The F. johnsoniae gliding motility adhesin SprB is delivered to the cell surface by the type IX secretion system (T9SS) and is rapidly propelled along the cell by the motility machinery. How this 6,497-amino-acid protein interacts with the secretion and motility machines is not known. Fusion of the C-terminal 218 amino acids of SprB to a foreign cargo protein resulted in its secretion, attachment to the cell surface, and rapid movement by the motility machinery. Efficient secretion of SprB required coexpression with the outer membrane protein SprF. Secreted proteins that have sequence similarity to SprB in their C-terminal regions are common in the phylum Bacteroidetes and may have roles in adhesion, motility, and virulence.
KEYWORDS: Bacteroidetes, Flavobacterium, gliding motility, protein secretion
ABSTRACT
Flavobacterium johnsoniae SprB moves rapidly along the cell surface, resulting in gliding motility. SprB secretion requires the type IX secretion system (T9SS). Proteins secreted by the T9SS typically have conserved C-terminal domains (CTDs) belonging to the type A CTD or type B CTD family. Attachment of 70- to 100-amino-acid type A CTDs to a foreign protein allows its secretion. Type B CTDs are common but have received little attention. Secretion of the foreign protein superfolder green fluorescent protein (sfGFP) fused to regions spanning the SprB type B CTD (sfGFP-CTDSprB) was analyzed. CTDs of 218 amino acids or longer resulted in secretion of sfGFP, whereas a 149-amino-acid region did not. Some sfGFP was secreted in soluble form, whereas the rest was attached on the cell surface. Surface-attached sfGFP was rapidly propelled along the cell, suggesting productive interaction with the motility machinery. This did not result in rapid cell movement, which apparently requires additional regions of SprB. Secretion of sfGFP-CTDSprB required coexpression with sprF, which lies downstream of sprB. SprF is similar in sequence to Porphyromonas gingivalis PorP. Most F. johnsoniae genes encoding proteins with type B CTDs lie immediately upstream of porP/sprF-like genes. sfGFP was fused to the type B CTD from one such protein (Fjoh_3952). This resulted in secretion of sfGFP only when it was coexpressed with its cognate PorP/SprF-like protein. These results highlight the need for extended regions of type B CTDs and for coexpression with the appropriate PorP/SprF-like protein for efficient secretion and cell surface localization of cargo proteins.
IMPORTANCE The F. johnsoniae gliding motility adhesin SprB is delivered to the cell surface by the type IX secretion system (T9SS) and is rapidly propelled along the cell by the motility machinery. How this 6,497-amino-acid protein interacts with the secretion and motility machines is not known. Fusion of the C-terminal 218 amino acids of SprB to a foreign cargo protein resulted in its secretion, attachment to the cell surface, and rapid movement by the motility machinery. Efficient secretion of SprB required coexpression with the outer membrane protein SprF. Secreted proteins that have sequence similarity to SprB in their C-terminal regions are common in the phylum Bacteroidetes and may have roles in adhesion, motility, and virulence.
INTRODUCTION
The gliding bacterium Flavobacterium johnsoniae secretes many proteins across its outer membrane via the type IX secretion system (T9SS) (1). T9SSs are common in but apparently confined to members of the phylum Bacteroidetes (2). They were first identified and have been most well studied in the nonmotile oral pathogen Porphyromonas gingivalis and in F. johnsoniae (3, 4). Many of the T9SS genes were originally discovered as F. johnsoniae genes required for cell movement over surfaces by gliding motility (5, 6). Some of the motility defects caused by mutations in these genes can be explained by failure to secrete the large mobile cell surface motility adhesins SprB and RemA (3, 7, 8). These adhesins are propelled rapidly along the surfaces of wild-type cells by other components of the gliding motility machinery. The adhesins follow a helical path along a cell, and their interactions with the substratum result in screw-like movement of the cell (9, 10).
The core components of the F. johnsoniae T9SS include GldK, GldL, GldM, GldN, SprA, SprE, and SprT (Fig. 1) (3, 5, 6, 8, 11), which are orthologs of P. gingivalis PorK, PorL, PorM, PorN, Sov, PorW, and PorT, respectively (3). Other components involved in but not essential for F. johnsoniae secretion include PorU and PorV. Mutations in porU and porV result in defects in secretion of some proteins that are targeted to the T9SS but not in that of others (1, 12). P. gingivalis porP, porZ, and PG1058 are also involved in secretion, and F. johnsoniae has orthologs of each of these. F. johnsoniae has multiple genes that are similar to P. gingivalis porP, and a requirement of these for secretion may be masked by redundancy. One of these genes, sprF, is required for secretion of SprB but not for secretion of other proteins that are targeted to the T9SS (7, 13). sprF lies immediately downstream of, and is cotranscribed with, sprB (13). PorP and SprF are predicted outer membrane β-barrel proteins (13), and outer membrane localization was confirmed for PorP (3). F. johnsoniae porZ and PG1058-like genes have not yet been examined for their roles in secretion. Most of the components of the T9SS are unique to members of the phylum Bacteroidetes, and none are similar in sequence to components of bacterial type I to type VIII secretion systems (3, 8).
FIG 1.
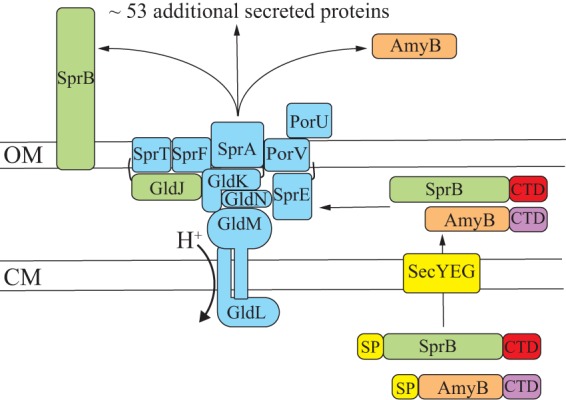
The F. johnsoniae type IX protein secretion system. Proteins required for secretion include the cytoplasmic membrane proteins GldL and GldM, which have been proposed to energize secretion, and the outer membrane-associated proteins GldK, GldN, SprA, SprE, and SprT. The gliding motility protein GldJ (green) is needed to stabilize the T9SS protein GldK (30). Many of the T9SS proteins may also have roles in motility in addition to their functions in secretion. SprF is required for the secretion of SprB but not for the secretion of other proteins by the T9SS (13). The secreted proteins have N-terminal signal peptides (SP) that target them for export across the cytoplasmic membrane by the Sec system before delivery to the T9SS. Proteins secreted by T9SSs have type A (purple) or type B (red) C-terminal domains (CTDs) required for secretion. PorV appears to be required for the secretion of proteins that have type A CTDs but is not needed for the secretion of SprB or for the secretion of some other proteins that have type B CTDs (1). Black lines on the lipoproteins GldJ, GldK, and SprE indicate lipid tails. OM, outer membrane; CM, cytoplasmic membrane.
Recent results have begun to reveal the structure and function of the T9SS. P. gingivalis PorK, PorL, PorM, and PorN (GldK, GldL, GldM, and GldN, respectively, in F. johnsoniae) appear to form a complex that spans the cell envelope (3, 14–16). The cytoplasmic membrane proteins PorL and PorM (GldL and GldM in F. johnsoniae) have been suggested to harvest the proton gradient and energize both secretion and motility (8, 15, 17). Studies of F. johnsoniae SprA (ortholog of P. gingivalis Sov) suggest that it forms the outer membrane pore through which secreted proteins pass (18).
The F. johnsoniae T9SS is required for secretion of dozens of proteins. In addition to the cell surface motility adhesins described above, it also secretes soluble extracellular enzymes such as the chitinase ChiA and the amylase AmyB (1, 8, 19). The P. gingivalis T9SS is involved in secretion of virulence factors such as the cell surface gingipain proteases (3). In both organisms, the secreted proteins have N-terminal signal peptides that allow export across the cytoplasmic membrane via the Sec machinery and conserved C-terminal domains (CTDs) that are required for T9SS-mediated secretion. The CTDs are typically cleaved during or after secretion (20–24). Most known T9SS CTDs belong to either protein domain family TIGR04183 (type A CTDs) or family TIGR04131 (type B CTDs). The roles of type A CTDs in secretion of F. johnsoniae and P. gingivalis proteins have been studied. Type A CTD regions of 70 to 100 amino acids are sufficient to target a foreign protein such as superfolder green fluorescent protein (sfGFP) for secretion from the periplasm across the outer membrane by the T9SS (12, 22). The structures of two type A CTDs have been determined (25, 26). Both have an Ig-like fold with seven β strands and are thought to interact with components of the T9SS. Some F. johnsoniae proteins with type A CTDs are released in soluble form, whereas others remain attached to the cell surface after secretion.
Type B CTDs are not similar in sequence to type A CTDs, and they have not been studied in detail. The 6,497-amino-acid protein SprB appears to require its type B CTD for secretion (12, 27). However, attachment of C-terminal regions of F. johnsoniae SprB to the foreign protein sfGFP failed to result in its secretion across the outer membrane (12).
Here we present evidence that type B CTDs target proteins for secretion by the T9SS and that specific PorP/SprF-like outer membrane proteins facilitate secretion of proteins carrying different type B CTDs. Moreover, the C-terminal 218-amino-acid region of SprB is sufficient to target a foreign protein for secretion and productive interaction (direct or indirect) with the motility machinery, resulting in rapid movement along the cell surface. The results highlight the diversity of proteins secreted by the T9SS and the roles of their C-terminal regions in secretion, cell surface localization, and interaction with the motility machinery.
RESULTS
Fusion of sfGFP to C-terminal regions of SprB results in its secretion.
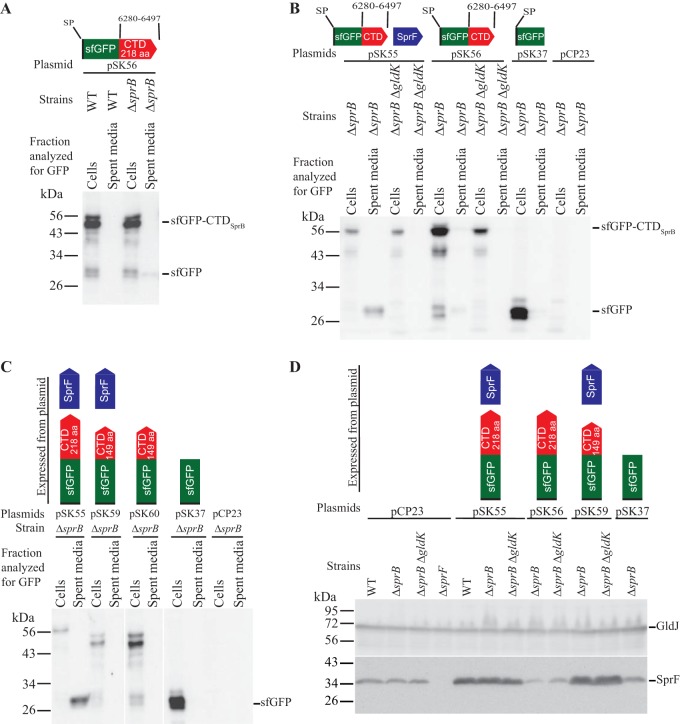
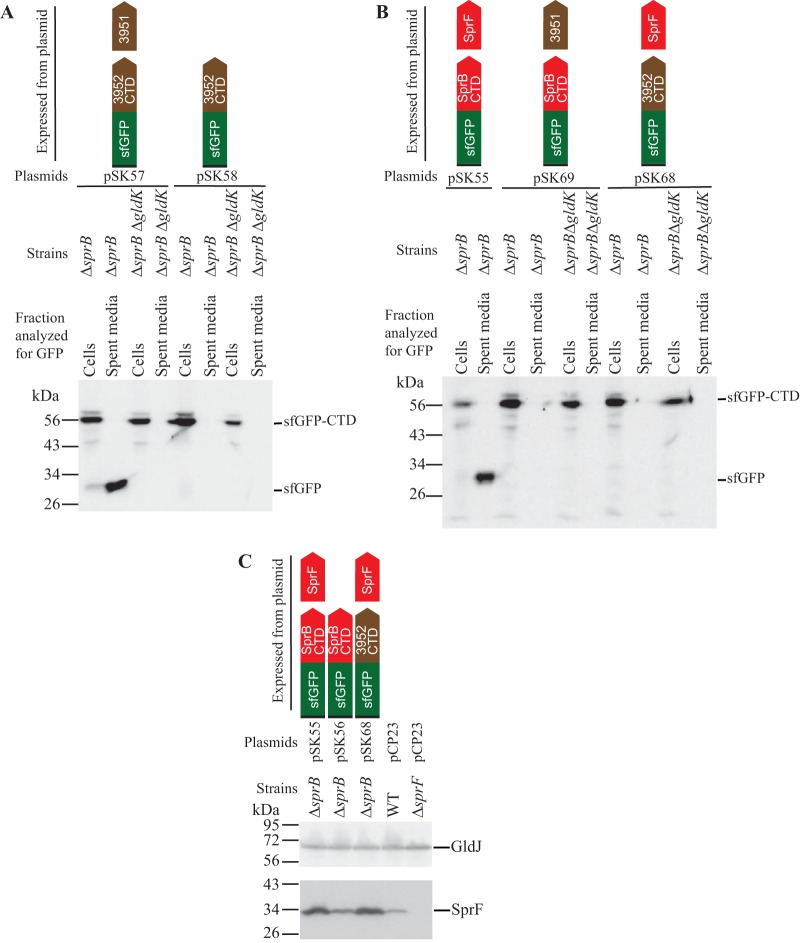
Many proteins secreted by T9SSs have type A CTDs, and the involvement of these in secretion has been demonstrated (1, 12, 21, 22, 28). Type B CTDs, such as the carboxy-terminal region of SprB, have been less well studied. Previous results indicated that sfGFP fused to a signal peptide at its amino terminus (SP-sfGFP) and to C-terminal regions of SprB of 99, 218, and 1,182 amino acids in length [SP-sfGFP-CTDSprB(99AA), SP-sfGFP-CTDSprB(218AA), and SP-sfGFP-CTDSprB(1182AA), respectively] was not secreted by wild-type cells (12). SprF is required for secretion of SprB but not for secretion of other proteins that are targeted to the T9SS (13). We previously suggested that SprF may play an adapter or chaperone-like function that facilitates secretion of SprB (13). One possible explanation for the failure of wild-type cells to secrete SP-sfGFP-CTDSprB is that the SprF in wild-type cells may have been fully engaged with its coexpressed partner, SprB, leaving little available to interact with SP-sfGFP-CTDSprB. To relieve this situation, we deleted sprB. ΔsprB mutants carrying pSK56, which expresses SP-sfGFP-CTDSprB(218AA), exhibited wild-type levels of SprF protein (Fig. 2D) and appeared to secrete a small amount of sfGFP (Fig. 2A and B). Coexpression of SP-sfGFP-CTDSprB and SprF from pSK55 resulted in higher levels of SprF (Fig. 2D) and increased secretion of sfGFP (Fig. 2B). Secretion did not occur in cells of the ΔsprB ΔgldK double mutant carrying the same plasmid (Fig. 2B), indicating that a functional T9SS was required. The presence of sfGFP in the spent medium was not likely the result of cell lysis or leakage, because coexpression of SP-sfGFP-CTDSprB (from pSK55) and SP-mCherry (from pJJ21) allowed secretion of sfGFP but not leakage of mCherry (see Fig. S2 in the supplemental material). A shorter region of the C terminus of SprB (149 amino acids) failed to result in secretion of sfGFP even when coexpressed with SprF (Fig. 2C). Regions of SprB spanning the C-terminal 368, 448, 663, and 1,182 amino acids were also fused to SP-sfGFP and examined for their ability to facilitate secretion of sfGFP. sprF was coexpressed on the same plasmids. Western immunoblot analyses of the strains expressing the fusion proteins demonstrated the accumulation of sfGFP in the spent medium (see Fig. S3 in the supplemental material). The apparent molecular mass of the soluble secreted sfGFP (approximately 30 kDa) did not change when the length of the C-terminal region of SprB fused to sfGFP was increased beyond 218 amino acids. This suggests processing of the fusion protein during or after secretion, with accumulation of stable sfGFP in the medium. F. johnsoniae produces many secreted proteases that could contribute to this partial digestion of the secreted protein (29). The results suggest that a CTD longer than 149 amino acids was required for secretion and that coexpression of SP-sfGFP-CTDSprB and SprF from the same plasmid resulted in enhanced secretion. Alignment of the C terminus of SprB with other F. johnsoniae proteins with type B CTDs indicates that regions of sequence identity extend only for the C-terminal 85 to 100 amino acids (see Fig. S4 in the supplemental material). Apparently, additional regions outside this conserved C-terminal region of SprB are required to effectively target SP-sfGFP-CTDSprB for secretion.
FIG 2.
T9SS-mediated secretion of sfGFP fused to the CTD of SprB, with or without coexpression with SprF. Cultures of wild-type (WT), ΔsprB mutant, ΔsprB ΔgldK T9SS-defective mutant, and ΔsprF mutant cells, carrying the plasmids indicated, were incubated in CYE at 25°C with shaking and harvested in the late exponential phase of growth. Strains with gldK deleted (ΔgldK) lack a functional T9SS. All strains except for the ΔsprF strain in panel D were wild type for sprF on the chromosome, but some expressed additional SprF from plasmid as indicated. (A to C) Cells and spent culture media were separated by centrifugation and analyzed by Western blotting using antiserum against GFP. Cell samples corresponded to 10 μg protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg protein before the cells were removed. The blot shown in panel C had four additional lanes that were removed. See Fig. S1 in the supplemental material for the full blot that includes these lanes. (D) Cells (20 μg protein per lane) were examined by Western blotting using antiserum against SprF and antiserum against GldJ (loading control). Plasmids used: pCP23 (empty vector); pSK37, which expresses sfGFP with an N-terminal signal peptide (SP); pSK56, which expresses SP-sfGFP fused to the 218-amino-acid CTD of SprB [SP-sfGFP-CTDSprB(218AA)]; pSK55, which coexpresses SP-sfGFP-CTDSprB(218AA) and SprF; pSK60, which expresses SP-sfGFP fused to the 149-amino-acid CTD of SprB [SP-sfGFP-CTDSprB(149AA)]; or pSK59, which coexpresses SP-sfGFP-CTDSprB149AA and SprF. Cartoons depicting the proteins expressed from the plasmids are indicated above the appropriate lanes.
Fusion of sfGFP to C-terminal regions of SprB results in its attachment to the cell surface and rapid movement.
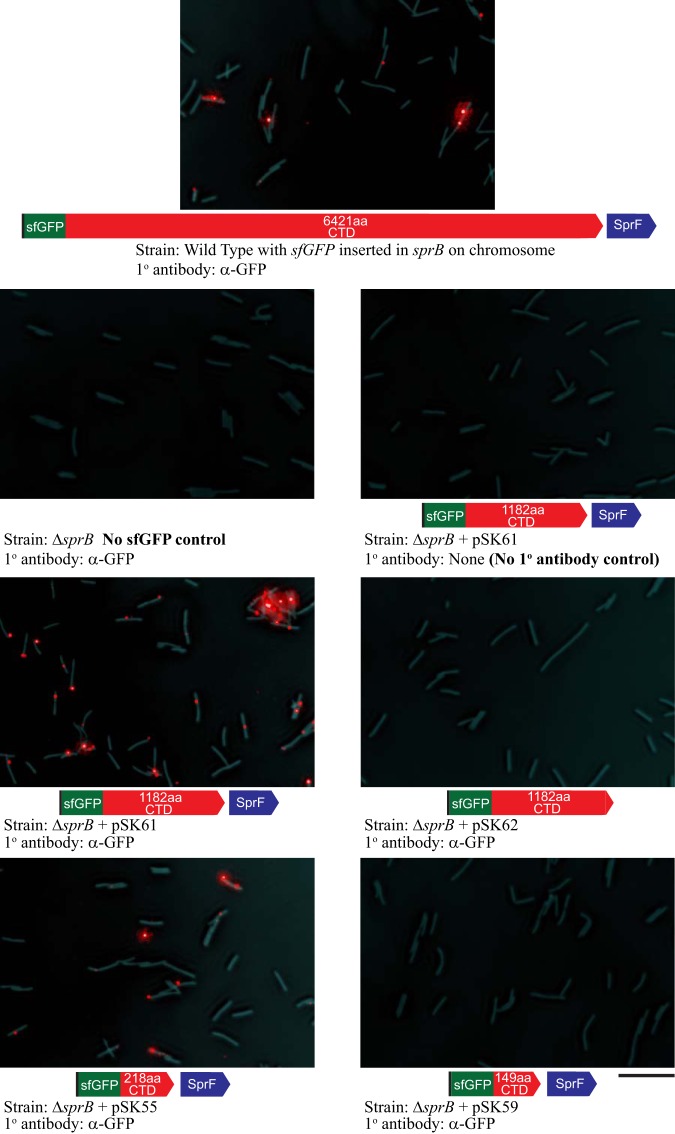
SprB attaches to the surfaces of cells after secretion by the T9SS (3, 9, 27). Cells that expressed SP-sfGFP-CTDSprB were examined for their ability to localize sfGFP to the cell surface by immunofluorescence microscopy using anti-GFP followed by secondary antibody conjugated to Alexa Fluor 594. Full-length SprB with sfGFP inserted after the N-terminal signal peptide [SP-sfGFP-CTDSprB(6421AA)] expressed from the chromosome localized to the cell surface (Fig. 3 and 4A; see Fig. S5 in the supplemental material). Similarly, cells that expressed SP-sfGFP-CTDSprB with CTDs of 218 amino acids or longer had sfGFP on their surfaces. In contrast, cells expressing SP-sfGFP-CTDSprB(149AA) did not have sfGFP on the cell surface (Fig. 3, 4A, and S5). Surface associated sfGFP was not observed above background levels unless the fusion protein was coexpressed with SprF (Fig. 3, 4, and S5). sfGFP did not interact with the cell surface unless it had the proper CTD. Fusion of SP-sfGFP to the CTDs of ChiA and AmyB resulted in secretion of sfGFP but not cell surface attachment (12), as confirmed here by immunofluorescence microscopy (see Fig. S6 and S7 in the supplemental material). SP-sfGFP-CTDAmyB and SP-sfGFP-CTDSprB had identical N-terminal signal peptides linked to sfGFP and differed only at their C termini, suggesting that the 218-amino-acid region of SprB was responsible for interaction with the cell surface. In all of the fluorescence microscopy experiments described above, cells were examined for red fluorescence. We also examined the same cells for green fluorescence to demonstrate that the filters used allowed detection of green fluorescence and red fluorescence without overlap (see Fig. S8 in the supplemental material). This also confirmed that sfGFP was present in cells that did not secrete it, either because of a T9SS mutation (ΔgldK) or because sfGFP was not fused to a C-terminal domain of SprB of 218 amino acids or longer.
FIG 3.
Detection of surface-localized sfGFP by immunofluorescence microscopy. Cells expressing SP-sfGFP fused to C-terminal regions (CTDs) of SprB ranging from 149 to 6,421 amino acids were exposed to anti-GFP (α-GFP) and secondary antibody fused to Alexa Fluor 594 and observed by fluorescence microscopy to detect sfGFP exposed on the cell surface. Exposure times for fluorescence images were all 33 ms. Phase-contrast images were overlaid to observe the relationship of the signals to cells. The top panel shows wild-type cells expressing full-length SprB from the chromosome, with sfGFP fused after the signal peptide. All remaining panels show ΔsprB mutant cells with or without plasmids expressing SP-sfGFP fused to CTDSprB of various lengths. All plasmids except pSK62 also express SprF to facilitate secretion. For additional controls and experimental results, see Fig. 4 and Fig. S5, S6, S7, and S8 in the supplemental material.
FIG 4.
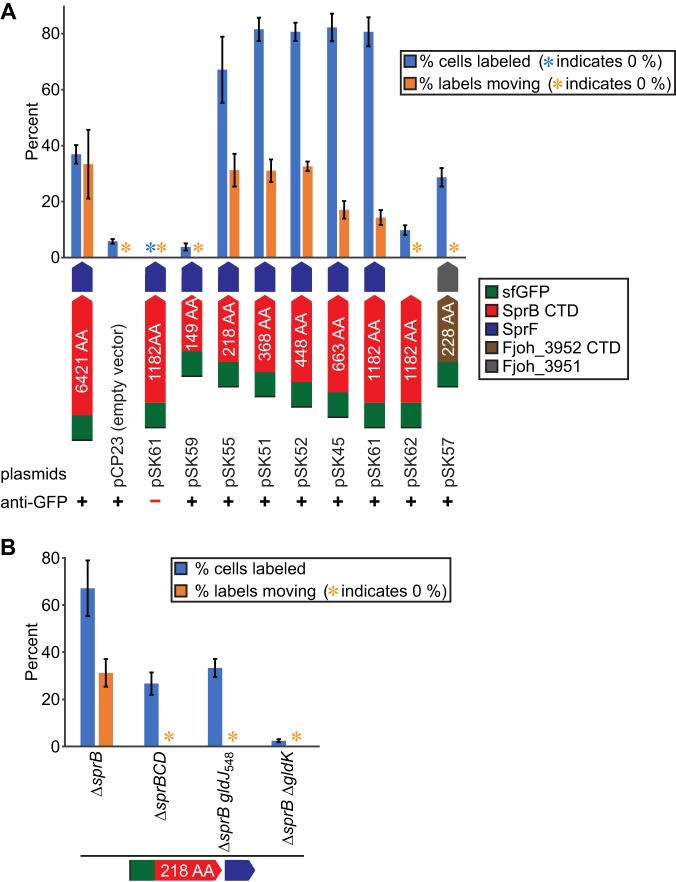
Labeling and movement of cell surface-localized sfGFP. Cells expressing SP-sfGFP fused to C-terminal regions (CTDs) of SprB and Fjoh_3952 were exposed to anti-GFP and secondary antibody fused to Alexa Fluor 594 and observed by fluorescence microscopy to detect sfGFP exposed on the cell surface. In both panels, the percentage of labeled cells and percentage of labels that moved were determined by examining three independent samples of 150 cells from each strain. For movement, cells were observed with simultaneous fluorescence excitation and low-light phase-contrast microscopy and examined for fluorescent labeling and propulsion of the label along the cell surface. Labels counted as moving traveled ≥2.5 μm during the 20-s observation period. Examples of moving and nonmoving labels are shown in Movies S1 to S3 in the supplemental material. Blue or orange asterisks indicate that 0 signals or 0 moving signals, respectively, were observed for these samples. Error bars indicate standard deviations from three measurements. (A) “6421 AA” indicates full-length SprB expressed from the chromosome, with sfGFP fused after the signal peptide. This strain also carried the empty vector, pCP23. All other sfGFP fusions were expressed from their respective plasmids in the ΔsprB mutant. (B) SP-sfGFP-CTDSprB(218AA) was expressed from pSK55 in ΔsprB, ΔsprBCD, ΔsprB gldJ548, and ΔsprB ΔgldK mutants.
Full-length SprB with sfGFP inserted after the N-terminal signal peptide was propelled along the cell surface (see Movie S1 in the supplemental material), as has been reported previously for wild-type SprB (9, 27). Similarly, sfGFP fused to CTDs of SprB of 218 amino acids or longer also moved rapidly along the cell surface, suggesting productive interaction of the C-terminal 218-amino-acid region of SprB with the gliding motility apparatus (Fig. 4 and 5; see Movie S2 in the supplemental material). Deletion of the region spanning sprC and sprD from the chromosome resulted in decreased localization of SP-sfGFP-CTDSprB(218AA) on the cell surface and no apparent movement (Fig. 4B), suggesting that SprC and/or SprD may be important for productive interaction with the rest of the motility machinery.
FIG 5.
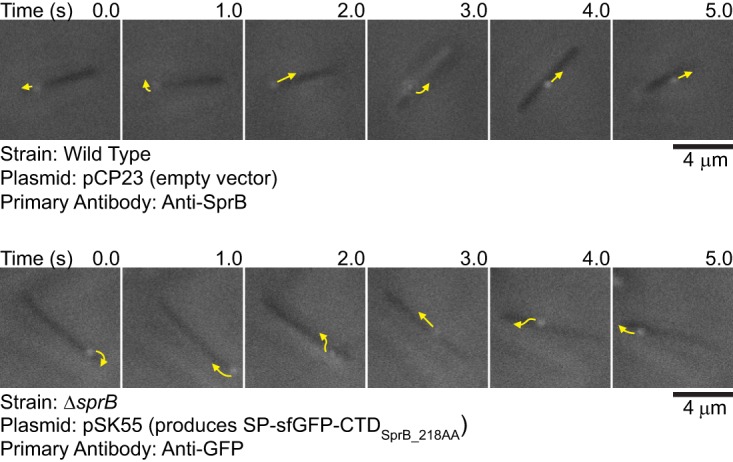
Movement of SprB and sfGFP on the cell surface. Cells that expressed SprB or that expressed SP-sfGFP fused to the C-terminal 218-amino-acid region of SprB were examined by simultaneous immunofluorescence and phase-contrast microscopy. Low-light phase contrast was used to detect the cells, and SprB and sfGFP were detected by exposure to appropriate antisera followed by exposure to goat anti-rabbit IgG conjugated to Alexa Fluor 594. Suspended cells were examined to avoid confusion resulting from the gliding movement of cells over surfaces. The sequences shown correspond to 5-s intervals from Movie S1 (top) and Movie S2 (bottom) in the supplemental material. Arrows indicate the movements of fluorescent signals in the next second. Controls demonstrating specificity of anti-SprB are given in references 27 and 30, and controls demonstrating specificity of anti-GFP are given in Fig. 3 and 4 and in Fig. S5, S7, and S8 in the supplemental material.
The motility protein GldJ is needed for gliding motility and for T9SS function (30). GldJ is required for stable accumulation of the T9SS protein GldK, explaining why GldJ is needed for secretion. The use of GldJ truncation mutants allows motility and secretion to be separated (30). gldJ mutant cells that express truncated GldJ548, lacking the C-terminal 13 amino acids of GldJ, accumulate GldK and have a functional T9SS, but they are nonmotile. Such cells secrete SprB to the cell surface but do not propel it along the cell surface. To demonstrate that the motility machinery was required for the observed movement of SP-sfGFP-CTDSprB(218AA) described above, we expressed this fusion protein in cells that produce truncated GldJ548. These cells secreted sfGFP and localized it to the cell surface, but they failed to propel it along the cell surface (Fig. 4B and Movie S2).
Expression of SP-sfGFP-CTDSprB in a ΔsprB mutant does not restore wild-type motility and colony spreading.
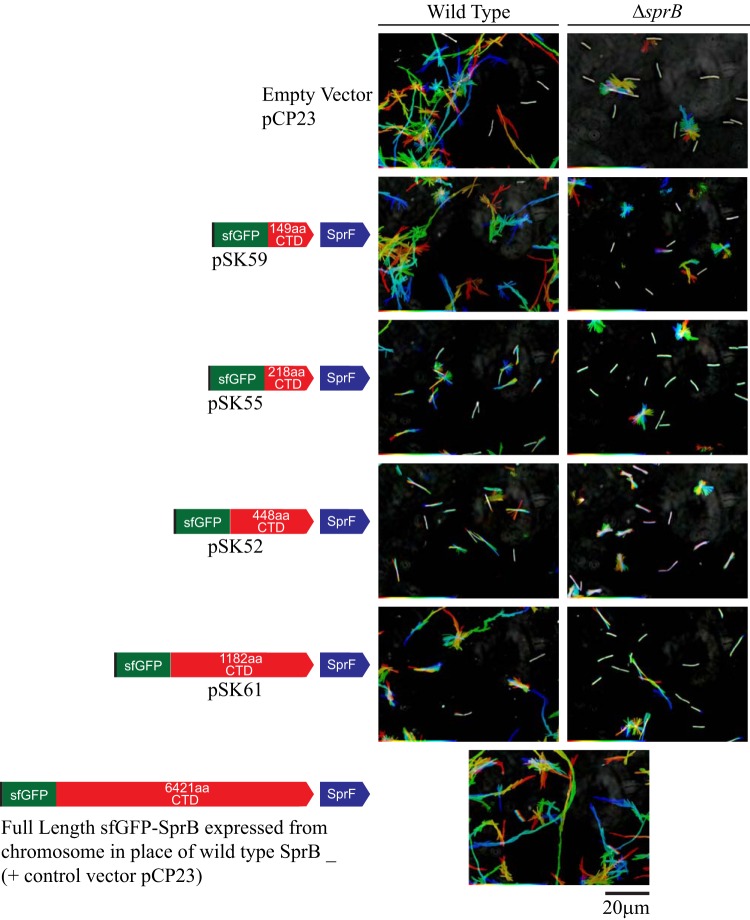
sprB null mutants are partially defective in motility (13, 27, 31). They form nonspreading colonies on agar and exhibit reduced but detectable motility on glass (Fig. 6 and 7). Whereas wild-type cells move in long paths on agar or glass, ΔsprB mutant cells are less active and also reverse direction frequently, thus making little net progress. The residual motility of ΔsprB mutant cells is explained by the presence of alternative motility adhesins, such as RemA (7, 27). We examined the ability of cell surface-localized sfGFP-CTDSprB to restore motility to ΔsprB mutant cells. ΔsprB mutants expressing SP-sfGFP-CTDSprB with C-terminal regions of SprB of 218, 448, and 1,182 amino acids failed to form spreading colonies on agar (Fig. 6) or to move effectively on glass (Fig. 7; see Movies S4 and S5 in the supplemental material), although some cells of the ΔsprB mutant that expressed sfGFP-CTDSprB(1182AA) moved slightly longer distances on glass than did cells of the ΔsprB mutant. As described above, sfGFP-CTDSprB with C-terminal regions of SprB greater than 218 amino acids interacted with the motility machinery, as indicated by their rapid propulsion along the cell surface. Additional regions of SprB may be required for productive interaction with the substratum, to allow the gliding motors to propel the cells effectively over agar or glass. The longest C-terminal region of SprB added to plasmid-encoded SP-sfGFP in this study (1,182 amino acids) spanned approximately 18% of full-length SprB. We failed to construct plasmids encoding SP-sfGFP fused to longer C-terminal regions of SprB, probably because of the extremely repetitive nature of the central region of sprB (27).
FIG 6.
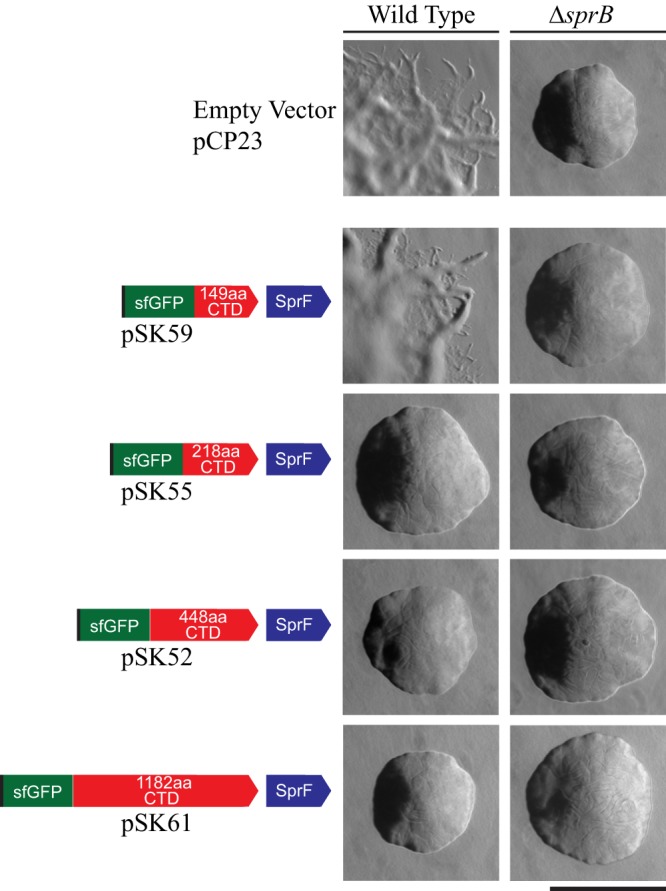
Colonies of wild-type and ΔsprB mutant F. johnsoniae cells expressing SP-sfGFP fused to C-terminal regions of SprB. Colonies were incubated at 25°C on PY2 agar (54) for 40 h. Photomicrographs were taken with a Photometrics Cool-SNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. The left column shows colonies of wild-type cells, and the right column shows colonies of ΔsprB mutant cells. Plasmids carried are indicated on the left, and the proteins encoded by the plasmids are shown by the cartoons. The scale bar indicates 1 mm and applies to all panels.
FIG 7.
Gliding of wild-type and ΔsprB mutant cells expressing SP-sfGFP fused to C-terminal regions of SprB. Cells carried either the empty vector pCP23 or the plasmids indicated that encode SP-sfGFP fused to C-terminal regions of SprB of various lengths. All plasmids except pCP23 also encode SprF. The bottom panel shows wild-type cells expressing full-length SprB from the chromosome, with sfGFP fused after the signal peptide. Cells were grown in MM overnight without shaking at 25°C, introduced into glass tunnel slides, and observed for motility at 25°C using a phase-contrast microscope. A series of images were taken for 120 s. Individual frames were colored from red (time zero) to yellow, green, cyan, and finally blue (120 s) and integrated into one image, resulting in rainbow traces of gliding cells. White cells correspond to those that exhibited little or no net movement. Multicolored “stars” indicate cells attached to the glass by one pole that rotated or flipped. The rainbow traces correspond to the sequences shown in Movies S4 and S5 in the supplemental material.
In addition to failing to restore full motility to a ΔsprB mutant as described above, expression of SP-sfGFP fused to C-terminal regions of SprB of 218, 448, and 1,182 amino acids in wild-type cells resulted in the formation of nonspreading colonies on agar (Fig. 6) and decreased motility on glass (Fig. 7). Overexpression of fragments of SprB that allow interaction with the motility machinery may interfere with the ability of full-length SprB to function properly. Expression of SP-sfGFP fused to the C-terminal 149 amino acids of SprB, which was not secreted from the cell, did not disrupt motility of wild-type cells (Fig. 6 and 7). Cells in which the wild-type chromosomal sprB was replaced with a modified and functional form of the gene that expresses full-length SprB with sfGFP inserted after the signal peptide [SP-sfGFP-CTDSprB(6421AA)] also moved similarly to the wild type on glass (Fig. 7).
SprF and the PorP/SprF-like protein Fjoh_3951 appear to be specific for their cognate secreted proteins.
F. johnsoniae is predicted to encode nine proteins that exhibit sequence similarity with and are similar in size to F. johnsoniae SprF and to the P. gingivalis T9SS protein PorP (see Fig. S9 in the supplemental material). Eight of the F. johnsoniae genes encoding these PorP/SprF-like proteins lie immediately downstream of genes encoding proteins with type B CTDs, similar to the relationship of sprF and sprB (see Fig. S10 in the supplemental material). The type B CTD-containing protein Fjoh_3952 and the PorP/SprF-like protein Fjoh_3951 were studied to determine if they behaved like SprB and SprF. Fjoh_3952 is produced by and secreted from wild-type cells in a process that requires the T9SS (1). The C-terminal 228-amino-acid region of Fjoh_3952 was fused to SP-sfGFP, and secretion was analyzed with or without coexpression of the porP/sprF-like gene, encoding Fjoh_3951. Coexpression of SP-sfGFP-CTDFjoh_3952 and Fjoh_3951 from pSK57 in cells that were wild type for secretion (the ΔsprB mutant was used here) resulted in sfGFP accumulation in the spent medium (Fig. 8A). In contrast, sfGFP was not found in the spent medium of cells that carried pSK58, which expressed SP-sfGFP-CTDFjoh_3952 without Fjoh_3951. Coexpression of SP-sfGFP-CTDFjoh_3952 and Fjoh_3951 from pSK57 in cells of a ΔgldK (T9SS-deficient) mutant did not result in sfGFP secretion (Fig. 8A). The results indicate that an intact T9SS and coexpression of the PorP/SprF-like protein Fjoh_3951 were required for secretion of SP-sfGFP-CTDFjoh_3952. Wild-type chromosomal Fjoh_3951 was present in all strains, but additional plasmid-encoded Fjoh_3951 protein was needed to foster secretion of SP-sfGFP-CTDFjoh_3952, in the same way that additional plasmid-encoded SprF was needed for efficient secretion of SP-sfGFP-CTDSprB (Fig. 2). Some of the secreted sfGFP-CTDFjoh_3952 attached to the cell surface (Fig. 4A; see Fig. S7 and S11 in the supplemental material), as was observed for sfGFP-CTDSprB. However, unlike sfGFP-CTDSprB, sfGFP-CTDFjoh_3952 did not move along the cell surface (Fig. 4A; see Movie S3 in the supplemental material), suggesting that Fjoh_3952 may not function as a motility adhesin as SprB does.
FIG 8.
Efficient secretion of SP-sfGFP fused to CTDSprB or to CTDFjoh_3952 requires coexpression with the appropriate PorP/SprF-like protein. Cultures of wild-type (WT), ΔsprB mutant, ΔsprB ΔgldK (T9SS-defective) mutant, and ΔsprF mutant cells, carrying the plasmids indicated, were incubated in CYE at 25°C with shaking and harvested in the late exponential phase of growth. (A and B) Cells and spent culture media were separated by centrifugation and analyzed by Western blotting using antiserum against GFP. Whole-cell samples corresponded to 10 μg protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg protein before the cells were removed. (C) Cells (20 μg protein per lane) were examined by Western blotting using antiserum against SprF and antiserum against GldJ (loading control). Plasmids used: pCP23 (empty vector); pSK58, which expresses SP-sfGFP fused to the 228-amino-acid CTD of Fjoh_3952 (SP-sfGFP-CTDFjoh_3952); pSK57, which coexpresses SP-sfGFP-CTDFjoh_3952 and Fjoh_3951; pSK56, which expresses SP-sfGFP-CTDSprB(218AA); pSK55, which coexpresses SP-sfGFP-CTDSprB(218AA) and SprF; pSK68, which coexpresses SP-sfGFP-CTDFjoh_3952 and SprF; and pSK69, which coexpresses SP-sfGFP-CTDSprB(218AA) and Fjoh_3951. Cartoons depicting the proteins expressed from the plasmids are indicated above the appropriate lanes.
To determine if SprF and Fjoh_3951 are interchangeable, we constructed pSK68, which expresses SP-sfGFP-CTDFjoh_3952 and SprF, and pSK69, which expresses SP-sfGFP-CTDSprB and Fjoh_3951. Cells carrying either plasmid failed to efficiently secrete soluble or cell surface-localized sfGFP (Fig. 8, S7, and S11), suggesting that the cognate PorP/SprF-like proteins were required for secretion of proteins carrying the CTDs of SprB and Fjoh_3952.
Analysis of the type B CTD of secreted protein Fjoh_1123.
Fjoh_1123 has a type B CTD and was shown to be secreted by the T9SS (1). Unlike most other F. johnsoniae genes encoding proteins with type B CTDs, the Fjoh_1123 gene is not located near a porP/sprF-like gene (Fig. S10). To determine if the Fjoh_1123 CTD would target sfGFP for secretion without coexpression with a porP/sprF-like gene, we constructed a plasmid that expressed SP-sfGFP fused to the C-terminal 238 amino acids of Fjoh_1123 (SP-sfGFP-CTDFjoh_1123). Western blot analysis and immunofluorescence microscopy demonstrated that sfGFP was not secreted by cells expressing this fusion protein (see Fig. S7, S11, and S12 in the supplemental material). Coexpression with one of the 10 F. johnsoniae PorP/SprF-like proteins may be required to facilitate secretion of Fjoh_1123 and SP-sfGFP-CTDFjoh_1123.
SprF outer membrane localization.
SprF might be an adapter or chaperone that aids secretion and surface localization of SprB. PSORTb and CELLO analyses predict that SprF is an outer membrane protein (13). Cell fractionation and Western blot analyses using anti-SprF antibodies were performed to test this prediction. SprF was detected in the outer membrane fraction of wild-type or ΔsprB mutant cells, whereas the known cytoplasmic membrane protein GldL localized to the cytoplasmic membrane (Fig. 9). SprF was not detected on the surfaces of intact wild-type or ΔsprB mutant cells using fluorescently labeled anti-SprF antibodies (data not shown). SprF was not susceptible to proteinase K treatment of intact wild-type cells, but SprF in ΔsprB cells was partially digested by proteinase K (Fig. 10). These results suggest that a limited portion of SprF is exposed on the external surface of the outer membrane but is protected from added proteinase K by SprB.
FIG 9.
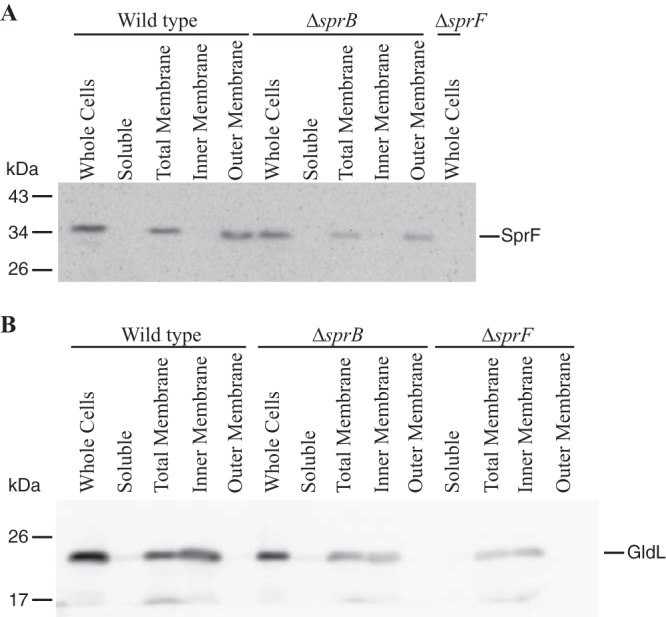
Localization of SprF. Wild-type and mutant cells were disrupted and separated into soluble and membrane fractions. Membranes were fractionated further by differential solubilization in Sarkosyl. Equal amounts of each fraction based on 20 μg protein of the starting material (whole cells) were loaded in each lane and separated by SDS-PAGE. Antisera against SprF (A) and GldL (B) were used to detect the respective proteins by Western blot analyses.
FIG 10.
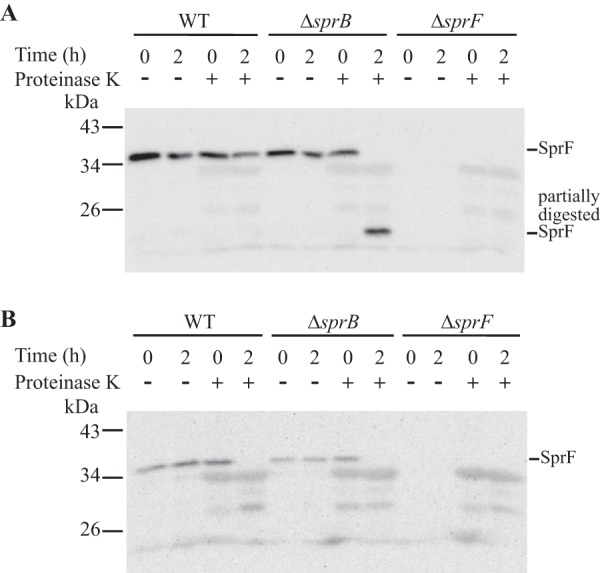
Proteinase K treatment to determine if SprF localizes to the cell surface. Wild-type, ΔsprB, and ΔsprF strains were analyzed. Proteinase K was added at a final concentration of 1 mg/ml to intact cells (A) and to cell extracts prepared by French pressure cell treatment (B), and cells and extracts were incubated at 25°C. Samples were removed at 0 h and 2 h for immunoblot analyses. Samples were separated by SDS-PAGE, and SprF was detected using antiserum against SprF. Samples not exposed to proteinase K (−) were also included.
Bioinformatic analysis of F. johnsoniae proteins predicted to be secreted by the T9SS.
Many proteins secreted by T9SSs have type A or type B CTDs (12). F. johnsoniae has 41 genes encoding proteins with type A CTDs and 12 encoding proteins with type B CTDs (see Tables S1 and S2 in the supplemental material). Many of these were shown to be secreted by wild-type cells, and they were not secreted by mutants that are deficient for the T9SS (1). Some proteins secreted by the T9SS, such as ChiA (19), lack regions of sequence similarity to either type A or type B CTDs. For this reason, the number of proteins secreted by the F. johnsoniae T9SS may be higher than the estimate given above. Of the 41 F. johnsoniae proteins with type A CTDs, 20 have predicted enzymatic activities (Table S1). In contrast, none of the 12 proteins with type B CTDs have predicted enzymatic activities, but 8 of them have features associated with some adhesins (Table S2). The 12 proteins with type B CTDs are very large, ranging from 78.5 to 672.0 kDa before processing, with a median of 239.6 kDa (Table S2). In contrast, F. johnsoniae proteins with type A CTDs have a median size of 94.3 kDa (Table S1).
Genomic context of F. johnsoniae porP/sprF-like genes.
F. johnsoniae is predicted to encode 10 PorP/SprF-like proteins (including SprF) that are similar in size (Fig. S9 and S10). The N-terminal 299-amino-acid region of the much larger protein SprD (1,588 amino acids) also exhibits limited similarity to SprF. Each of the 10 PorP/SprF-like proteins has an N-terminal signal peptide, and each is predicted to be an outer membrane protein by CELLO analysis (32). Nine of the genes encoding these proteins lie immediately downstream of genes encoding proteins with type B CTDs (Fig. S10). The PorP/SprF-like proteins encoded by these genes presumably aid the secretion of the product of the upstream gene. Of these nine porP/sprF-like genes, six are located immediately upstream of, and transcribed in the same direction as, genes encoding predicted outer membrane proteins that are similar in sequence to PG1058 (see Fig. S10 and S13 in the supplemental material), which is required for P. gingivalis T9SS function (33). The C-terminal regions of each of these PG1058-like proteins are similar in sequence to the C-terminal peptidoglycan-interacting domain of Escherichia coli OmpA (34, 35). Unlike P. gingivalis PG1058, which was predicted to be a lipoprotein (33), each of the F. johnsoniae PG1058-like proteins lack the conserved cysteine adjacent to the amino-terminal signal peptide that is required of bacterial lipoproteins. The synteny of porP/sprF-like genes and PG1058-like genes suggests possible functional links between the encoded proteins in T9SS-mediated secretion.
Predicted secreted proteins and porP/sprF-like genes in other members of the Bacteroidetes.
Examination of the completed genome sequences of 104 members of the phylum Bacteroidetes revealed the presence of core genes of the T9SS in 90 of these (12). Each of these has proteins with type A CTDs, and 88 also have proteins with type B CTDs. In most cases they encode multiple proteins with type A CTDs and multiple proteins with type B CTDs (see Table S3 in the supplemental material). Of those examined, Fluviicola taffensis DSM 16823 had the most predicted secreted proteins: 180 proteins with type A CTDs and 50 with type B CTDs. Members of the class Bacteroidia are unusual among the Bacteroidetes in encoding few proteins with type B CTDs. Among 17 genomes from members of the class Bacteroidia with T9SS genes, 14 encode only a single type B CTD-containing protein, and another has none.
Each of the organisms that have genes encoding type B CTDs also has one or more porP/sprF-like genes (Table S3). The number of predicted proteins with type B CTDs often reflects the number of predicted PorP/SprF-like proteins. For example, F. johnsoniae has 12 type B CTD-containing proteins and 11 PorP/SprF-like proteins, whereas P. gingivalis has a single type B CTD-containing protein, PG1035, and a single PorP/SprF-like protein, PorP (Table S3). Type B CTD-encoding genes and porP/sprF-like genes are absent in all but one of the organisms that lack core T9SS genes.
Similar to the situation for F. johnsoniae, porP/sprF-like genes in other Bacteroidetes are often located immediately downstream of genes encoding proteins with type B CTDs. Of 527 porP/sprF-like genes identified, 314 (60%) lie immediately downstream of and transcribed in the same direction as genes encoding type B CTD-containing proteins (Table S3). In some cases, multiple type B CTD-encoding genes are found immediately upstream of individual porP/sprF-like genes. In the most extreme case (Cytophaga hutchinsonii CHU_3434), five type B CTD-encoding genes are immediately upstream. Our analysis underestimates the association of porP/sprF-like genes with type B CTD-encoding genes for two reasons. First, some porP/sprF-like genes are immediately upstream (instead of downstream) of genes encoding type B CTDs, and these were excluded. Some of these are transcribed in the same direction as the genes encoding type B CTDs, whereas others are oriented in the opposite direction. Second, some porP/sprF-like genes are not immediately adjacent to genes encoding type B CTD-containing proteins but instead are separated by one gene and were thus not counted in our analysis.
In addition to the porP/sprF-like genes described above that are located near genes encoding predicted T9SS-secreted proteins, other porP/sprF-like genes are adjacent to genes encoding components of the T9SS secretion machine. Of the 527 porP/sprF-like genes in Table S3, 42 are immediately upstream and oriented in the same direction as gldK, gldL, gldM, and gldN, which encode core components of the T9SS. These include P. gingivalis porP (3) and Cytophaga hutchinsonii sprP (36). gldK, gldL, gldM, and gldN form an operon (8) and are arranged in this order in each organism that has these genes, whether or not they have a porP/sprF-like gene upstream. In addition to the porP/sprF-like genes associated with gldK, 12 porP/sprF-like genes lie immediately upstream of gldJ, which is similar in sequence to gldK and has been linked to both gliding motility and T9SS function (30). The association of porP/sprF-like genes with gldJ and gldK suggests that the gliding motility and T9SS proteins GldJ and GldK may have a functional relationship with PorP/SprF-like proteins.
DISCUSSION
T9SSs are prevalent in members of the phylum Bacteroidetes and secrete tens to hundreds of different proteins from a single strain. These proteins have type A or type B CTDs that target them to the secretion system (1, 8, 27, 28, 37). The features of type A CTDs have been functionally studied in F. johnsoniae and P. gingivalis (12, 21, 22), but type B CTDs have received little attention. The results presented here demonstrate that type B CTDs are common in the phylum Bacteroidetes and suggest that they target proteins for secretion by the T9SS.
Type B CTDs differ in sequence from type A CTDs (12, 19), and they also appear to differ functionally. Regions longer than 149 amino acids of the SprB type B CTD were required to target sfGFP for secretion. An additional protein, SprF, was also required for secretion. Wild-type levels of SprF were not sufficient to facilitate secretion of SP-sfGFP-CTDSprB expressed from a plasmid. However, efficient secretion occurred when SP-sfGFP-CTDSprB and SprF were expressed together from the same plasmid. F. johnsoniae encodes nine PorP/SprF-like proteins in addition to SprF. Many of the porP/sprF-like genes are arranged like sprF, immediately downstream of genes encoding proteins with type B CTDs. Expression of one of these, Fjoh_3951, was required for efficient secretion of sfGFP linked to the type B CTD of Fjoh_3952. SprF and Fjoh_3951 were not interchangeable. Expression of SprF did not facilitate secretion of SP-sfGFP-CTDFjoh_3952, and expression of Fjoh_3951 did not facilitate secretion of SP-sfGFP-CTDSprB. It appears that proteins carrying each of these CTDs required coexpression with the appropriate cognate PorP/SprF-like protein for secretion.
P. gingivalis has only a single protein, PorP, that is similar to F. johnsoniae SprF. P. gingivalis PorP is essential for T9SS-mediated secretion, including secretion of proteins with type A CTDs (3). In contrast, the individual F. johnsoniae PorP/SprF proteins that have been examined have more limited roles in secretion. SprF, for example, is only known to be required for secretion of SprB (7, 13). Further studies are needed to determine if any of the F. johnsoniae PorP/SprF-like proteins are critical for T9SS function or if collectively they perform a critical role that is masked by redundancy.
The exact functions of PorP, SprF, or any of the PorP/SprF-like proteins in secretion are not known. They are all predicted outer membrane β-barrel proteins, and outer membrane localization was confirmed for PorP (3) and SprF (Fig. 9 and 10). Recent results indicate that P. gingivalis PorP interacts directly or indirectly with the T9SS components PorK and PorM, both of which have large periplasmic domains (15). PorK, PorL, PorM, and PorN are each essential components of the T9SS, and they appear to form a complex (3, 15). The interactions of P. gingivalis PorP with PorK and PorM and the genetic arrangement of porP immediately upstream of the porK-porL-porM-porN operon (15) suggest that PorP may interact with this T9SS complex to perform its role in secretion.
After secretion by the T9SS, SprB is attached on the cell surface (3, 9, 27). The C-terminal 218-amino-acid region of SprB was sufficient to allow this attachment. We do not know how SprB interacts with the cell surface, but SprF and other outer membrane motility proteins such as SprC and SprD may be involved. Wild-type cells have large amounts of SprB on the cell surface, and only a small amount appears to be released from the cells (27). In contrast, we observed that cells expressing SP-sfGFP-CTDSprB not only had sfGFP on the cell surface but also released substantial amounts of soluble sfGFP into the spent medium (Fig. 2; see Fig. S3 in the supplemental material). F. johnsoniae produces many secreted proteases that could explain the release of sfGFP from the cell surface, although other explanations are possible.
SprB is a motility adhesin that moves rapidly along the cell surface, propelled by other components of the motility machinery (9, 27). The C-terminal 218-amino-acid region of SprB was sufficient to allow productive interaction with the motility machinery, resulting in movement. Identification of proteins that interact with this C-terminal region of SprB may help determine how it is linked to the gliding machinery. Since SprF is required for secretion of proteins carrying the C-terminal region of SprB, it may interact with this region. It may also remain attached to SprB after its secretion and perform additional roles in motility, although this has not been demonstrated. sprC, sprD, sprB, and sprF are part of an operon, and nonpolar mutations in each gene cause similar motility defects (13). The N-terminal region of the large motility protein SprD is similar in sequence to SprF (see Fig. S9 and S10 in the supplemental material), suggesting that SprD may also interact with SprB. Deletion of sprC and sprD resulted in decreased localization of SP-sfGFP-CTDSprB to the cell surface and lack of movement along the cell surface (Fig. 4), further suggesting that SprC or SprD may be involved in these processes. In addition to interacting with the motility machinery, SprB is thought to interact with the substratum to provide the traction needed for cell movement. Expression of sfGFP fused to C-terminal regions of SprB ranging from 218 to 1,182 amino acids failed to substantially enhance cell movement beyond the limited movement already exhibited by the ΔsprB mutant cells. Additional studies are needed to dissect the 6,497-amino-acid SprB protein and determine the regions required for productive interaction with the substratum.
The importance of T9SSs in the nutrition, physiology, motility, and virulence of members of the phylum Bacteroidetes has been demonstrated (1, 3, 19, 36, 38–40). Most of the proteins with easily predicted functions that are secreted by T9SSs have type A CTDs. In contrast, with the exception of the motility protein SprB, the functions of the many proteins with type B CTDs are not known. Many of these are large and have features that are found in some adhesins (see Table S2 in the supplemental material). They may allow cells to interact with each other and with surfaces. The results presented demonstrate that these proteins are common among members of the phylum Bacteroidetes and highlight the roles of type B CTDs and their associated outer membrane PorP/SprF-like proteins in T9SS-mediated secretion across the outer membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
F. johnsoniae ATCC 17061 (UW101) and its rpsL2 streptomycin-resistant derivative CJ1827 were the wild-type strains used in this study (29, 31, 41, 42). Unless indicated otherwise, F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (43). Escherichia coli strains were grown in lysogeny broth (LB) at 37°C (44). Strains and plasmids used in this study are listed in Table 1, and primers are listed in Table S4 in the supplemental material. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; erythromycin 100 μg/ml; kanamycin, 30 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| DH5αmcr | Strain used for general cloning | Life Technologies (Grand Island, NY) |
| HB101 | Strain used with pRK2013 for triparental conjugation | 52, 53 |
| F. johnsoniae strains | ||
| UW101 | Wild-type F. johnsoniae ATCC 17061T | 29, 42 |
| CJ1584 | Δ(sprC sprD sprB) | 13 |
| CJ1827 | rpsL2 Smr “wild-type” F. johnsoniae strain used in construction of deletion mutants | 31 |
| CJ1922 | ΔsprB rpsL2 (Smr) | 31 |
| CJ2122 | ΔgldK rpsL2 (Smr) | 8 |
| CJ2518 | ΔsprF rpsL2 (Smr) | This study |
| CJ2386 | gldJ548 rpsL2 (Smr) | 30 |
| CJ2491 | sfGFP::sprB rpsL2 (Smr) | This study |
| CJ2737 | ΔsprB ΔgldK rpsL2 (Smr) | This study |
| CJ2839 | ΔsprB gldJ548 rpsL2 (Smr) | This study |
| Plasmids | ||
| pCB3 | Encodes SP-sfGFP-CTDChiA(105AA); Apr (Tcr) | 12 |
| pCP11 | E. coli-F. johnsoniae shuttle plasmid; Apr (Emr) | 43 |
| pCP23 | E. coli-F. johnsoniae shuttle plasmid; Apr (Tcr) | 54 |
| pJJ01 | Construct used to delete gldK; Apr (Emr) | 8 |
| pJJ21 | Expresses periplasmic mCherry from pCP11; Apr (Emr) | This study |
| pRK2013 | Helper plasmid for triparental conjugation; IncP Tra+ Kmr | 53 |
| pRR51 | rpsL-containing suicide vector used for constructing deletion mutants; Apr (Emr) | 31 |
| pRR67 | Construct used to delete sprB; Apr (Emr) | 31 |
| pSK37 | sfGFP with stop codon cloned into pYT40; Apr (Tcr) | 12 |
| pSK45 | 1,992-bp region encoding 663 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(663AA) and SprF; Apr (Tcr) | This study |
| pSK50 | 1,992-bp region encoding 663 amino acids of CTDSprB inserted into pYT179; encodes SP-sfGFP-CTDSprB(663AA); Apr (Tcr) | This study |
| pSK51 |
1,107-bp region encoding 368 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(368AA) and SprF; Apr (Tcr) |
This study |
| pSK52 | 1,347-bp region encoding 448 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(448AA) and SprF; Apr (Tcr) | This study |
| pSK53 | 1,107-bp region encoding 368 amino acids of CTDSprB inserted into pYT179; encodes SP-sfGFP-CTDSprB(368AA); Apr (Tcr) | This study |
| pSK54 | 1,347-bp region encoding 448 amino acids of CTDSprB inserted into pYT179; encodes SP-sfGFP-CTDSprB(448AA); Apr (Tcr) | This study |
| pSK55 | 657-bp region encoding 218 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(218AA) and SprF; Apr (Tcr) | This study |
| pSK56 | Encodes SP-sfGFP-CTDSprB(218AA); Apr (Tcr) | 12 |
| pSK57 | 687-bp region encoding 228 amino acids of CTDFjoh_3952, 16-bp intergenic region and 1,032-bp region encoding Fjoh_3951 inserted into pYT179; encodes SP-sfGFP-CTDFjoh_3952(228AA) and Fjoh_3951; Apr (Tcr) | This study |
| pSK58 | 687-bp region encoding 228 amino acids of CTDFjoh_3952 inserted into pYT179; encodes SP-sfGFP-CTDFjoh_3952(228AA); Apr (Tcr) | 12 |
| pSK59 | 450-bp region encoding 149 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(149AA) and SprF; Apr (Tcr) | This study |
| pSK60 | 450-bp region encoding 149 amino acids of CTDSprB inserted into pYT179; encodes SP-sfGFP-CTDSprB(149AA); Apr (Tcr) | This study |
| pSK61 | 3,549-bp region encoding 1,182 amino acids of CTDSprB, 16-bp intergenic region and 1,178-bp region encoding SprF inserted into pYT179; encodes SP-sfGFP-CTDSprB(1182AA) and SprF; Apr (Tcr) | This study |
| pSK62 | 3,549-bp region encoding 1,182 amino acids of CTDSprB inserted into pYT179; encodes SP-sfGFP-CTDSprB(1182AA); Apr (Tcr) | 12 |
| pSK64 | 762-bp region encoding 238 amino acids of Fjoh_1123 inserted into pYT179; encodes SP-sfGFP-CTDFjoh_1123(238AA); Apr (Tcr) | This study |
| pSK68 | 1,294-bp region encoding SprF inserted into pSK58; encodes SP-sfGFP-CTDFjoh_3952(228AA) and SprF; Apr (Tcr) | This study |
| pSK69 | 1,032-bp region encoding Fjoh_3951 inserted into pSK56; encodes SP-sfGFP-CTDSprB(218AA) and Fjoh_3951; Apr (Tcr) | This study |
| pSK82 | Encodes SP-sfGFP-CTDAmyB(99AA); Apr (Tcr) | 12 |
| pTB263 | Expresses sfGFP; Apr | 55 |
| pYT40 | remA promoter, start codon, and N-terminal signal peptide-encoding region inserted into pCP23; Apr (Tcr) | 12 |
| pYT108 | Plasmid used to construct pYT296; Apr (Emr) | This study |
| pYT112 | Plasmid used to construct pYT296; Apr (Emr) | This study |
| pYT179 | sfGFP amplified without stop codon and cloned into pYT40; encodes SP-sfGFP; Apr (Tcr) | 12 |
| pYT296 | Plasmid used to insert the gene encoding sfGFP into chromosomal sprB to construct CJ2491, which produces full-length SprB with sfGFP inserted after the signal peptide; Apr (Emr) | This study |
| pYT316 | Construct used to delete F. johnsoniae sprF; Apr (Emr) | 45 |
Antibiotic resistance phenotypes are as follows: ampicillin, Apr; erythromycin, Emr; kanamycin, Kmr; streptomycin, Smr; tetracycline, Tcr. The antibiotic resistance phenotypes given in parentheses are those expressed in F. johnsoniae but not in E. coli. The antibiotic resistance phenotypes without parentheses are those expressed in E. coli but not in F. johnsoniae.
Construction of deletion mutants.
The sprF deletion mutant CJ2518 was generated using pYT316 as described previously (45) except that the deletion was introduced into F. johnsoniae CJ1827. The ΔsprB ΔgldK double mutant CJ2737 was constructed using pJJ01 to delete gldK as described previously (8) except that the deletion was introduced into the ΔsprB mutant CJ1922 (31). The ΔsprB gldJ548 double mutant was constructed using pRR67 to delete sprB as described previously (31) except that the deletion was introduced into the gldJ548 truncation mutant CJ2386 (30).
Insertion of the sfGFP gene into chromosomal sprB.
The sfGFP-encoding gene was inserted into chromosomal sprB essentially as previously described for myc tag insertions (7). A 2.3-kbp fragment starting upstream of sprB and extending through base 228, numbered from the A of the start codon, was amplified by PCR using Phusion DNA polymerase (New England BioLabs, Ipswich, MA) and primers 1356 (engineered XbaI site) and 1302 (engineered BamHI site). This fragment and pRR51 were digested with XbaI and BamHI and ligated together to form pYT108. A second fragment starting at base 229 of sprB and extending 1.9 kbp downstream was amplified using primers 1358 (engineered SalI site) and 1305 (engineered SphI site), digested with SalI and SphI, and ligated into pYT108 that had been digested with the same enzymes to generate pYT112. A 75- bp region encoding sfGFP with an upstream linker (peptide sequence LEGPAGL) and a downstream linker (peptide sequence GGSGGGSG) was amplified from pTB263 using primer 1749 (engineered XbaI site) and primer 1366 (engineered SalI site). This fragment and pYT112 were digested with XbaI and SalI and ligated together, resulting in insertion of the sfGFP-encoding fragment between the sprB fragments as pYT296. Plasmid pYT296 was introduced into F. johnsoniae strain CJ1827 by triparental conjugation, and a mutant that encoded full-length SprB with sfGFP inserted after the signal peptide was isolated as previously described (7) and confirmed by PCR amplification and DNA sequencing.
Generation of plasmids that express fluorescent proteins with signal peptides at the N termini and with regions of SprB CTDs at the C termini.
Regions of DNA encoding the C terminus of SprB (CTDSprB) were introduced into plasmid pYT179, which expressed the N-terminal signal peptide of RemA fused to sfGFP (SP-sfGFP) (12), resulting in plasmids that produce SP-sfGFP-CTDSprB. These include pSK56 [expresses SP-sfGFP-CTDSprB(218AA)] and pSK62 [expresses SP-sfGFP-CTDSprB(1182AA)], which have been previously described (12). Additional plasmids expressing SP-sfGFP-CTDSprB(149AA) (pSK60), SP-sfGFP-CTDSprB(368AA) (pSK53), SP-sfGFP-CTDSprB(448AA) (pSK54), and SP-sfGFP-CTDSprB(663AA) (pSK50) were constructed in a similar way using the primers listed in Table S4. In each case a region encoding the C terminus of SprB was amplified and inserted into the XbaI and SphI sites of pYT179 to generate a construct encoding SP-sfGFP-CTDSprB. Plasmids that also included sprF downstream from each of the SP-sfGFP-CTDSprB-encoding constructs were also prepared. For example, a 657-bp fragment encoding 218 amino acids of CTDSprB, the 17-bp intergenic region, and a 1,294-bp fragment carrying the sprF gene was amplified by PCR primers 1843 (engineered XbaI site) and 955 (engineered SphI site). This fragment was introduced into XbaI- and SphI-digested pYT179 to generate pSK55. Plasmids encoding SP-sfGFP-CTDSprB with 149, 368, 448, 663, and 1,182 amino acids from the C terminus of SprB followed by and coexpressed with sprF (pSK59, pSK51, pSK52, pSK45, and pSK61, respectively) were constructed similarly using the primers listed in Table S4. A plasmid encoding mCherry as a periplasmic marker was generated by cloning the chiA promoter and N-terminal signal peptide region fused to the mCherry gene from pSSK54 (19) into pCP11. A 1,254-bp fragment was amplified from pSSK54 with primers 2429 (engineered XbaI site) and 2430 (engineered SalI site). This fragment and pCP11 were digested using XbaI and SalI and ligated together to form pJJ21.
Generation of plasmids that express SP-sfGFP fused to regions of the CTDs of Fjoh_3952 and Fjoh_1123.
A 687-bp fragment encoding the C-terminal 228 amino acids of the type B CTD-containing protein Fjoh_3952 and the entire porP/sprF-like gene product Fjoh_3951 was amplified and cloned into pYT179 using primers 1868 (engineered XbaI site) and 1869 (engineered SphI site) to generate plasmid pSK57.
Plasmid pSK69, which encodes both SP-sfGFP-CTDSprB and Fjoh_3951, was also constructed, as was plasmid pSK68, which encodes both SP-sfGFP-CTDFjoh_3952 and SprF. Fjoh_3951 was amplified using primers 1892 (engineered SphI site) and 1969 (engineered SphI site) and cloned into the SphI site of pSK56, generating pSK69. Similarly, sprF was amplified using primers 1883 (engineered SphI site) and 955 (engineered SphI site) and cloned into the SphI site of pSK58 (12) to generate pSK68. Plasmids were confirmed by sequencing. A region spanning 762 bp of CTDFjoh_1123 was also cloned into pYT179 using primers 1881 (engineered XbaI site) and 1182 (engineered SphI site), generating plasmid pSK64, which encodes SP-sfGFP-CTDFjoh_1123.
Western blot analyses.
F. johnsoniae cells were grown to early stationary phase in CYE at 25°C with shaking as previously described (12). Cells were pelleted by centrifugation at 22,000 × g for 15 min, and the culture supernatant (spent medium) was separated. For whole-cell samples, the cells were suspended in the original culture volume of phosphate-buffered saline (PBS), consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 2 mM KH2PO4 (pH 7.4). Equal amounts of spent media and whole cells were boiled in SDS-PAGE loading buffer for 10 min. Proteins were separated by SDS-PAGE, and Western blot analyses were performed as previously described (46) except that polyvinylidene difluoride (PVDF) membranes were used instead of nitrocellulose membranes. Equal amounts of each sample based on the starting material were loaded in each lane. For cell extracts this corresponded to 10 μg protein, whereas for spent medium this corresponded to the equivalent volume of spent medium that contained 10 μg cell protein before the cells were removed. Anti-GFP (0.5 mg per ml; GenScript, Piscataway, NJ) was used at a dilution of 1:3,000 to detect sfGFP in Western blots, and anti-mCherry (BioVision Incorporated, Milpitas, CA) was used at a dilution of 1:5,000 to detect mCherry. Affinity-purified polyclonal antibodies against the SprF peptide QSISNCPCTQSPVHD were produced by Biomatik Corporation (Cambridge, ON, Canada) and were used at a dilution of 1:3,000. Polyclonal antisera against GldL and GldJ were previously described (8, 47).
Localization of SprF.
Wild-type and mutant cells of F. johnsoniae were grown to early stationary phase in MM medium (48) at 25°C with shaking. Cells were harvested and washed once in PBS by centrifugation for 10 min at 4,000 × g, suspended in PBS, and adjusted to an optical density at 600 nm (OD600) of 1.5, and EDTA-free Halt protease inhibitor cocktail was added (Thermo Fisher Scientific, Rockford, IL). The cells were disrupted by using a French pressure cell and fractionated into soluble, inner membrane (Sarkosyl-soluble), and outer membrane (Sarkosyl-insoluble) fractions essentially as described previously (49). Equal amounts of each fraction based on 20 μg protein of the starting material (whole cells) were separated by SDS-PAGE, and Western blotting was performed as described above.
Proteinase K treatment of cells to determine the localization of SprF.
Cells of F. johnsoniae were grown in CYE at 25°C with shaking. Cells were collected, washed and suspended in 20 mM sodium phosphate–10 mM MgCl2 (pH 7.5), and diluted to an OD600 reading of 1.5. To examine SprF, proteinase K was added to the intact cells to a final concentration of 1 mg/ml and incubated at 25°C with gentle mixing. In each case, an identical sample was lysed using a French pressure cell, unbroken cells and debris were removed by centrifugation, and then proteinase K was added as described above. At 0 and 2 h, 150 μl of cells or lysed cells was sampled, 10 mM phenylmethylsulfonyl fluoride was added, and the samples were boiled for 1 min to stop digestion. SDS-PAGE loading buffer was added, and the samples were boiled for another 7 min. Control samples that were not exposed to proteinase K were also included. Equal volumes were separated by SDS-PAGE and transferred to PVDF membranes, and proteins were detected with antiserum against SprF.
Microscopic observation of cell movement.
The movement of F. johnsoniae cells on glass was examined by phase-contrast microscopy at 25°C. Cells were grown in MM at 25°C without shaking until late exponential phase. Motility on glass was analyzed using liquid-filled tunnel slides prepared as described previously (7), using Nichiban NW-5 double-sided tape (Nichiban Co, Tokyo, Japan) to hold a glass coverslip over a glass slide. Cells suspended in MM medium were introduced into tunnel slides, incubated for 5 min, and observed for motility using an Olympus BH-2 phase-contrast microscope. Images were recorded using a Photometrics Cool-SNAPcf2 camera and analyzed using MetaMorph software (Molecular Devices, Downingtown, PA) and ImageJ version 1.52i (http://rsb.info.nih.gov/ij/). Rainbow traces of cell movements were made using ImageJ and the macro Color FootPrint (9).
Analysis of wild-type and mutant cells for surface localization and movement of sfGFP.
Cell surface localization of sfGFP was examined by immunofluorescence microscopy. Cells were grown in MM at 25°C without shaking until late exponential phase. One milliliter of cell culture was centrifuged at 1,000 × g for 10 min, and the cell pellet was suspended and washed 2 times in 500 μl of TC buffer (43). The cells were pelleted by centrifugation and suspended in 50 μl of TC plus 2% bovine serum albumin (BSA). Cells were exposed to 1 μl of polyclonal anti-GFP rabbit IgG (0.5 mg/ml; GenScript, Piscataway, NJ) for 30 min at room temperature and then pelleted (1,000 × g for 10 min), washed 1 time with 500 μl TC buffer, suspended in 50 μl TC plus 2% BSA, and exposed to 1 μl of F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa Fluor 594 (2 mg/ml; Invitrogen, Carlsbad, CA). Cells were incubated for 30 min in the dark and collected by centrifugation (1,000 × g for 10 min). Cells were washed once with 500 μl of TC, suspended in 200 μl of TC, and kept in the dark until analysis (no more than 1 h after labeling). Samples were spotted on glass slides previously coated with a thin layer of 1.0% agarose dissolved in TC and allowed to sit 30 s before applying coverslips. Samples were observed using a Nikon Eclipse 50i microscope (Nikon Instruments, Inc., Melville, NY) with an ExFo XL120 mercury lamp, a Y-2E/C TR filter to detect red fluorescence, and an Endow 41017 GFP filter to detect green fluorescence (Chroma Technology Corp., Bellows Falls, VT). Epifluorescence and phase-contrast images were recorded separately with a Hamamatsu ORCA-Flash4.0 LT+ camera through a Nikon CFI60 Plan Fluor phase-contrast DLL 100×/1.3-numerical-aperture objective using NIS-Elements Advanced Research software (Nikon Instruments, Inc., Melville, NY) and ImageJ. Exposure times for all fluorescence images were 33 ms.
GIMP (https://www.gimp.org/) was used to generate overlay images showing red fluorescence over phase contrast. Briefly, the phase-contrast images were inverted and the contrast adjusted so the cells were light gray on a black background. These were then colorized to dark turquoise (RGB 12, 28, 28) using the “colorize” command. Red fluorescence was similarly processed without inversion and colorized red (RGB 204, 0, 0). The black background of red fluorescence images was then made transparent and overlaid on the corresponding colorized phase-contrast images. Surface localization statistics were based on examination of overlay images of 150 cells in three replicate experiments (450 cells total).
To examine cells for movement of cell surface-localized sfGFP, cells labeled with antibodies as described above were introduced into a tunnel slide and incubated for 3 min before imaging. Cells were imaged by simultaneous low-light phase-contrast microscopy and fluorescence microscopy, and images were recorded as described above. Suspended cells were examined to avoid confusion resulting from the gliding movement of cells over surfaces. A total of 450 cells for each strain were observed for 20 s each in three replicate experiments (150 cells per experiment). Cells were counted as labeled if a fluorescent signal remained associated with the cell for the entire 20-s observation period. Signals were considered to have moved if they were displaced by 2.5 μm or more with respect to a cell pole at any time during observation.
Bioinformatic analyses.
Genome sequences were analyzed for T9SS genes that encode proteins that belong to appropriate TIGRFAM multiple-sequence alignment families (50). This was accomplished using the Integrated Microbial Genomes (IMG version 4.0.1 [https://img.jgi.doe.gov/]) Function Profile Tool to examine the genomes for sequences predicted to encode orthologs of GldK (TIGR03525), GldL (TIGR03513), GldM (TIGR03517), GldN (TIGR03523), and SprA (TIGR04189). The genomes were also examined for genes encoding proteins with type A CTDs (TIGR04183) or type B CTDs (TIGR04131) and for genes encoding PorP/SprF-like proteins (TIGR03519) in the same way. In each case, the trusted cutoffs assigned by the J. Craig Venter Institute (JCVI) were used to identify family members. These cutoffs allow identification of the vast majority of family members, with vanishingly few false positives (50). F. johnsoniae proteins related to P. gingivalis W83 PG1058 were identified by BLASTP analysis (51).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grant MCB-1516990 from the National Science Foundation.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00218-19.
REFERENCES
- 1.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 5.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson SS, Glocka PP, Agarwal S, Grimm DP, McBride MJ. 2007. Flavobacterium johnsoniae SprA is a cell-surface protein involved in gliding motility. J Bacteriol 189:7145–7150. doi: 10.1128/JB.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. 2012. Flavobacterium johnsoniae RemA is a mobile cell-surface lectin involved in gliding. J Bacteriol 194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell-surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava A, Roland T, Berg HC. 2016. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys J 111:1008–1013. doi: 10.1016/j.bpj.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. doi: 10.1128/JB.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. 2017. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J Bacteriol 199:e00884-16. doi: 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. doi: 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent MS, Canestrari MJ, Leone P, Stathopulos J, Ize B, Zoued A, Cambillau C, Kellenberger C, Roussel A, Cascales E. 2017. Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J Biol Chem 292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leone P, Roche J, Vincent MS, Tran QH, Desmyter A, Cascales E, Kellenberger C, Cambillau C, Roussel A. 2018. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat Commun 9:429. doi: 10.1038/s41467-017-02784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivastava A, Lele PP, Berg HC. 2015. A rotary motor drives Flavobacterium gliding. Curr Biol 25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharade SS, McBride MJ. 2014. The Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen KA, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol 189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J Bacteriol 193:132–142. doi: 10.1128/JB.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds E. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thøgersen IB, Gao J, Kwan AH, Trewhella J, Dubin G, Gomis-Rüth FX, Nguyen K-A, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6:23123. doi: 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thøgersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen K-A, Potempa J, Gomis-Rüth FX. 2016. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci Rep 6:37708. doi: 10.1038/srep37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification and cell surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 29.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston JJ, Shrivastava A, McBride MJ. 2018. Untangling Flavobacterium johnsoniae gliding motility and protein secretion. J Bacteriol 200:e00362-17. doi: 10.1128/JB.00362-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes RG, Pucker HG, McBride MJ. 2011. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant motility genes remF, remG, remH, and remI. J Bacteriol 193:2418–2428. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu CS, Chen YC, Lu CH, Hwang JK. 2006. Prediction of protein subcellular localization. Proteins 64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 33.Heath JE, Seers CA, Veith PD, Butler CA, Nor Muhammad NA, Chen YY, Slakeski N, Peng B, Zhang L, Dashper SG, Cross KJ, Cleal SM, Moore C, Reynolds EC. 2016. PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11:e0164313. doi: 10.1371/journal.pone.0164313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Suarez ML, Samsudin F, Piggot TJ, Bond PJ, Khalid S. 2016. Full-length OmpA: structure, function, and membrane interactions predicted by molecular dynamics simulations. Biophys J 111:1692–1702. doi: 10.1016/j.bpj.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samsudin F, Ortiz-Suarez ML, Piggot TJ, Bond PJ, Khalid S. 2016. OmpA: a flexible clamp for bacterial cell wall attachment. Structure 24:2227–2235. doi: 10.1016/j.str.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. doi: 10.1007/s00253-013-5355-2. [DOI] [PubMed] [Google Scholar]
- 37.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 38.Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, Barbier P, Gullstrand CW, Roets JE, Powers JL, Kulkarni SS, Erbes DH, Garcia JC, Nie P, McBride MJ. 2017. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium columnare. Appl Environ Microbiol 83:e01769-17. doi: 10.1128/AEM.01769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo YQ, Hu D, Guo J, Wang T, Xiao YC, Wang XL, Li SW, Liu M, Li ZL, Bi DR, Zhou ZT. 2017. Riemerella anatipestifer type IX secretion system is required for virulence and gelatinase secretion. Front Microbiol 8:2553. doi: 10.3389/fmicb.2017.02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kita D, Shibata S, Kikuchi Y, Kokubu E, Nakayama K, Saito A, Ishihara K. 2016. Involvement of the type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl Environ Microbiol 82:1756–1766. doi: 10.1128/AEM.03452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang LYE, Pate JL, Betzig RJ. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol 159:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride MJ, Braun TF. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol 186:2295–2302. doi: 10.1128/jb.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride MJ, Kempf MJ. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol 178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 45.Zhu Y, Thomas F, Larocque R, Li N, Duffieux D, Cladiere L, Souchaud F, Michel G, McBride MJ. 2017. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ Microbiol 19:2164–2181. doi: 10.1111/1462-2920.13699. [DOI] [PubMed] [Google Scholar]
- 46.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun TF, McBride MJ. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol 187:2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, McBride MJ, Subramaniam S. 2007. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunnicutt DW, McBride MJ. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J Bacteriol 182:911–918. doi: 10.1128/jb.182.4.911-918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolivar F, Backman K. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol 68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 53.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci U S A 94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uehara T, Dinh T, Bernhardt TG. 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.