Abstract
The microtubule-associated protein tau undergoes aberrant modification resulting in insoluble brain deposits in various neurodegenerative diseases, including frontotemporal dementia (FTD), progressive supranuclear palsy, and corticobasal degeneration. Tau aggregates can form in different cell types of the central nervous system (CNS) but are most prevalent in neurons. We have previously recapitulated aspects of human FTD in mouse models by overexpressing mutant human tau in CNS neurons, including a P301S tau variant in TAU58/2 mice, characterized by early-onset and progressive behavioral deficits and FTD-like neuropathology. The molecular mechanisms underlying the functional deficits of TAU58/2 mice remain mostly elusive. Here, we employed functional genomics (i.e. RNAseq) to determine differentially expressed genes in young and aged TAU58/2 mice to identify alterations in cellular processes that may contribute to neuropathy. We identified genes in cortical brain samples differentially regulated between young and old TAU58/2 mice relative to nontransgenic littermates and by comparative analysis with a dataset of CNS cell type–specific genes expressed in nontransgenic mice. Most differentially-regulated genes had known or putative roles in neurons and included presynaptic and excitatory genes. Specifically, we observed changes in presynaptic factors, glutamatergic signaling, and protein scaffolding. Moreover, in the aged mice, expression levels of several genes whose expression was annotated to occur in other brain cell types were altered. Immunoblotting and immunostaining of brain samples from the TAU58/2 mice confirmed altered expression and localization of identified and network-linked proteins. Our results have revealed genes dysregulated by progressive tau accumulation in an FTD mouse model.
Keywords: Tau protein (Tau), neurodegenerative disease, mouse, gene expression, neurobiology, transgenic mice
Introduction
The microtubule-associated protein tau (generally termed “tau”) is encoded by the MAPT gene on human chromosome 17, which gives rise to different isoforms by alternative splicing (1). The tau protein binds to the microtubule network throughout neurons. Although it is enriched in the axon shaft, we have shown that under physiological conditions it is also present at post-synaptic sites (2). Tau harbors a particularly high number of 81 known and putative phosphorylation sites that regulate many of its functions, including binding to microtubules and other partners (1, 3, 4). In diseases commonly referred to as tauopathies, tau becomes increasingly phosphorylated due to an imbalance of kinase and phosphatase activity, resulting in hyperphosphorylation (5). Hyperphosphorylated tau accumulates in the soma and dendrites of neurons and is prone to form fibrillar aggregates (= paired helical filaments; PHFs),2 eventually presenting as neurofibrillary tangles (NFTs) (6). Tau hyperphosphorylation and deposition is also found in other CNS cell types, including oligodendrocytes and astrocytes in certain tauopathies (7). Tauopathies comprise a significant number of neurodegenerative diseases, including Alzheimer's disease (AD) and frontotemporal dementia (FTD). FTD belongs to a collection of neurodegenerative conditions clinically characterized by variable behavioral, language, motor, and cognitive deficits (8, 9). These conditions furthermore include Pick's disease, corticobasal degeneration, progressive supranuclear palsy, and globular glial tauopathy (7). Familial forms of FTD have been linked to mutations in a range of gene loci, including MAPT (10). Pathogenic MAPT mutations have been instrumental in developing transgenic mouse models of tauopathies that present with both FTD and AD-like neuropathology and functional deficits (11, 12). Although these mouse models have made significant contributions to the understanding of processes underlying neurodegeneration and to the testing and development of potential therapeutic approaches (13, 14), the mechanistic insights gained remain incomplete.
We have recently reported a new P301S tau transgenic mouse line, TAU58/2, that shows progressively increasing tau pathology together with early-onset functional deficit, including FTD-like behavioral changes and compromised motor performance (15, 16). The TAU58/2 model of FTD has since been used by others (17, 18). It would therefore be of general interest to provide insight into systems changes in TAU58/2 mice and other P301S tau transgenic mouse models of FTD.
Here, we provide insight into system changes in TAU58/2 mice and report CNS cell type–annotated differential gene expression in young and aged animals. Validation at the protein level revealed previously unknown changes to pre-synaptic factors, indicating that progressive alterations of this neuronal compartment is associated with tau pathology in this mouse model of FTD.
Results
Differential gene expression in young and old TAU58/2 mice
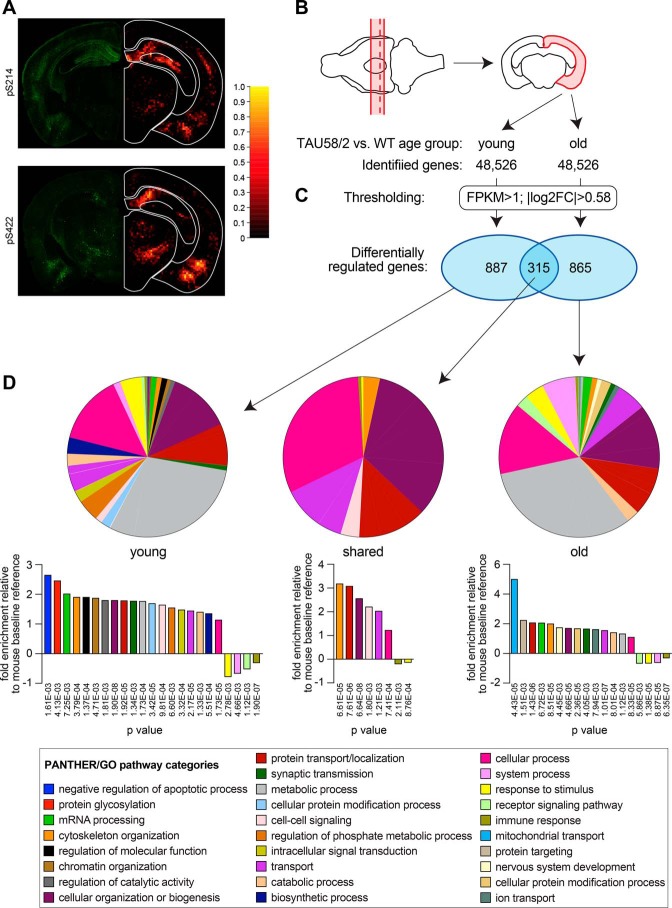
Transgenic P301S tau expression is associated with hyperphosphorylation of tau in several brain regions in TAU58/2 mice (Fig. 1A). We have previously shown that neuropathology, including the hyperphosphorylation of tau and presence of NFTs, progressively increases as TAU58/2 mice age (16). Similarly, functional deficits such as disinhibition and motor impairments progress with time (15, 16). The underlying molecular mechanisms of the functional deficits associated with a progressive tau pathology in TAU58/2 mice remain unknown. To obtain an unbiased insight into the molecular changes associated with transgenic P301S tau expression over time, we performed quantitative polyadenylated RNAseq (RNAseq) of cortical brain samples from both young (3 months: early pathology) and old (10 months: progressed pathology) TAU58/2 mice and their nontransgenic littermates (Fig. 1B). This allowed us to compare differential gene regulation at individual ages, between ages, and between genotypes (i.e. TAU58/2 versus nontransgenic mice). 48,526 genes were identified for subsequent quantitative comparison in each group. After thresholding (fragments per kilobase million (FPKM) >1 and ABS (log2FC) >0.58) for three complete sequences per gene and group, we found 1,202 differentially-regulated genes in young TAU58/2 mice compared with their nontransgenic littermate controls (Fig. 1C and Fig. S1 and Table S1). For comparison, 1,180 genes were differentially regulated between old TAU58/2 and nontransgenic control mice. There was an overlap of 315 differentially-regulated genes between young and old TAU58/2 gene sets.
Figure 1.
mRNA expression profiling in P301S tau transgenic TAU58/2 mice. A, left, immunofluorescence staining with phosphorylation site-specific tau antibodies to serine 214 (pS214; green) and 422 (pS422; green) in brains from old TAU58/2 mice. Right, heat maps of staining intensity from six different brains. B, schematic of dissection procedure to obtain corresponding cortical brain samples (red outlines) from old and young TAU58/2 mice and nontransgenic littermate controls for mRNA sequencing. Red broken line indicates level of brain section shown in A. C, thresholding of pairwise expression of identified genes (FPKM >1, |log2FC| >0.58, p < 0.05) identified unique and shared differentially-regulated genes (blue ellipses) in young and old TAU58/2 mice. D, PANTHER pathway mapping of differentially-regulated genes in young and old TAU58/2 mice revealed a broad range of potentially altered cellular and molecular processes, including their representation relative to a C57Bl/6 baseline reference.
Instead of analyzing individual differentially deregulated genes, we were interested in system changes associated with tau pathology in young and old TAU58/2 mice. Therefore, we subjected the differentially-regulated genes from either young or old TAU58/2 or those found in both age groups to gene ontology analysis using the online PANTHER software version 13.1 to predict altered pathways (Fig. 1D) (19). In addition to gene annotation to distinct pathways, PANTHER also provides the statistical representation of each pathway relative to a mouse baseline dataset. Differentially regulated genes from both young and old TAU58/2 mice were annotated to a wide range of cellular pathways with over-representations of several pathways relative to the PANTHER reference dataset. For young TAU58/2 mice, this included synaptic transmission and cell–cell signaling, and vesicle-mediated transport pathways were over-represented relative to the PANTHER reference dataset, while the immune response pathways were under-represented. For old TAU58/2 mice, the mitochondrial transport pathway was most prominently over-represented. The overlapping and differentially-regulated genes between young and old TAU58/2 mice were over-represented by cellular organization and transport pathways.
A similar complex picture of gene annotations to GOTERMS and pathways was obtained when we subjected the gene data sets of differentially-regulated genes from TAU58/2 mice to gene ontology analysis using the online DAVID software version 6.8 (data not shown).
Taken together, we found a large number of differentially-regulated genes in young and old TAU58/2 mice with overlap in the genes of the cytoskeleton organization, protein transport, cellular organization, cell–cell signaling, transport, cellular process, immune response, and stimulus response pathways. Accordingly, gene ontology analysis provided a wide spectrum of predicted changed cellular/molecular pathways. Given the broad spectrum of predicted systems affected in both age groups, further stratification was undertaken to obtain increased granularity while keeping bias to a minimum. Note that a separate analysis of differentially-regulated genes that reached significance (p < 0.05; Fig. S1) did not reveal deregulation of relevant distinct pathways or networks (data not shown). Hence, we explored an alternative approach for reducing complexity as outlined below.
CNS cell type–specific gene annotation of differentially expressed genes in TAU58/2 mice
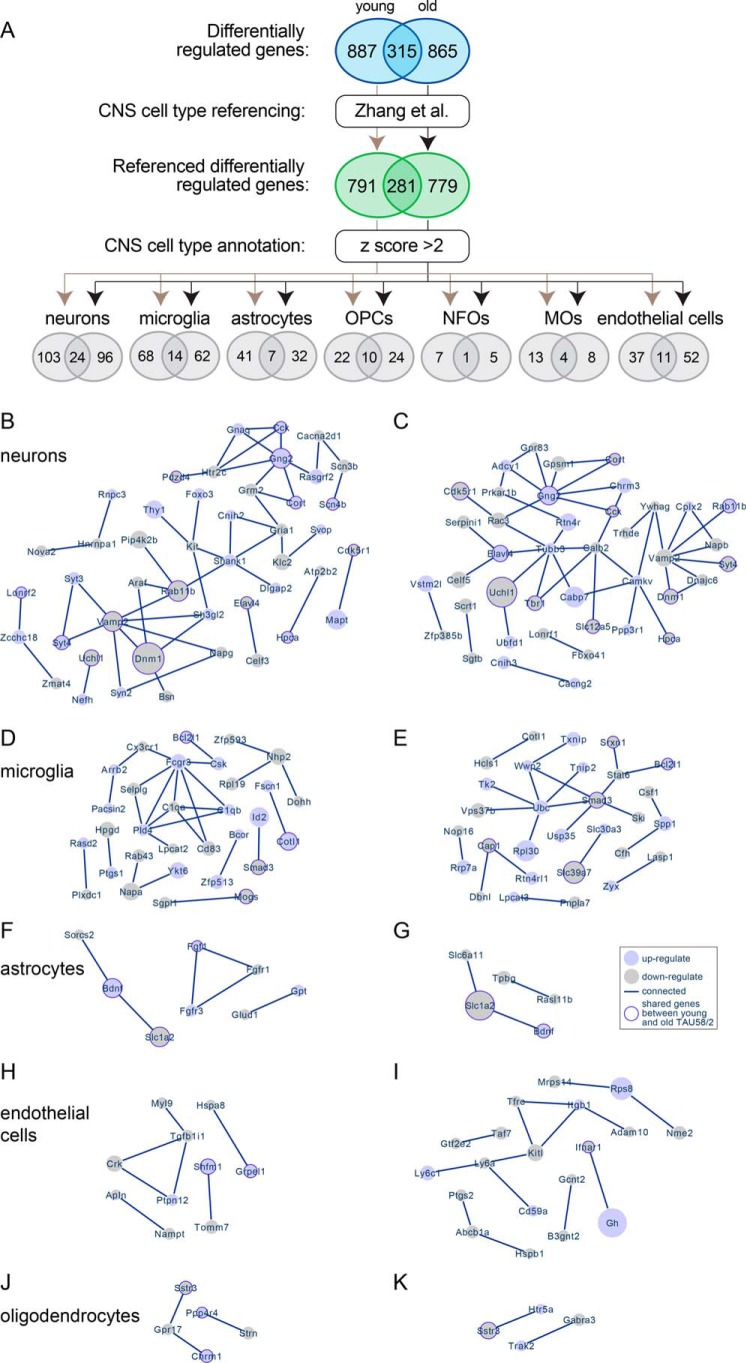
Given that the RNA for sequencing was extracted from tissue homogenates, our analysis may have suffered from differentially-regulated genes being annotated to common pathways despite the possibility that these genes are not expressed in the same cell type. Therefore, we decided to reference our differentially-regulated genes from young and old TAU58/2 mice to the dataset of CNS cell type–specific gene expression (20). 1,072 (89%) and 1,060 (90%) of the differentially-regulated genes from young and old TAU58/2 mice were covered by the CNS cell type–specific gene expression dataset, respectively (Fig. 2A). This included 281 (89%) differentially-regulated genes that overlapped between young and old TAU58/2 mice.
Figure 2.
Annotation of differentially-regulated genes to CNS cell types. A, process schematic for annotation of differentially-regulated genes from young and old TAU58/2 mice to CNS cell types by referencing to a previously published dataset (Zhang et al. (20)). Ellipses indicate number of genes at each step, including overlapping differentially expressed genes between young and old TAU58/2 mice. B and C, predicted networks of differentially expressed genes annotated to neurons in young (B) and old (C) TAU58/2 mice. Light-blue–filled circles indicate up-regulated genes, and light-gray–filled circles indicate down-regulated genes. The size of filled circles indicates relative degree of deregulation (large = highly deregulated and small = moderate deregulation as compared with gene expression in nontransgenic controls). Dark-blue borders indicate deregulation in both young and old TAU58/2 mice. Lines indicate connection predicted by STRING analysis. D and E, predicted networks of differentially-regulated genes annotated to microglia in young (D) and old (E) TAU58/2 mice. F and G, predicted networks of differentially expressed genes annotated to astrocytes in young (F) and old (G) TAU58/2 mice. H and I, predicted networks of differentially-regulated genes annotated to endothelial cells in young (H) and old (I) TAU58/2 mice. J and K, predicted networks of differentially expressed genes annotated to oligodendrocytes in young (J) and old (K) TAU58/2 mice.
Next, we sorted the referenced differentially-regulated genes from both age groups against CNS cell-specific data sets (neurons, astrocytes, microglia, and oligodendrocyte precursor cells (OPCs), newly-formed oligodendrocytes (NFOs), myelinating oligodendrocytes (MOs), and endothelial cells) and applied a cutoff criterion of a z score of >2 (Fig. 2A) (20). This resulted in cell type–specific expression lists of 271 (25%) and 279 (26%) from the differentially-regulated genes of young and old TAU58/2 mice, respectively (Fig. 2A). In both young and old TAU58/2 mice, the highest number of cell-specific differentially-regulated genes were annotated to neurons, consistent with cell-intrinsic effects of neuronal transgene expression. This was followed by differentially-regulated genes annotated to microglia, consistent with a microgliosis in TAU58/2 mice (16). Interestingly, Iba1 staining showed increased numbers of microglia only in aged but not young TAU58/2 mice (Fig. S2), suggesting differential gene regulation in microglia prior to overt microgliosis. For comparison, microgliosis was more prominent in the brainstem of TAU58/2 mice. Numbers of differentially-regulated genes in astrocytes, oligodendrocytes (= combined OPCs, NFOs, and MOs), and endothelial cells in young TAU58/2 mice were similar. The number of differentially-regulated genes annotated to astrocytes and endothelial cells were 22 and 41% lower, respectively, in old versus young TAU58/2 mice. Accordingly, TAU58/2 mice showed moderate, progressive astrocytosis, in contrast to strong astrocytosis in the brainstem (Fig. S2). Taken together, differentially-regulated genes from young and old TAU58/2 mice were predominantly expressed in specific CNS cell types.
CNS cell type–specific gene clusters indicate explicitly altered functional pathways
Next, we subjected the annotated differentially-regulated genes for each CNS cell type and age group to STRING protein–protein interaction network analysis for prediction of altered molecular pathways and gene clusters (21). For neurons, STRING analysis revealed one large network of interconnected genes in young TAU58/2 mice (Fig. 2B and Fig. S3A). This network contained two major components: genes associated with synaptic vesicle trafficking (Vamp2, Syt3/4, Syn2, Rab11b, and Dnm1) and with glutamate receptor signaling (Gria1, Grm2, Gng2, and Shank1). Similarly, STRING analysis displayed one large network in old TAU58/2 mice, sharing association with synaptic vesicle trafficking (Vamp2, Syt3/4, Rab11b, and Dnm1) with young TAU58/2 mice (Fig. 2C and Fig. S3B). In contrast to young TAU58/2 mice, glutamate receptor signaling was no longer prominent (Gng2), but other receptor signaling-associated (Gpr83, Prkar1b, and Gpsm1) and tubulin-associated (Tubb3) genes were part of the network in old TAU58/2 mice.
For microglia, STRING analysis identified one prominent yet different network of interacting genes in young and old TAU58/2 mice, as well as some nonoverlapping and less connected gene clusters (Fig. 2, D and E, and Fig. S4). In young TAU58/2 mice, the main microglial cluster of differentially-regulated genes was associated with immune (Cx3cr1, Fcgr3, Selplg, and Csk) or complement (C1qa and C1qb) activation. Notably, we found complement factors C1q and C3 stained phosphorylated tau-harboring neurons in the brains of young and aged TAU58/2 mice, showing initial signs of C3 foci in young TAU58/2 and marked cytoplasmic C1q and C3 accumulation in aged TAU58/2 (Fig. S5). The prominent gene cluster in old TAU58/2 mice was associated with ubiquitination (Ubc and Wwp2) and cytokine signaling (Smad3, Stat6, and Bcl2l1). We noted that Ubc took a central position in the large gene cluster in old TAU58/2 mice. Because of its central role in protein regulation, this may contribute to a clustering of otherwise functionally unrelated proteins. Differential regulation of Smad3 is shared between microglia-annotated genes of young and old TAU58/2 but was only part of the prominent cluster in old mice.
For astrocytes, STRING analysis revealed less prominent gene clusters in both young and old TAU58/2 mice (Fig. 2, F and G, and Fig. S6). Notably, a similar cluster with up-regulated Bdnf and down-regulated Slc1a2 was present in young and old TAU58/2 mice. An Fgf-related gene cluster was only up-regulated in young TAU58/2 mice, whereas only Fgf1 was down-regulated in old TAU58/2 mice.
For endothelial cells, STRING analysis showed a prominent and differentially-regulated gene cluster in old TAU58/2 mice associated with surface receptors (Tfrc, Ly6a, Ly6c1, Cd59a, and Itgb1) and their regulation (Kitl and Adam10) (Fig. 2I and Fig. S7A). A minor gene cluster of genes regulating focal adhesion (Crk, Tgfb1i1, and Myl9) was present in young TAU58/2 mice (Fig. 2H and Fig. S7B).
No significant gene clusters were identified for oligodendrocytes, even when combining differentially-regulated genes from OPCs, NFOs, and MOs (Fig. 2, J and K, and Fig. S8). Taken together, STRING analysis of differentially-regulated genes annotated to specific CNS cell types identified both overlapping and distinct gene clusters in young and old TAU58/2 mice.
Post-translational validation of differentially regulated candidates and associated proteins
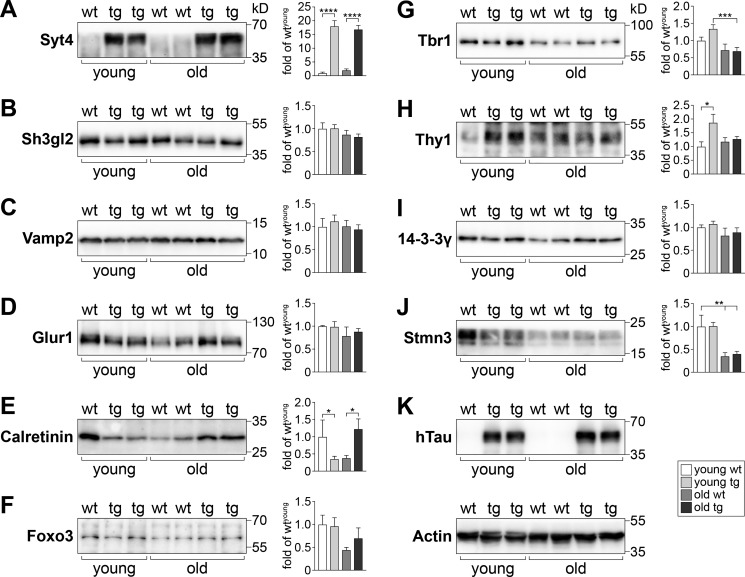
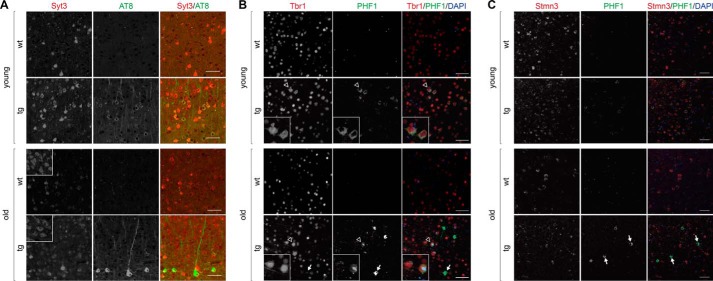
Focusing on the differentially-regulated genes annotated to neurons in young and old TAU58/2 mice, we selected candidates for determining their protein translation levels in cortex extracts from independent cohorts of TAU58/2 and nontransgenic mice of both age groups. Both antibody availability and assay performance were criteria for selection. Among the tested proteins linked to synaptic vesicle trafficking, we found a marked increase in the protein levels of synaptotagmin 4 (Syt4) in both young and old TAU58/2 mice as compared with their nontransgenic littermate controls (Fig. 3A). Similarly, cortical neurons showed markedly increased staining for the related synaptotagmin 3 (Syt3) in young and old TAU58/2 brains compared with nontransgenic controls (Fig. 4A). For comparison, endophilin A1 (Sh3gl2) levels were unchanged in young and old TAU58/2 mice when compared with the age-matched controls (Fig. 3B). Similarly, vesicle-associated membrane protein 2 (Vamp2) levels were found to be unchanged in both age groups, irrespective of transgenic P301S tau expression (Fig. 3C). The α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit glutamate receptor 1 (Glur1 (also known as GluA1, Gria1)) was expressed at comparable levels in young and old TAU58/2 and nontransgenic mice (Fig. 3D). Calretinin (Calb2) expression was reduced levels in young TAU58/2 compared with nontransgenic mice. In contrast, calretinin levels were markedly increased in old TAU58/2 mice (Fig. 3E). The transcription factors forkhead box protein O3 (Foxo3) and T-box brain protein 1 (Tbr1) levels were comparable in young TAU58/2 and nontransgenic mice. In contrast, Foxo3 showed a trend toward increased levels in old TAU58/2 compared with nontransgenic mice (Fig. 3F). There was an age-dependent decrease of Tbr1 levels (Fig. 3G). In contrast to predominant nuclear localization of Tbr1 in the cortex of nontransgenic mice, neurons harboring hyperphosphorylated tau showed cytoplasmic mis-localization of Tbr1 in young TAU58/2 brains (Fig. 4B). Similar mis-localizations of Tbr1 in tau-positive neurons were present in aged TAU58/2 brains, whereas NFTs have lost Tbr1 expression. Levels of the Thy-1 membrane protein (Thy1) were markedly increased in young but not old TAU58/2 mice compared with respective nontransgenic controls (Fig. 3H). The scaffolding 14-3-3 protein γ (Ywhag, 14-3-3γ) was unchanged in young and old TAU58/2 mice as compared with nontransgenic controls (Fig. 3I). The microtubule-destabilizing factor stathmin 3 (Stmn3) was unchanged in young TAU58/2 mice. In old mice, Stmn3 levels were lower than in young mice with no difference between old TAU58/2 and nontransgenic mice (Fig. 3J). Interestingly, Stmn3 co-localized with hyperphosphorylated tau in cortical neurons of young TAU58/2 mice, whereas NFTs in the cortex of aged TAU58/2 brains lost Stmn3 staining (Fig. 4C). Probing Western blotting membranes for human tau confirmed the expression of P301S mutant transgenic tau in TAU58/2 mice, and actin was used as loading control (Fig. 3K). In summary, Western blotting confirmed changes to transcript products of differentially-regulated genes annotated to neurons in both young and old TAU58/2 mice.
Figure 3.
Validation of translation products from differentially-regulated genes in young and old TAU58/2 brains. A–J, Western blotting of cortical brain extracts from young and old TAU58/2 (transgenic (tg)) and nontransgenic (wt) mice with specific antibodies and subsequent quantification of band intensities relative to levels detected for young WT samples from multiple blots (sample sizes: n = 3 (young wt); n = 7 (young tg); n = 4 (old wt); n = 6 (old tg); *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; one-way ANOVA (Fisher LSD test)). K, probing with the human tau (hTau)-specific tau13 antibody-confirmed genotypes. Representative blot of actin levels confirmed equal loading.
Figure 4.
Altered expression of Syt3, Tbr1, and Stmn3 in tau harboring neurons of TAU58/2 mice. A, staining of young and old TAU58/2 and nontransgenic control brains with antibodies to Syt3 (red) and phosphorylated tau (AT8). Insets show Syt3 staining of dorsal thalamic nucleus as a reference area of same intensity in TAU58/2 and nontransgenic mice. Scale bar, 50 μm. B, staining of young and old TAU58/2 and nontransgenic control brains with antibodies to Tbr1 (red) and phosphorylated tau (PHF1). Merged images include nuclear DAPI (blue) staining. Arrows indicate NFTs with marked accumulation of phosphorylated tau and absence of Tbr1. Insets show higher resolution images of neurons indicated by open arrowheads in larger images. Scale bar, 50 μm. C, staining of young and old TAU58/2 and nontransgenic control brains with antibodies to Stmn3 (red) and phosphorylated tau (PHF1). Merged images include nuclear DAPI (blue) staining. Arrows indicate NFTs with marked accumulation of phosphorylated tau and absence of Stmn3. Scale bar, 50 μm.
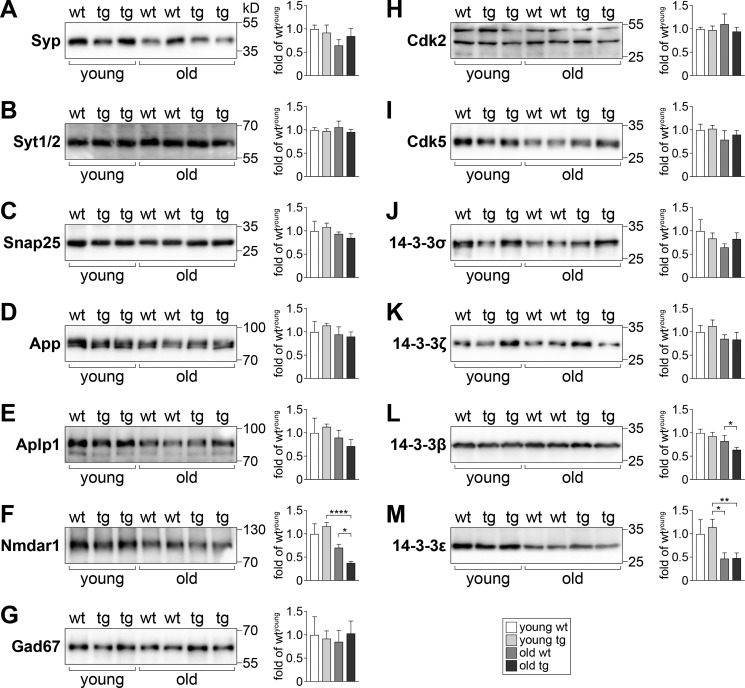
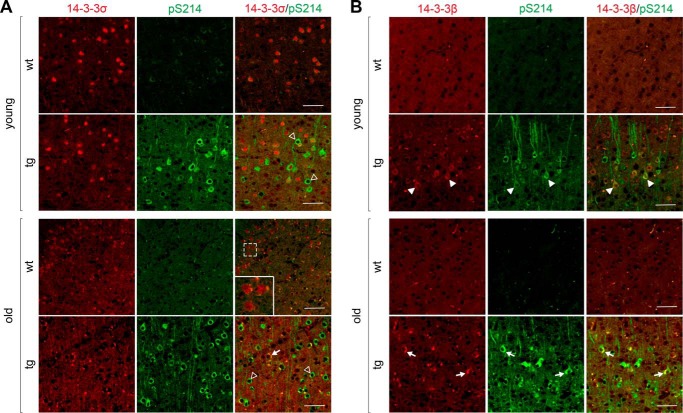
Focusing further on neuron-specific and differentially-regulated genes in young and old TAU58/2 mice, we conducted an additional analysis via the MouseNet functional prediction server (version 2). MouseNetV2 is an algorithm-based tool to predict new pathway members based on established networks of input datasets (i.e. guide genes) (22). Both guide gene datasets of neuron-annotated differentially-regulated genes in young and old TAU58/2 mice returned a significant area under the curve input analysis, indicating closeness of genes (area under receiver operating characteristic curve: young = 0.5963 (p < 0.0001); old = 0.6445 (p < 0.0001)). Several proteins were predicted to be linked to clusters of guide genes (Table S2). These predictions were validated by Western blotting of brain extracts from young and old TAU58/2 mice using commercial antibodies. Candidates for validation were selected by MouseNet score, antibody availability, and assay performance. Of the synaptic proteins tested, synaptophysin (Syp) showed unchanged levels in young and old TAU58/2 mice compared with nontransgenic mice (Fig. 5A). In contrast to Syt4, Syt1/2 showed no change in expression levels in young and old TAU58/2 mice (Fig. 5B). Similarly, the synaptosomal-associated protein 25 (Snap25) was unchanged between ages and genotypes (Fig. 5C). The (vesicular) membrane proteins amyloid-β precursor protein (App) and the related APP-like protein 1 (Aplp1) were also expressed at similar levels in young and old TAU58/2 and nontransgenic mice (Fig. 5, D and E). The postsynaptic N-methyl-d-aspartate (NMDA) receptor subunit 1 (Nmdar1) was found to be expressed at higher levels in young compared with old mice, with a trend toward higher levels in young TAU58/2 mice compared with nontransgenic controls (Fig. 5F). In contrast, Nmdar1 levels were significantly reduced in old TAU58/2 mice compared with old nontransgenic controls. Glutamate decarboxylase 1 (Gad1 = Gad67) was expressed at comparable levels in young and old TAU58/2 and nontransgenic mice (Fig. 5G). Next, we probed for levels of cyclin-dependent–like kinase 2 and 5 (Cdk2 and Cdk5). Both Cdk2 and Cdk5 levels were unchanged in young and old mice irrespective of their genotype (Fig. 5, H and I). Finally, we tested for the expression of different 14-3-3 isoforms; 14-3-3σ (Ywhas) showed a trend toward an age-dependent decrease with lower levels in old mice compared with young nontransgenic mice (Fig. 5J). In young TAU58/2 and nontransgenic mice, levels of 14-3-3σ were comparable, but there was a trend toward increased expression in old TAU58/2 mice compared with old nontransgenic controls. Staining of young nontransgenic brain sections showed 14-3-3σ was predominantly nuclear, whereas in young TAU58/2 mice a subset of phosphorylated tau-harboring neurons lacked nuclear 14-3-3σ (Fig. 6A). In old TAU58/2 brains, nuclear straining was largely absent together with frequent large cytoplasmic focal 14-3-3σ staining that co-localized with phosphorylated tau. Old nontransgenic brains maintained nuclear 14-3-3σ staining. Interestingly, few smaller 14-3-3σ foci in the nontransgenic cortex co-labeled with phosphorylated tau, possibly reflecting a process of brain aging. 14-3-3ζ (Ywhaz) showed similar expression levels in young and aged TAU58/2 and nontransgenic mice (Fig. 5K). 14-3-3β (Ywhab) levels remained unchanged in young TAU58/2 mice but were lower in old TAU58/2 mice compared with nontransgenic controls (Fig. 5L). Although the staining of brain sections from young TAU58/2 showed cytoplasmic accumulation of 14-3-3β that co-labeled with phosphorylated tau, 14-3-3β localized to the neuropil in nontransgenic mice (Fig. 6B). For comparison, the old TAU58/2 cortex harbored frequent and large cytoplasmic aggregates of 14-3-3β that co-labeled with phosphorylated tau. Small and infrequent foci of 14-3-3β in old nontransgenic brains similarly co-labeled with phosphorylated tau, possibly reflecting aging. Finally, 14-3-3ϵ (Ywhae) was lower in old than in young mice, irrespective of their genotype (Fig. 5M).
Figure 5.
Expression of candidate proteins predicted by MouseNet in young and old TAU58/2 brains. A–M, Western blotting of cortical brain extracts from young and old TAU58/2 (tg) and nontransgenic (wt) mice with specific antibodies and subsequent quantification of band intensities relative to levels detected for young WT samples from multiple blots (sample sizes: n = 3 (young wt); n = 7 (young tg); n = 4 (old wt); n = 6 (old tg); *, p < 0.05; **, p < 0.01; ****, p < 0.0001; one-way ANOVA (Fisher LSD test)). Representative blot of actin levels is shown in Fig. 3K.
Figure 6.
Co-aggregation of 14-3-3 proteins with phosphorylated tau in neurons in aged TAU58/2 mice. A, staining of young and old TAU58/2 and nontransgenic (nontg) control brains with antibodies to 14-3-3σ (red) and phosphorylated tau (pS214; green). Open arrowheads indicated neurons with lack of neuronal 14-3-3σ staining. Arrows indicated focal staining of 14-3-3σ aggregates. Inset, magnification of neurons in aged nontransgenic staining. Scale bar, 50 μm. B, staining of young and old TAU58/2 and nontransgenic (nontg) control brains with antibodies to 14-3-3β (red) and phosphorylated tau (pS214; green). Arrowheads indicate cytoplasmic accumulation of 14-3-3β. Arrows indicated focal staining of 14-3-3β aggregates. Scale bar, 50 μm.
Discussion
In this study, we report comparative gene expression analysis between young and old human P301S transgenic TAU58/2 mice. Cell-type–specific gene network analysis revealed distinct and shared pathway clusters in both young and old TAU58/2 mice, indicating different molecular mechanisms during disease progression.
Analysis of the entire gene expression data sets from young and old TAU58/2 mice, including shared differentially-regulated genes, provided limited insight into underlying molecular/cellular mechanisms due to the complexity of the data. The only pathway that showed a notable over-representation of altered gene expression was associated with mitochondrial transport, which may indicate perturbations of cell respiration. It is well-established in cells and mice that increased levels of pathological tau compromise the transport of mitochondria (and other cargos) along the microtubule network (23–25). Furthermore, we have previously shown impaired mitochondrial respiration in mutant tau transgenic strains (26, 27). To this end, it remains to be shown whether mitochondrial distribution and respiration are changed over time in TAU58/2 mice.
However, considering both data sets of most significantly-deregulated genes in young and old TAU58/2 mice as well as outputs from different pathway analysis, no obvious candidate genes or pathways could be identified that warranted targeted investigation. This may be due to the very broad impact of transgenic P301S mutant tau expression on many different pathways or, more likely, reflects that current pathways prediction tools do not consider differential expression of genes in independent cell types. In other words, different dysregulated genes that map to the same pathway may well be expressed in different cell populations, hence preventing direct physical interaction or connection at the molecular level. Therefore, we have referenced the differentially-regulated genes from young and old TAU58/2 mice against a comprehensive gene expression data set of distinct cells types from mouse brains (20). This significantly reduced the complexity of the data and identified distinct cell type–specific gene networks.
The differentially-regulated genes annotated to neurons depicted changes to larger gene clusters in both young and old TAU58/2 mice. Both age groups shared a network of altered expression of genes associated with synaptic vesicle trafficking, suggesting early and persistent functional changes of the pre-synapse in TAU58/2 mice. This is in line with recently reported pre-synaptic deficits in other tau transgenic mouse models (28–30). Alterations in glutamate receptor signaling was prominent in young TAU58/2 mice but did not contribute to the differentially-regulated gene cluster in old mice. This may indicate that aberrant glutamatergic activation contributes to early deficits in TAU58/2, whereas other receptor-signaling pathways and changes associated with the tubulin cytoskeleton contribute to deficits as mice age. Accordingly, hyperexcitability has been described in different tau transgenic mouse models (31, 32). Interestingly, the mRNA-encoding astrocyte-annotated Slc1a2 (also known as Glt1 or Eaat2) that removes glutamate from synapses (33) was consistently down-regulated in both young and old TAU58 mice, possibly contributing to increased glutamatergic signaling. We have previously shown earlier, and confirmed in this study, progressive astrocytosis and microgliosis in TAU58/2 mice (16). Consistently, young TAU58/2 mice showed clusters of differentially regulated microglia-annotated genes associated with immune- and complement activation, indicative of an inflammatory response. Accordingly, the microglial checkpoint protein Cx3cr1 was down-regulated (34). In old TAU58/2 mice, the differential gene signature for microglia was less specific and limited to general cytokine response molecules and ubiquitination. Interestingly, our analysis suggests that endothelial cells and oligodendrocytes were also compromised when human mutant tau is expressed in neurons. This suggests a rather complex cellular response to neuronal tau pathology in TAU58/2 mice that changes with progression of pathology.
Validation of differentially-regulated genes annotated to neurons by Western blotting revealed distinct changes to protein levels in the presence of transgenic P301S mutant tau in TAU58/2 mice and between age groups. Consistent with an absence of differential regulation, the majority of proteins linked to guide genes by MouseNet showed no change in expression levels in TAU58/2 mice. Nevertheless, we identified changed protein expression levels for Nmdar1 and 14-3-3 isoforms in TAU58/2 mice. Together, this shows that subjecting transcriptional data to a variety of available analysis tools and referencing to cell-specific gene expression data sets can identify deregulated pathways and proteins in mouse models. However, we also show that differential regulation of individual genes does not necessarily mean that protein levels are also changed. Hence, validating the differential mRNA datasets at the translational level remains necessary. Although Western blotting has not revealed protein changes of the differentially-regulated genes or age groups, this does not exclude changes to protein expression or activity in a subpopulation of cells.
Probing brain extracts for expression levels revealed changes in a range of proteins associated with synaptic vesicle trafficking, including Syt4, Syp, and Sh3gl2, whereas others were not changed, such as Syt1/2, SNAP25, and Vamp2. Furthermore, staining showed increased Syt3 labeling of cortical neurons in TAU58/2 brains. This is in line with the prediction of a network of differentially-regulated genes associated with synaptic vesicle trafficking in young and old TAU58/2 mice. Here, pathological tau may affect specific factors in synaptic vesicle trafficking rather than blocking this process per se. Similarly, we have previously shown that axonal transport is disrupted by pathological tau by specific interference with motor proteins, rather than clogging the passage of all cargos in axons (35). Syt4 levels are significantly increased following neuronal hyperexcitation (36). Neuronal hyperactivity has been reported for tau transgenic mice (31, 32). Accordingly, increased Syt4 and possibly Syt3 levels in TAU58/2 may be in response to neuronal hyperexcitation. Interestingly, we have recently shown that Syt1 interacts with human tau via a specific motif at its N terminus (37). Furthermore, Syt and other synaptic proteins may interact differentially with distinct tau isoforms (38). Syt1 levels were increased in the CSF of AD patients (39). Conversely, Syt2 levels were decreased in synapses in FTD and AD brains (40).
We found a trend toward increased expression of Nmdar1 in young TAU58/2 mice, although levels were significantly reduced in old TAU58/2 mice. This may explain, at least in part, the predicted cluster of differentially-regulated genes associated with glutamatergic signaling in young but not old TAU58/2 mice. Furthermore, this is consistent with early hyperexcitation phenotypes reported for P301L tau transgenic mice (31, 32). We have previously shown that tau is directly involved in the regulation of NMDA receptor signaling at the post-synapse (2, 4, 41, 42). Interestingly, the molecular modulator of neuronal excitation and long-term potentiation, calretinin (43), was significantly decreased in young and increased in aged TAU58/2 mice, possibly contributing to early hyperexcitation. Conversely, its late up-regulation may reflect compensatory mechanisms. Whether TAU58/2 mice present with hyperexcitation and neuronal network synchronicity similar to AD mouse models (44, 45) and how these molecular processes contribute to functional deficits of these mice remain to be shown.
Previous reports have linked different 14-3-3 protein isoforms to NFT pathology in humans (46–48) and tau aggregation (49). Furthermore, increased CSF 14-3-3 levels have been reported for a range of neurodegenerative diseases, including FTD and AD (50), and may aid in diagnosing underlying conditions (51). 14-3-3 directly interacts with tau, which is regulated by and mediates phosphorylation of tau (52–54). This interaction may contribute to regulation of the neuronal microtubule network (53, 55) and contribute to tau aggregation (56, 57). Here, we found reduced Ywhag mRNA expression in aged TAU58/2 mice. However, this did not translate to altered 14-3-3γ protein levels. Interestingly, protein levels of other 14-3-3 family members were changed in aged TAU58/2 mice. Specifically, 14-3-3σ and 14-3-3β protein levels were decreased and increased in old TAU58/2 mice, respectively. Both 14-3-3σ and 14-3-3β aggregated in the tau pathology-harboring neurons in aged TAU58/2 mice, possibly contributing to changes in levels. Interestingly, 14-3-3σ also lost its normal nuclear localization as TAU58/2 mice aged, whereas 14-3-3β accumulated in the cytoplasm already in young TAU58/2 mice, which both may be due to their interaction with phosphorylated tau in the cytoplasm (52–54). Furthermore, 14-3-3ϵ levels were found to be age-dependently reduced. Given the diverse functions and roles of 14-3-3 proteins in multiple pathways, changes to their expression levels in the response to transgenic P301S tau expression could potentially affect many physiological processes. Notably, 14-3-3 has been found to regulate the dynamics of neurofilaments (58). Hence, altered 14-3-3 protein expression may contribute to the neurofilament pathology found in TAU58/2, other tau transgenic mice, and human tauopathies (16). Although infrequent, we found neuronal foci of 14-3-3σ and 14-3-3β that co-labeled with phosphorylated tau in aged nontransgenic mice, suggesting an age-related underlying process that may be accelerated in TAU58/2 mice by transgenic tau expression and pathology. To this end, 14-3-3 may play a central role in neurodegenerative diseases that warrants further investigation in tau transgenic mouse models of FTD.
Finally, we found significantly increased levels of the surface protein Thy1, which is also known as CD90 antigen, in young TAU58/2 mice. Notably, the P301S tau transgene is driven by the murine Thy1.2 promoter. Although it cannot be fully excluded that this affects the expression of endogenous Thy1, transcription factor dilution would rather result in reduced expression levels. Therefore, increased Thy1 levels are more likely linked to the presence of pathogenic tau in TAU58/2 mice, but its roles in the brain in health and disease remain elusive.
In summary, reducing the complexity of whole transcriptome data by annotating genes to specific CNS cell types has identified several new proteins that were found to be changed in tau transgenic TAU58/2 mice. Although it is unlikely that this information would have been obtained from interrogating the entire data set for expression changes or by pathway analysis, we do acknowledge that the exclusion of differentially-regulated genes during the data analysis possibly excluded other relevant genes. Nevertheless, our study provides new insight into the complex effects of human mutant tau accumulation in an in vivo model of FTD. Finally, our differential gene data set may prove useful for interrogation of specific genes of interest by the wider dementia research community.
Materials and methods
Mice
TAU58/2 mice express P301S tau under the neuronal mThy1.2 promoter as reported previously (16). Mice were maintained on a pure C57Bl/6J background and held in littermate groups on a 12-h light/dark cycle with access to standard chow and water ad libitum. Male transgenic and nontransgenic littermates were used for this study. Mice were killed by cervical dislocation, and brains were rapidly removed, dissected, and tissue snap-frozen for mRNA and protein expression. Three mice per age and genotype were used for RNAseq. All experiments were approved by the Animal Ethics Committees of the University of New South Wales and Macquarie University.
RNAseq
Extraction of mRNA and mRNA sequencing were done by Macrogen (Korea). Briefly, RNA extraction was performed using Maxwell 16 LEV simplyRNA tissue kits (Promega) and library construction performed using TruSeq-stranded total RNA with Ribo-Zero Gold sample preparation kit and TruSeq rapid SBS kit. Sequencing was subsequently performed on the Illumina HiSeq 2500 platform following the HiSeq 2500 System User Guide Document no. 15035786 version 01 HCS 2.2.70 protocol and using HCS version 2.2 for sequencing control. All samples generated >3 billion bp reads of 100-bp paired-ends reads which corresponds to 50× coverage. The HISAT2, StringTie, and Ballgown pipeline was used for analysis as described previously (59). Reads were mapped against the mm10 Mus musculus reference genome using HISAT2 version 2.0.4 with the “–dta” option and sorted using samtools version 0.0.19. The subsequently sorted alignment data were compared with the GRCm38.85 assembly release to estimate transcript abundance and table counts using StringTie version 1.3.0 with options “-B -f 0.1 -m 200 -g 50.” Output from StringTie was used for differential expression analysis in Ballgown version 2.2.0 on the RStudio platform using R version 3.4.2. FPKM for between-group comparison was performed using the stattest() function in Ballgown.
CNS cell type–annotation of genes
For gene expression data sets from both young and old TAU58/2 mice, raw FPKM values with a threshold of mean WT FPKM >1 and ABS(log2FC) >0.58 were sorted ascendingly. Genes with 0 FPKM were ignored for the analysis, unless all reads were 0 for all WT or transgenic samples. Then genes were sorted according to a CNS cell-specific gene expression data set (20). The z scores were determined for each cell type and age group. Exclusion lists with a z score of >2 showed no overlap between cell types. These genes were therefore annotated as uniquely/preferentially expressed in distinct cell types and used for further analysis by online tools PANTHER and STRING.
Gene cluster and pathway analysis
All differentially-regulated genes from young, old, and overlapping genes were individually used for pathway analysis with the online PANTHER software version 13.1 to predict altered pathways (19). Related pathways were grouped for presentation purposes. A representation of pathway-associated genes relative to a PANTHER reference dataset from naïve mice were furthermore determined.
Gene product lists from each young and old murine brain cell types were translated into networks using STRING version 10.5 (21). Data source settings for nodal associations were set to default (“on” for text-mining, databases, experiments, co-expression, neighborhood, gene fusion, and co-occurrence) with “medium confidence” (≥0.400) interaction score minimums. Networks were visualized using Cytoscape version 3.6.1 (60). Each node corresponds to a protein and each edge to a STRING-based physical and/or functional interaction. Node size is proportional to FPKM fold-change in transgenic relative to nontransgenic samples. Blue and gray nodes represent up- and down-regulation, respectively. Solid border nodes are differentially regulated cell-specific gene products found in both young and old transgenic mice.
Western blotting
Western blotting was done as described previously (61, 62). Primary antibodies were to the following: Syt4 (catalog no. MABN109, 1:1K; Merck-Millipore); Sh3gl2 (catalog no. ab169762; 1:1K, Abcam); Vamp2 (catalog no. ab3347, 1:1K, Abcam); Glur1 (catalog no. MAB2263, 1:1K, Merck-Millipore); calretinin (catalog no. sc-5043, 1:1K, Santa Cruz Biotechnology); Foxo3 (catalog no. 07-702, Merck-Millipore); Tbr1 (catalog no. ab31940, 1:1K, Abcam); Thy1 (catalog no. 550571, 1:1K, BD Biosciences); 14-3-3γ (catalog no. sc-731, 1:1K, Santa Cruz Biotechnology); Stmn3 (catalog no. ab171625, 1:1K, Abcam); hTau (catalog no. sc-21796, 1:5K, Santa Cruz Biotechnology); actin (catalog no. A2066, 1:2K, Sigma); Syp (catalog no. ab52636, 1:1K, Abcam); Syt1/2 (catalog no. sc-12466, 1:1K, Santa Cruz Biotechnology); Snap25 (catalog no. ab5666, 1:1K, Abcam); App (catalog no. ab32136, 1:1K, Abcam),;Aplp1 (catalog no. 171615, 1:1K, Calbiochem); Nmdar1 (catalog no. MAB363, 1:1K, Merck-Millipore); Gad67 (catalog no. MAB5406, 1:1K, Merck-Millipore); Cdk2 (catalog no. sc-748, 1:1K, Santa Cruz Biotechnology); Cdk5 (catalog no. ab40773, 1:1K, Abcam); 14-3-3σ (catalog no. sc-100638, 1:1K, Santa Cruz Biotechnology); 14-3-3ζ (catalog no. ab51129, 1:1K, Abcam); 14-3-3β (catalog no. sc-25276, 1:1K, Santa Cruz Biotechnology); and 14-3-3ϵ (catalog no. sc-1020, 1:1K, Santa Cruz Biotechnology). Protein bands were visualized using HRP-coupled secondary antibodies, goat anti-rabbit (catalog no. sc-2004, 1:5K, Santa Cruz Biotechnology); goat anti-mouse (catalog no. 115-035-166, 1:5K, Jackson ImmunoResearch); mouse anti-goat (catalog no. sc-2354, 1:5K, Santa Cruz Biotechnology); and the HRP Luminata Crescendo substrate (Merck Millipore). Quantification was done with the Fiji software version 1.0 (National Institutes of Health).
Tissue staining
Immunohistochemistry was done as described previously (16). Primary antibodies were against the following: Gfap (catalog no. G9269, 1:200, Sigma); Iba1 (catalog no. WEE4506, 1:200, Wako); PHF1 (1:250, gift from P. Davies); pS214 (catalog no. AB170892, 1:1K, Abcam); Tbr1 (catalog no. AB31940, 1:100, Abcam); AT8 (catalog no. MN1020, 1:250, Pierce); pS422 (catalog no. AB79415, 1:500, Abcam); Stmn3 (catalog no. AB171625, 1:100, Abcam); 14-3-3σ (catalog no. sc-100638, 1:50, Santa Cruz Biotechnology); 14-3-3β (catalog no. sc-25276, 1:100, Santa Cruz Biotechnology); Syt3 (catalog no. orb13704, 1:100, Biorbyt); C1q (catalog no. AB71940, 1:25, Abcam); and C3 (catalog no. AB11862, 1:25, Abcam). All images were done on a BX51 fluorescence microscope (Olympus) using the Cell Sens software. Iba1 and Gfap-positive cells were counted on one section per brain and an area of 1,000 × 1,000 μm.
Statistical analysis
Statistical analysis was done with the Prism 7 software (GraphPad) using the indicated statistical tests. All values are given as mean and standard error of the mean.
Author contributions
Y. D. K., G. T. S., and L. M. I. conceptualization; Y. D. K., G. C., K. S., C. A., M. P., and A. F. F. data curation; Y. D. K., G. C., K. S., C. A., M. B., J. M., E. P., and A. I. formal analysis; Y. D. K., G. T. S., and L. M. I. supervision; Y. D. K., G. M. H., O. P., M. C. K., M. K., J. R. H., C. T. L., J. S. M., A. I., J. J. K., G. T. S., and L. M. I. funding acquisition; Y. D. K. and L. M. I. writing-original draft; Y. D. K., J. J. K., G. T. S., and L. M. I. project administration; Y. D. K., G. C., K. S., M. B., J. M., M. P., A. F. F., E. P., G. M. H., O. P., M. C. K., M. K., J. R. H., C. T. L., J. S. M., A. I., J. J. K., G. T. S., and L. M. I. writing-review and editing; Y. D. K., G. C., M. B., and L. M. I. visualization.
Supplementary Material
This work was supported by National Health and Medical Research Council Grants 1081916, 1103258, 1107657, 1132524, 1136241, 1143848, and 1143978, Australian Research Council Grants DP170100781, DP170100843, and CE110001021, CurePSP Grant 609-2015-07, and a generous donation by the Crescitelli family. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S2.
- PHF
- paired helical filament
- NFT
- neurofibrillary tangle
- FTD
- frontotemporal dementia
- CNS
- central nervous system
- AD
- Alzheimer's disease
- OPC
- oligodendrocyte precursor cell
- NFO
- newly formed oligodendrocyte
- MO
- myelinating oligodendrocyte
- HRP
- horseradish peroxidase
- NMDA
- N-methyl-d-aspartate
- FPKM
- fragment per kilobase million
- DAPI
- 4,6-diamidino-2-phenylindole
- CSF
- cerebrospinal fluid
- ANOVA
- analysis of variance
- Syp
- synaptophysin
- App
- amyloid-β precursor protein.
References
- 1. Buée L., Bussière T., Buée-Scherrer V., Delacourte A., and Hof P. R. (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 33, 95–130 10.1016/S0165-0173(00)00019-9 [DOI] [PubMed] [Google Scholar]
- 2. Ittner L. M., Ke Y. D., Delerue F., Bi M., Gladbach A., van Eersel J., Wölfing H., Chieng B. C., Christie M. J., Napier I. A., Eckert A., Staufenbiel M., Hardeman E., and Götz J. (2010) Dendritic function of Tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell 142, 387–397 10.1016/j.cell.2010.06.036 [DOI] [PubMed] [Google Scholar]
- 3. Maas T., Eidenmüller J., and Brandt R. (2000) Interaction of tau with the neural membrane cortex is regulated by phosphorylation at sites that are modified in paired helical filaments. J. Biol. Chem. 275, 15733–15740 10.1074/jbc.M000389200 [DOI] [PubMed] [Google Scholar]
- 4. Ittner A., Chua S. W., Bertz J., Volkerling A., van der Hoven J., Gladbach A., Przybyla M., Bi M., van Hummel A., Stevens C. H., Ippati S., Suh L. S., Macmillan A., Sutherland G., Kril J. J., et al. (2016) Site-specific phosphorylation of tau inhibits amyloid-beta toxicity in Alzheimer's mice. Science 354, 904–908 10.1126/science.aah6205 [DOI] [PubMed] [Google Scholar]
- 5. Ballatore C., Lee V. M., and Trojanowski J. Q. (2007) Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 10.1038/nrn2194 [DOI] [PubMed] [Google Scholar]
- 6. Goedert M., Spillantini M. G., Jakes R., Rutherford D., and Crowther R. A. (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3, 519–526 10.1016/0896-6273(89)90210-9 [DOI] [PubMed] [Google Scholar]
- 7. Forrest S. L., Kril J. J., Stevens C. H., Kwok J. B., Hallupp M., Kim W. S., Huang Y., McGinley C. V., Werka H., Kiernan M. C., Götz J., Spillantini M. G., Hodges J. R., Ittner L. M., and Halliday G. M. (2018) Retiring the term FTDP-17 as MAPT mutations are genetic forms of sporadic frontotemporal tauopathies. Brain 141, 521–534 10.1093/brain/awx328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burrell J. R., Halliday G. M., Kril J. J., Ittner L. M., Götz J., Kiernan M. C., and Hodges J. R. (2016) The frontotemporal dementia-motor neuron disease continuum. Lancet 388, 919–931 10.1016/S0140-6736(16)00737-6 [DOI] [PubMed] [Google Scholar]
- 9. Warren J. D., Rohrer J. D., and Rossor M. N. (2013) Clinical review. Frontotemporal dementia. BMJ 347, f4827 10.1136/bmj.f4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., et al. (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- 11. Götz J., and Ittner L. M. (2008) Animal models of Alzheimer's disease and frontotemporal dementia. Nat. Rev. Neurosci. 9, 532–544 10.1038/nrn2420 [DOI] [PubMed] [Google Scholar]
- 12. Ittner L. M., Halliday G. M., Kril J. J., Götz J., Hodges J. R., and Kiernan M. C. (2015) FTD and ALS-translating mouse studies into clinical trials. Nat. Rev. Neurol. 11, 360–366 10.1038/nrneurol.2015.65 [DOI] [PubMed] [Google Scholar]
- 13. Brunden K. R., Trojanowski J. Q., and Lee V. M. (2009) Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat. Rev. Drug Discov. 8, 783–793 10.1038/nrd2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Eersel J., Ke Y. D., Liu X., Delerue F., Kril J. J., Götz J., and Ittner L. M. (2010) Sodium selenate mitigates tau pathology, neurodegeneration, and functional deficits in Alzheimer's disease models. Proc. Natl. Acad. Sci. U.S.A. 107, 13888–13893 10.1073/pnas.1009038107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Przybyla M., Stevens C. H., van der Hoven J., Harasta A., Bi M., Ittner A., van Hummel A., Hodges J. R., Piguet O., Karl T., Kassiou M., Housley G. D., Ke Y. D., Ittner L. M., and Eersel J. (2016) Disinhibition-like behavior in a P301S mutant tau transgenic mouse model of frontotemporal dementia. Neurosci. Lett. 631, 24–29 10.1016/j.neulet.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 16. van Eersel J., Stevens C. H., Przybyla M., Gladbach A., Stefanoska K., Chan C. K., Ong W. Y., Hodges J. R., Sutherland G. T., Kril J. J., Abramowski D., Staufenbiel M., Halliday G. M., and Ittner L. M. (2015) Early-onset axonal pathology in a novel P301S-Tau transgenic mouse model of frontotemporal lobar degeneration. Neuropathol. Appl. Neurobiol. 41, 906–925 10.1111/nan.12233 [DOI] [PubMed] [Google Scholar]
- 17. Van der Jeugd A., Vermaercke B., Halliday G. M., Staufenbiel M., and Götz J. (2016) Impulsivity, decreased social exploration, and executive dysfunction in a mouse model of frontotemporal dementia. Neurobiol. Learn. Mem. 130, 34–43 10.1016/j.nlm.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 18. Yin Z., Valkenburg F., Hornix B. E., Mantingh-Otter I., Zhou X., Mari M., Reggiori F., Van Dam D., Eggen B. J. L., De Deyn P. P., and Boddeke E. (2017) Progressive motor deficit is mediated by the denervation of neuromuscular junctions and axonal degeneration in transgenic mice expressing mutant (P301S) Tau protein. J. Alzheimers Dis. 60, S41–S57 10.3233/JAD-161206 [DOI] [PubMed] [Google Scholar]
- 19. Mi H., and Thomas P. (2009) PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 563, 123–140 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O'Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim E., Hwang S., Kim H., Shim H., Kang B., Yang S., Shim J. H., Shin S. Y., Marcotte E. M., and Lee I. (2016) MouseNet v2: a database of gene networks for studying the laboratory mouse and eight other model vertebrates. Nucleic Acids Res. 44, D848–D854 10.1093/nar/gkv1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishihara T., Hong M., Zhang B., Nakagawa Y., Lee M. K., Trojanowski J. Q., and Lee V. M. (1999) Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron 24, 751–762 10.1016/S0896-6273(00)81127-7 [DOI] [PubMed] [Google Scholar]
- 24. Stamer K., Vogel R., Thies E., Mandelkow E., and Mandelkow E. M. (2002) Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol. 156, 1051–1063 10.1083/jcb.200108057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ittner L. M., Fath T., Ke Y. D., Bi M., van Eersel J., Li K. M., Gunning P., and Gotz J. (2008) Parkinsonism and impaired axonal transport in a mouse model of frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 105, 15597–16002 10.1073/pnas.0808084105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. David D. C., Ittner L. M., Gehrig P., Nergenau D., Shepherd C., Halliday G., and Götz J. (2006) Beta-amyloid treatment of two complementary P301L tau-expressing Alzheimer's disease models reveals similar deregulated cellular processes. Proteomics 6, 6566–6577 10.1002/pmic.200600634 [DOI] [PubMed] [Google Scholar]
- 27. Rhein V., Song X., Wiesner A., Ittner L. M., Baysang G., Meier F., Ozmen L., Bluethmann H., Dröse S., Brandt U., Savaskan E., Czech C., Götz J., and Eckert A. (2009) Amyloid-beta and Tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc. Natl. Acad. Sci. U.S.A. 106, 20057–20062 10.1073/pnas.0905529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Decker J. M., Krüger L., Sydow A., Zhao S., Frotscher M., Mandelkow E., and Mandelkow E. M. (2015) Pro-aggregant Tau impairs mossy fiber plasticity due to structural changes and Ca(2+) dysregulation. Acta Neuropathol. Commun. 3, 23 10.1186/s40478-015-0193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McInnes J., Wierda K., Snellinx A., Bounti L., Wang Y. C., Stancu I. C., Apóstolo N., Gevaert K., Dewachter I., Spires-Jones T. L., De Strooper B., De Wit J., Zhou L., and Verstreken P. (2018) Synaptogyrin-3 mediates presynaptic dysfunction induced by Tau. Neuron 97, 823–835.e8 10.1016/j.neuron.2018.01.022 [DOI] [PubMed] [Google Scholar]
- 30. Zhou L., McInnes J., Wierda K., Holt M., Herrmann A. G., Jackson R. J., Wang Y. C., Swerts J., Beyens J., Miskiewicz K., Vilain S., Dewachter I., Moechars D., De Strooper B., Spires-Jones T. L., et al. (2017) Tau association with synaptic vesicles causes presynaptic dysfunction. Nat. Commun. 8, 15295 10.1038/ncomms15295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. García-Cabrero A. M., Guerrero-López R., Giráldez B. G., Llorens-Martín M., Avila J., Serratosa J. M., and Sánchez M. P. (2013) Hyperexcitability and epileptic seizures in a model of frontotemporal dementia. Neurobiol. Dis. 58, 200–208 10.1016/j.nbd.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 32. Liu S., Shen Y., Shultz S. R., Nguyen A., Hovens C., Adlard P. A., Bush A. I., Chan J., Kwan P., O'Brien T. J., and Jones N. C. (2017) Accelerated kindling epileptogenesis in Tg4510 tau transgenic mice, but not in tau knockout mice. Epilepsia 58, e136–e141 10.1111/epi.13847 [DOI] [PubMed] [Google Scholar]
- 33. Kirschner M. A., Copeland N. G., Gilbert D. J., Jenkins N. A., and Amara S. G. (1994) Mouse excitatory amino acid transporter EAAT2: isolation, characterization, and proximity to neuroexcitability loci on mouse chromosome 2. Genomics 24, 218–224 10.1006/geno.1994.1609 [DOI] [PubMed] [Google Scholar]
- 34. Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., David E., Baruch K., Lara-Astaiso D., Toth B., Itzkovitz S., Colonna M., Schwartz M., and Amit I. (2017) A unique microglia type associated with restricting development of Alzheimer's disease. Cell 169, 1276–1290.e17 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 35. Ittner L. M., Ke Y. D., and Götz J. (2009) Phosphorylated Tau interacts with c-Jun N-terminal kinase-interacting protein 1 (JIP1) in Alzheimer disease. J. Biol. Chem. 284, 20909–20916 10.1074/jbc.M109.014472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferguson G. D., Thomas D. M., Elferink L. A., and Herschman H. R. (1999) Synthesis degradation, and subcellular localization of synaptotagmin IV, a neuronal immediate early gene product. J. Neurochem. 72, 1821–1831 10.1046/j.1471-4159.1999.0721821.x [DOI] [PubMed] [Google Scholar]
- 37. Stefanoska K., Volkerling A., Bertz J., Poljak A., Ke Y. D., Ittner L. M., and Ittner A. (2018) An N-terminal motif unique to primate tau enables differential protein-protein interactions. J. Biol. Chem. 293, 3710–3719 10.1074/jbc.RA118.001784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C., Song X., Nisbet R., and Götz J. (2016) Co-immunoprecipitation with Tau isoform-specific antibodies reveals distinct protein interactions and highlights a putative role for 2N Tau in disease. J. Biol. Chem. 291, 8173–8188 10.1074/jbc.M115.641902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Öhrfelt A., Brinkmalm A., Dumurgier J., Brinkmalm G., Hansson O., Zetterberg H., Bouaziz-Amar E., Hugon J., Paquet C., and Blennow K. (2016) The pre-synaptic vesicle protein synaptotagmin is a novel biomarker for Alzheimer's disease. Alzheimers Res. Ther. 8, 41 10.1186/s13195-016-0208-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goetzl E. J., Kapogiannis D., Schwartz J. B., Lobach I. V., Goetzl L., Abner E. L., Jicha G. A., Karydas A. M., Boxer A., and Miller B. L. (2016) Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer's disease. FASEB J. 30, 4141–4148 10.1096/fj.201600816R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ittner L. M., and Gotz J. (2011) Amyloid-beta and tau - a toxic pas de deux in Alzheimer's disease. Nat. Rev. Neurosci. 12, 65–72 10.1038/nrn2967 [DOI] [PubMed] [Google Scholar]
- 42. Ittner A., and Ittner L. M. (2018) Dendritic Tau in Alzheimer's disease. Neuron 99, 13–27 10.1016/j.neuron.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 43. Camp A. J., and Wijesinghe R. (2009) Calretinin: modulator of neuronal excitability. Int. J. Biochem. Cell Biol. 41, 2118–2121 10.1016/j.biocel.2009.05.007 [DOI] [PubMed] [Google Scholar]
- 44. Palop J. J., Chin J., Roberson E. D., Wang J., Thwin M. T., Bien-Ly N., Yoo J., Ho K. O., Yu G. Q., Kreitzer A., Finkbeiner S., Noebels J. L., and Mucke L. (2007) Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer's disease. Neuron 55, 697–711 10.1016/j.neuron.2007.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ittner A. A., Gladbach A., Bertz J., Suh L. S., and Ittner L. M. (2014) p38 MAP kinase-mediated NMDA receptor-dependent suppression of hippocampal hypersynchronicity in a mouse model of Alzheimer inverted question marks disease. Acta Neuropathol. Commun. 2, 149 10.1186/s40478-014-0149-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Layfield R., Fergusson J., Aitken A., Lowe J., Landon M., and Mayer R. J. (1996) Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci. Lett. 209, 57–60 10.1016/0304-3940(96)12598-2 [DOI] [PubMed] [Google Scholar]
- 47. Qureshi H. Y., Li T., MacDonald R., Cho C. M., Leclerc N., and Paudel H. K. (2013) Interaction of 14-3-3zeta with microtubule-associated protein tau within Alzheimer's disease neurofibrillary tangles. Biochemistry 52, 6445–6455 10.1021/bi400442d [DOI] [PubMed] [Google Scholar]
- 48. Umahara T., Uchihara T., Tsuchiya K., Nakamura A., Ikeda K., Iwamoto T., and Takasaki M. (2004) Immunolocalization of 14-3-3 isoforms in brains with Pick body disease. Neurosci. Lett. 371, 215–219 10.1016/j.neulet.2004.08.079 [DOI] [PubMed] [Google Scholar]
- 49. Hernández F., Cuadros R., and Avila J. (2004) Zeta 14-3-3 protein favours the formation of human tau fibrillar polymers. Neurosci. Lett. 357, 143–146 10.1016/j.neulet.2003.12.049 [DOI] [PubMed] [Google Scholar]
- 50. Burkhard P. R., Sanchez J. C., Landis T., and Hochstrasser D. F. (2001) CSF detection of the 14-3-3 protein in unselected patients with dementia. Neurology 56, 1528–1533 10.1212/WNL.56.11.1528 [DOI] [PubMed] [Google Scholar]
- 51. Leitão M. J., Baldeiras I., Almeida M. R., Ribeiro M. H., Santos A. C., Ribeiro M., Tomás J., Rocha S., Santana I., and Oliveira C. R. (2016) Sporadic Creutzfeldt-Jakob disease diagnostic accuracy is improved by a new CSF ELISA 14-3-3gamma assay. Neuroscience 322, 398–407 10.1016/j.neuroscience.2016.02.057 [DOI] [PubMed] [Google Scholar]
- 52. Sluchanko N. N., and Gusev N. B. (2011) Probable participation of 14-3-3 in tau protein oligomerization and aggregation. J. Alzheimers Dis. 27, 467–476 10.3233/JAD-2011-110692 [DOI] [PubMed] [Google Scholar]
- 53. Sadik G., Tanaka T., Kato K., Yamamori H., Nessa B. N., Morihara T., and Takeda M. (2009) Phosphorylation of tau at Ser214 mediates its interaction with 14-3-3 protein: implications for the mechanism of tau aggregation. J. Neurochem. 108, 33–43 10.1111/j.1471-4159.2008.05716.x [DOI] [PubMed] [Google Scholar]
- 54. Qureshi H. Y., Han D., MacDonald R., and Paudel H. K. (2013) Overexpression of 14-3-3z promotes tau phosphorylation at Ser262 and accelerates proteosomal degradation of synaptophysin in rat primary hippocampal neurons. PLoS ONE 8, e84615 10.1371/journal.pone.0084615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Joo Y., Schumacher B., Landrieu I., Bartel M., Smet-Nocca C., Jang A., Choi H. S., Jeon N. L., Chang K. A., Kim H. S., Ottmann C., and Suh Y. H. (2015) Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau. FASEB J. 29, 4133–4144 10.1096/fj.14-265009 [DOI] [PubMed] [Google Scholar]
- 56. Li T., and Paudel H. K. (2016) 14-3-3zeta mediates Tau aggregation in human neuroblastoma M17 cells. PLoS ONE 11, e0160635 10.1371/journal.pone.0160635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Papanikolopoulou K., Grammenoudi S., Samiotaki M., and Skoulakis E. M. C. (2018) Differential effects of 14-3-3 dimers on Tau phosphorylation, stability and toxicity in vivo. Hum. Mol. Genet. 27, 2244–2261 10.1093/hmg/ddy129 [DOI] [PubMed] [Google Scholar]
- 58. Miao L., Teng J., Lin J., Liao X., and Chen J. (2013) 14-3-3 proteins interact with neurofilament protein-L and regulate dynamic assembly of neurofilaments. J. Cell Sci. 126, 427–436 10.1242/jcs.105817 [DOI] [PubMed] [Google Scholar]
- 59. Pertea M., Kim D., Pertea G. M., Leek J. T., and Salzberg S. L. (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ittner L. M., Koller D., Muff R., Fischer J. A., and Born W. (2005) The N-terminal extracellular domain 23–60 of the calcitonin receptor-like receptor in chimeras with the parathyroid hormone receptor mediates association with receptor activity-modifying protein 1. Biochemistry 44, 5749–5754 10.1021/bi048111o [DOI] [PubMed] [Google Scholar]
- 62. Koller D., Ittner L. M., Muff R., Husmann K., Fischer J. A., and Born W. (2004) Selective inactivation of adrenomedullin over calcitonin gene-related peptide receptor function by the deletion of amino acids 14–20 of the mouse calcitonin-like receptor. J. Biol. Chem. 279, 20387–20391 10.1074/jbc.M313058200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.