Abstract
The successful assembly and regulation of the kinetochore are critical for the equal and accurate segregation of genetic material during the cell cycle. CENP-C (centromere protein C), a conserved inner kinetochore component, has been broadly characterized as a scaffolding protein and is required for the recruitment of multiple kinetochore proteins to the centromere. At its C terminus, CENP-C harbors a conserved cupin domain that has an established role in protein dimerization. Although the crystal structure of the Saccharomyces cerevisiae Mif2CENP-C cupin domain has been determined, centromeric organization and kinetochore composition vary greatly between S. cerevisiae (point centromere) and other eukaryotes (regional centromere). Therefore, whether the structural and functional role of the cupin domain is conserved throughout evolution requires investigation. Here, we report the crystal structures of the Schizosaccharomyces pombe and Drosophila melanogaster CENP-C cupin domains at 2.52 and 1.81 Å resolutions, respectively. Although the central jelly roll architecture is conserved among the three determined CENP-C cupin domain structures, the cupin domains from organisms with regional centromeres contain additional structural features that aid in dimerization. Moreover, we found that the S. pombe Cnp3CENP-C jelly roll fold harbors an inner binding pocket that is used to recruit the meiosis-specific protein Moa1. In summary, our results unveil the evolutionarily conserved and unique features of the CENP-C cupin domain and uncover the mechanism by which it functions as a recruitment factor.
Keywords: centromere, kinetochore, X-ray crystallography, analytical ultracentrifugation, meiosis, CENP-C, cupin domain, fission yeast, Moa1
Introduction
Faithful chromosome segregation during cell division is essential for survival in eukaryotes. During cell division, duplicated chromosomes must segregate in an accurate and equal manner to ensure the proper separation of genetic material. If chromosomes fail to divide evenly, the resulting consequences are severe and include aneuploidy, birth defects, and cancer (1–3).
The centromere and kinetochore are two vital components that are required for the successful distribution of genetic material during the cell cycle. The centromere is a unique region on the chromosome that serves as the site of kinetochore assembly and subsequent microtubule attachment. Budding yeasts, such as Saccharomyces cerevisiae, possess a unique point centromere that contains a genetically well-defined 125-bp DNA sequence, which is essential for kinetochore assembly (4–7). However, most other eukaryotes, including humans, Schizosaccharomyces pombe, and Drosophila melanogaster, possess regional centromeres that are larger (kilobases to megabases long) and are epigenetically specified (8).
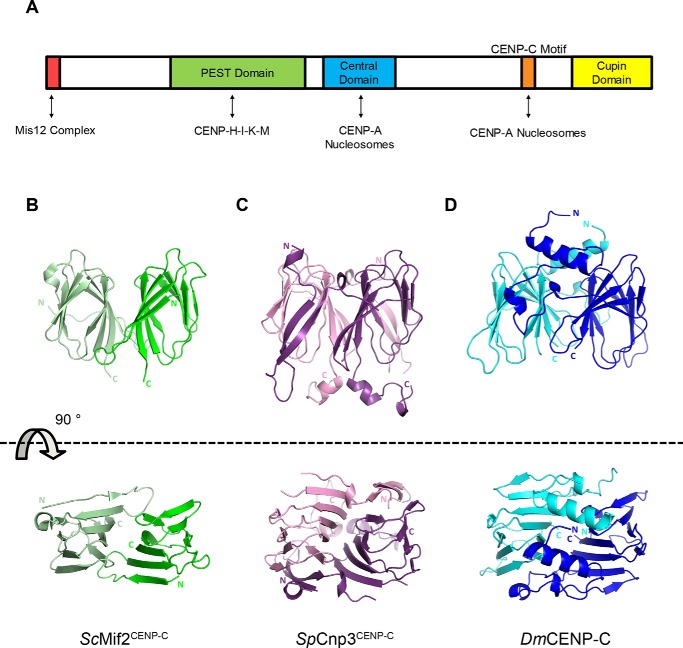
The kinetochore is a multicomponent protein complex that assembles on the centromere and facilitates a physical connection between the chromosome and the microtubules during cell division. Early EM images showed that the kinetochore has a trilaminar structure with two clear electron-dense regions (9). These two electron-dense regions are now known as the inner and outer kinetochores, named the constitutive centromere-associated network (CCAN)3 and the Knl1–Mis12–Ndc80 network, respectively (10, 11). CENP-C, one of the first centromere-associated proteins to be discovered, is a vital CCAN component and functions as a scaffolding protein by specifically recognizing centromere-specific CENP-A nucleosomes and subsequently recruiting other CCAN components to the centromere (12–16). The functional significance of CENP-C is apparent from its remarkable evolutionary conservation. Notably, D. melanogaster and Caenorhabditis elegans kinetochores lack the vast majority of known CCAN components, yet CENP-C homologs are clearly present in these organisms (17, 18). The N-terminal region of CENP-C serves as a recruitment platform for multiple kinetochore proteins including, the Mis12, the CENP-H-I-K-M, and the CENP-L-N complexes, which are critical for building a strong connection to the outer kinetochore and microtubules (see Fig. 1A) (16, 19–24). Although the protein sequence of the CENP-C N-terminal region has diverged among organisms, the CENP-C C-terminal region features two conserved domains: the CENP-C motif and the cupin domain. The CENP-C motif is the hallmark of CENP-C homologs and is required for CENP-A nucleosome recognition (15). The C-terminal end of the protein contains the cupin domain, which is known to homodimerize (25).
Figure 1.
Side and aerial views of CENP-C cupin domain crystal structures. A, classified domains of human CENP-C and their respective binding partners indicated by double-headed arrows. B, crystal structure of the ScMif2CENP-C cupin domain (PDB code 2VPV) (25). Monomers are colored green and light green. C, crystal structure of the SpCnp3CENP-C cupin domain (PDB code 6O2D). Monomers are colored in purple and pink. D, crystal structure of the DmCENP-C cupin domain (PDB code 6O2K). Monomers are colored in blue and cyan. Illustrations of protein structures used in all figures were generated with PyMOL (Delano Scientific, LLC). Both N and C termini are labeled.
The crystal structure of the S. cerevisiae Mif2CENP-C (hereafter referred to as ScMif2CENP-C) cupin domain revealed a homodimeric complex where each monomer is comprised of a nine-stranded jelly roll fold (see Fig. 1B) (25). However, the structural significance of the cupin jelly roll fold is yet to be addressed. In particular, S. cerevisiae point centromeres are genetically specified by conserved DNA elements (CDEI, CDEII, and CDEIII) that are essential for establishing a platform for kinetochore assembly. The composition of the S. cerevisiae inner kinetochore significantly differs from that of higher eukaryotes with no apparent homolog for the CBF3 complex identified at regional centromeres (26). Interestingly, even organisms with regional centromeres have dramatically different inner kinetochore compositions, because D. melanogaster inner kinetochores only seem to consist of CENP-C, with no orthologs for any other CCAN proteins identified to date (18). Therefore, although CENP-C is highly conserved in eukaryotes, its structure and physiological role may vary to accommodate differences in centromere and kinetochore composition. By comparing CENP-C cupin domain structures from organisms with point centromeres (S. cerevisiae) against those with regional centromeres and differing kinetochore compositions (S. pombe and D. melanogaster), we sought to delineate the conserved and unique structural features among cupin domains and further elucidate their functional relevance in kinetochore assembly.
In addition to its role in promoting dimerization, the CENP-C cupin domain can function as a centromere-recruiting factor for key kinetochore components. A former yeast two-hybrid study has demonstrated that the S. pombe Cnp3CENP-C (hereafter referred to as SpCnp3CENP-C) cupin domain interacts with Moa1, a key meiosis I–specific protein and functional equivalent of human MEIKIN, and recruits it to centromeres (23, 27, 28). It is known that kinetochores behave differently in mitosis and meiosis. During meiosis I, sister kinetochores attach to microtubules emanating from the same spindle pole, and centromeric cohesion is maintained to ensure the separation of homologous chromosomes. Moa1 is responsible for monopolar attachment during meiosis I and indirectly recruits Shugoshin to ensure consequent protection of centromeric cohesion (27, 29). The exact details of the interaction between the SpCnp3CENP-C cupin domain and Moa1, including the binding interface and relevant residues, have remained elusive.
Here, we present the crystal structures of the SpCnp3CENP-C and D. melanogaster CENP-C (hereafter referred to as DmCENP-C) cupin domains at 2.52 and 1.81 Å resolution, respectively. Although we find a conserved dimeric core structure within each cupin domain, we find that both the SpCnp3CENP-C and DmCENP-C cupin domains possess additional and unique structural features. Particularly, the SpCnp3CENP-C cupin domain features a distinct β-hairpin at the N-terminal side of the jelly roll core, which is essential for maintaining the stability of the dimeric state in vitro and has mitotic roles in vivo. Additionally, the SpCnp3CENP-C cupin domain features a binding pocket for the meiosis-specific protein Moa1. Thus, our results further reveal a structural role for the SpCnp3CENP-C C terminus in meiosis.
Results
Structural determination of the S. pombe and D. melanogaster CENP-C cupin domains
To elucidate the evolutionarily conserved and/or differentiating features of the CENP-C cupin domain between organisms with point and regional centromeres, we determined the crystal structures of CENP-C cupin domains belonging to two organisms with regional centromeres: S. pombe (SpCnp3CENP-C) and D. melanogaster (DmCENP-C). The crystal structure of the SpCnp3CENP-C cupin domain (residues 489–643) was determined by single-wavelength anomalous diffraction (SAD) with selenomethionine-substituted protein to calculate initial phases. The electron density map at 2.52 Å resolution was used for model building, and the final model was refined at Rwork/Rfree values of 0.217/0.251, respectively (Fig. 1C and Table 1). Residues 489–493 of the SpCnp3CENP-C cupin domain structure were not visible because of their flexible nature. The initial model of DmCENP-C cupin domain (residues 1244–1411) was built based on the 2.63 Å resolution electron density map calculated from SAD phases using selenomethionine-labeled protein. This model was then used for molecular replacement against a 1.81 Å native data set collected from crystals of a longer DmCENP-C cupin domain construct (residues 1190–1411). The electron density for additional residues was visible after molecular replacement. After building the additional residues, the final model was refined at Rwork/Rfree values of 0.196/0.232 (Fig. 1D and Table 1).
Table 1.
Data collection and refinement statistics
The values in parentheses are for the highest-resolution shell. NA, not available.
|
SpCnp3CENP-C cupin domain |
DmCENP-C cupin domain |
||
|---|---|---|---|
| Cnp3 cupin (489–643) | SeMet CENP-C cupin (1244–1411) | Native CENP-C cupin (1190–1411) | |
| Data collection | |||
| Wavelength (Å) | 0.9786 | 0.9786 | 0.9786 |
| Space group | P41212 | P3121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 55.16, 55.16, 206.64 | 86.24, 86.24, 112.24 | 51.93, 61.72, 87.92 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution | 34.44–2.52 (2.59–2.52) | 40.25–2.63 (2.70–2.63) | 30.86–1.81 (1.86–1.81) |
| Rmerge | 0.056 (0.578) | 0.257 (2.327) | 0.041 (0.469) |
| Rpim | 0.016 (0.156) | 0.056 (0.502) | 0.020 (0.257) |
| I/σI | 32.9 (5.0) | 11.8 (1.9) | 21.6 (2.8) |
| Completeness (%) | 99.6 (99.3) | 99.9 (100.0) | 99.6 (99.4) |
| Redundancy | 13.2 (14.5) | 21.8 (22.3) | 4.8 (4.2) |
| Refinement | |||
| Resolution (Å) | 33.07–2.52 | 30.86–1.81 | |
| No. reflections | 11,475 | 25,138 | |
| Rwork/Rfree | 0.217/0.251 | 0.196/0.232 | |
| No. atoms | |||
| Protein | 2246 | 2224 | |
| Ligand/ion | NA | NA | |
| Water | 5 | 128 | |
| B-factors | |||
| Protein (average B-factor) | 71.68 | 29.59 | |
| Ligand/ion | NA | NA | |
| Water | 62.60 | 28.27 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | |
| Bond angles (°) | 1.248 | 0.902 | |
| Ramachandran plot (%) | |||
| Favored | 97.43 | 98.14 | |
| Allowed | 2.57 | 1.86 | |
| Outliers | 0.00 | 0.00 | |
| PDB code | 6O2D | 6O2K | |
Structural comparison of CENP-C cupin domains at point and regional centromeres
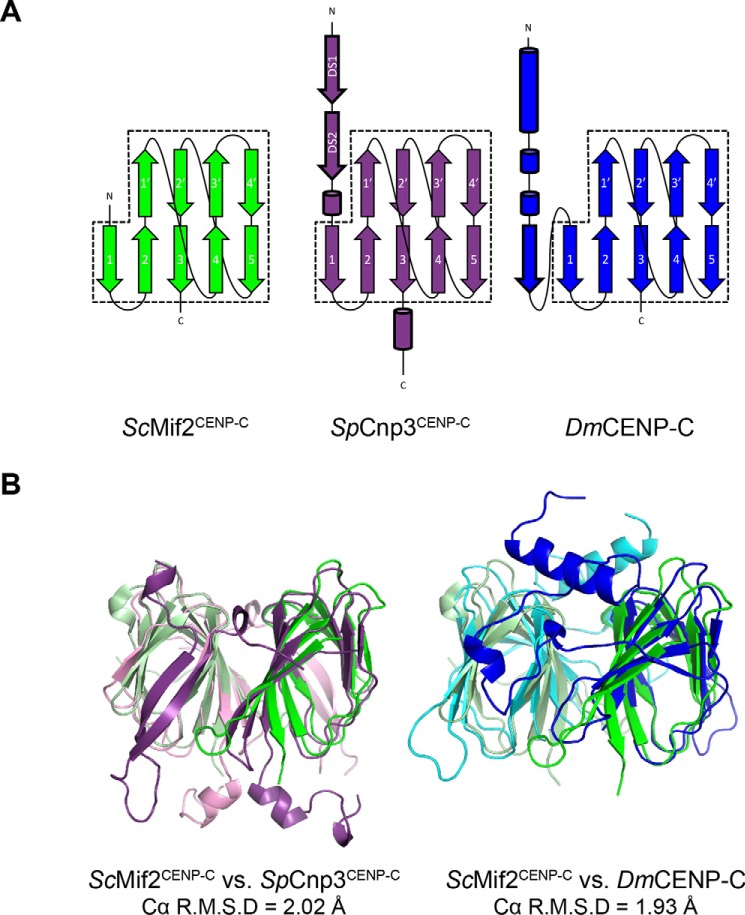
The structural core of both the SpCnp3CENP-C and DmCENP-C cupin domains shows a dimeric complex where each monomer forms a jelly roll fold consisting of nine β-strands that form two antiparallel β sheets (Fig. 2A). The five-stranded sheet (β1–β5) participates in dimerization, whereas the remaining four strands (β1′–β4′) complete the jelly roll fold to form the characteristic shape of the cupin domain. We find that the core nine-stranded fold of the CENP-C cupin domains from organisms possessing point centromeres (ScMif2CENP-C cupin domain; PDB code 2VPV) is well-preserved evolutionarily with those with regional centromeres (SpCnp3CENP-C and DmCENP-C), showing Cα R.M.S. differences of 2.02 Å (ScMif2CENP-C versus SpCnp3CENP-C (532–625; 158 Cα residues)) and 1.93 Å (ScMif2CENP-C versus DmCENP-C (1320–1411; 161 Cα residues)) (Fig. 2B). In both SpCnp3CENP-C and DmCENP-C cupin domains, the two β-sheets (β1–β5 and β1′–β4′) within the jelly roll fold create a pocket-like surface that is often used as a site for metal ion binding in other cupin proteins (30–32).
Figure 2.
CENP-C cupin domains from point and regional centromeres share a core jelly roll fold. A, secondary structure representations of the ScMif2CENP-C, SpCnp3CENP-C, and DmCENP-C cupin domains. The conserved jelly roll fold is comprised of nine β-strands. β-Strands 1–5 participate in dimerization, whereas β-strands 1′–4′ complete the rest of the conserved jelly roll fold (dashed boxes). The regional centromere CENP-C cupin domains of S. pombe and D. melanogaster possess additional secondary structures shown in cylinders (α-helix) and arrows (β-strand) with bold outlines. B, the core jelly roll fold of the ScMif2CENP-C cupin domain (residues 437–530) overlays well with the core fold of the SpCnp3CENP-C cupin domain (residues 532–625, Cα R.M.S. deviation = 2.02 Å) as well as with the core fold of the DmCENP-C cupin domain (residues 1320–1411, Cα R.M.S. deviation = 1.93 Å).
Although the core jelly roll folds are conserved, the CENP-C cupin domains from organisms with regional centromeres exhibit additional features at their N- and C-terminal ends. The SpCnp3CENP-C cupin domain features a β-hairpin and a short α-helix at its N-terminal side and an additional α-helix on the C-terminal end of the jelly roll (Fig. 2). The DmCENP-C cupin domain possesses two extra α-helices and an extra β-strand at the N-terminal side of the jelly roll (Fig. 2). These additional features have not been observed in the ScMif2CENP-C cupin domain. Because the ScMif2CENP-C cupin domain construct that was used for crystallization (residues 365–530) was longer than that within the determined structure (residues 437–530), residues 365–436 of the ScMif2CENP-C cupin domain are likely to be disordered.
Additionally, despite the high level of conservation in the overall architecture of the jelly roll fold, the primary sequence of residues within the dimer interface is not well-preserved among the three CENP-C cupin domain structures (Fig. S1). The ScMif2CENP-C cupin domain mainly relies on hydrophobic interactions along the interface for dimer formation. In contrast, the SpCnp3CENP-C and DmCENP-C cupin domains primarily rely on hydrogen bond interactions formed by their additional secondary structures outside of the jelly roll fold to maintain their dimeric state.
Taken together, although the overall structural features of the ScMif2CENP-C (point centromere) cupin domain are well-preserved in the SpCnp3CENP-C and DmCENP-C (regional centromere) cupin domains, the interactions at the dimer interface are not conserved. The cupin domains from organisms with regional centromeres also have additional structural components. We propose that these additional features may further reinforce the dimerization in the CENP-C cupin domains at regional centromeres.
Additional secondary structures in the CENP-C cupin domains from organisms with regional centromeres contribute to dimerization and protein stability
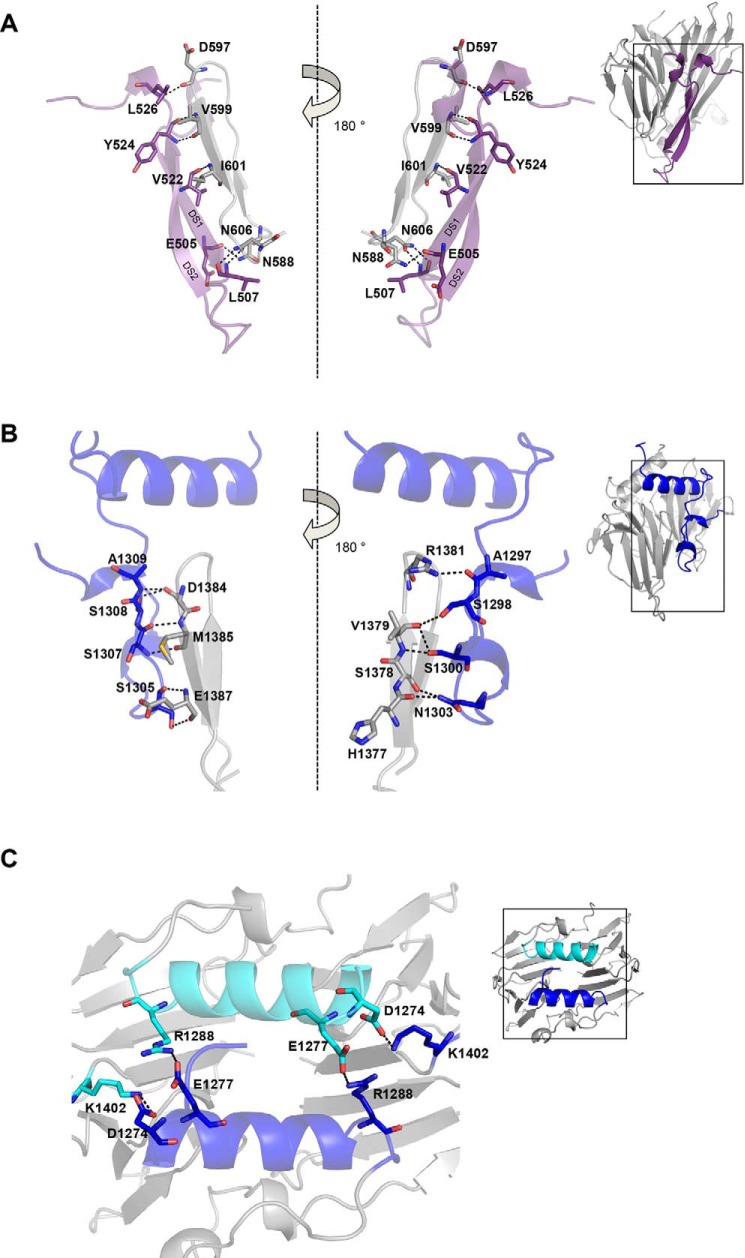
The structural comparison of ScMif2CENP-C cupin domain with the SpCnp3CENP-C and DmCENP-C cupin domains revealed that organisms with regional centromeres encode CENP-C cupin domains that possess secondary structures in addition to the jelly roll folds (Fig. 2). Two β-strands at the N terminus of the SpCnp3CENP-C cupin domain form a β-hairpin structure and then participate in domain swapping between monomers. This unexpected domain-swapped region (DS) is likely to further enforce the dimerization of the SpCnp3CENP-C cupin domain. This is evidenced by the formation of multiple hydrogen bonds between the backbones of the DS2 β-strand and the adjacent β5-strand of the opposing chain (Fig. 3A). Specifically, the backbones of residues Tyr-524 and Val-522 in DS2 hydrogen bond to the backbones of residues Val-599 and Ile-601 in β5-strand, respectively. DS1 residues Glu-505 and Leu-507 also hydrogen bond with nearby loops via the side chains of Asn-588 and Asn-606, respectively. Additionally, the backbones of Leu-526 and Asp-597, residues that are located in adjacent loops, also participate in hydrogen bonding (Fig. 3A). Thr-628, Asp-627, and Arg-634, residues within the C-terminal helical region of the SpCnp3CENP-C cupin domain, further contribute to the dimeric state by interacting with residues along the opposite chain.
Figure 3.
CENP-C cupin domains from organisms with regional centromeres possess additional interactions mediated by their extra secondary structure features. A, zoom in of the SpCnp3CENP-C domain-swapped region to highlight the interactions (dashes) and interacting residues (sticks) between the β hairpin (purple) and the β-strands of the opposite chain (gray). B, zoom in of the DmCENP-C N-terminal region to highlight the interactions (dashes) and interacting residues (sticks) between the loop region (blue) and the β-strands of the opposite chain (gray). C, zoom in of the DmCENP-C N-terminal helices to highlight the residues (sticks) and interactions (dashes) between the two chains (blue and cyan).
A similar domain-swapping feature is also observed within the DmCENP-C cupin domain as a loop in place of the β-hairpin at its corresponding position. Although not as structurally well-defined as the SpCnp3CENP-C β-hairpin, this loop structure may similarly contribute to stabilizing dimerization with several hydrogen bond interactions through a combination of side chain and backbone interactions between the overlapping loop and the adjacent β-strands of the opposing chain. Ser-1305 and Ser-1307 respectively hydrogen bond with Glu-1387 and Met-1385 of β5 of the opposing chain. Asn-1303 hydrogen bonds with His-1377 and Ser-1378 of strand β4′, whereas Ser-1298 and Ser-1300 both interact with Val-1379 of strand β4′. Ala-1297, Ser-1308, and Ala-1309 all form hydrogen bonds with loop regions adjacent to strands β5 and β4′ (Fig. 3B). Lastly, residues Asp-1274, Glu-1277, and Arg-1288, from the N-terminal DmCENP-C helical region contribute to dimerization through interactions with surrounding loops via Lys-1402, Arg-1288, and Glu-1277, respectively (Fig. 3C).
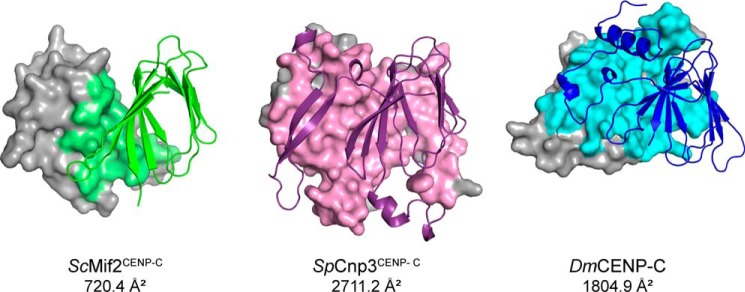
The calculated surface area of the dimer interface of the ScMif2CENP-C cupin domain is 720.4 Å2 per monomer (33). The corresponding surface areas within the SpCnp3CENP-C and DmCENP-C cupin domains are 2711.2 and 1804.9 Å2 per monomer, respectively (Fig. 4) (33). The differences in interface surface area between CENP-C cupin domains from organisms with point versus regional centromeres strongly indicate that the dimerization of CENP-C cupin domains at regional centromeres needs to be further strengthened by the additional secondary structure components.
Figure 4.
CENP-C cupin domains at regional centromeres (SpCnp3CENP-C and DmCENP-C) possess more expansive dimer interfaces than that at point centromeres (ScMif2CENP-C). One monomer in each structure is represented as a cartoon, and interfacing residues are colored accordingly within the gray surface representation of the opposite monomer. Dimer interface areas were calculated using PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html).4
The dimeric state of the CENP-C cupin domain is evolutionarily conserved in vitro
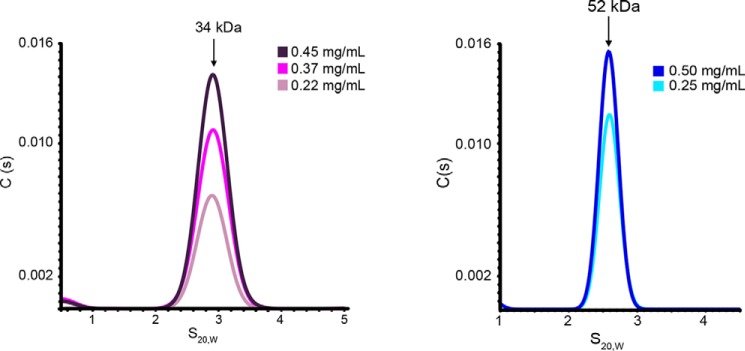
To analyze the oligomeric state of the SpCnp3CENP-C and DmCENP-C cupin domains, sedimentation velocity analytical ultracentrifugation (SV-AUC) was employed. SV-AUC analysis of purified SpCnp3CENP-C and DmCENP-C cupin domains each showed a single peak with estimated molecular masses of 34 kDa (expected molecular mass of the monomer is 16.8 kDa) and 52 kDa (expected molecular mass of the monomer is 24.9 kDa), respectively (Fig. 5). This demonstrates that both cupin domains form homodimers in solution, similarly to the ScMif2CENP-C cupin domain (25).
Figure 5.
CENP-C cupin domains encoded by organisms with regional centromeres are dimeric in vitro. SV-AUC analysis of the SpCnp3CENP-C (left panel) and DmCENP-C (right panel) cupin domains shows that both are dimeric in vitro. The colors represent samples of differing concentrations. The expected molecular mass of the SpCnp3CENP-C cupin monomer is 16.8 kDa. The expected molecular mass of the DmCENP-C cupin monomer is 24.9 kDa.
The domain-swapped region is essential to maintain the dimeric state and structural integrity of the SpCnp3CENP-C cupin domain
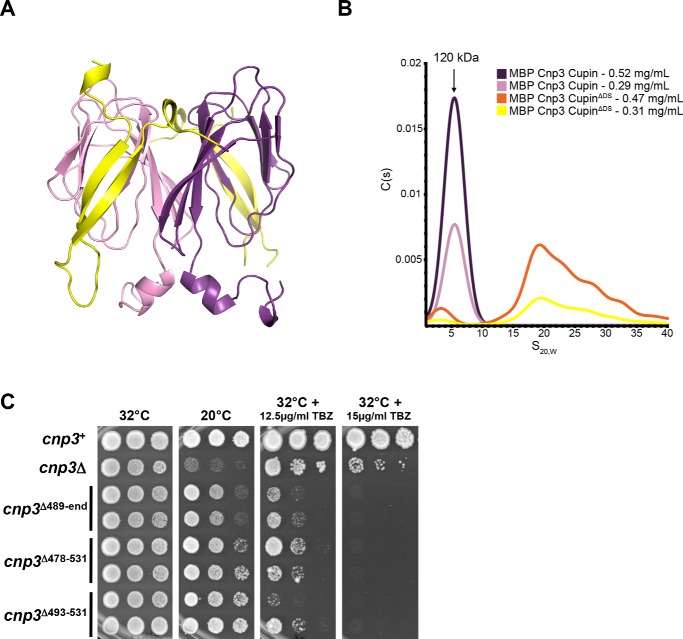
To further investigate the functional role of the unique SpCnp3CENP-C β-hairpin structure, an N-terminally truncated construct of the SpCnp3CENP-C cupin domain was generated by removing the domain-swapped region (Cnp3 CupinΔDS; residues 532–643) and subsequently subjected to SV-AUC analysis (Fig. 6A). Analysis of maltose-binding protein (MBP)–tagged Cnp3 CupinΔDS shows distinct peak shifts compared with MBP-tagged WT SpCnp3CENP-C cupin domain (residues 489–643) with a lower molecular mass population as well as a higher molecular mass population (Fig. 6B). The latter is likely to be higher molecular mass aggregates. Notably, cleavage of the MBP tag from MBP-tagged Cnp3 CupinΔDS for further purification resulted in protein aggregation that was unable to be further purified. This is in stark contrast to the tag cleavage and purification of the WT SpCnp3CENP-C cupin domain (residues 489–643) that could be purified in large amounts and crystallized without any issues with protein stability.
Figure 6.
The DS region (β-hairpin) is required for maintaining the dimeric state in vitro and promotes SpCnp3CENP-C function in vivo. A, the MBP-tagged SpCnp3CENP-C CupinΔDS construct (residues 532–643) does not contain the N-terminal β hairpin (yellow) of the SpCnp3CENP-C cupin domain. This construct was used for SV-AUC analysis to determine the significance of the SpCnp3CENP-C DS region. B, SV-AUC analysis of the purified MBP-tagged SpCnp3CENP-C CupinΔDS protein (orange, yellow) reveals that it sediments as higher molecular mass aggregates. MBP-tagged SpCnp3CENP-C cupin (purple, pink) continues to sediment as a dimeric population. Expected molecular mass of the MBP-tagged SpCnp3CENP-C cupin monomer is 62.4 kDa. C, cnp3Δ478–531 and cnp3Δ493–531 DS truncation mutants display hypersensitivity to TBZ but show little cold sensitivity compared with cnp3Δ cells. 5-fold serial dilutions of cells of the indicated genotypes were spotted on YES medium supplemented with or without the indicated concentrations of TBZ and incubated at the indicated temperatures for 3–7 days. Two independent isolates of each genotype (cupin domain truncation) are shown. Also see Fig. S2.
To investigate whether the domain-swapped region is required for SpCnp3CENP-C function in vivo, we generated S. pombe strains expressing internally truncated mutants of SpCnp3CENP-C, from the cnp3 endogenous locus. cnp3Δ493–531 cells expressed the otherwise full-length SpCnp3CENP-C protein selectively missing the domain-swapped region alone, i.e. residues 493–531, which form part of the N terminus within the solved SpCnp3CENP-C cupin domain structure. Similarly, cnp3Δ478–531 cells expressed the otherwise full-length SpCnp3CENP-C protein missing residues 478–531, which we predicted would additionally eliminate the disordered region preceding the domain-swapped region. A strain expressing a C-terminally truncated mutant cnp3Δ489-end eliminating the entire cupin domain of SpCnp3CENP-C, i.e. residues 489–643, was also generated as a control. Although cnp3Δ493–531 and cnp3Δ478–531 cells displayed very little cold sensitivity when compared with cnp3Δ cells, they showed hypersensitivity to the microtubule depolymerizing drug thiabendazole (TBZ), much like cnp3Δ cells, suggesting that mitotic chromosome segregation is disrupted in the absence of the domain-swapped region within SpCnp3CENP-C (Fig. 6C). This effect could at least in part be attributed to the significantly diminished ability of Cnp3Δ493–531 and Cnp3Δ478–531 to localize to fission yeast centromeres, as found by live cell imaging and co-immunofluorescence assays (Fig. S2). These results collectively suggest that the domain-swapped region within the cupin domain is essential for SpCnp3CENP-C function in vivo.
Taking these results together, we conclude that unlike the ScMif2CENP-C cupin domain, the jelly roll fold architecture alone is not sufficient to maintain the stability of the SpCnp3CENP-C cupin dimer. The domain-swapped region at the N terminus of the SpCnp3CENP-C cupin domain plays an important role in stabilizing and maintaining the dimeric state of the SpCnp3CENP-C cupin domain and is essential for its function in vivo.
Moa1 binds to the inner pocket of the SpCnp3CENP-C cupin domain
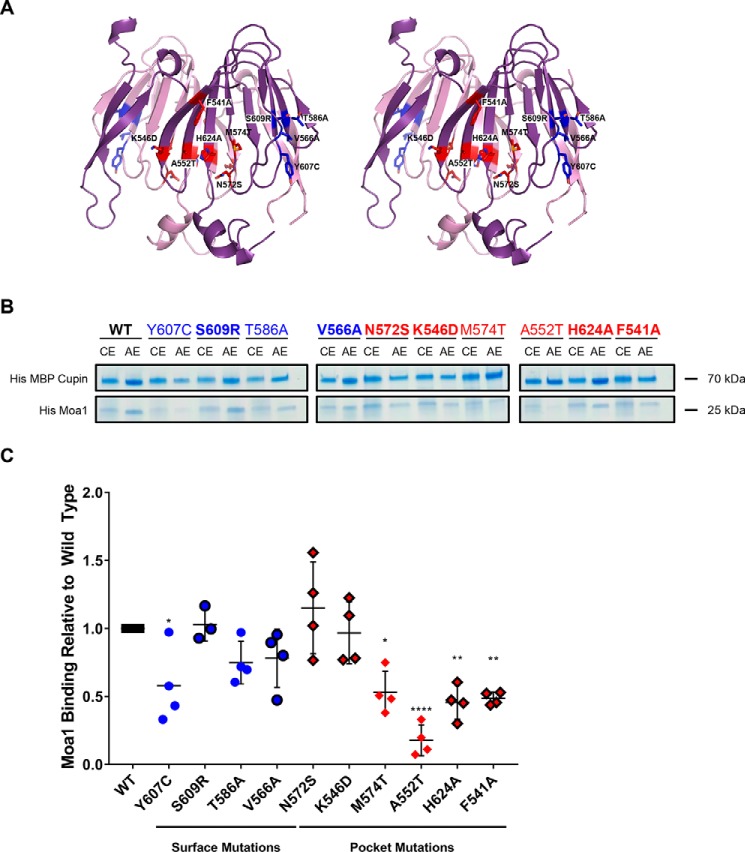
A previous study has demonstrated an association between the SpCnp3CENP-C cupin domain and the meiosis-specific protein Moa1 (23). To elucidate the structural role of the cupin fold and map the binding region required for Moa1 recruitment to centromeres, we introduced 10 point mutations within the SpCnp3CENP-C cupin domain that can be broadly classified into two categories: those within the pocket (N572S, K546D, M574T, A552T, H624A, and F541A) and those on the surface (Y607C, S609R, T586A, and V566A) of the cupin domain (Fig. 7A). Mutations Y607C, T586A, M574T, and A552T were used as controls for the binding experiments, because they have already been shown to inhibit SpCnp3CENP-C interaction with Moa1 in vivo (23). His-MBP-tagged SpCnp3CENP-C cupin (WT or mutated) and His-tagged Moa1 were co-expressed using a polycistronic construct and subsequently subjected to amylose-resin affinity pulldowns to determine the relative amount of mutant complex formation compared with WT protein (Fig. 7B). Moa1 was only stably expressed and remained soluble when it was co-expressed with the SpCnp3CENP-C cupin domain. Five SpCnp3CENP-C cupin domain mutations were found to significantly reduce Moa1 binding: Y607C, M574T, A552T, H624A, and F541A, and notably the latter four mutations are all positioned within the cupin domain–binding pocket (Fig. 7C). Therefore, the affinity pulldown experiments indicate that the pocket within the SpCnp3CENP-C cupin domain is responsible for Moa1 interaction.
Figure 7.
Moa1 associates with the inner binding pocket of the SpCnp3CENP-C cupin domain. A, point mutations were designed to determine the Moa1 binding interface in the SpCnp3CENP-C cupin domain. The mutations (stick side chains) were split into two categories: pocket mutations (red) and surface mutations (blue). B, amylose affinity pulldowns were performed with the His-MBP-tagged WT and mutant SpCnp3CENP-C cupin domains to determine the binding interface used for His-tagged Moa1 binding. Cobalt affinity column elutions (CE) show the initial expression levels of both components of the complex. Amylose affinity column elutions (AE) show the amount of Moa1 that was able to form a complex with the SpCnp3CENP-C cupin domain within each sample. Mutations that have been previously shown to disrupt Moa1 recruitment in vivo are not shown in bold (23). C, quantification of the AE pulldown band intensity ratios of each mutant relative to WT. Five SpCnp3CENP-C cupin domain point mutations, Y607C, M574T, A552T, H624A, and F541A, significantly reduce complex formation between His-MBP Cupin and His-Moa1. Four of these mutations are located within the cupin domain inner pocket (red). Points not shown in bold are controls that have been shown to disrupt Moa1 recruitment in vivo (23). One-way analysis of variance (n = 4, means with S.D.) was used. *, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001. One outlier was removed from S609R analysis using Grubbs's test.
Differential scanning calorimetry (DSC) experiments were performed to confirm that disrupted Moa1 complex formation observed in a subset of SpCnp3CENP-C cupin mutants is due to the mutation of critical residues required for the interaction and not due to improper cupin domain folding. DSC experiments show that WT cupin domain has a denaturation temperature of 48.34 °C, whereas the mutations that significantly disrupted Moa1 binding, Y607C, M574T, A552T, H624A, and F541A, have denaturation temperatures of 47.34, 42.08, 48.62, 51.13, and 47.14 °C, respectively (Fig. S3). Thus, the DSC experiments indicate that mutations that significantly disrupt the association of the SpCnp3CENP-C cupin domain with Moa1 do not disturb the structural integrity of the cupin domain, suggesting that they exclusively affect Moa1 binding. Notably, the M574T mutant does exhibit a lower denaturation temperature compared with the WT protein and other point mutants. We have verified that this mutant maintains a dimeric state (data not shown). However, we cannot rule out the possibility that this pocket mutation may destabilize the cupin domain in another manner.
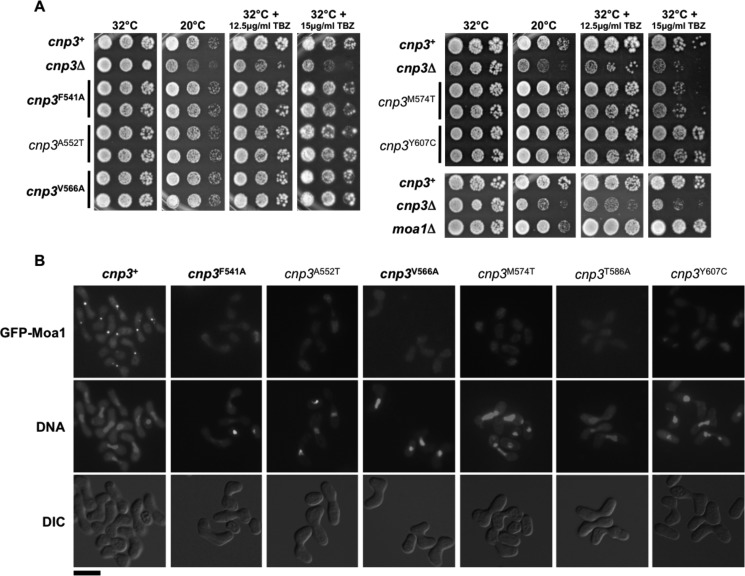
A subset of the 10 mutations listed above were also tested for their effects on SpCnp3CENP-C function and Moa1 localization in vivo, because Cnp3CENP-C has been shown to recruit Moa1 to centromeres during S. pombe meiosis (23). Mutations F541A, A552T, M574T (pocket), or Y607C, V566A (surface) were introduced into the endogenous cnp3 genomic locus. A552T and Y607C served as controls because they have been previously shown to specifically disrupt SpCnp3CENP-C function in meiosis but not mitosis. Cells expressing Cnp3A552T or Cnp3Y607C display neither cold sensitivity nor TBZ sensitivity while being unable to recruit Moa1 to centromeres during meiosis I (Fig. 8) (23). We observed very similar phenotypes for cells expressing Cnp3F541A (pocket) and Cnp3V566A (surface), despite V566A only mildly affecting Moa1 binding in vitro. Cnp3M574T-expressing cells additionally displayed mild TBZ sensitivity as has been previously demonstrated (Figs. 7C and 8 and Fig. S4) (23). Together, these results suggest that mutations in the SpCnp3CENP-C cupin domain, mainly in the pocket region, disrupt Moa1 association in vitro, and consequently Moa1 recruitment to centromeres during meiosis in vivo. On the other hand, mutations in the cupin domain have largely no effects on nonmeiotic functions of SpCnp3CENP-C, thus highlighting a largely meiosis-specific role for the SpCnp3CENP-C cupin fold pocket in S. pombe.
Figure 8.
Point mutations within the SpCnp3CENP-C cupin domain exclusively affect its function in meiosis, through disruption of Moa1 recruitment to centromeres. A, SpCnp3CENP-C cupin domain point mutants (pocket and surface) largely display no TBZ or cold sensitivity when compared with cnp3Δ cells (note that moa1Δ cells also display no TBZ or cold sensitivity). 5-fold serial dilutions of cells of the indicated genotypes were spotted on YES medium supplemented with or without the indicated concentrations of TBZ and incubated at the indicated temperatures for 3–7 days. Two independent isolates of each genotype (cupin domain mutant) are shown. Point mutants unique to this study are in bold, whereas previously described mutants are not (23). B, point mutations within the SpCnp3CENP-C cupin pocket (F541A, A552T, and M574T) and surface (V566A, T586A, and Y607C) result in failure to recruit Moa1 to centromeres during meiosis I. Homothallic (h90) fission yeast cells of the indicated genotypes expressing GFP-tagged Moa1 were induced into meiosis at 32 °C and arrested in meiotic prophase I using a mei4Δ allele. DNA was stained with 4′,6′-diamino-2-phenylindole, following which cells were imaged live. Point mutants unique to this study are in bold, whereas previously described mutants are not (23). Scale bar, 10 μm. Also see Fig. S4. DIC, differential interference contrast.
Discussion
Here, we present and analyze the structures of CENP-C cupin domains from two evolutionarily distinct organisms with regional centromeres: S. pombe Cnp3CENP-C and D. melanogaster CENP-C. Along with the previously published crystal structure of the S. cerevisiae Mif2CENP-C cupin domain, these three structures originate from organisms with diverse centromere architecture and kinetochore composition. We sought to define and distinguish the conserved and unique structural features of the CENP-C cupin domain at two different types of centromeres, point and regional, to gain a better understanding of the evolutionary conservation of CENP-C cupin domain function.
Through structural comparison, we found that the core nine-stranded jelly roll fold is conserved among these structures. However, the mechanisms of dimerization differ between cupin domains associating with regional versus point centromeres because of the former relying on secondary structures additional to the jelly roll fold and thus creating a larger dimerization interface.
We further discovered that the cupin domain contains an inner pocket, which plays an important role in recruiting key binding partners such as Moa1 in S. pombe. Through structure-guided mutagenesis of the SpCnp3CENP-C cupin domain, we revealed that the inner binding pocket is essential for association with Moa1. We further demonstrated that mutation of key pocket residues in vivo leads to failure in recruiting Moa1 to centromeres in meiosis I.
Comparison of the CENP-C cupin domain dimer interface in organisms with point and regional centromeres
One clear distinction between CENP-C cupin domains at point and regional centromeres is the interactions at the dimer interface. The ScMif2CENP-C (point centromere) cupin domain forms a dimer through the jelly roll fold, which is likely sufficient to form the ScMif2CENP-C dimer (Fig. 1B). Although it is possible that the ScMif2CENP-C cupin domain contains extra structural features at its C-terminal end, it is unlikely that the ScMif2CENP-C cupin domain contains any additional secondary structure features at its N-terminal region. The construct used for crystallization of the ScMif2CENP-C cupin domain consisted of extra residues on the N-terminal side that were not present in the final structure and are therefore, likely to be unstructured. CENP-C cupin domains at regional centromeres, however, seem to require additional secondary structures to form a stable dimer, with a larger dimer interface surface (Figs. 1, B–D, and 4). The SpCnp3CENP-C cupin domain possesses an additional secondary structure element, namely a β-hairpin (domain-swapped region) N-terminal to the jelly roll fold. Our experiments both in vitro and in vivo demonstrate that this additional secondary structure element is essential for the stability and function of SpCnp3CENP-C. Deletion of the Mif2CENP-C cupin domain in S. cerevisiae results in a temperature-sensitive phenotype (25). Analogously, deletion of the Cnp3CENP-C cupin domain (cnp3Δ489-end) in S. pombe results in mild cold sensitivity and hypersensitivity to thiabendazole, suggesting that dimerization mediated by the cupin domain is essential for SpCnp3CENP-C function in vivo (Fig. 6C).
Similar to SpCnp3CENP-C, the D. melanogaster CENP-C cupin domain also contains additional loop and helical structures to aid in dimerization. The comparison of dimer interfaces across the three analyzed orthologous CENP-C cupin domains further supports our observation that the cupin domains associated with regional centromeres possess dimerization interfaces over twice the size of the ScMif2CENP-C interface (point centromere) (Fig. 4). Additional structural analyses of CENP-C cupin domains from other organisms will be required to determine whether extra secondary structures and a larger dimerization interface are consistent features of CENP-C cupin domains found at regional centromeres.
The point centromere has a well-defined centromeric DNA sequence bound by a single CENP-ACse4 nucleosome, whereas the regional centromere is composed of a larger centromeric DNA sequence (several kilobases to several megabases) with multiple CENP-A nucleosomes sporadically incorporated together with canonical histone H3 nucleosomes (34–36). In fact, CENP-A nucleosomes have been shown to cluster toward the centromeric surface to maximize their accessible surface area for inner kinetochore recognition (35, 37). Accordingly, several models have been proposed to explain how CENP-A and H3 centromeric nucleosomes are organized (38). The potential role of CENP-C in organizing CENP-A nucleosomes at the regional centromere has been proposed in an earlier study (14). In this model, CENP-C utilizes its CENP-A recognition domain and cupin domain to function as a bridge to connect neighboring CENP-A nucleosomes. Compared with the point centromere, bridging of CENP-A nucleosomes at the regional centromere may require a larger and stronger dimerization interface. Indeed, former studies have shown that CENP-C depletion in HeLa cells affects the size, shape, and structural integrity of the inner kinetochore plate in electron micrographs (39).
The S. pombe Cnp3CENP-C cupin fold forms a functional binding pocket for Moa1
Using the SpCnp3CENP-C cupin domain, we further demonstrate that the characteristic cupin fold has structural significance beyond dimerization, by forming a pocket for binding partners. Although the majority of residues within the pocket are not structurally conserved among determined CENP-C cupin domain structures, they all possess a similar pocket that is internally lined with predominantly hydrophobic and aromatic residues. Notably, the crystal packing observed in the SpCnp3CENP-C cupin domain revealed that the C-terminal tail of an adjacent symmetry mate fits within the pocket opening, likely mimicking the pocket binding with its partner (Fig. S5A). The utilization of the cupin pocket is not unique to SpCnp3CENP-C and has been shown to bind metals and sugars in other cupin proteins (Fig. S5B) (32, 40). It is therefore likely that the residues along the interior of the cupin pocket may be specifically tailored to its respective binding partner(s) to optimize partner binding.
Here, we show that the interaction between the SpCnp3CENP-C cupin domain and meiosis-specific protein Moa1 occurs at this internal cupin domain pocket. Our in vitro pulldowns as well as in vivo recruitment studies demonstrate that Moa1 no longer interacts with the cupin domain and fails to localize to the centromere in meiosis I when key residues within the cupin inner pocket are mutated (Figs. 7 and 8 and Fig. S4). However, when these pocket mutants are monitored during mitosis in vivo, they display no abnormal phenotypes, further suggesting that the SpCnp3CENP-C cupin-binding pocket is specific for Moa1 recruitment during meiosis I (Fig. 8). A former study already showed that two mutations A552T and Y607C in the SpCnp3CENP-C cupin domain severely affect meiotic chromosome segregation (23). Interestingly, in our pulldown studies, Y607C, a surface mutation, significantly decreased Moa1 binding. Because Y607C is the only surface mutation tested to significantly affect Moa1 binding in vitro, it is likely that Moa1 uses this residue as a secondary contact point for additional interactions. Interestingly, another tested surface mutation, V566A, significantly disrupted centromere localization of Moa1 in vivo while only mildly affecting Moa1 interaction in vitro, much like T586A as previously demonstrated (Figs. 7 and 8B) (23). These observations further support the likelihood of the cupin domain surface being involved in mediating secondary interactions between SpCnp3CENP-C and Moa1.
The CENP-C cupin pocket is likely to mediate crucial interactions in metazoans
The SpCnp3CENP-C cupin–Moa1 interaction may be conserved in vertebrates because MEIKIN, the functional mouse homolog of Moa1, has been shown to bind to the C-terminal region of mouse CENP-C (28). Therefore, it is likely that MEIKIN binds to the cupin domain of CENP-C via the conserved cupin pocket during meiosis. Additionally, there is evidence to suggest that the D. melanogaster CENP-C cupin pocket likely binds Cal1, an evolutionarily distinct CENP-A chaperone with functions in both mitosis and meiosis (41).
In summary, our study uncovers crucial details about the structure and function of the conserved cupin domain of CENP-C in organisms with point and regional centromeres. Our findings provide insights into how CENP-C proteins might have evolved to interact with and recruit binding partners to centromeres in a wide range of eukaryotes. Our results have important implications for understanding how CENP-C, one of the mostly highly conserved centromere proteins, regulates chromosome segregation in both mitosis and meiosis.
Experimental procedures
Purification of S. pombe Cnp3 cupin domain
N-terminal His-MBP tagged Cnp3 489–643 was cloned into the pET3a vector and expressed in Escherichia coli Rosetta (DE3) cells using PA-5052 (native protein) or PASM-5052 (selenium-labeled protein) auto-inducible medium (42). Harvested cells were resuspended in 30 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 3 mm β-mercaptoethanol with protease inhibitor cocktails. After sonication on ice for 2 min, soluble lysate was recovered by centrifugation at 34,541 × g for 1 h. Lysate was applied to a cobalt affinity column (Takara) pre-equilibrated with buffer A (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 3 mm β-mercaptoethanol). The resin was subsequently washed with buffer A, a high salt buffer (30 mm Tris-HCl, pH 8.0, 1 m NaCl, 3 mm β-mercaptoethanol), and once more with buffer A before eluting with the elution buffer (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 300 mm imidazole, and 3 mm β-mercaptoethanol). The N-terminal His-MBP tags were cleaved by tobacco etch virus protease (1:100 ratio) while dialyzing in 30 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm DTT at 4 °C overnight. The cleaved tags were removed with secondary cobalt and amylose (New England Biolabs) affinity columns. The amylose column flow through fraction was applied to the HiTrap Q HP anion exchange column (GE Healthcare) with a NaCl gradient (50 mm to 1 m NaCl), and fractions containing the S. pombe Cnp3 cupin domain were pooled. Pooled fractions were concentrated using an Amicon centrifugal filter (Millipore Sigma) and applied to a HiLoad 16/600 Superdex 200-pg size exclusion chromatography column (GE Healthcare) pre-equilibrated with 30 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine (TCEP). Desired fractions were pooled and applied to a final amylose column to remove any remaining MBP, and the collected flow through was concentrated for crystallization and subsequent biochemical assays.
Purification of D. melanogaster CENP-C cupin domain
The overall cloning and protein expression procedure of native and selenomethionine-substituted D. melanogaster CENP-C cupin domains (residues 1244–1411 and 1190–1411) was the same as the S. pombe Cnp3 cupin domain, described above. The native CENP-C 1190–1411 protocol did not require the HP-Q column before being applied to the Superdex 200 column.
Crystallization and structure determination of the S. pombe Cnp3 cupin domain
Crystals of the S. pombe Cnp3 cupin domain were grown using the hanging-drop diffusion method at room temperature. Plate-shaped crystals were obtained by mixing purified proteins (12.7 mg/ml) with 0.1 m HEPES (pH 7.0), 20% PEG 3350, and 6% (w/v) trimethylamine N-oxide dihydrate in a 1:1 ratio (v/v). The reservoir solution was made up of 0.25 m potassium fluoride, 0.125 m HEPES (pH 7.0), and 25% PEG 3350. Crystals were cryo-protected in reservoir solution with a final concentration of 35% PEG 3350 and flash-frozen in liquid nitrogen. A 2.52 Å data set of a selenomethionine-substituted S. pombe Cnp3 cupin domain crystal was collected at Advanced Photon Source (APS) on Beamline 21-ID-G (LS-CAT) at the wavelength of the selenium anomalous peak position (λ = 0.9786 Å). The data set was indexed and scaled using XDS, and it belonged to the space group of P 41212 with the unit cell size of a = 55.16 Å, b = 55.16 Å, c = 206.64 Å, α = β = γ = 90° (43, 44). The electron density map was generated via PHENIX.autosol (45). The initial model was built using PHENIX.autobuild, and the model building and structure refinement were done using the programs COOT and PHENIX.refine, respectively (46–48). The final refined model has Rwork/Rfree values of 0.217/0.251, and the Ramachandran analysis was done using MolProbity with the result of 97.43 (favored), 2.57 (allowed), and 0.00% (outlier) (49).
Crystallization and structure determination of D. melanogaster CENP-C cupin domains
Purified native D. melanogaster CENP-C cupin domain protein (residues 1244–1411) at 19.7 mg/ml was used for crystallization using the hanging-drop diffusion method. Ridged oval-shaped crystals were obtained by mixing proteins with 0.2 m NaCl, 0.1 m MES (pH 6.0), 15% (v/v) pentaerythritol propoxylate (5/4 PO/OH) in a 1:1 ratio (v/v). Selenomethionine-substituted crystals of this short CENP-C cupin domain were grown in the same condition by providing native crystals as microseeds. These crystals were then cryo-protected in 0.2 m NaCl, 0.1 m MES (pH 6.0), 15% (v/v) pentaerythritol propoxylate (5/4 PO/OH) containing 20% glycerol as a cryo-protectant and quickly frozen in liquid nitrogen. The 2.63 Å SAD data set of the D. melanogaster CENP-C cupin domain (residues 1244–1411) was collected at APS using Beamline 21-ID-G at the wavelength of 0.9786 Å. The collected data were further processed using Mosflm and Aimless (50, 51). The diffracted crystal belonged to the space group P3121 with the unit cell dimensions of a = 86.24 Å, b = 86.24 Å, c = 112.24 Å, α = γ = 90°, and β = 120°. The initial SAD map was generated using PHENIX.autosol (45). Using PHENIX.autobuild, an initial model was obtained and was further manually built and refined using the program COOT and PHENIX.refine, respectively (46–48). The obtained model was used for molecular replacement against the native data set of D. melanogaster CENP-C cupin domain (residues 1190–1411), which diffracted to 1.81 Å resolution.
The native crystals of the D. melanogaster CENP-C cupin domain (residues 1190–1411) were grown by mixing purified proteins (21 mg/ml) and the precipitation solution (0.1 m MOPS (pH 7.0) and 12% (w/v) PEG 4000) in a 1:1 ratio (v/v) using the hanging-drop diffusion method at room temperature. The crystals were further cryo-protected with a final concentration of 35% (w/v) PEG 4000 and frozen in the liquid nitrogen. The data set was collected at a 0.9786 Å wavelength under cryogenic conditions at APS using Beamline 21-ID-G. The 1.81 Å native data set of the space group P212121 and unit cell dimensions of a = 51.93 Å, b = 61.72 Å, c = 87.92 Å, α = β = γ = 90° was indexed and scaled using the program XDS (43, 44). The initial phases were calculated by molecular replacement (Phaser) using the model built from the short construct of the D. melanogaster CENP-C cupin domain (residues 1190–1411) as a search model (52). Further model building and the refinement were done using the programs COOT and PHENIX.refine, respectively (47, 48). Residues 1190–1269 were likely proteolytically cleaved during the crystallization process and were not visible within our structure. The Rwork and Rfree values of the final refined model of D. melanogaster CENP-C cupin domain (residues 1190–1411) were 0.196 and 0.232, respectively. The final model has the Ramachandran plot of favored/allowed/disallowed with 98.14/1.86/0.00% based on MolProbity (49).
Cupin Moa1 binding assays
N-terminal tagged His-MBP Cupin and His Moa1 were cloned as a polycistronic construct into the pET3a vector using ligation-independent cloning (53). Point mutations were introduced to the cupin domain via QuikChange II XL site-directed mutagenesis kit (Agilent). The proteins were subsequently expressed in 1 liter of E. coli Rosetta (DE3) cells using PA-5052 auto-inducible medium (42). Harvested cells were resuspended in 30 ml of buffer consisting of 30 mm Tris-HCl (pH 8.0), 500 mm NaCl, 5% glycerol, and 3 mm β-mercaptoethanol with protease inhibitor cocktails. After sonication and subsequent spin down, the lysate was applied to affinity columns. Both columns were equilibrated with buffer B (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5% glycerol, 3 mm β-mercaptoethanol) before sample loading. 15 ml of cell lysate was loaded onto 3 ml of cobalt resin (Takara) and allowed to drain through. The resin was then washed with 20 ml of buffer B, and then bound protein was eluted with 10 ml of elution buffer (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5% glycerol, 300 mm imidazole, and 3 mm β-mercaptoethanol). The remaining 15 ml of cell lysate was rocked with 2 ml of amylose resin (New England Biolabs) at room temperature for 1 h. The resin was subsequently washed with 20 ml of buffer B, and protein-eluted with 5 ml of elution buffer (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 20 mm maltose, 5% glycerol, 3 mm β-mercaptoethanol). The assay was replicated four times for WT cupin domain and each point mutant. Amylose elution gel band intensities (AEIs) were quantified using ImageJ (54). His MBP Cupin AEI/His Moa1 AEI ratios were calculated for each sample. WT AEI ratios were then divided by the AEI ratio for each sample, and this value was subsequently used in GraphPad Prism 7.00 for Windows (GraphPad Software, La Jolla, CA) to run statistical analysis.
Differential scanning calorimetry
The denaturation temperature of WT Cnp3 cupin domain and each significant point mutant was measured using a Nano DSC (TA Instruments). Nano DSC was first conditioned with 0.3 ml of buffer made of 30 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 1 mm TCEP in both reference and sample cells. Program for conditioning run consisted of a temperature range of 25 °C to 80 °C, a scan rate of 2 °C/min, pressure set at 3 atm, and a 60-s equilibration. The same buffer samples were used for a subsequent baseline run with a temperature range of 25 to 80 °C, a scan rate of 1 °C/min, pressure set at 3 atm, and a 60-s equilibration. Protein sample concentrations ranged from 0.66 to 3.17 mg/ml in a buffer consisting of 30 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 1 mm TCEP. 1 ml of protein sample was degassed under vacuum for 15 min before loading. Sample cell was loaded with 0.3 ml of protein, and the same program conditions were used as baseline run. The data were processed using NanoAnalyze Software (TA Instruments).
SV-AUC
SV-AUC was carried out using 420 μl of sample loaded into two-sector Epon centerpieces with a 1.2-cm path-length (Beckman Coulter, Indianapolis, IN) in an An60Ti rotor in a Beckman Optima Xl-I analytical ultracentrifuge and run at 22 or 6 °C. The sedimentation was initiated after at least 2 h of temperature equilibration. Measurement was completed in intensity mode. Sedimentation was monitored by absorbance at 280 nm at 42,000 rpm. The samples were prepared as mentioned.
All SV-AUC data were analyzed using UltraScan III software, version 4.0, and fitting procedures were completed on XSEDE clusters at the Texas Advanced Computing Center (Lonestar, Stampede, jetstream) through the UltraScan Science Gateway (https://www.xsede.org/web/guest/gateways-listing)4 (55). The partial specific volume (vbar) of each species was estimated within UltraScan III based on the protein sequence. Raw intensity data were converted to pseudo-absorbance by using the intensity of the air above the meniscus as a reference and edited. Next, 2D sedimentation spectrum analysis was performed to subtract time-invariant noise, and the meniscus was fit using 10 points in a 0.05-cm range (56). A sedimentation coefficient distribution of 1–20 for the DmCENP-C cupin domain, 1–50 for MBP SpCnp3CENP-C Cupin, or 0.5–5 for the untagged SpCnp3CENP-C cupin domain was fitted. The arrays were fitted using an S range of 1–20, 1–50, or 0.5–5 S, an f/f0 range of 1–4 with 64 grid points for each, 10 uniform grid repetitions, and 400 simulation points. 2D sedimentation spectrum analysis was then repeated at the determined meniscus to fit radially invariant and time-invariant noise together using 10 iterations.
His-MBP tagged Cnp3 CupinΔDS purification
N-terminal His-MBP tagged Cnp3 489–643 was cloned into the pET3a vector and expressed in E. coli Rosetta (DE3) cells using PA-5052 (42). Harvested cells were resuspended in 30 mm Tris-HCl (pH 8.0), 500 mm NaCl, and 3 mm β-mercaptoethanol with protease inhibitor cocktails. After sonication on ice for 1.5 min, soluble lysate was recovered by centrifugation at 34,541 × g for 1 h. The lysate was applied to a cobalt affinity column (Takara) pre-equilibrated with buffer A (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 3 mm β-mercaptoethanol). The resin was subsequently washed with buffer A, a high salt buffer (30 mm Tris-HCl, pH 8.0, 1 m NaCl, 3 mm β-mercaptoethanol) and once more with buffer A before eluting with the elution buffer (30 mm Tris-HCl, pH 8.0, 500 mm NaCl, 300 mm imidazole, and 3 mm β-mercaptoethanol). The elution was concentrated using an Amicon centrifugal filter (Millipore Sigma) and applied to a HiLoad 16/600 Superdex 200-pg size exclusion chromatography column (GE Healthcare) pre-equilibrated with 30 mm Tris-HCl (pH 8.0), 500 mm NaCl, 1 mm TCEP. Desired fractions were diluted to differing A280 values for AUC analysis. The purification protocol was identical for the MBP Cnp3 CupinΔDS samples.
S. pombe strains and growth assays
Standard methods were used for fission yeast growth, genetics, and manipulation (57). Gene deletion, tagging, and mutagenesis were carried out by either the lithium acetate transformation method or electroporation. Mutations within the SpCnp3CENP-C cupin domain (truncations and point mutants), as well as C-terminally GFP-tagged alleles of SpCnp3CENP-C, were generated by PCR-based methods and integrated at the endogenous cnp3 genomic locus using either a ura4, natMX, or kanMX selection marker targeted to the 3′-UTR of cnp3 (58). 5-fold serial dilutions of the indicated strains were spotted onto YES medium supplemented with or without the indicated concentrations of TBZ and incubated at the indicated temperatures for 3–7 days. Genotypes of S. pombe strains used in this study are listed in Table S1.
Western analyses of GFP-tagged Cnp3 mutants expressed in vivo
Whole-cell extracts from cells expressing GFP-tagged WT or truncation mutants of SpCnp3CENP-C were prepared by TCA precipitation as previously described (59). Proteins in whole-cell extracts were analyzed on Western blots probed with monoclonal anti-GFP antibody (7.1/B.1, Roche). Anti-cdc2 antibody (y100.4, Abcam) was used for loading control.
Cytology
For fluorescence imaging of GFP-tagged SpCnp3CENP-C, the indicated S. pombe strains were grown in liquid PMG medium overnight. The cells were imaged live using a Deltavision Elite microscope (GE Healthcare) equipped with a sCMOS camera and solid-state light-emitting diodes controlled by Softworx. A FITC filter was used for imaging GFP fluorescence. Deconvolution was performed using Softworx software. 10 Z sections for GFP (Cnp3) signals were converted into single two-dimensional images by projecting the maximum signal at each pixel position using Icy software (Institut Pasteur). For immunofluorescence (IF), cells were fixed in 3.7% formaldehyde for 15 min at room temperature. Immunolocalization was performed as previously described (60). Primary antibodies used for IF were anti-GFP A11122 (1:200) (Thermo Fisher/Invitrogen) and anti-Cnp1CENP-A antiserum (1:2000) (gift from R. Allshire). Alexa Fluor 488– and 594–coupled secondary antibodies were used at 1:1000 dilution (A21441 and A11016 from Thermo Fisher/Invitrogen). To induce meiosis, homothallic fission yeast cells (h90) expressing GFP-tagged Moa1 in WT or cnp3 mutant backgrounds were grown to exponential phase in YES, resuspended in 20 g/liter leucine, and spotted onto SPAS medium. Following incubation at 32 °C for 24 h to allow the mei4Δ mutation to arrest cells in meiotic prophase I (27), the cells were either imaged live using Vectashield with 4′,6′-diamino-2-phenylindole (Vector Laboratories) as mounting medium or processed for IF. Fluorescence imaging of GFP-Moa1 (live and IF) and Cnp3-GFP (IF) was performed using a Zeiss Axioimager Z2 microscope (Carl Zeiss AG, Germany) equipped with a Zeiss Colibri LED illumination system and Hamamatsu digital camera C11440. Ten Z sections for GFP (Moa1 or Cnp3) and RFP (Cnp1CENP-A) signals acquired using Zen software (Zeiss), as applicable, were converted into single two-dimensional images by projecting the maximum signal at each pixel position using Icy software (Institut Pasteur).
Author contributions
J. K. C., B. G. M., L. S., and U.-S. C. conceptualization; J. K. C., V. M., P. K. G., B. A. M., P. K., S. A., L. S., and U.-S. C. data curation; J. K. C., V. M., P. K. G., B. A. M., P. K., S. A., and U.-S. C. software; J. K. C., V. M., P. K. G., B. A. M., P. K., S. A., L. S., and U.-S. C. formal analysis; J. K. C., B. A. M., B. G. M., L. S., and U.-S. C. validation; J. K. C., V. M., P. K. G., B. A. M., P. K., S. A., L. S., and U.-S. C. visualization; J. K. C., P. K., S. A., B. G. M., L. S., and U.-S. C. methodology; J. K. C. and U.-S. C. writing-original draft; J. K. C., L. S., and U.-S. C. writing-review and editing; B. G. M., L. S., and U.-S. C. supervision; B. G. M., L. S., and U.-S. C. funding acquisition; L. S. and U.-S. C. resources; L. S. and U.-S. C. investigation; L. S. and U.-S. C. project administration.
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Photon Source LS-CAT beamlines for advice and assistance with data collection, and Kathleen Wisser and Joseph Schauerte for their guidance in setting up DSC experiments. We thank Dr. James C. A. Bardwell for the use of his analytical ultracentrifuge and Dr. Peter Thorpe for advice, as well as kindly allowing us access to his fluorescence microscope. We are grateful to the Yeast Genetic Resource Center, National Bioresource Project (Japan) for S. pombe strains and Prof. Robin Allshire for the anti-Cnp1CENP-A antibody.
This work was supported by National Institutes of Health Grant DK111465, March of Dimes Foundation Grant N019154-00 (to U.-S. C.), and a Queen Mary University of London start-up grant and Biotechnology and Biological Sciences Research Council Grant BB/R00868X/1 (to L. S.). This work was also supported in part by National Institutes of Health Grant R01GM-108829 (to B. G. M.), National Institutes of Health Cellular and Molecular Biology Training Grant T-32-GM007315 (to J. K. C.), and Rackham Graduate Student Precandidate and Candidate Research Grants (to J. K. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 6O2D and 6O2K) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CCAN
- constitutive centromere-associated network
- R.M.S.
- root mean square
- PDB
- Protein Data Bank
- SAD
- single-wavelength anomalous diffraction
- DS
- domain-swapped region
- SV-AUC
- sedimentation velocity analytical ultracentrifugation
- MBP
- maltose-binding protein
- TBZ
- thiabendazole
- DSC
- differential scanning calorimetry
- TCEP
- tris(2-carboxyethyl)phosphine
- APS
- Advanced Photon Source
- AEI
- amylose elution gel band intensity
- IF
- immunofluorescence.
References
- 1. Torres E. M., Williams B. R., and Amon A. (2008) Aneuploidy: cells losing their balance. Genetics 179, 737–746 10.1534/genetics.108.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hassold T., and Hunt P. (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- 3. Yuen K. W., Montpetit B., and Hieter P. (2005) The kinetochore and cancer: what's the connection? Curr. Opin. Cell Biol. 17, 576–582 10.1016/j.ceb.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 4. Clarke L., and Carbon J. (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–509 10.1038/287504a0 [DOI] [PubMed] [Google Scholar]
- 5. Fitzgerald-Hayes M., Clarke L., and Carbon J. (1982) Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29, 235–244 10.1016/0092-8674(82)90108-8 [DOI] [PubMed] [Google Scholar]
- 6. Cottarel G., Shero J. H., Hieter P., and Hegemann J. H. (1989) A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell Biol. 9, 3342–3349 10.1128/MCB.9.8.3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lechner J., and Carbon J. (1991) A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–725 10.1016/0092-8674(91)90501-O [DOI] [PubMed] [Google Scholar]
- 8. McKinley K. L., and Cheeseman I. M. (2016) The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brinkley B. R., and Stubblefield E. (1966) The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma 19, 28–43 10.1007/BF00332792 [DOI] [PubMed] [Google Scholar]
- 10. Cheeseman I. M., and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 11. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., and Desai A. (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 12. Earnshaw W. C., and Rothfield N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- 13. Moroi Y., Peebles C., Fritzler M. J., Steigerwald J., and Tan E. M. (1980) Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. U.S.A. 77, 1627–1631 10.1073/pnas.77.3.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carroll C. W., Milks K. J., and Straight A. F. (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato H., Jiang J., Zhou B. R., Rozendaal M., Feng H., Ghirlando R., Xiao T. S., Straight A. F., and Bai Y. (2013) A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKinley K. L., Sekulic N., Guo L. Y., Tsinman T., Black B. E., and Cheeseman I. M. (2015) The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere–kinetochore interface. Mol. Cell 60, 886–898 10.1016/j.molcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Przewloka M. R., and Glover D. M. (2009) The kinetochore and the centromere: a working long distance relationship. Annu. Rev. Genet. 43, 439–465 10.1146/annurev-genet-102108-134310 [DOI] [PubMed] [Google Scholar]
- 18. Drinnenberg I. A., Henikoff S., and Malik H. S. (2016) Evolutionary turnover of kinetochore proteins: a ship of theseus? Trends Cell Biol. 26, 498–510 10.1016/j.tcb.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Screpanti E., De Antoni A., Alushin G. M., Petrovic A., Melis T., Nogales E., and Musacchio A. (2011) Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391–398 10.1016/j.sbi.2011.03.007,10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Przewloka M. R., Venkei Z., Bolanos-Garcia V. M., Debski J., Dadlez M., and Glover D. M. (2011) CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21, 399–405 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 21. Klare K., Weir J. R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., and Musacchio A. (2015) CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 10.1083/jcb.201412028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H., and Fukagawa T. (2015) Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 26, 3768–3776 10.1091/mbc.E15-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka K., Chang H. L., Kagami A., and Watanabe Y. (2009) CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 24. Hinshaw S. M., and Harrison S. C. (2013) An Iml3–Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 5, 29–36 10.1016/j.celrep.2013.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen R. L., Espelin C. W., De Wulf P., Sorger P. K., Harrison S. C., and Simons K. T. (2008) Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell 19, 4480–4491 10.1091/mbc.e08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meraldi P., McAinsh A. D., Rheinbay E., and Sorger P. K. (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7, R23 10.1186/gb-2006-7-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yokobayashi S., and Watanabe Y. (2005) The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123, 803–817 10.1016/j.cell.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 28. Kim J., Ishiguro K., Nambu A., Akiyoshi B., Yokobayashi S., Kagami A., Ishiguro T., Pendas A. M., Takeda N., Sakakibara Y., Kitajima T. S., Tanno Y., Sakuno T., and Watanabe Y. (2015) Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517, 466–471 10.1038/nature14097 [DOI] [PubMed] [Google Scholar]
- 29. Miyazaki S., Kim J., Yamagishi Y., Ishiguro T., Okada Y., Tanno Y., Sakuno T., and Watanabe Y. (2017) Meikin-associated polo-like kinase specifies Bub1 distribution in meiosis I. Genes Cells 22, 552–567 10.1111/gtc.12496 [DOI] [PubMed] [Google Scholar]
- 30. Pang H., Bartlam M., Zeng Q., Miyatake H., Hisano T., Miki K., Wong L. L., Gao G. F., and Rao Z. (2004) Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J. Biol. Chem. 279, 1491–1498 10.1074/jbc.M310022200 [DOI] [PubMed] [Google Scholar]
- 31. Simmons C. R., Liu Q., Huang Q., Hao Q., Begley T. P., Karplus P. A., and Stipanuk M. H. (2006) Crystal structure of mammalian cysteine dioxygenase: a novel mononuclear iron center for cysteine thiol oxidation. J. Biol. Chem. 281, 18723–18733 10.1074/jbc.M601555200 [DOI] [PubMed] [Google Scholar]
- 32. Rajavel M., Mitra A., and Gopal B. (2009) Role of Bacillus subtilis BacB in the synthesis of bacilysin. J. Biol. Chem. 284, 31882–31892 10.1074/jbc.M109.014522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 34. Furuyama S., and Biggins S. (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 104, 14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blower M. D., Sullivan B. A., and Karpen G. H. (2002) Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergmann J. H., Rodríguez M. G., Martins N. M., Kimura H., Kelly D. A., Masumoto H., Larionov V., Jansen L. E., and Earnshaw W. C. (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30, 328–340 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marshall O. J., Marshall A. T., and Choo K. H. (2008) Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J. Cell Biol. 183, 1193–1202 10.1083/jcb.200804078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukagawa T., and Earnshaw W. C. (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu S. T., Rattner J. B., Jablonski S. A., and Yen T. J. (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175, 41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soisson S. M., MacDougall-Shackleton B., Schleif R., and Wolberger C. (1997) Structural basis for ligand-regulated oligomerization of AraC. Science 276, 421–425 10.1126/science.276.5311.421 [DOI] [PubMed] [Google Scholar]
- 41. Schittenhelm R. B., Althoff F., Heidmann S., and Lehner C. F. (2010) Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 123, 3768–3779 10.1242/jcs.067934 [DOI] [PubMed] [Google Scholar]
- 42. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 43. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 10.1107/S0907444909012098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmid-Burgk J. L., Xie Z., and Benenson Y. (2014) Hierarchical ligation-independent assembly of PCR fragments. Methods Mol. Biol. 1116, 49–58 10.1007/978-1-62703-764-8_4 [DOI] [PubMed] [Google Scholar]
- 54. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Demeler B. (2005) UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. In Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., and Rowe A. J., eds) pp. 210–230, Royal Society of Chemistry, London [Google Scholar]
- 56. Brookes E., Cao W., and Demeler B. (2010) A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 39, 405–414 10.1007/s00249-009-0413-5 [DOI] [PubMed] [Google Scholar]
- 57. Moreno S., Klar A., and Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- 58. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. 3rd, Steever A. B., Wach A., Philippsen P., and Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in 1Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 59. Salas-Pino S., Gallardo P., Barrales R. R., Braun S., and Daga R. R. (2017) The fission yeast nucleoporin Alm1 is required for proteasomal degradation of kinetochore components. J. Cell Biol. 216, 3591–3608 10.1083/jcb.201612194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Subramanian L., Toda N. R., Rappsilber J., and Allshire R. C. (2014) Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 4, 140043 10.1098/rsob.140043 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.