Abstract
Asparaginase is an amino acid–depleting agent used to treat blood cancers. Metabolic complications due to asparaginase affect liver function in humans. To examine how the liver response to asparaginase changes during maturity to adulthood, here we treated juvenile (2-week), young adult (8-week), and mature adult (16-week) mice with drug or excipient for 1 week and conducted RNA-Seq and functional analyses. Asparaginase reduced body growth and liver mass in juveniles but not in the adult animals. Unbiased exploration of the effect of asparaginase on the liver transcriptome revealed that the integrated stress response (ISR) was the only molecular signature shared across the ages, corroborating similar eukaryotic initiation factor 2 phosphorylation responses to asparaginase at all ages. Juvenile livers exhibited steatosis and iron accumulation following asparaginase exposure along with a hepatic gene signature indicating that asparaginase uniquely affects lipid, cholesterol, and iron metabolism in juvenile mice. In contrast, asparaginase-treated adult mice displayed greater variability in liver function, which correlated with an acute-phase inflammatory response gene signature. Asparaginase-exposed adults also had a serine/glycine/one-carbon metabolism gene signature in liver that corresponded with reduced circulating glycine and serine levels. These results establish the ISR as a conserved response to asparaginase-mediated amino acid deprivation and provide new insights into the relationship between the liver transcriptome and hepatic function upon asparaginase exposure.
Keywords: eukaryotic initiation factor 2 (eIF2), amino acid, triglyceride, mRNA, development, age-dependent response, asparaginase, hepatic metabolism, integrated stress response, integrated stress response (ISR)
Introduction
Asparaginase is a drug used in the treatment of acute lymphoblastic leukemia in humans and various lymphoid malignancies in dogs and cats. It kills tumor cells via extracellular depletion of the amino acids glutamine and asparagine. Adverse metabolic events including hepatotoxicity compromise treatment success of asparaginase-based chemotherapy regimens in humans (1). Our previous efforts to understand the molecular events induced by asparaginase in livers of cancer-free mice (2, 3) show that in nongrowing adults, asparaginase triggers phosphorylation of eukaryotic translation initiation factor 2 (eIF2)2 by the eIF2 kinases general control nonderepressible 2 (GCN2) and protein kinase R–like endoplasmic reticulum kinase (PERK) (2, 4–6). The phosphorylation of eIF2 slows down global protein synthesis and promotes translation of select mRNAs, including the basic leucine transcription factor, activating transcription factor 4 (ATF4). This cytoprotective signal cascade, coined the integrated stress response (ISR), results in changes in expression levels of ATF4 target genes to regain homeostasis and adapt to cellular stress (7, 8). Genetic disruption of the GCN2-eIF2-ATF4 axis predisposes to adverse complications such as hepatic dysfunction, immunosuppression, and pancreatitis (3, 9, 10). Pre-existing obesity activates PERK upon asparaginase exposure and when in combination with Gcn2 deletion aggravates asparaginase-associated liver dysfunction (6).
In the current study, we sought to understand age-associated liver responses to asparaginase. We hypothesized, based on a published report showing a decline in total eIF2 protein with age (11), that ISR activation would differ according to age. Comparative analysis of livers from mice treated with asparaginase instead showed that ISR activation was similar at all ages and associated with a conserved ATF4 gene target signature. Age-specific differences in the liver transcriptome included inhibition of lipid and cholesterol biosynthesis and iron bioavailability in juveniles, whereas adult livers displayed increased serine/glycine/one-carbon metabolism and inflammation. These results provide insight into the relationship between age and the resources available to support recovery from amino acid depletion by asparaginase.
Results
Asparaginase reduces body growth and liver mass in juvenile mice only
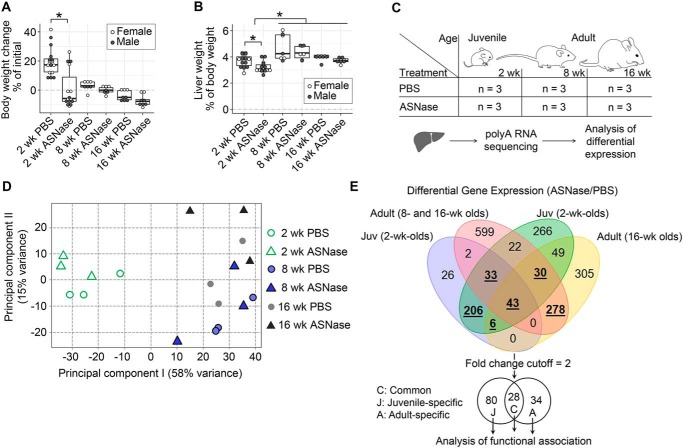
To identify age-specific hepatic responses following chronic asparaginase exposure, we utilized 2-, 8-, and 16-week-old mice representing preweaned juveniles, young adults, and mature adults, respectively. Asparaginase stunted the growth of juvenile mice and decreased their liver masses relative to body weight (Fig. 1, A and B). In contrast, body and liver weights were not significantly altered by drug in adults.
Figure 1.
Age affects hepatic response to ASNase. ASNase exposure stunts body growth (A) and reduces relative liver mass (B) in juveniles but not in adults following 1 week of daily intraperitoneal injections as compared with control groups that were injected with vehicle (PBS). The box plots show median values, top and bottom hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the whiskers extend to the smallest and the largest value within 1.5 distance of the interquartile range. *, p < 0.05 by Wilcoxon rank-sum test. C, schematic diagram of the experimental setup to analyze hepatic transcriptomes of the mice; n indicates group number. D, principal component analysis clusters juvenile mice separately from adult mice regardless of treatment. E, Venn diagram shows numbers of commonly and uniquely altered transcript abundances in the murine hepatic transcriptomes discovered in the two comparison sessions: (i) juveniles (blue set) versus combined 8- and 16-week-old adult group (red set) and (ii) juveniles (green set) versus 16-week-old mice (yellow set). Boldface underlined numbers of the colored Venn diagram were further filtered using the 2-fold change cutoff in gene expression induced by ASNase, resulting in 28 common, 80 juvenile-specific, and 34 adult-specific elements for further analysis of functional association.
Age influences the liver transcriptome in response to asparaginase
We next sought to examine how asparaginase affects the liver transcriptome across the three age groups (Fig. 1C). Total RNA was isolated from liver, enriched for polyadenylated RNA, and sequenced to a depth of 30 million reads (7). Principal component analysis clustered juvenile transcriptomes separately from adults regardless of treatment (Fig. 1D), suggesting dominance of age over asparaginase exposure on the hepatic transcriptome. Quantitative analysis of differentially expressed genes confirmed that juveniles differed considerably from adults, whereas young and mature adults were largely similar.
Further examination of adult versus juvenile transcriptomes revealed increased variability in adult transcript abundances as compared with juveniles (12, 13). With this in mind, we constructed a Venn analysis to compare our differentially expressed gene lists: (i) juveniles were compared with 8- and 16-week-old mice combined into a single group of adults, and (ii) juveniles were compared only with the 16-week-old adults (Fig. 1E). Genes that were identified in both comparisons and showed greater than 2-fold difference in expression were selected for further consideration. These criteria ensure 87–100% statistical power given the sequencing depth and the number of replicates (14). Using this filtering scheme, we identified 28 genes altered by asparaginase regardless of age. In addition, 80 genes were altered by asparaginase in juveniles only, and 34 genes were altered by asparaginase only in adults.
Asparaginase activates the ISR in liver regardless of age
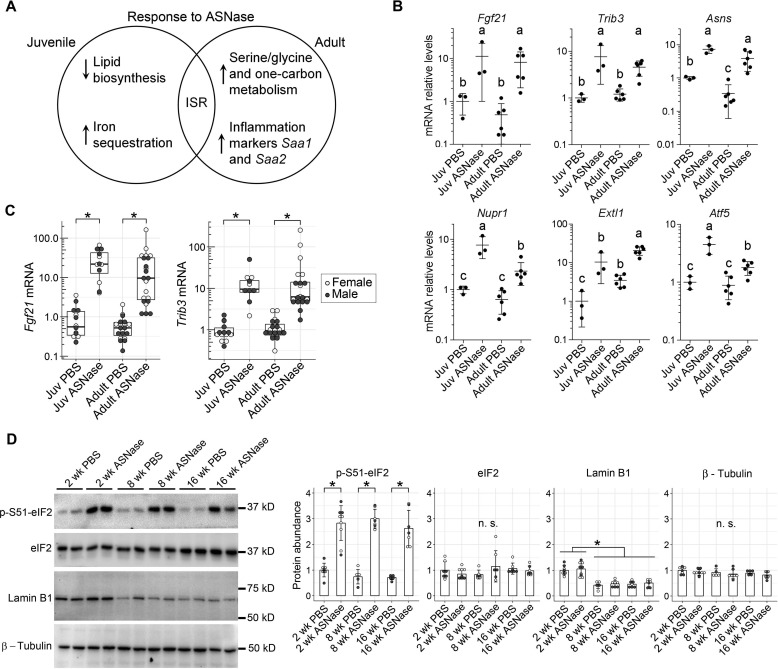
Analysis of the resulting gene lists using DESeq2 (15), DAVID gene annotation enrichment analysis, KEGG pathway mapping (16), and STRING (search tool for the retrieval of interacting genes/proteins) (17) revealed unique pathways or processes by age. The 28 genes found in common between juvenile and adult mice treated with asparaginase did not associate with annotated processes. Instead, visual examination of the gene list revealed an activated ISR (Fig. 2A) (8, 9, 18, 19). ATF4 target genes present in this common signature included (i) Fgf21, a major hepatokine that regulates whole-body metabolism according to nutritional status (9); (ii) Trib3, a pseudokinase that binds and interferes with the activity of ATF4 and Akt/protein kinase B (20–22); and (iii) Asns, asparagine synthetase that replenishes intracellular levels of asparagine and other amino acids (8) (Fig. 2B). In addition, Nupr1 (also known as p8), a gene transcript responsive to amino acid starvation and dietary protein dilution (23–25), was increased 7-fold in juveniles and 4-fold in adults, whereas Extl1, which encodes an N-glycosyltransferase (26), showed 10-fold increase by drug in juveniles and 6-fold in adults, and Atf5, a well-known responder to ER stress and amino acid starvation (6, 7), was increased 4-fold in juveniles and 2-fold in adults. A full list of the asparaginase-responsive genes independent of age is summarized in Table S1A. One gene worth noting in this supplemental list is Rho/Rac guanine nucleotide exchange factor 2 (Arhgef2), reportedly responsive to either leucine- or cysteine-deficient conditions (27), which increased 3–4-fold in both juveniles and adults exposed to asparaginase. In almost all instances, the level of induction was greater in juveniles relative to adults.
Figure 2.
The ISR is a conserved gene signature in the livers of juvenile and adult mice following 1 week of daily ASNase exposure. A, Venn diagram representing functional annotations of the common and unique gene signatures in juveniles (2 weeks old) and adults (8 and 16 weeks old) exposed to ASNase daily for 1 week. B, the shared or common transcriptional signature in liver following chronic ASNase exposure includes the activating transcription factor 4 (ATF4) target genes Fgf21, Trib3, Asns, Nupr1, Exlt1, Atf5, and other genes from Table S1A. Data are represented as means normalized to the juvenile (Juv) PBS control group ± S.D. (error bars); means not sharing a common letter are significantly different, padj < 0.1. C, transcript levels of Fgf21 and Trib3 in the livers of juvenile and adult mice were confirmed by RT-qPCR. D, immunoblot analysis of phosphorylation of eIF2 α-subunit at serine 51 (p-S51-eIF2) substantiates similar ISR induction to ASNase across ages. Lamin B1, but not eIF2 or β-tubulin, decreases with age. Densitometry of blots are shown to the right of the representative immunoblots. Bar graphs display means normalized to the juvenile (Juv) PBS control group ± S.D. Data displayed in C and D were evaluated by Wilcoxon rank-sum test. *, p < 0.05. n.s., not significant.
The RNA-Seq results were validated on a larger cohort of animals by examining gene expression in juvenile (2-week) and adult (8–16-week) mice using RT-qPCR. We confirmed that both Fgf21 and Trib3 were similarly induced by asparaginase and found no influence of either age or sex on transcript abundances (Fig. 2C). This gene signature agreed with eIF2 phosphorylation levels in which the response to asparaginase was similar across ages (Fig. 2D). Total eIF2 levels were also not affected by age or asparaginase, similar to β-tubulin, whereas lamin B1, a protein known to decrease with age (28), showed the expected age-related decrease in protein levels. Taken together, these results indicate that the ISR is a conserved hepatic stress response to asparaginase regardless of age.
Age influences hepatic lipid metabolism in response to asparaginase
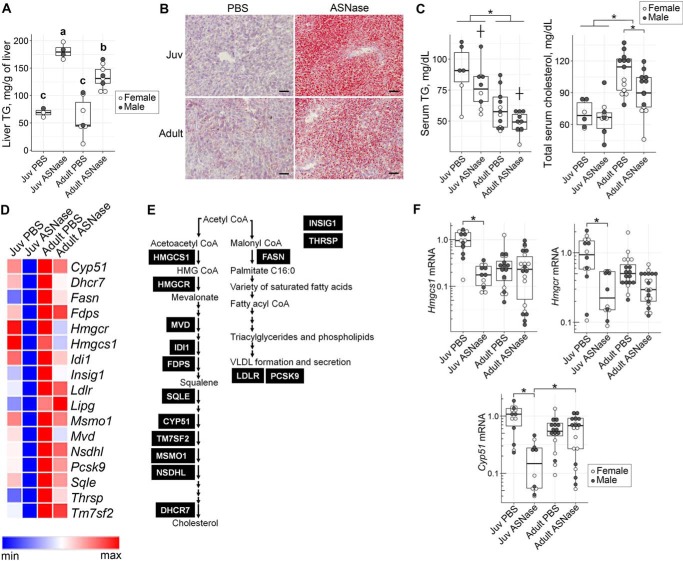
Asparaginase-induced liver steatosis was greatest in juveniles, displaying the highest liver triglycerides biochemically alongside intense Oil Red O staining in liver sections (Fig. 3, A and B). Asparaginase-treated adults showed modest changes in liver triglycerides, similar to that previously reported by our group (9). Circulating triglycerides were highest in excipient-treated juveniles, reducing both with age and by asparaginase exposure (Fig. 3C). On the other hand, total serum cholesterol was highest in excipient-treated adults and was reduced by asparaginase in adults only.
Figure 3.
Age affects lipid metabolism in response to ASNase. Hepatic triglyceride (TG) measurement (A) and Oil Red O staining (B) of liver (scale bar, 100 μm) show larger accumulation of red droplets representing hepatic TG in juveniles (Juv) as compared with adults. Data in A were evaluated by two-factor ANOVA with ASNase and age as independent factors. Means not sharing a common letter are different, reflecting an ASNase × age interaction, p < 0.05. C, serum TG and total cholesterol in sera of mice exposed to either ASNase or to PBS vehicle. Data in C were evaluated by two-factor ANOVA. *, significant effect of age (p < 0.05); †, significant effect of ASNase (p < 0.05). D, heat map showing genes of the lipid biosynthesis pathway that were significantly altered in expression by ASNase in the juveniles (padj < 0.1), but not the adults. The blue-to-red color scale represents mRNA abundances obtained from RNA-Seq data across the four treatment groups. For exact expression values and corresponding protein function, see Table S1B. E, diagram indicates enzymes of the lipid biosynthesis pathway encoded by genes from D. F, validation of Hmgcs1, Hmgcr, and Cyp51 mRNA abundances by RT-qPCR. Values are normalized to the ones of the juvenile PBS control group. The box plots show median values, top and bottom hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the whiskers extend to the smallest and the largest value within 1.5 distance of the interquartile range. *, p < 0.05 by Wilcoxon rank-sum test.
Although circulating triglycerides and cholesterol were both reduced by asparaginase in adults, the liver transcriptome showed minimal changes in lipid metabolism genes. Instead, the transcriptome of juveniles uniquely displayed down-regulation of lipid biosynthesis upon asparaginase exposure (Fig. 2A). Specifically, more than 60% of genes encoding proteins that function in the cholesterol and fatty acid biosynthesis were down-regulated roughly 3–5-fold in juveniles but were largely unresponsive in adults (Fig. 3 (D and E) and Table S1B). Confirmation by RT-qPCR on a larger cohort of animals showed 5-fold reductions in Hmgcs1, Hmgcr, and Cyp51 in juveniles, but not in adults (Fig. 3F). These reductions are interpreted to function as a homeostatic response attempting to correct the hepatic steatosis.
Age influences iron metabolism
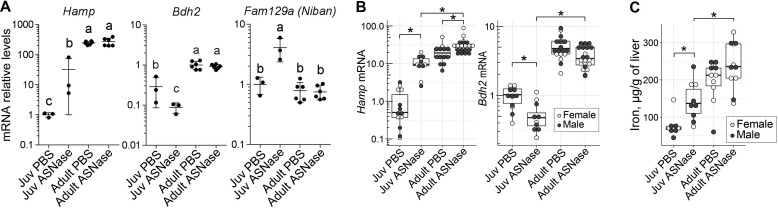
Examination of the juvenile-specific transcriptional changes in response to asparaginase also indicated increased iron sequestration (Figs. 2A and 4A). Specifically, chronic asparaginase exposure corresponded with 10-fold induction of Hamp, a gene encoding the small peptide hepcidin, which functions to block iron export from cells under conditions of high iron, inflammation, or ER stress. There was also a 3-fold reduction in Bdh2, which encodes 3-hydroxybutyrate dehydrogenase 2, an enzyme catalyzing a rate-limiting step in the synthesis of the iron-chelating compound 2,5-dihydroxybenzoic acid siderophore (29, 30), suggestive of reductions in iron bioavailability for extrahepatic tissues. We also noted a 4-fold increased expression of Fam129a (also known as Niban), an ATF4 target gene induced in cell culture models of ER stress (31) and shown to increase in response to a low-iron diet in mouse models (32).
Figure 4.
Age affects hepatic iron metabolism in response to ASNase. A, transcript abundances for genes pertaining to iron metabolism (Hamp, Bdh2, and Fam129a) that were significantly altered by chronic ASNase exposure in RNA-Seq data set. Data are represented as means normalized to the juvenile (Juv) PBS control group ± S.D. (error bars). Means not sharing a common letter are different (padj < 0.1). B, validation of Hamp and Bdh2 mRNA abundances by RT-qPCR. Values are normalized to the ones of the juvenile PBS control group. C, total iron content in livers of juveniles and adults exposed to either ASNase or vehicle control (PBS). The box plots in B and C show median values, top and bottom hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the whiskers extend to the smallest and the largest value within 1.5 distance of the interquartile range. *, p < 0.05 by Wilcoxon rank-sum test.
Age by itself induced Hamp 10–20-fold over untreated juvenile levels such that asparaginase failed to further increase transcript levels in adult mice (Fig. 4A). Age-related changes in Hamp expression were observed in aging rat tissues (33), suggesting that hepcidin plays a role in mitigating age-related iron accumulation (34). The inability of adults to increase Hamp expression in response to asparaginase suggests a ceiling effect to amino acid depletion. To confirm this, we assessed Hamp and Bdh2 expression by RT-qPCR on a larger cohort of animals (Fig. 4B). Hamp abundance increased 10-fold upon asparaginase exposure in juveniles and only 1.5-fold in adults, whereas Bdh2 mRNA abundance decreased 2-fold in juveniles with asparaginase and did not change in adults (Fig. 4B), fully confirming the RNA-Seq data. Because this transcriptional signature predicted increased iron sequestration by liver, we measured total iron content. A 2-fold increase in total hepatic iron was induced by asparaginase in juvenile livers, whereas no significant change was observed in adults (Fig. 4C), agreeing with changes in Hamp expression. Of note, total iron content of adult livers was at least 2-fold higher than that of juvenile ones, directionally correlating with hepatic Hamp expression and reflecting age-related iron accumulation.
Asparaginase provokes inflammation and alters one-carbon metabolism in adult liver
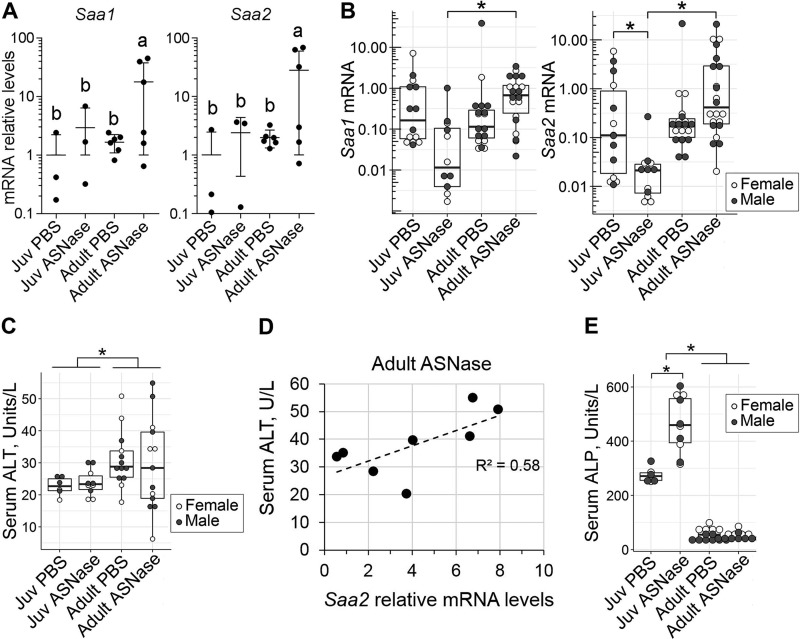
The adult-specific transcriptional response to asparaginase showed elevations in Saa1 and Saa2 transcripts, which encode serum amyloid A1 and A2 proteins (Fig. 5A). SAA1 and SAA2 are principally produced by the liver under acute inflammatory conditions. We confirmed these findings by RT-qPCR in a larger cohort of animals and found that adults displayed 2–3-fold increases in Saa1 and Saa2 in response to asparaginase, whereas juveniles showed the opposite, up to 10-fold reductions in these transcripts (Fig. 5B). To check whether increased Saa1 and Saa2 mRNAs coincided with alterations in liver function, we measured levels of alanine aminotransferase (ALT) in the sera of mice (Fig. 5C). Notably, serum ALT levels positively correlated with the hepatic Saa2 mRNA levels in adult mice treated with asparaginase (Fig. 5D). No change in the serum alkaline phosphatase (ALP) levels was observed in adults. Basally elevated ALP levels in untreated juveniles (Fig. 5E) reflect the developmental phase of active growth of the juvenile mice and are also reported for humans (35). Further elevation of ALP levels by asparaginase exposure is common among pediatric patients and usually resolves on its own after completion of the asparaginase protocol (36).
Figure 5.
Hepatic transcriptional response of adults to chronic ASNase exposure includes markers of acute-phase response. A, mRNA levels of Saa1 and Saa2, encoding serum amyloid A1 and A2, obtained from RNA-Seq data. Data are represented as means normalized to the juvenile (Juv) PBS control group ± S.D. (error bars) Means not sharing a common letter are different (padj < 0.1). B, validation of Saa1 and Saa2 mRNA abundances by RT-qPCR. Values are normalized to the ones of the juvenile PBS control group. C, serum ALT levels in mice exposed to ASNase or vehicle control (PBS). D, correlation plot delineating levels of the Saa2 mRNA (from B) to the levels of serum ALT (from C) in adult mice treated with ASNase (Spearman correlation coefficient r = 0.761, p < 0.58). E, serum ALP levels. The box plots show median values, top and bottom hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the whiskers extend to the smallest and the largest value within 1.5 distance of the interquartile range. *, p < 0.05 by Wilcoxon rank-sum test.
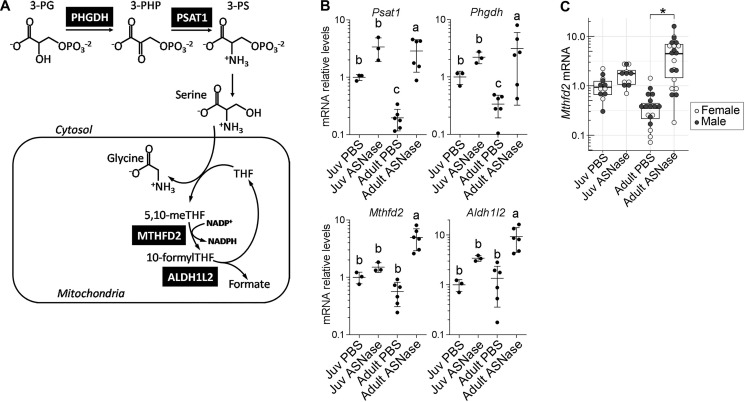
The other identified adult-specific response to asparaginase encoded key steps in serine/glycine metabolism (Phgdh and Psat1) and the mitochondrial folate cycle (Mthfd2 and Aldh1l2) (Fig. 6 (A–C) and Table S1C). To determine whether these changes corresponded with differential effects on circulating amino acids, we measured serum concentrations of amino acids by HPLC (Table 1). In both juveniles and adults, asparaginase significantly reduced concentrations of asparagine, aspartate, and glutamine and significantly increased concentrations of alanine, arginine, glutamate, glycine, lysine, serine, and threonine. Notably, age independently reduced serine and glycine concentrations substantially. Thus, elevated abundances of Phgdh and Psat1 transcripts suggest that adults under amino acid stress have reduced availability of serine as a substrate to help drive either mitochondrial NADPH production (37) or purine nucleotide synthesis, the ultimate products of the folate cycle (38). Validation of the Mthfd2 expression on a larger cohort of animals confirmed reduced transcript abundance in excipient-treated adults and up to 10-fold increased abundance of the transcript in adults following asparaginase regardless of sex (Fig. 5C).
Figure 6.
Age affects one-carbon metabolism in response to asparaginase. A, schematic depiction of the serine/glycine and one-carbon metabolism pathway illustrates reactions catalyzed by the enzymes (black boxes). B, transcript levels of genes encoding the enzymes of serine/glycine and one-carbon metabolism; values are obtained from RNA-Seq data. Data are represented as means normalized to the juvenile (Juv) PBS control group ± S.D. (error bars). Means not sharing a common letter are different (padj < 0.1). C, validation of Mthfd2 mRNA levels by RT-qPCR. The box plots show median values, top and bottom hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the whiskers extend to the smallest and the largest value within 1.5 distance of the interquartile range. *, p < 0.05 by Wilcoxon rank-sum test.
Table 1.
Amino acids concentrations in the sera of mice exposed to eight daily injections of either ASNase or PBS excipient
Data were analyzed by two-factor ANOVA with ASNase and age as independent factors. Main effects of ASNase and age are shown in columns to the right of the treatment groups. Means in a row not sharing a common letter are different, reflecting an ASNase × age interaction (*, p < 0.05).
| Amino acids in serum | Average values (μm) |
p values |
||||
|---|---|---|---|---|---|---|
| Juv PBS (n = 9) | Juv ASNase (n = 9) | Adult PBS (n = 6) | Adult ASNase (n = 8) | Effect of ASNase | Effect of age | |
| Alanine | 357 ± 29 | 511 ± 56 | 348 ± 24 | 451 ± 104 | 0.002* | 0.351 |
| Arginine | 138 ± 9 | 173 ± 22 | 96 ± 21 | 107 ± 26 | 0.047* | 0.000* |
| Asparagine | 42.4 ± 31.3 | 5.7 ± 0.4 | 40.1 ± 33.1 | 5.6 ± 0.6 | 0.004* | 0.915 |
| Aspartate | 40 ± 24 | 21 ± 11 | 36 ± 21 | 20 ± 9 | 0.042* | 0.746 |
| Cystine | 6.3 ± 3.2 | 3.4 ± 0.3 | 3.5 ± 3.7 | 4.0 ± 6.8 | 0.628 | 0.655 |
| Glutamate | 81 ± 6 (b) | 168 ± 74 (b) | 57 ± 38 (b) | 330 ± 77 (a) | 0.000* | 0.034* |
| Glutamine | 405 ± 16 | 242 ± 49 | 597 ± 159 | 263 ± 120 | 0.001* | 0.090 |
| Glycine | 783 ± 47 (c) | 1151 ± 129 (a) | 449 ± 61 (b) | 512 ± 53 (b) | 0.000* | 0.000* |
| Histidine | 60 ± 3 | 62 ± 6 | 59 ± 10 | 56 ± 14 | 0.879 | 0.581 |
| Isoleucine | 54 ± 4 | 68 ± 10 | 69 ± 21 | 75 ± 18 | 0.246 | 0.208 |
| Leucine | 69 ± 4 | 93 ± 21 | 112 ± 36 | 120 ± 30 | 0.269 | 0.027* |
| Lysine | 289 ± 54 | 449 ± 95 | 188 ± 38 | 237 ± 64 | 0.003* | 0.000* |
| Methionine | 65 ± 6 | 74 ± 10 | 74 ± 21 | 76 ± 26 | 0.612 | 0.594 |
| Proline | 304 ± 12 | 312 ± 23 | 304 ± 168 | 285 ± 62 | 0.918 | 0.791 |
| Serine | 241 ± 11 | 344 ± 51 | 124 ± 17 | 178 ± 43 | 0.000* | 0.000* |
| Threonine | 203 ± 11 | 288 ± 26 | 156 ± 25 | 244 ± 81 | 0.007* | 0.117 |
| Tryptophan | 87 ± 5 | 76 ± 2 | 109 ± 23 | 106 ± 20 | 0.455 | 0.012* |
| Valine | 168 ± 21 | 225 ± 33 | 205 ± 58 | 235 ± 59 | 0.109 | 0.380 |
Age alters liver gene expression independent of asparaginase
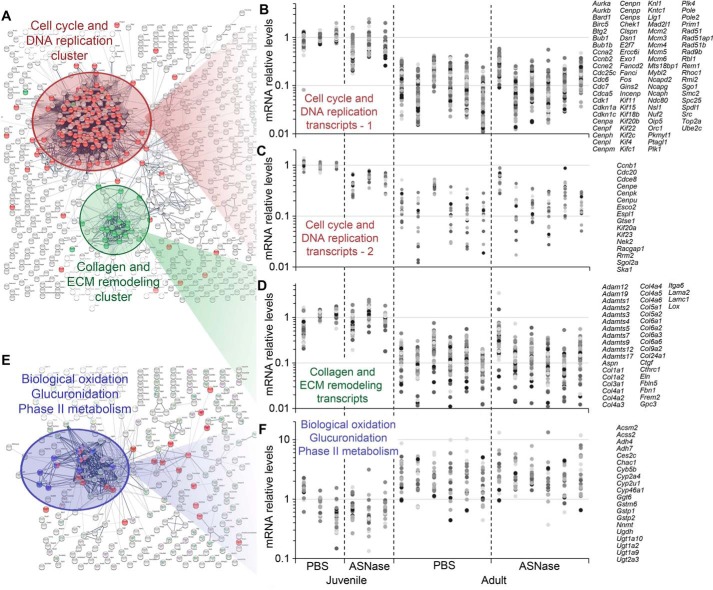
In this analysis, more genes were differentially altered by age versus asparaginase (Table S2). Therefore, we utilized unbiased STRING analysis to identify major pathways that changed in the liver during maturity to adulthood. Our analysis revealed two large clusters of genes that were increased in young mice relative to adults (Fig. 7, A–D). First, a cell cycle/DNA replication cluster was identified, which contained genes encoding regulatory and structural proteins involved in chromosome maintenance and mitosis. This cluster is indicative of high numbers of replicating cells that become reduced when animals reach maturity. Roughly 16% of the genes contained within the cell cycle and DNA replication cluster had 2–3-fold decreased mRNA abundance after asparaginase treatment in the preweaned juvenile group. A second gene cluster included collagen and extracellular matrix remodeling, also consistent with a higher growth state in preweaned juveniles.
Figure 7.
Functional association analysis of genes affected by maturity to adulthood revealed three large clusters. A, genes whose expression is down-regulated during maturity to adulthood encode proteins that regulate cell cycle/DNA replication (red) and proteins that constitute extracellular matrix (ECM) and its remodelers (green). B and C, graphs showing abundances of transcripts functionally associated with the cell cycle/DNA replication cluster. B, transcripts with abundances decreased at least 2–3-fold by ASNase in the juveniles; C, transcripts with abundances not affected by ASNase. D, transcripts of the ECM remodeling cluster decreased in abundance 2–10-fold during maturity to adulthood. E, genes whose expression is up-regulated during maturity to adulthood encode proteins catalyzing oxidation reactions, glucuronidation, and other reactions of the phase II metabolism. F, graphs showing abundances of transcripts functionally associated with the collagen and ECM remodeling cluster increased 2–10-fold during maturity to adulthood. B, C, D, and F share a common x axis comprising individual animals within each treatment group for a total of 18 animals whose transcriptomes were sequenced. Dots colored in different shades of gray represent mRNA abundances for genes from the corresponding clusters. All of the expression values in these panels are normalized to the average value of the juvenile (Juv) PBS control group. Lists of genes are given on the right side of each panel. For exact expression values and corresponding protein functions, see Table S2.
Older mice showed specific enrichment (up-regulated 3–10-fold) in genes encoding enzymes that function in biological oxidation, glucuronidation, and phase II liver metabolism reactions (Fig. 7, E and F). These data suggest an increased demand for inactivation of a variety of xenobiotics and metabolites in adult liver that may reflect increased metabolic demand following full transition from dam's milk to solid food (39, 40).
Discussion
This study aimed to explore the mechanistic basis for age-related differences to asparaginase in liver, complementing our portfolio of work detailing the metabolic and tissue-specific effects of asparaginase (2–4, 6, 7, 9, 18). A major finding of this study is that the ISR is a core response triggered in liver by asparaginase both in the juvenile period and in adulthood. This is an important finding because it emphasizes the conserved nature of this homeostatic program to protect and adapt to stress during development to adulthood.
Adult mice deficient in genes encoding ISR components respond poorly to asparaginase and suffer a variety of metabolic problems related to liver dysfunction (6, 9). Adult WT mice in our previous studies showed healthy adaptation to asparaginase only if they were lean. As such, we did not expect the adult mice in this study, who were also lean, to demonstrate frank liver toxicity. Instead, based on clinical reports suggesting a lower incidence of asparaginase-associated toxicities in younger patients (41), we surmised that activation of the ISR by asparaginase may be influenced by age. The similarity in ISR activation to asparaginase independent of age emphasizes that complications may arise at any age if this pathway is defective or otherwise compromised. The finding that age does not substantially alter the ISR to asparaginase also stands in contrast to that of pre-existing obesity, which promotes a maladaptive ISR to asparaginase and significantly aggravates liver steatosis and dysfunction in WT mice (6). Thus, we conclude that genetic background and metabolic state are more relevant factors to consider versus age in understanding the basis for asparaginase-associated hepatotoxicity. Indeed, human patients with polymorphisms in the ATF4 gene targets Asns and Atf5 demonstrate increased toxicity and lower event-free survival (42), and obesity increases the relative risk for hepatotoxicity in human patients (43).
Juvenile-specific responses to amino acid stress by asparaginase were (i) inhibition of hepatic lipid biosynthesis transcripts alongside hepatic steatosis and (ii) increased iron sequestration. We interpret the down-regulation of genes encoding triglyceride and cholesterol synthesis pathways to be a homeostatic response to increased triglyceride accumulation in juvenile livers. The mechanism by which asparaginase promotes hepatic steatosis in intact mice is unclear, but mice lacking Gcn2 show massive hepatic steatosis due to reduced triglyceride secretion in association with loss of ApoB100 protein expression (6, 9). Liver protein synthesis is reduced by asparaginase (4), and this reduction may affect lipoprotein production more severely in younger mice because of their rapid growth phase. The development of steatosis may also provoke oxidative stress, stimulating cellular defenses to prevent tissue damage. Reductions in hepatic protein synthesis likely impact iron–sulfur cluster protein synthesis and heme biosynthesis in juveniles, provoking iron sequestration as an antioxidant response. The coordinated regulation of amino acid and iron metabolism is conserved throughout eukaryotes (44) and occurs through activation of GCN2, the main responder to asparaginase in mouse liver.
Adult-specific changes in the transcriptome to amino acid stress by asparaginase included (i) acute-phase pro-inflammatory response and (ii) up-regulation of one-carbon metabolism. The presence of an acute-phase pro-inflammatory response is corroborated by clinical observations of increased SAA protein in the serum of high-risk older acute lymphoblastic leukemia pediatric patients (>10 years old) treated with asparaginase as opposed to younger low-risk patients (<9 years old) (45). These results collectively indicate a pro-inflammatory response unique to older patients. The identified pro-inflammatory response in adult mice did not correlate with iron levels or hepcidin gene expression. This contrasts with juvenile mice, which showed a relationship between iron levels and hepcidin in liver after asparaginase exposure. Hepcidin is a liver-produced hormone that acts to regulate iron absorption and distribution across tissues. Infection and inflammation induce hepcidin production, reducing intestinal iron uptake and hepatic iron export (46). The relationship between hepcidin, iron, and inflammation in the context of aging is complex (34). Future studies exploring these themes as a basis for liver dysfunction during asparaginase treatment are warranted.
The other transcriptional signature in adults reflects heightened capacity for detoxification and metabolic maintenance rather than de novo production of biomaterial. Maturity to adulthood is accompanied by 4–5-fold reduced pool of free tissue nucleotides as compared with juveniles (47). The current study supports this by showing adults have significantly reduced circulating concentrations of the amino acids that serve as precursors to nucleotide synthesis. Thus, adults must employ a stress remediation strategy that is fundamentally different from juveniles. Our data suggest that adults up-regulate serine/glycine and one-carbon metabolism perhaps to sustain or support de novo nucleotide synthesis via elevated Mthfd2 expression. MTHFD2 supplies formate as a source of carbon for purine nitrogenous bases (38, 48). Supporting de novo production of nucleotides is one of several functions of the MTHFD2 enzyme. Specifically, MTHFD2 also functions to support mitochondria with NADPH during stress (37) and regulates ribosome biogenesis from inside the nucleus (49).
In summary, the hepatic response to asparaginase differs with age although activation of the ISR is conserved during maturity to adulthood. Whereas lipid metabolism, oxidative stress defenses, amino acid metabolism, and inflammation are all processes regulated by the ISR, questions remain regarding how this regulation occurs during asparaginase exposure and whether activation of ISR by asparaginase is important for metabolic control by promoting ATF4 synthesis or via repression of mTOR complex 1 signaling as we recently detailed (18). Future studies should explore the relationship between ISR activation and the pathways and processes that are functionally altered at each age.
Experimental procedures
Animals and care
All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee at Rutgers University, and all animals received humane care according to the criteria outlined in the Guide for the Care and use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institute of Health (NIH publication 86-23, revised 1985) and ARRIVE) C57Bl/6J mice of both sexes were provided free access to food (5001 Laboratory Rodent Diet, LabDiet) and water and maintained on a 12-h light/dark cycle (7 a.m./7 p.m.) with same-sex littermates until the experimental group assignment, wherein mice were housed in individual plastic cages. Cages contained soft bedding and enrichment. Information about sexes of the animals utilized for the analysis of transcriptome is contained in Tables S1 and S2. For validation of major findings of the global RNA-Seq by RT-qPCR, a total of 68 mice were used: juvenile PBS (7 females and 7 males), juvenile asparaginase (ASNase) (6 females and 6 males), adult PBS (9 females and 11 males), adult ASNase (11 females and 11 males).
Injections and sample collection
Three age groups of mice, preweaned juvenile (2 weeks old), young adults (8 weeks old), and nongrowing adults (16 weeks old), were assigned to receive 7–8 daily intraperitoneal injections of either asparaginase (3 IU/g body weight, Elspar, Merck) or PBS excipient. Asparaginase was reconstituted from powder, and the reconstituted solution was analyzed for asparaginase activity as described (4). Animals were killed by decapitation ∼6 h following the final injection. Whole livers were rapidly dissected and frozen in liquid nitrogen for the subsequent biochemical analyses.
Histology
Liver tissues were fixed in 4% paraformaldehyde, cryoprotected in 15 and 30% sucrose PBS-based solutions, and frozen for cryosectioning. Cryosections (∼10 μm) were stained with Oil Red O to visualize lipid content.
Triglyceride measurements
Triglycerides were measured in frozen liver tissue samples (∼40 mg) using the Colorimetric Triglyceride Quantification kit (BioVision, catalog no. K622, Mountain View, CA) according to the manufacturer's instructions.
Immunoblotting
The frozen tissue powder was lysed in 1:40 ratio with the lysis buffer (25 mm HEPES, pH 7.5, 10 mm DTT, 0.1% SDS, 1× protease inhibitor mixture (Sigma; P8340), 1 mm sodium orthovanadate, 0.5% deoxycholate, 50 mm β-glycerophosphate, 2 mm EDTA, 1 mm microcystin, 50 mm NaF, 3 mm benzamidine) using the Polytron bench top homogenizer followed by heating the lysates for 5 min in Laemmli buffer. Primary antibodies used were as follows: anti-phospho-(Ser-51)-eIF2 (Cell Signaling Technology, CST 3597), anti-eIF2α (Santa Cruz Biotechnology, Inc., sc-11386), anti-lamin B1 (Cell Signaling Technology, catalog no. 9087), anti-tubulin β (Bioss, bs-4511R). Secondary antibody was peroxidase-AffiniPure goat anti-rabbit TgC (H+L) from Jackson ImmunoResearch (111-035-003). The images were taken with Fluorchem M imager (ProteinSimple), and band densities were quantified using AlphaView software.
Iron measurement
Iron content of frozen murine livers was determined as described (50), using reagents from the QuantiChrom Iron Assay Kit (BioAssay Systems, catalog no. DIFE-250, Hayward, CA). Briefly, all of the glassware was washed with soap and water, rinsed with milliQ water three times, and then soaked overnight in 20% nitric acid to ensure removal of iron traces. About 50 mg of powdered frozen liver (unflushed) per biological replicate was placed in the clean glass tubes and dried in a Savant SpeedVac at 80 °C for 30 min. The dried pellets were digested with 100 μl of 70% nitric acid at 90 °C for 15–20 min, neutralized with 160 μl of 10 m NaOH, and further diluted with 0.4 m ammonium acetate (pH 4.2) up to 1 ml of the final volume. Aliquots of 100 μl of the neutralized diluted digestant were pipetted to 96-well plate in triplicates and then mixed with 25 μl of the colorimetric solution (water: Solution B/ascorbic acid:Solution C/chelating fluorophore, 3:1:1). For cloudy samples, 5 μl of 10% SDS per well were added to dissolve the white flaky organic precipitate. Absorbance at 600 nm was measured within 10 min. A standard curve was generated using an iron chloride solution according to the manufacturer's instructions.
Serum analysis
Serum alanine aminotransferase, alkaline phosphatase, serum triglyceride, and total cholesterol were measured using commercially available kits (Pointe Scientific, Canton, MI). Amino acid composition of the serum was determined as described previously (18).
RNA-Seq
Libraries for RNA-Seq were prepared and sequenced at the JP Sulzberger Columbia Genome Center (Columbia University, New York) as described (7). The reads were mapped to the mouse genome (mm10) with Bowtie 2 and TopHat version 2.0. Sequencing data for the 8-week cohort of mice were taken from (7). Differences in gene expression were determined using DEseq2 approach and evaluated according to treatment and age of the mice. Expression values in Tables S1 and S2 are given in transcripts per million (TPM), which was calculated according to the following formula,
| (Eq. 1) |
where Xi is the number of reads mapped to gene i; li is the length of the longest transcript of gene i; and N is the total number of genes identified per sample.
RT-qPCR validation
RNA was isolated from 10–20 mg of frozen tissue powder using TriReagent (Sigma-Aldrich, catalog no. T9424). Quality of the isolated RNA was assessed by measuring A260/280 and A260/230 ratios; integrity was assessed by visualization of the rRNA on agarose gel. cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, catalog no. 4368814) following the manufacturer's instructions with 1 μg of input RNA for 20 μl of reverse transcription reaction. The resulting cDNA libraries were diluted 20 times with water and used in a SYBR Green–based protocol (Thermo Fisher Scientific, catalog no. 4309155) with the following primers: Fgf21 (forward, 5′-AGCATACCCCATCCCTGACT-3′; reverse, 5′-AGGAGACTTTCTGGACTGCG-3′), Trib3 (forward, 5′-CAGCAACTGTGAGAGGACGA-3′; reverse, 5′-TGGAATGGGTATCTGCCAGC-3′), Mthfd2 (forward, 5′-AGTGCGAAATGAAGCCGTTG-3′; reverse, 5′-GACTGGCGGGATTGTCACC-3′), Cyp51 (forward, 5′-GTTTCAGGCGCAGGGATAGA-3′; reverse, 5′-CATCTGTTAGAGGACGCCCG-3′), Hmgcr (forward, 5′-ATCCTGACGATAACGCGGTG-3′; reverse, 5′-AAGAGGCCAGCAATACCCAG-3′), Hmgcs1 (forward, 5′-TGATCCCCTTTGGTGGCTGA-3′; reverse, 5′-AGGGCAACGATTCCCACATC-3′), Hamp (forward, 5′-AGGGCAGACATTGCGATACC-3′; reverse, 3′-GCAACAGATACCACACTGGGA-3′), Bdh2 (forward, 5′-CAAAAGGACTCGGAGGGGAC-3′; reverse, 3′-ACCAGTTGTGCAGGTGGTAA-3′), Saa1 (forward, 5′-ACCAGATCTGCCCAGGAGAC-3′; reverse, 5′-CCCTTGGAAAGCCTCGTGAAC-3′), Saa2 (forward, 5′-ATGGAGACAAATACTTCCATGCT-3′; reverse, 5′-GCCGAAGAATTCCTGAAAGCTC-3′).
Statistical analysis
Global transcriptome analyses (n = 3 per group) were conducted using the DEseq2 R-script. Differential expression of a gene was considered statistically significant between treatment groups when the adjusted probability (padj) value was <0.1. Principal component analysis was performed on the log2-transformed counts normalized for library size as described in the DESeq2 vignette. Serum amino acids and all lipid measurements were analyzed by two-factor ANOVA with ASNase and age as independent factors. Statistically significant comparisons (main or interaction effects) were further assessed using Tukey's HSD. All other measurements were analyzed with Wilcoxon rank-sum test through pairwise comparisons. Correlation between Saa2 mRNA levels and serum ALT levels was established by Spearman's rank correlation hypothesis testing.
Author contributions
T. G. A. and I. A. N. conceptualization; I. A. N., E. T. M., B. K., W. O. J. and T. G. A. animal handling and biochemical investigation; I. A. N., C. C. S., Q. Z., B. K., W. O. J. and W. J. B. data curation; I. A. N. validation; I. A. N. and W. O. J. visualization; I. A. N., E. T. M., Q. Z., and T. G. A. methodology; I. A. N. writing original draft; I. A. N., W. J. B., and T. G. A. writing, review, and editing; T. G. A. funding acquisition; W. J. B., G. L. G. and T. G. A. resources; T.G.A. supervision; T. G. A. project administration.
Supplementary Material
Acknowledgments
We thank Yongping Wang and Rana Al Baghdadi for expert technical assistance and Ronald Wek, Premal Shah, and Elizabeth Snyder for fruitful discussions of the manuscript.
This work was supported by National Institutes of Health Grants HD070487 (to T. G. A.), DK109714 (to T. G. A.), K12GM093854 (to I. A. N.), R21ES028258 (to G. L. G.), China Scholarship Council grant 201406270118 (Q. Z.), VA-BX002741 (to G. L. G.) and National Institute of Food and Agriculture Grant NE1439 (to W. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editor's Picks.
This article contains Tables S1 and S2.
Data sets analyzed in this study are available at the Gene Expression Omnibus under accession number GSE122243.
- eIF2
- eukaryotic initiation factor 2
- GCN2
- general control nonderepressible 2
- PERK
- protein kinase R–like endoplasmic reticulum kinase
- ATF4
- activating transcription factor 4
- ISR
- integrated stress response
- qPCR
- quantitative PCR
- ER
- endoplasmic reticulum
- ALT
- alanine aminotransferase
- STRING
- search tool for the retrieval of interacting genes/proteins
- ANOVA
- analysis of variance
- ASNase
- asparaginase
- ALP
- alkaline phosphatase
- ECM
- extracellular matrix.
References
- 1. Christ T. N., Stock W., and Knoebel R. W. (2018) Incidence of asparaginase-related hepatotoxicity, pancreatitis, and thrombotic events in adults with acute lymphoblastic leukemia treated with a pediatric-inspired regimen. J. Oncol. Pharm. Pract. 24, 299–308 10.1177/1078155217701291 [DOI] [PubMed] [Google Scholar]
- 2. Phillipson-Weiner L., Mirek E. T., Wang Y., McAuliffe W. G., Wek R. C., and Anthony T. G. (2016) General control nonderepressible 2 deletion predisposes to asparaginase-associated pancreatitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G1061–G1070 10.1152/ajpgi.00052.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilson G. J., Bunpo P., Cundiff J. K., Wek R. C., and Anthony T. G. (2013) The eukaryotic initiation factor 2 kinase GCN2 protects against hepatotoxicity during asparaginase treatment. Am. J. Physiol. Endocrinol. Metab. 305, E1124–E1133 10.1152/ajpendo.00080.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reinert R. B., Oberle L. M., Wek S. A., Bunpo P., Wang X. P., Mileva I., Goodwin L. O., Aldrich C. J., Durden D. L., McNurlan M. A., Wek R. C., and Anthony T. G. (2006) Role of glutamine depletion in directing tissue-specific nutrient stress responses to l-asparaginase. J. Biol. Chem. 281, 31222–31233 10.1074/jbc.M604511200 [DOI] [PubMed] [Google Scholar]
- 5. Bunpo P., Dudley A., Cundiff J. K., Cavener D. R., Wek R. C., and Anthony T. G. (2009) GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent l-asparaginase. J. Biol. Chem. 284, 32742–32749 10.1074/jbc.M109.047910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nikonorova I. A., Al-Baghdadi R. J. T., Mirek E. T., Wang Y., Goudie M. P., Wetstein B. B., Dixon J. L., Hine C., Mitchell J. R., Adams C. M., Wek R. C., and Anthony T. G. (2017) Obesity challenges the hepatoprotective function of the integrated stress response to asparaginase exposure in mice. J. Biol. Chem. 292, 6786–6798 10.1074/jbc.M116.768408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Baghdadi R. J. T., Nikonorova I. A., Mirek E. T., Wang Y., Park J., Belden W. J., Wek R. C., and Anthony T. G. (2017) Role of activating transcription factor 4 in the hepatic response to amino acid depletion by asparaginase. Sci. Rep. 7, 1272 10.1038/s41598-017-01041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lomelino C. L., Andring J. T., McKenna R., and Kilberg M. S. (2017) Asparagine synthetase: function, structure, and role in disease. J. Biol. Chem. 292, 19952–19958 10.1074/jbc.R117.819060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson G. J., Lennox B. A., She P., Mirek E. T., Al Baghdadi R. J. T., Fusakio M. E., Dixon J. L., Henderson G. C., Wek R. C., and Anthony T. G. (2015) GCN2 is required to increase fibroblast growth factor 21 and maintain hepatic triglyceride homeostasis during asparaginase treatment. Am. J. Physiol. Endocrinol. Metab. 308, E283–E293 10.1152/ajpendo.00361.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bunpo P., Cundiff J. K., Reinert R. B., Wek R. C., Aldrich C. J., and Anthony T. G. (2010) The eIF2 kinase GCN2 is essential for the murine immune system to adapt to amino acid deprivation by asparaginase. J. Nutr. 140, 2020–2027 10.3945/jn.110.129197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimball S. R., Vary T. C., and Jefferson L. S. (1992) Age-dependent decrease in the amount of eukaryotic initiation factor 2 in various rat tissues. Biochem. J. 286, 263–268 10.1042/bj2860263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Somel M., Khaitovich P., Bahn S., Pääbo S., and Lachmann M. (2006) Gene expression becomes heterogeneous with age. Curr. Biol. 16, R359–R360 10.1016/j.cub.2006.04.024 [DOI] [PubMed] [Google Scholar]
- 13. White R. R., Milholland B., MacRae S. L., Lin M., Zheng D., and Vijg J. (2015) Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16, 899 10.1186/s12864-015-2061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M. W., Gaffney D. J., Elo L. L., Zhang X., and Mortazavi A. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 17. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nikonorova I. A., Mirek E. T., Signore C. C., Goudie M. P., Wek R. C., and Anthony T. G. (2018) Time-resolved analysis of amino acid stress identifies eIF2 phosphorylation as necessary to inhibit mTORC1 activity in liver. J. Biol. Chem. 293, 5005–5015 10.1074/jbc.RA117.001625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carraro V., Maurin A.-C., Lambert-Langlais S., Averous J., Chaveroux C., Parry L., Jousse C., Ord D., Ord T., Fafournoux P., and Bruhat A. (2010) Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2α/ATF4 pathway. PLoS One 5, e15716 10.1371/journal.pone.0015716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du K., Herzig S., Kulkarni R. N., and Montminy M. (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 10.1126/science.1079817 [DOI] [PubMed] [Google Scholar]
- 21. Ord D, and Ord T. (2003) Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp. Cell Res. 286, 308–320 10.1016/S0014-4827(03)00070-3 [DOI] [PubMed] [Google Scholar]
- 22. Jousse C., Deval C., Maurin A. C., Parry L., Chérasse Y., Chaveroux C., Lefloch R., Lenormand P., Bruhat A., and Fafournoux P. (2007) TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J. Biol. Chem. 282, 15851–15861 10.1074/jbc.M611723200 [DOI] [PubMed] [Google Scholar]
- 23. Maida A., Zota A., Sjøberg K. A., Schumacher J., Sijmonsma T. P., Pfenninger A., Christensen M. M., Gantert T., Fuhrmeister J., Rothermel U., Schmoll D., Heikenwälder M., Iovanna J. L., Stemmer K., Kiens B., et al. (2016) A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J. Clin. Invest. 126, 3263–3278 10.1172/JCI85946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Averous J., Lambert-Langlais S., Cherasse Y., Carraro V., Parry L., B'chir W., Jousse C., Maurin A. C., Bruhat A., and Fafournoux P. (2011) Amino acid deprivation regulates the stress-inducible gene p8 via the GCN2/ATF4 pathway. Biochem. Biophys. Res. Commun. 413, 24–29 10.1016/j.bbrc.2011.08.028 [DOI] [PubMed] [Google Scholar]
- 25. Jin H. O., Seo S. K., Woo S. H., Choe T. B., Hong S. I., Kim J. I., and Park I. C. (2009) Nuclear protein 1 induced by ATF4 in response to various stressors acts as a positive regulator on the transcriptional activation of ATF4. IUBMB Life 61, 1153–1158 10.1002/iub.271 [DOI] [PubMed] [Google Scholar]
- 26. Kim B.-T., Kitagawa H., Tamura J., Saito T., Kusche-Gullberg M., Lindahl U., and Sugahara K. (2001) Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode 1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/heparin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 7176–7181 10.1073/pnas.131188498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sikalidis A. K., Lee J.-I., and Stipanuk M. H. (2011) Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids 41, 159–171 10.1007/s00726-010-0571-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dreesen O., Ong P. F., Chojnowski A., and Colman A. (2013) The contrasting roles of lamin B1 in cellular aging and human disease. Nucleus 4, 283–290 10.4161/nucl.25808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Z., Ciocea A., and Devireddy L. (2014) Endogenous siderophore 2,5-dihydroxybenzoic acid deficiency promotes anemia and splenic iron overload in mice. Mol. Cell. Biol. 34, 2533–2546 10.1128/MCB.00231-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zughaier S. M., Stauffer B. B., and McCarty N. A. (2014) Inflammation and ER stress downregulate BDH2 expression and dysregulate intracellular iron in macrophages. J. Immunol. Res. 2014, 140728 10.1155/2014/140728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evstafieva A. G., Kovaleva I. E., Shoshinova M. S., Budanov A. V., and Chumakov P. M. (2018) Implication of KRT16, FAM129A and HKDC1 genes as ATF4 regulated components of the integrated stress response. PLoS One 13, e0191107 10.1371/journal.pone.0191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi M., Kato H., Hada H., Itoh-Nakadai A., Fujiwara T., Muto A., Inoguchi Y., Ichiyanagi K., Hojo W., Tomosugi N., Sasaki H., Harigae H., and Igarashi K. (2017) Iron-heme-Bach1 axis is involved in erythroblast adaptation to iron deficiency. Haematologica 102, 454–465 10.3324/haematol.2016.151043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Y., Fuscoe J. C., Zhao C., Guo C., Jia M., Qing T., Bannon D. I., Lancashire L., Bao W., Du T., Luo H., Su Z., Jones W. D., Moland C. L., Branham W. S., et al. (2014) A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 5, 3230 10.1038/ncomms4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu J., Knutson M. D., Carter C. S., and Leeuwenburgh C. (2008) Iron accumulation with age, oxidative stress and functional decline. PLoS One 3, e2865 10.1371/journal.pone.0002865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schiele F., Henny J., Hitz J., Petitclerc C., Gueguen R., and Siest G. (1983) Total bone and liver alkaline phosphatases in plasma: biological variations and reference limits. Clin. Chem. 29, 634–641 [PubMed] [Google Scholar]
- 36. Hijiya N., and van der Sluis I. M. (2016) Asparaginase-associated toxicity in children with acute lymphoblastic leukemia. Leuk. Lymphoma 57, 748–757 10.3109/10428194.2015.1101098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., and Rabinowitz J. D. (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 10.1038/nature13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ben-Sahra I., Hoxhaj G., Ricoult S. J. H., Asara J. M., and Manning B. D. (2016) mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 10.1126/science.aad0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fu Z. D., Csanaky I. L., and Klaassen C. D. (2012) Effects of aging on mRNA profiles for drug-metabolizing enzymes and transporters in livers of male and female mice. Drug Metab. Dispos. 40, 1216–1225 10.1124/dmd.111.044461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kwekel J. C., Desai V. G., Moland C. L., Branham W. S., and Fuscoe J. C. (2010) Age and sex dependent changes in liver gene expression during the life cycle of the rat. BMC Genomics 11, 675 10.1186/1471-2164-11-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boissel N., and Sender L. S. (2015) Best practices in adolescent and young adult patients with acute lymphoblastic leukemia: a focus on asparaginase. J. Adolesc. Young Adult Oncol. 4, 118–128 10.1089/jayao.2015.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ben Tanfous M., Sharif-Askari B., Ceppi F., Laaribi H., Gagné V., Rousseau J., Labuda M., Silverman L. B., Sallan S. E., Neuberg D., Kutok J. L., Sinnett D., Laverdière C., and Krajinovic M. (2015) Polymorphisms of asparaginase pathway and asparaginase-related complications in children with acute lymphoblastic leukemia. Clin. Cancer Res. 21, 329–334 10.1158/1078-0432.CCR-14-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orgel E., Sposto R., Malvar J., Seibel N. L., Ladas E., Gaynon P. S., and Freyer D. R. (2014) Impact on survival and toxicity by duration of weight extremes during treatment for pediatric acute lymphoblastic leukemia: a report from the Children's Oncology Group. J. Clin. Oncol. 32, 1331–1337 10.1200/JCO.2013.52.6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caballero-Molada M., Planes M. D., Benlloch H., Atares S., Naranjo M. A., and Serrano R. (2018) The Gcn2–eIF2α pathway connects iron and amino acid homeostasis in Saccharomyces cerevisiae. Biochem. J. 475, 1523–1534 10.1042/BCJ20170871 [DOI] [PubMed] [Google Scholar]
- 45. Braoudaki M., Lambrou G. I., Vougas K., Karamolegou K., Tsangaris G. T., and Tzortzatou-Stathopoulou F. (2013) Protein biomarkers distinguish between high- and low-risk pediatric acute lymphoblastic leukemia in a tissue specific manner. J. Hematol. Oncol. 6, 52 10.1186/1756-8722-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt P. J. (2015) Regulation of iron metabolism by hepcidin under conditions of inflammation. J. Biol. Chem. 290, 18975–18983 10.1074/jbc.R115.650150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bolla R. I., and Miller J. K. (1980) Endogenous nucleotide pools and protein incorporation into liver nuclei from young and old rats. Mech. Ageing Dev. 12, 107–118 10.1016/0047-6374(80)90087-1 [DOI] [PubMed] [Google Scholar]
- 48. Pedley A. M., and Benkovic S. J. (2017) A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 42, 141–154 10.1016/j.tibs.2016.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koufaris C., and Nilsson R. (2018) Protein interaction and functional data indicate MTHFD2 involvement in RNA processing and translation. Cancer Metab. 6, 12 10.1186/s40170-018-0185-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hedayati M., Abubaker-Sharif B., Khattab M., Razavi A., Mohammed I., Nejad A., Wabler M., Zhou H., Mihalic J., Gruettner C., DeWeese T., and Ivkov R. (2018) An optimised spectrophotometric assay for convenient and accurate quantitation of intracellular iron from iron oxide nanoparticles. Int. J. Hyperthermia 34, 373–381 10.1080/02656736.2017.1354403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.