Abstract
TP53 is the most frequently mutated tumor suppressor gene in many cancers, yet biochemical characterization of several of its reported mutations with probable biological significance have not been accomplished enough. Specifically, missense mutations in TP53 can contribute to tumorigenesis through gain-of-function of biochemical and biological properties that stimulate tumor growth. Here, we identified a relatively rare mutation leading to a proline to leucine substitution (P152L) in TP53 at the very end of its DNA-binding domain (DBD) in a sample from an Indian oral cancer patient. Although the P152Lp53 DBD alone bound to DNA, the full-length protein completely lacked binding ability at its cognate DNA motifs. Interestingly, P152Lp53 could efficiently tetramerize, and the mutation had only a limited impact on the structure and stability of full-length p53. Significantly, when we expressed this variant in a TP53-null cell line, it induced cell motility, proliferation, and invasion compared with a vector-only control. Also, enhanced tumorigenic potential was observed when P152Lp53-expressing cells were xenografted into nude mice. Investigating the effects of P152Lp53 expression on cellular pathways, we found that it is associated with up-regulation of several pathways, including cell-cell and cell-extracellular matrix signaling, epidermal growth factor receptor signaling, and Rho-GTPase signaling, commonly active in tumorigenesis and metastasis. Taken together, our findings provide a detailed account of the biochemical and cellular alterations associated with the cancer-associated P152Lp53 variant and establish it as a gain-of-function TP53 variant.
Keywords: carcinogenesis, tumor suppressor gene, tumor cell biology, gene expression, DNA-binding protein, gain-of-function mutant, P152Lp53, p53 tetramerization, transcriptional regulator
Introduction
p53 is the most well-studied mammalian transcription factor (1). It is biologically active as a homotetramer (2, 3) and cooperatively binds to its target DNA in a sequence-specific manner (4). It is involved in eliciting multifaceted response upon DNA damage in terms of transcriptional output that decides various cellular processes like DNA repair (5), cell cycle arrest (6), and apoptosis (7), thereby maintaining the genomic integrity of the cell (8).
TP53 mutations have been reported in almost every type of cancer with varying rates (9). At least 50% of all tumors exhibit mutation of TP53 (10). Unlike the majority of tumor suppressor genes, which are usually inactivated during cancer progression by deletions or truncating mutations, the vast majority (75%) of the cancer-associated mutations in TP53 are missense mutations (11, 12). Although missense mutations are diverse in their location within the p53 coding sequence, a great majority (>90%) of these missense mutations are clustered within the central, highly conserved DNA-binding domain (DBD)4 region (13, 14) and among these mutated residues are a small number (approximately six) of residues where mutations are found with unusually high frequency. These residues are termed as “hot spot residues” (15).
Mutations have an impact on p53 protein at two different yet interconnected levels. p53 mutants that arise due to the mutations in amino acid residues that make direct contact with the target DNA sequence are “DNA contact” or Class I mutants (such as mutation in Arg-273, Arg-248), whereas “conformational” or Class II mutants (such as Arg-175, Gly-245, Arg-282) arise due to the mutations in residues responsible for maintaining the tertiary structure or conformation of the protein and hence cause disruption in the structure of p53 protein at either a local or global level (16–18).
Mutations in p53 can have various consequences that are not mutually exclusive. Apart from losing WT functions due to loss in DNA-binding activity of p53 and dominant-negative effects where mutant p53 forms mixed tetramer with coexpressed WT p53 and renders it incapable of DNA binding and transactivation (19); missense mutations in p53 also contribute to tumorigenesis through their novel “gain-of-function” effects where mutant p53 acquire new biochemical and biological properties (20, 21) and drives tumor growth through increased cell migration, invasion, proliferation, genomic instability, antiapoptosis, antineoplastic, therapy resistance etc. (22). High frequency of missense mutations, high expression levels and prolonged t½ of mutant p53 (23) in the cell conspicuously supports the gain-of-function hypothesis (15).
Oral squamous cell carcinoma (OSCC) is the most frequent cancer type of head and neck squamous cell carcinoma (HNSCC) (24). TP53 mutations are etiologically associated with OSCC. One study reports tobacco users with a significantly higher evidence of TP53 mutation (25). R248Q, R175H, R273H, and R282W are the most frequent missense mutations observed in OSCC (26). The P152L mutation, although, not a hot spot mutation, still ranks 36th among the 50 most frequent p53 cancer mutants (27). Moreover, the P152L mutation has also been reported in OSCC at the tongue and in the gingivobuccal site (28). However, surprisingly, biochemical and functional characterization of this mutant protein has not been carried out yet.
The proline to leucine mutation, at the 152nd position, resides in the S3/S4 turn opposite to the DNA-binding surface of the mutant p53. Here, we first performed biochemical assays to assess the impact of the P152L mutation on the cognate site DNA-binding ability. Our data clearly indicates the limited but detrimental effect of this mutation on the critical structural conformation essential for DNA binding of the full-length protein, which is ineffective in the context of isolated DBD. The stable expression of P152Lp53 in a p53-null cell line (H1299) significantly induces the tumorigenicity in a xenograft mouse model. RNA-Seq analysis of tumor cell-derived RNA samples reveals unique gain-of-function pathways induced by P152Lp53.
Results
Identification of P152Lp53 missense mutation and its zygosity in an oral cancer patient sample
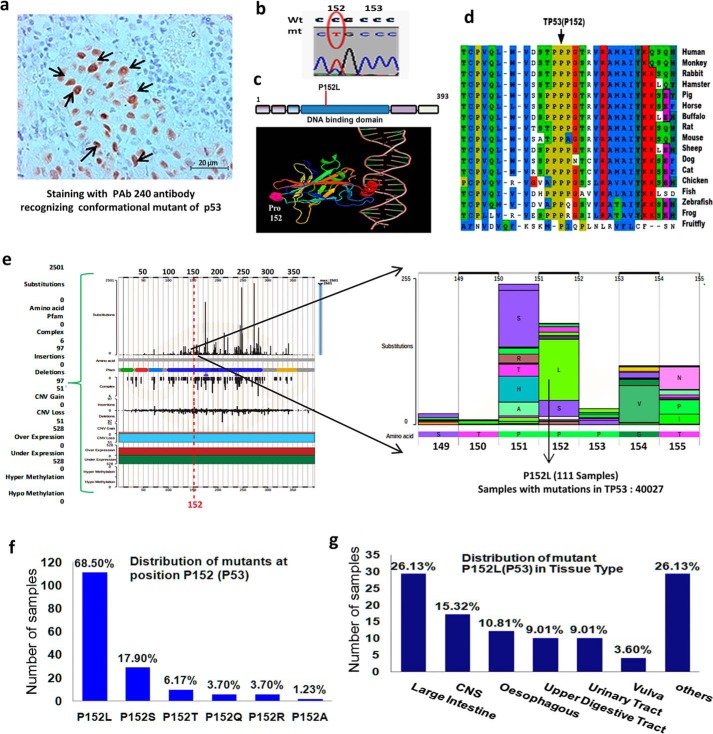
To screen TP53 mutations in oral cancer samples, we performed immunohistochemistry with PAb240 antibody, which recognizes only conformational p53 mutants. From the tumor samples showing high expression levels of mutant p53 in the nucleus, we isolated genomic DNA from the stained region (Fig. 1a). The samples were sequenced for the region spanning the DNA-binding domain using specific primers from exons 4 to 9. In one of the oral cancer samples, we found C > T nucleotide substitution, which leads to a change in the amino acid from proline to leucine at position 152 in the DNA-binding domain of p53 (Fig. 1b). To predict a possible model of P152Lp53 interaction with the cognate DNA-binding site through the PyMOL-generated image, the coordinates of tumor suppressor p53 complexed with DNA (RCSB code 1TUP) were used. It was observed that Pro-152 is far and opposite to the surface of the protein, which is in contact with the cognate DNA sequence site (Fig. 1c). The zygosity of the mutant allele in the patient sample was determined by an allelic discrimination assay. Due to certain limitations of the software, calls were made manually by assessing the calculated normalized dye fluorescence (ΔRn) as a function of cycle number and the cycle threshold (CT) values, from which the mutant allele from the patient sample was found to be a heterozygote for P152L mutation (Table S5).
Figure 1.
Identification of P152Lp53 mutation in a patient sample and prevalence of somatic P152Lp53 missense mutations in human cancers. a, immunohistochemistry for mutant p53 by PAb240 antibody (indicated by →) on an oral cancer sample; b, sequence chromatogram showing C > T substitution at 152 position; c, location of Pro-152 (shown in pink) in 3D structure of the p53 core domain bound to DNA visualized by PyMOL (molecular visualization software); d, multiple sequence alignment of human TP53 protein sequence across various organisms. The Pro-152 residue is indicated by a black arrow. e, prevalence of missense mutations in a stretch of 150–155 amino acids of p53 analyzed from COSMIC database version 86; f, bar plot representation of number and frequency of various missense mutations present at TP53 (Pro-152) loci out of 111 samples of available data at COSMIC. The percentage frequency of various amino acid substitutions are shown above the bars. g, bar plot representation of number and frequency distribution of P152Lp53 mutation occurrence over various cancer tissue types out of 111 samples of P152L mutations. CNS, central nervous system, others: adrenal gland, breast, hematopoietic and lymphoid tissue, liver, lung, pancreas, skin, soft tissue, and stomach tissue.
Prevalence and range of P152Lp53 missense mutation in human cancer
The P152L mutation in p53 has earlier been identified in a patient with esophageal cancer (29), oral squamous cell carcinoma (28), and Li-Fraumeni syndrome having pediatric adrenocortical carcinoma (30). To the best of our knowledge, the detailed functional characterization of P152Lp53 is yet to be done. We performed multiple sequence alignment of p53 protein sequences and observed the conservation of Pro-152 across the organisms during evolution suggesting its structural and functional importance (Fig. 1d). Upon analyzing the prevalence of the P152Lp53 mutation in cancer samples using COSMIC database, it was observed that, most of the mutation at the Pro-152 locus (73%) were missense rather than insertion or deletion mutations (Fig. S1a). Within a sequence stretch of 150–155 amino acids in the DNA-binding domain, frequency of the P152L missense mutation was relatively high as compared with other amino acid substitutions and is comparable with P151S substitution (Fig. 1e), which has already been found to confer gain-of-function activity in head and neck cancer cells (31). The most prevalent missense amino acid change at Pro-152 was found to be P152L (68.5%, Fig. 1f), indicating a higher occurrence of leucine substitution at Pro-152 loci in cancers as compared with other amino acid substitutions. The P152L mutation in p53 occurs in several solid cancers and its total distribution across different cancer tissue types is as follows: large intestine (26.13%), central nervous system (15.32%), esophagus (10.81%), and so on (Fig. 1g). An overall analysis suggested high prevalence of the TP53 (P152L) allele in human cancers, among all other type of mutations at the Pro-152 codon.
Biochemical characterization of P152L p53
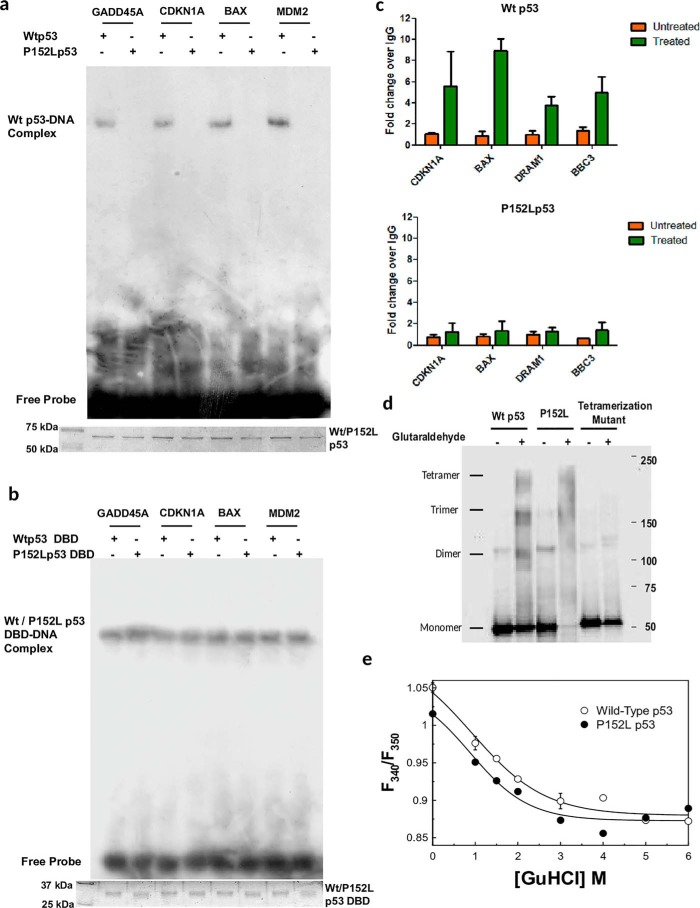
During DNA damage activated p53 gets overexpressed and stabilized and causes activation of a number of p53 downstream target genes through its direct binding to its cognate sites on respective gene promoters. Hence, to investigate impact of the P152L mutation on the DNA-binding ability of the p53 protein we cloned and expressed both the full-length and the DNA-binding domain (Fig. S1b) of WT p53 and P152Lp53 and performed electrophoretic mobility shift assay (EMSA) with recombinant FLAG-tagged full-length Wtp53 and P152Lp53 protein using several γ-32P–labeled oligonucleotides containing p53 responsive elements of the following genes, GADD45A, CDKN1A, BAX, and MDM2. We observed that the full-length P152Lp53 protein could not bind to the p53 cognate sequences at all, as compared with WT protein (Fig. 2a). To ensure the sequence specific binding of p53 to its oligonucleotides, point mutations in the GADD45A consensus sequence was created and subjected to EMSA. As expected, both the WT and mutant p53 could not form any complex with the GADD45A mutant consensus sequence (Fig. S1c, lanes 4 and 8). These observations suggest that P152L missense mutations leads to abrogation of the DNA-binding ability of p53.
Figure 2.
Biochemical characterization of P152Lp53. a, EMSA showing the abrogation of the DNA-binding property of recombinant full-length P152Lp53; b, EMSA showing the DNA binding property of DBD of WT and P152Lp53 (protein loading is shown in Coomassie-stained SDS-PAGE gel image); c, ChIP-qPCR to determine the occupancy of Wtp53 and P152Lp53 at p53 responsive gene promoters. The mean relative fold-change of p53 occupancy over IgG ± S.D. have been plotted (n = 2). d, tetramer formation assay by glutaraldehyde cross-linking resolved in 3–12% gradient SDS-PAGE followed by Western blotting. e, equilibrium GuHCl denaturation of wildtype (○) and P152L (●) p53. The fitted curve is to a two-state equation. Each data point is an average of three independent biological replicates.
Pro-152 is located in a very intriguing position in the DBD of p53 (Fig. 1c). Mutation at this position should not affect the DNA binding of p53, at least of the DBD. Therefore, the effect of the P152L mutation on the DNA-binding ability of the DBD of p53 was assessed. For this purpose EMSA was performed using bacterially expressed untagged WT and P152Lp53 DBD protein. Remarkably, we observed that both WT and P152Lp53 DBD were able to form DNA-protein complex with the GADD45A, CDKN1A, BAX, and MDM2 promoter consensus sequence (Fig. 2b). Likewise, they could not bind to the GADD45A mutant consensus sequence suggesting that DBD-DNA complexes are a result of sequence-specific binding (Fig. S1d, lanes 2 and 3 versus lanes 5 and 6). We validated this result further with cold competition assay for mutant p53 DBD binding to the GADD45A promoter consensus sequence (Fig. S1e, lanes 2–5 and 6–8). These results collectively show that the P152L mutation does not alter the DNA-binding ability of p53 DBD, although it abrogates full-length p53 DNA binding. To study the mutational effect on the transcriptional outcome in the cellular context, ChIP-qPCR for various p53 responsive genes (CDKN1A, BAX, DRAM1, and BBC3) was performed in the H1299 doxycycline-inducible WT and P152Lp53 cell line (Fig. S2a) to assess the enrichment of P152Lp53 on several p53 promoter sequences. Consistent with the in vitro data, a significant decrease in promoter binding was observed for P152Lp53 as against wtp53 (Fig. 2c). In addition, a luciferase assay was performed using transient transfection and expression of WT and P152Lp53 in the H1299 (p53−/−) cell line. The p53 expressing vectors (either mutant or WT) were co-transfected with reporter vector containing p53 responsive elements. Upon transfection of expression vectors, P152Lp53 showed very minimal promoter activation as compared with WT (Fig. S2b). The protein expression was confirmed by Western blotting (Fig. S2c). This observation is consistent with the result obtained in a similar assay (32) where a significant reduction of TP53 function was observed upon P152L mutation.
Functionally p53 binds to the cognate DNA site as a tetramer (2, 33). Several of the mutations in the oligomerization domain of p53, such as L344P or L330H, abolish its ability to tetramerize and hence these mutants exist as monomers and do not bind to DNA. Although a few exceptions do exist: for example, L344A and R337C mutants, which retain the ability to form dimer and tetramer, and hence either show partial or no loss of DNA-binding ability, respectively (Table 1, top). Intriguingly, there are studies where it has been reported that few conformational mutants (V143A, G245S), which have their DNA-binding ability abrogated, still form tetramers upon glutaraldehyde treatment (Table 1, top). To examine the impact of the P152L mutation on its tetramer formation ability, we performed glutaraldehyde cross-linking followed by Western blotting where we observed that P152Lp53 retains the ability to form a tetramer (Fig. 2d, fourth lane). The tetramerization mutant (L344P), as expected, did not show any tetramer formation, rather a dimer was observed. The glutaraldehyde percentage (0.005%) used was standardized as the minimum concentration required to form a tetramer of WT p53.
Table 1.
The DNA binding and oligomerization status of p53 mutants (information retrieved from TP53 mutant database (http://p53.fr/TP53Mutload/database_access/search.php), and the DNA binding property of full-length and DBD of p53 mutants
| p53 mutants | Binding and oligomerization status |
||
|---|---|---|---|
| Class | DNA binding | Oligomerization status | |
| V143A | Conformational | − − | Tetramer |
| G245S | Conformational | − − | Tetramer |
| L344P | Tetramerization mutant | − − | Monomer |
| L344A | Tetramerization mutant | Partial | Dimer |
| L330H | Tetramerization mutant | − − | Monomer |
| R337C | Tetramerization mutant | + + | Tetramer |
The observation that the P152Lp53 tetramer is devoid of DNA-binding property, whereas the DBD retains significant sequence-specific DNA-binding ability, indicates altered subunit-subunit association. The mutant protein may also be aggregated or inherently unstable. To better understand the effect of the mutation on the protein structure, we first measured the hydrodynamic radii of the WT and mutant p53 by dynamic light scattering. Fig. S2d shows the dynamic light scattering profile of the WT and P152Lp53. The measured hydrodynamic radii are 35.5 and 40.9 Å, respectively, for the WT and P152Lp53. The measured radii suggest that neither of the proteins is aggregated to any significant extent and the P152Lp53 protein is modestly enlarged. This is consistent with the idea that the subunits are arranged in a different mode in the mutant protein. GuHCl (guanidine hydrochloride) denaturation of the WT and mutant tetramers was performed to measure the stability of the proteins. Fig. 2e shows the GuHCl-induced equilibrium denaturation profile of the WT and P152Lp53 as measured by increased solvent exposure of tryptophan residues (F340/F350) as a function of GuHCl. The fitted curve is a two-state equation as described in Ref. 34. The fluorescence emission spectra of the mutant are modestly red-shifted compared with that of the WT (reflected in the lower F340/F350 value of the mutant). The midpoints of the transition of the WT and mutant protein are around 1.1 m GuHCl, indicating the stability is not impacted by the mutation in a major way. The change of exposure recorded here most likely reflects the unfolding of the proteins as CD spectra recorded at 3 m GuHCl is devoid of any protein-like bands (data not shown).
Gain-of-function properties of P152Lp53
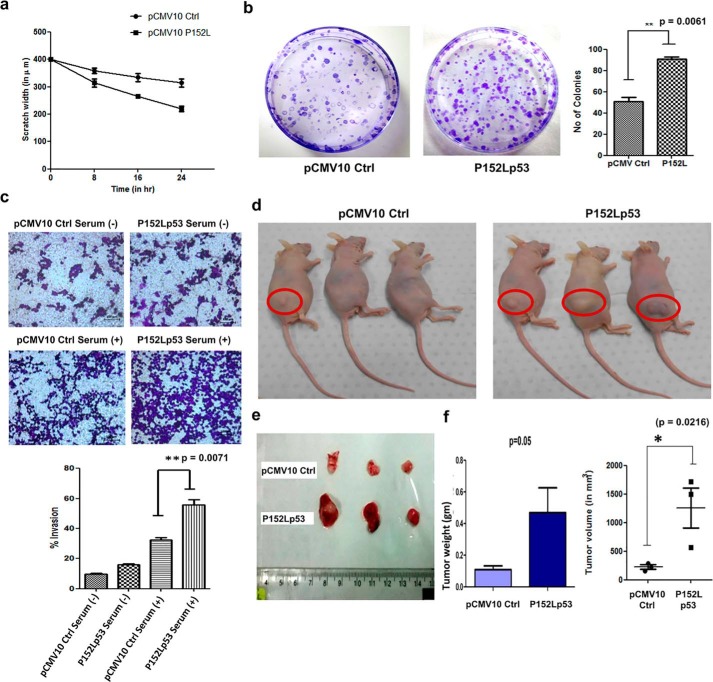
To delineate whether P152Lp53 manifests any gain-of-function properties, we generated a stable cell line expressing P152Lp53 in H1299 (p53−/−) cells. The expression of the mutant protein was confirmed by Western blot analysis and immunofluorescence showed that P152Lp53 localizes in the nucleus as that of WT p53 (Fig. S3, a and b). To ascertain whether expression of mutant p53 alters the migration potential of H1299 stable cell lines, a wound-healing assay was performed. We observed that H1299 cells expressing P152Lp53 migrated faster as compared with the vector control cell line (Fig. 3a, Fig. S3c). Furthermore, we also tested the proliferation ability of P152Lp53 expressing cells through a colony formation assay in which the number of colonies obtained determines the proliferative potential of the cell. It was observed that the number of colonies were significantly higher (1.8 times) in P152Lp53 expressing cells, as compared with the vector control cells (p = 0.0061) (Fig. 3b). Interestingly, the size of colonies was found to be predominantly larger for P152Lp53 expressing cells compared with vector control. To examine the effect of P152Lp53 on the invasive potential, a Transwell-invasion assay was performed. The H1299 cell line stably expressing P152Lp53 displayed an increased percentage invasion as compare with the vector control cell line (p = 0.0071) (Fig. 3c).
Figure 3.
Gain-of-function properties of P152Lp53 in H1299 cells. a, wound-healing assay showing the migration rate of the H1299 cell line stably expressing P152Lp53 compared with pCMV10 vector control (n = 2); b, colony formation assay of pCMV10 Ctrl and the P152Lp53 stable cell line to assess cell proliferation (n = 3); c, Transwell assay showing increased invasive potential of cells expressing P152Lp53 (n = 3); d, non-orthotopic nude mice showing enhanced tumor formation ability of H1299 cells stably expressing P152Lp53 injected in the right flank region; e, tumor extracted after the 38th day of injection showing the difference in size of the tumor of P152Lp53 mice as compared with vector control mice; f, statistical analysis of weight and volume of the tumor extracted from three pCMV10 Ctrl and P152Lp53 mice.
To delineate the tumorigenic potential of P152Lp53 in vivo, the H1299 cell line stably expressing P152Lp53 and vector control cell line were injected subcutaneously into the right flank region of nude mice. After 38 days, upon the appearance of a visible tumor, mice were sacrificed. We observed that the size of the tumors formed was significantly large in nude mice injected with P152Lp53 expressing cells as compared with vector control (Fig. 3, d and e). The difference in both the tumor weight (p = 0.05) and volume (p = 0.0216) between P152Lp53 and vector control mice groups were found to be statistically significant (Fig. 3f). These results collectively suggest that due to P152L missense mutation in the p53 gene, mutant p53 protein have acquired several gain-of-functions effects such as increased cell migration, proliferation, invasion, and tumorigenicity.
Mechanism of gain-of-function of P152Lp53
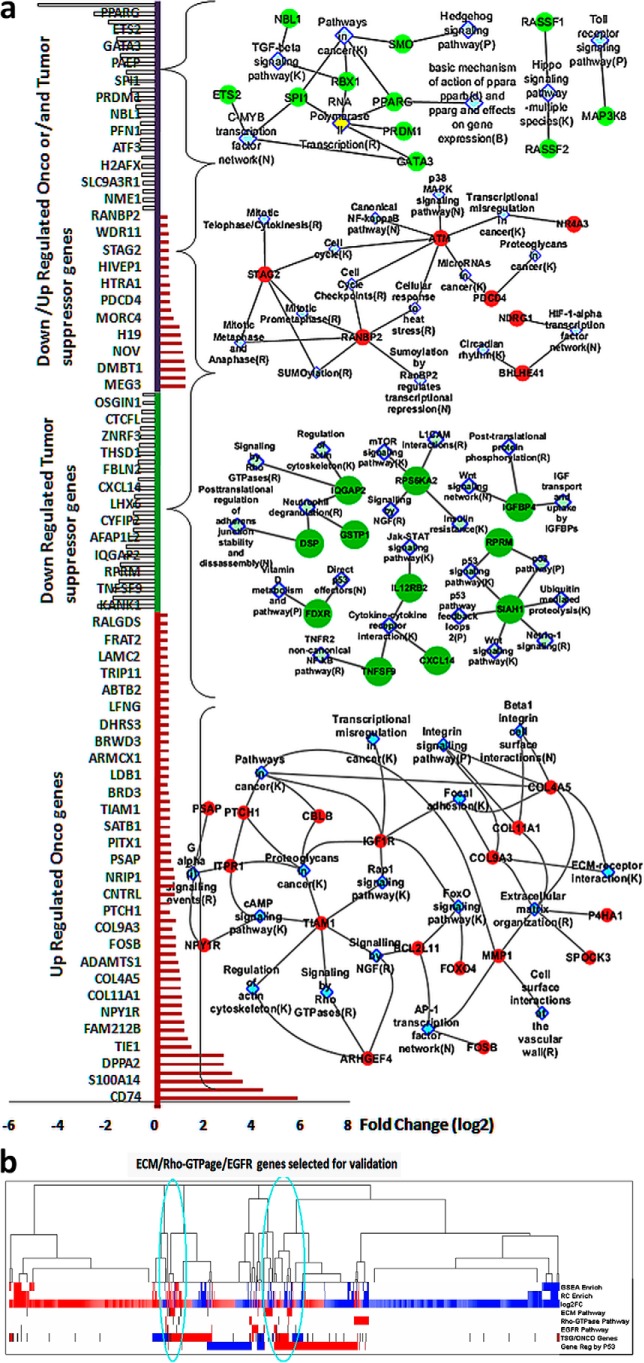
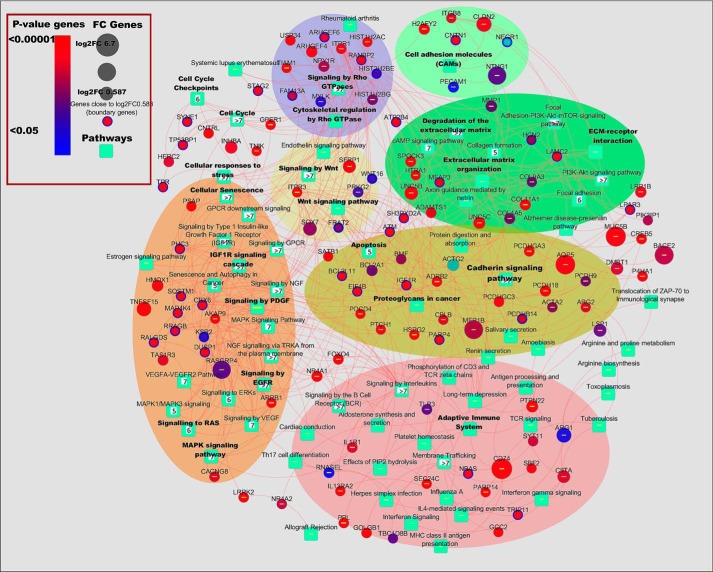
To gain the mechanistic insights of the gain-of-function effects elicited by P152L p53, RNA-Seq was performed for RNA isolated (Fig. S3d) from the xenografted tumor samples (pCMV10 Ctrl and P152Lp53). A total of 661 genes (295 up-regulated and 366 down-regulated) were obtained with a p value ≤0.05 and FC ≥ 1.5 (File S1). Among these, many known and reported oncogenes were found to be up-regulated and many reported tumor suppressor genes (TSG) were down-regulated (Fig. 4a). To get accurate insight of the underlying biological mechanisms and instigated pathways, we performed a multistage enrichment pathway and various comparative analyses. First we did pathway enrichment analysis on all up-regulated and down-regulated genes using 3 methods: GSEA, Reactome FI, and an in-house developed pathway portal. The enriched pathways (up/down) obtained through an in-house developed pathway portal and based upon a significant p value (≤ 0.05) and are tabulated in Files S2 and S3. In addition, several other pathways were found to be enriched when we considered the percentage of differentially up-regulated genes' share belonging to several pathways that contribute to tumorigenesis (File S4). An integrated pathway network of all up-regulated DEG genes obtained from the significantly up-regulated enriched pathways (p < 0.05, fold-change ≥ 1.5) clearly shows that P152Lp53 assists in tumor formation and progression through significant up-regulation of several pathways such as cell-cell/cell-ECM signaling pathway, EGFR signaling pathway, PI3K-Akt signaling pathway, Rho-GTPase signaling pathway, and the Wnt signaling pathway (Fig. 5). Some of the boundary genes (1.5 < FC > 1.4 and p value ≤ 0.055) were also considered during pathway enrichment and network analysis of up-regulated genes (File S5). Through pathway enrichment analysis we investigated the involvement of the various already reported oncogenes, TSG, and genes with dual roles such as ONCO/TSG, which were differentially expressed in our data (Fig. 4a, Files S6–S9).
Figure 4.
Selection of enriched pathways and genes and their involvement in various tumorigenic pathways. a, Onco, tumor suppressor, and dual role of DEG and their enriched pathway networks showing various pathways in metastasis and proliferation. b, hierarchical clustering of DEG, GSEA, and Reactome FI enrichment and other comparisons. DEG up-regulated (log2FC) and in TSG/ONCO data sets (TSG/ONCO genes), the ONCO genes and the literature reported P53 up-regulated genes as shown in red. DEG down-regulated (log2FC) and in TSG/ONCO data sets (TSG/ONCO genes), the TSG (tumor suppressor) genes, and the literature reported P53 down-regulated genes as shown in blue. The black lines are the genes selected for ECM, Rho-GTPase, and EGFR pathways, which mostly fall in the densely clustered region of the genes that are involved in various P53-regulated mechanisms.
Figure 5.
Network of pathways and genes induced by P152Lp53 expression. RNA-seq analysis of the tumor (P152Lp53 versus pCMV10 Ctrl) showing the pathways and genes up-regulated (fold-change ≥ 1.5, p value ≥ 0.05). Some of the boundary genes (fold-change ≥ 1.41 but <1.5) are also considered in the pathway network.
Various oncogenes involved in proliferation and metastasis in various cancer types were found to be up-regulated. Notably among them were COL11A1, PECAM1, LAMC2, MMP1, BCL2L11, TIAM2, and CBLB. Likewise, several TSG were found to be down-regulated such as SIAH1, FDXR, TNFSF9, IL12RB2, FBLN2, and PRICKLE1. Down-regulation of several of these TSG reportedly enhances tumorigenesis. We also classified several differentially expressed genes, which have dual functionality of ONCO or TSG depending on the context and their differential expression further supported our proliferative and metastatic claims. For instance, NDRG1 and HTRA1 were up-regulated, whereas ETS2 was down-regulated (Fig. 4a). We then subjected these selected enriched pathways and DEG to various comparisons and hierarchical clustering (Fig. 4b) and the densely clustered genes were selected for the qPCR validations.
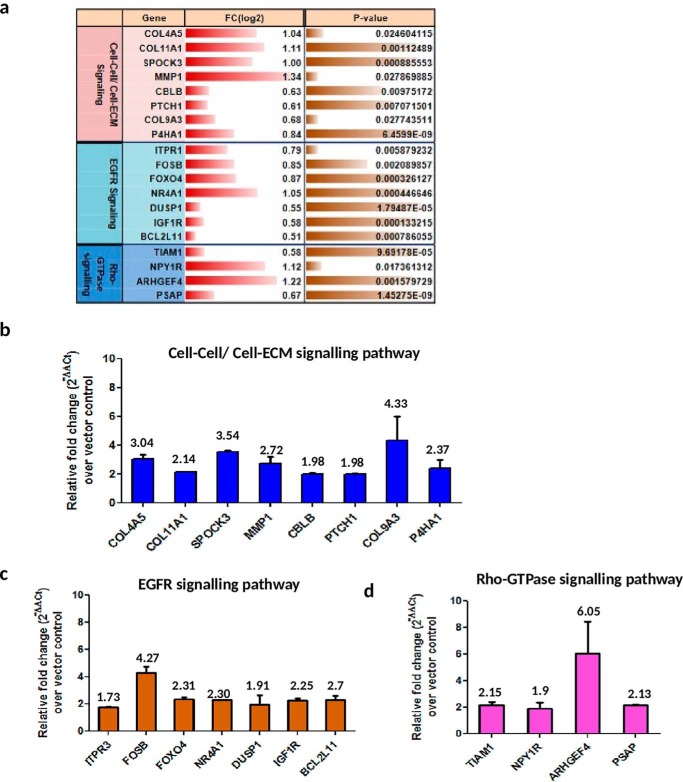
To validate the RNA-Seq analysis, tumor-isolated RNA was subjected to qPCR verification for a few genes belonging to the three pathways: cell-cell/cell-ECM signaling pathway, EGFR signaling pathway, and Rho-GTPase signaling pathway being selected as per the analysis in Fig. 4b. Of the 19 genes selected for validation (Fig. 6a), all genes from the P152L p53 tumor were found to have relative fold-change above 1.5-fold over vector control tumor and 14/19 genes had relative fold-change of above 2 (Fig. 6, b–d). Many of the collagens (essential component of ECM), such as COL4A5, COL11A1 and COL9A3, were up-regulated. Collagens have been implicated in various aspects of neoplastic transformation, for example, COL11A1 is overexpressed in several cancers and has been shown to promote cell proliferation, migration, invasion, and drug resistance in lung cancer (35). Matrix metalloproteinase 1 (MMP1), an endopeptidase, which is involved in tissue remodeling, angiogenesis, and metastatic progression through its proteolytic activity of hydrolyzing the components of ECM (36) was up-regulated. Prolyl 4-hydroxylases (P4Hs) are involved in collagen biogenesis and have been found to be essential for breast cancer metastasis (37). One of the isoforms of P4Hs, i.e. P4HA1, was found to be up-regulated. Several other genes belonging to the EGFR signaling pathway, such as ITPR3, NR4A1, IGF1R etc. were significantly up-regulated. An increased level of ITPR3 is associated with growth and aggressiveness of different types of tumors. NR4A1, a transcription factor, has been found to be a potent activator of transforming growth factor-β/SMAD signaling thereby promoting breast cancer invasion and metastasis (38). Various studies have established that cancer cells express insulin and insulin-like growth factor 1 receptors, and that their expression is critical for anchorage independent growth (characteristic of malignant cells) and are important activators of the Akt and mitogen-activated protein kinase signaling pathways in neoplastic tissue (39). Furthermore, several Rho-GTPase signaling genes such as TIAM1 were also found up-regulated. Increased TIAM1 expression is associated with increased invasiveness and epithelial-mesenchymal transition in several cancers (40). Collectively, these up-regulated genes and pathways involved might work in concert and contribute to proliferation, adhesion, migration, invasion to neighboring tissues, tumor-associated angiogenesis, and eventually metastasis.
Figure 6.
Validation of RNA-Seq analysis by qPCR. a, selected up-regulated pathways and genes (with fold-change (log2) and p value) from RNA-Seq analysis. Verification of b, cell-cell/cell-ECM signaling pathway; c, EGFR signaling pathway; and d, Rho-GTPase signaling pathway was performed by qPCR. Data are plotted as the relative fold-change increase in genes of P152Lp53 tumor over pCMV10 vector control tumor (n = 2). Individual fold-change values are indicated above the bar. All error bars are calculated using standard deviation.
Discussion
To date many of the missense mutations in p53 are found to have GOF effects. Not all p53 missense mutations are GOF mutations. As the likelihood of p53 hot spot mutants to be reported in cancer is more than other p53 mutants, our knowledge pertaining to the GOF effects of p53 mutants has largely been acquired from studies that tested for the functional properties of hot spot mutants. For instance, GOF properties for some of the mutants, such as R175H, R273H, R248W and R248Q, have extensively been studied as they are among the most frequent mutants in many cancers. This has rendered many of the potentially oncogenic p53 mutants that are relatively rare or less frequent in sporadic cancers, structurally or functionally uncharacterized. Moreover, no association between the GOF properties and frequency of reported p53 mutants could be established because several rare mutants such as V143A and D281G were also found to possess GOF properties (26).
To add to the complexity, different p53 mutants have intrinsic differences in their biological activities and behave differently depending upon the cell type and genetic background of the cell. Even different amino acid substitutions at the same position in the p53 protein can have dramatically different phenotypic effects. For example R248Q but not R248W, is able to confer invasive ability when overexpressed in p53 null cells (41). Hence our knowledge is still limited so as to how a single point mutation or a specific mutation in p53 can be very detrimental to cause havoc inside the cell in terms of transforming a normal cell to a tumor promoting cell.
This study for the first time carried out the biochemical characterization of the P152Lp53 mutant and established it as a gain-of-function mutant. P152L mutation resulted in loss of DNA-binding ability of full-length p53 protein but not that of the DNA-binding domain. This observation is interesting in light of the fact that very few (for example V143A) p53 mutants have been reported, where DBD of the mutant has the propensity of binding to DNA but the full-length mutant is unable to bind to DNA (Table 1, lower), indicative of the global impact of the mutation rather than local. Presumably P152L mutation does not lead to any significant local structural alterations in its DBD region rather it changes the global conformation of full-length p53 protein in a way that the DNA binding is lost. Because the P152L mutant also retains the ability to form a tetramer we hypothesize that the alteration of structure due to the mutation is not to an extent that the mutant p53 monomer cannot form tetramers, rather a conformationally altered tetramer is formed that is unable to bind to the p53 consensus binding sites. Slightly enlarged hydrodynamic radii of P152Lp53 as compared with WT p53 with no major effect in its stability further strengthen our hypothesis.
Remarkably it also displayed many of the gain-of-function properties in the form of increased migration, proliferation, invasion, and tumorigenicity, which clearly designates it into the GOF mutants' category of p53 mutants. The identification of this mutation from the oral cancer patient also consolidates its gain-of-function property.
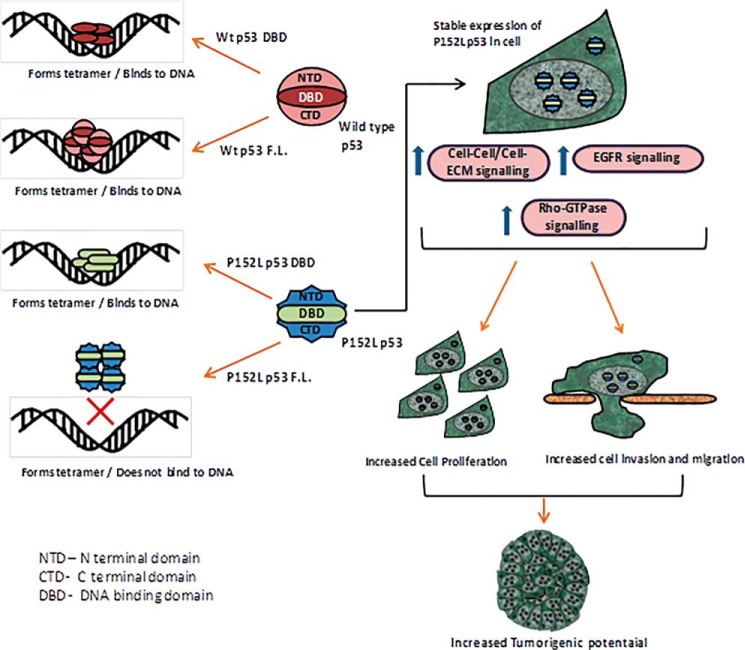
In conclusion, the P152L substitution likely results in tertiary structural alterations leading to significant functional changes in the p53 protein with loss of p53 transcriptional activity and several effects demonstrating P152Lp53 as a new gain-of-function mutant, which is conformationally altered. Remarkably, its mutational consequence appear to be a global phenomenon in the protein conformation, rather than local structural changes in the DNA-binding domain (Fig. 7).
Figure 7.
Summarized cartoon representation of the biochemical and functional characterization of P152L p53.
Experimental procedures
Immunohistochemistry and mutation detection
Oral tumor samples for this investigation were collected from patients at Bangalore Institute of Oncology (BIO), Bangalore, Karnataka, and Sri Devaraj Urs Academy of Higher Education and Research (SDUAHER), Kolar, Karnataka. Ethical clearance and approval for human studies was obtained from the Ethical Review Board at BIO, Central Ethics Committee at SDUAHER, Kolar (reference number SDUAHER/KLR/R&D/79/2014–15) and Institutional Human Bioethics and Biosafety Review Committee at JNCASR (reference number JNC/IBSC/2013/07–1495). Written informed consent was obtained from all patients before collection of samples and the sample collection procedures abide by the Declaration of Helsinki principles. 5-μm paraffin sections of oral cancers were kept overnight on a hot plate at 55 °C and 1 h at 60 °C. After deparaffinization and rehydration, sections were subjected to antigen retrieval in sodium citrate buffer (pH 7.2) for 20 min and then cooled down to room temperature for further processing. Sections were blocked in 5% skimmed milk solution for 2 h at room temperature. Primary antibody (PAb240 mutant p53 specific antibody, Millipore) was then added and kept in a humid chamber for overnight incubation at room temperature. Slides were developed using a streptavidin-avidin biotin kit (Dako Inc.) and used diaminobenzidine as a substrate and counterstained by diluted hematoxylin. Slides were mounted with DPX. The positive region was excised by using the Pin Point DNA isolation kit (Zymo Research Corp., CA) and genomic DNA was isolated. Using specific primers (Table S1) spanning the whole DNA-binding domain of the p53 gene, PCR was done and the amplified DNA products were given for Sanger sequencing. Mutations were analyzed by BioEdit software.
Luciferase assay
200 ng of PG13-luc plasmid containing p53 responsive elements and 300 ng of pCMV2-β-gal plasmid were co-transfected with 400 ng of pCMV2, pCMV2-wtp53, or P152Lp53 mammalian expression vectors in the H1299 cell line. 24 h after transfection, cells were lysed in the 1× reporter lysis buffer provided with the Luciferase Assay Kit (Promega). After a brief vortexing, whole cell lysates were centrifuged at 13,000 rpm (4 °C) for 5 min. Supernatant was collected in a fresh tube and 5 μl was added to the luciferase assay substrate (5 μl). Luminescence was measured as relative light units by a WALLAC 1409 liquid scintillation counter and normalized with β-gal count.
Allelic discrimination assay
TaqMan-MGB genotyping assay mix (×40), contained forward and reverse primers, one probe matching the WT sequence variant labeled with VIC and another probe matching to the mutant (SNP) sequence variant labeled with FAM. A working master mix (5 μl) contained 0.125 μl of TaqMan-genotyping assay mix (×40), 2.5 μl of TaqMan Genotyping Master Mix (Applied Biosystems, Carlsbad, CA), and 50 ng of human genomic DNA. Applied Biosystems StepOnePlus Real-Time PCR Systems and its own in build software was used for the genotyping assay experiment. The AB standard PCR protocol was 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min, repeating steps 2–3 for 40 cycles. The software analyzed the before and after florescence level and calculated normalized dye fluorescence (ΔRn) as a function of cycle number for Allele1 (WT) or Allele2 (mutant). Based on this number the software made an automatic call of either Allele1 (homozygous1/1), Allele2 (homozygous2/2), or heterozygous (1/2). Due to certain limitation of the software, calls were made manually by assessing (ΔRn) and the cycle threshold (CT) values.
Multiple sequence alignment of p53 protein sequence and TP53 mutation statistics at Pro-152 location
Multiple sequence alignment was done to check conservation of “P” amino acid at the 152th position across phylogenetically close organisms. After retrieving the protein sequences of the selected organisms from UniProt database, the BLAST was done using pBLAST. The multiple sequence alignment was performed using CLASTALX (42) and visualized using AliView (43). To access the impact of the reported mutations and it's statistics in TP53 at the location Pro-152, the data from COSMIC, version 86 (https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=TP53)5 was taken and analyzed as described under “Results.”
Modeling proline to leucine substitution at 152nd amino acid position in p53
To model the location of proline at position 152 of p53, coordinates of the human p53 DNA-binding domain bound to DNA with accession code 1TUP, tumor suppressor p53 complexed with DNA were taken from RCSB, and analyzed with the structural analysis program PyMOL (DeLano Scientific, Palo Alto, CA).
Recombinant protein expression and purification
FLAG-tagged full-length WT p53 and P152Lp53 and untagged DBD of WT p53 and P152Lp53 were expressed and purified from BL21(DE3)pLysS Escherichia coli strain. The recombinant FLAG-tagged p53 proteins were purified through affinity chromatography using M2-agarose (Sigma) binding resin. Untagged p53 DBD proteins were purified through cation exchange chromatography using SP-Sepharose binding resin (GE Healthcare).
Gel mobility shift assay
Various radiolabeled oligonucleotides (Table S6) containing p53 responsive-elements (GADD45A, CDKN1A, BAX, and MDM2) were incubated in a reaction volume of 40 μl containing 8 μl of 5× EMSA buffer (100 mm HEPES, pH 7.9, 125 mm KCl, 0.5 mm EDTA, 50% glycerol, 10 mm MgCl2), 2 μl of 60 μg/ml of double stranded poly(dI-dC), 4 μl of BSA (1 mg/ml), 2 μl of Nonidet P-40, 2 μl of DTT with the proteins as indicated for 30 min at 30 °C. Samples were analyzed on 6% native PAGE containing 0.5 × Tris borate-EDTA buffer and electrophoresed at 4 °C for 2 h (200 V). The gel was dried and an autoradiogram was then developed. For cold competition experiments, an increasing concentration of cold oligo (GADD45A/GADD45A mutant consensus sequence) was added 15 min prior to the addition of radiolabeled GADD45A CS oligonucleotide.
Chromatin immunoprecipitation
ChIP followed by qPCR at p53 responsive gene promoters was performed as described in Ref. 44.
Dynamic light scattering
Dynamic light scattering measurements were recorded at room temperature for both FL-WT and FL-P152L p53 in 20 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl and 100 μm ZnCl2 at a concentration of 1 μm with a Zetasizer Nano ZS (Malvern Instruments, UK) equipped with 4 milliwatt He-Ne laser source (λ = 632.8 nm) and a detector positioned at a scattering angle of 173°. Scattering data were collected as an average of three measurements with 20 scans for each measurement.
Denaturation experiment
Purified proteins were dialyzed against 20 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl and 100 μm ZnCl2 overnight at 4 °C and further fractionated by size exclusion chromatography with a 16/600 Superdex-200 pg (GE Healthcare) 4 °C pre-equilibrated with buffer A. A peak fraction was collected and concentrated with Amicon® Ultra-15 100-kDa Centrifugal Filter Units up to 2 μm for each FL-WT and FL-P152L p53.
For denaturation experiments, protein samples were incubated overnight (∼18 h) at 25 °C at increasing concentrations of GuHCl (0.25–6 m) (G3272, Sigma). Intrinsic tryptophan fluorescence intensity was measured at 25 °C using an Hitachi F-7000 FL spectrophotometer (excitation 295 nm; emission 310–450 nm; excitation/emission slit width 5 nm; at 2400 nm/min). The fluorescence intensities of corresponding solutions without protein samples were subtracted as background from each reading. The measurements were performed in triplicates, with each dataset consisting of an average of four individual scans.
Glutaraldehyde cross-linking
200 ng of recombinant protein was incubated for 30 min at room temperature in Buffer H containing 20 mm HEPES (pH 7.9), 100 mm KCl, 0.5 mm EDTA, 10% glycerol, 0.5 mm phenylmethylsulfonyl fluoride with 0.005% glutaraldehyde in a final reaction volume of 10 μl. The reaction was terminated by adding SDS gel loading dye and the reaction products were analyzed by gradient gel (3–12%) followed by Western blotting with p53 antibody DO1 (Millipore).
Cell culture and stable transfection
Human lung carcinoma H1299 (p53 null) cell line (ATCC) was cultured in RPMI 1640 media supplemented with 2 mm glutamine (Himedia, India) and 10% (v/v) FCS (Gibco). The pEBTetD p53 H1299 cells and pEBTetD P152Lp53 H1299 cells were cultured in supplemented RPMI 1640 under 1.2 μg/ml of puromycin antibiotic selection. The pFLAG-CMV-10 H1299 and pFLAG-CMV-10-P152Lp53 H1299 cells were cultured in supplemented RPMI 1640. Proliferating cell cultures were maintained in a 5% CO2 humidified incubator at 37 °C. Cells were checked for Mycoplasma contamination before carrying out the experiments by Mycoplasma PCR detection kit (Applied Biological Materials). Stable transfection of 1.5 μg of WT p53, P152Lp53, or vector control plasmids were performed using 3 μl of Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. 48 h after transfection cells were kept in RPMI supplemented with 10% (v/v) FCS and antibiotic selection pressure for about 2 weeks after which cells were examined for protein expression by Western blotting and immunofluorescence.
Colony formation assay
Harvested cells were counted using a hemocytometer and diluted such that 500 cells were seeded into 60-mm dishes that were incubated for 2 weeks for colony formation. Colonies were then fixed with 5 ml of ice-cold 70% methanol for 30 min and stained with 5 ml of 0.01% (w/v) crystal violet in distilled water for 1 h. Excess staining was removed and dishes were allowed to dry and colonies were counted.
In vitro wound-healing assay
Cells were seeded in 30-mm dishes to create a confluent monolayer. After the confluence was achieved, cell monolayer was scrapped in a straight line to create a “scratch” of equal width in both assessed and control cells, with a p20 micropipette tip. Cell debris was removed and the dish was refreshed with RPMI medium with 10% FBS containing mitomycin (0.02 mg/ml). Images were acquired using a phase-contrast microscope.
Non-orthotopic xenograft mouse model study
Animal experiments were carried out with the approval from the Institutional Animal Ethics Committees (IAEC) of IISc and JNCASR. The nude mice (NU/NU mice) used for the animal experiments were purchased from Jackson Laboratories (USA). The mice were housed in an accredited facility under a 12-h light/day cycle and provided food and water ad libitum. H1299 cells (5 million cells in serum-free media) were subcutaneously injected into 6-week-old female Balb/c nude mice in the right flank region. Growth in tumor was observed for about 1 month followed by mice sacrifice by cervical dislocation, tumor extraction (38th day), and measurement of tumor weight and size by vernier caliper.
RNA isolation and RT-qPCR
Total RNA was isolated from the mice tumor using the TRIzol reagent (Invitrogen) as per the manufacturer's instructions. Further purification and cDNA preparation was carried out as described in Ref. 44. qPCR was performed using SYBR Green (Sigma) and mRNA-specific primers (Table S4).
RNA sequencing and analysis
The library preparation and sequencing was carried out at Macrogen Inc. (Seoul, South Korea), where the mRNA libraries using Illumina TruSeq library preparation kit were generated from 1 μg of total RNA and then sequenced using Illumina HiSeq2500 to generate the RNA-seq fastaq paired end data. About 40 million paired end reads of each, two control (pCMV10 Ctrl) and two mutant (P152Lp53) samples were generated. FASTQ files of these 4 samples were first quality checked using FastQC-0.11.6. The quality of the sequencing data are above 30 (Phred quality Score) with the read length 101 for all the control and mutant samples (Table S2). All four samples were then aligned using STAR Aligner-2.5 (45) with human reference genome version GRCH38p10. The alignment statistics is as shown in Table S3. The read alignment counts were generated using QoRTs-v1.3.0 (46) and then Deseq2 (version 1.14.1) (47) was used to obtain differentially expressed genes. The data were submitted to GEO (accession no. GSE119654). Pathways enrichment and network analysis was performed with in-house developed perl scripts and pathway databases with pathways data taken from KEGG, REACTOME, WIKI, PANTHER, NCI-NATURE, BIOCARTA, HUMANcyc. The differentially expressed genes were further classified into Onco, tumor suppressor, or with dual role (Onco and TSG). The selected genes and pathways for each category (Onco, TSG, DUAL) are shown in Fig. 4a (analyses data in Files S6–S9). Three strategies were used to decipher and annotate the functional mechanisms of P152Lp53. First the GSEA and Reactome FI based enrichment (as described in detail in Ref. 48) with the in-house pathway database. The hierarchical clustering of these differentially expressed genes, pathways, and reported P53-regulated genes and Onco/TSG reported genes were used to decipher the major pathways and genes being affected in the present study (Fig. 4b and Files S6–S9). The network analysis and representation was done using Cytoscape (49).
Statistical analysis
The statistical analysis (for cell line based GOF experiments) was done by applying unpaired, two-tailed, equal variance Student's t test. p value less than 0.05 was considered statistically significant. All the tests were carried out by Prism 5 (GraphPad software).
Author contributions
S. Singh, M.K., and T.K.K. conceptualization; S. Singh, M.K., S. Sen, P.U., S.B., and V.S.T. data curation; S. Singh, M.K., S.K., S.R., A.D., and T.K.K. investigation; S. Singh, M.K., S.K., S. Sen, P.U., S.B., N.M., V.S.T., S.R., A.D., and T.K.K. methodology; S. Singh, M.K., S.K., S. Sen, P.U., A.D., and T.K.K. writing-original draft; S. Singh, S.K., S. Sen, S.B., S.R., and T.K.K. writing-review and editing; S.K., and N.M., formal analysis; S. Singh, S.K., S. Sen validation; V.S.T., S.R., A.D., and T.K.K. resources; S.R., A.D., and T.K.K. supervision; T.K.K., funding acquisition; T.K.K. project administration.
Supplementary Material
Acknowledgments
We are grateful to Prof. Kumaravel Somasundaram (Chairman), Central Animal Facility, Indian Institute of Science, for help in the mice experiment and very fruitful discussions during the progress of this work. We thank B. S. Suma, Confocal Facility, JNCASR, for technical help. We thank the anonymous reviewers for critical and insightful comments and Transcription and Disease Lab members for their valuable suggestions.
This work was supported by Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR), Department of Biotechnology (DBT), Government of India, Virtual National Oral Cancer Institute (VNOCI) Grant (BT/PRI7576/MED/30/1690/2016) and Life Science Education and Training at JNCASR Grant (DBT/INF/22/SP27679/2018), Department of Science and Technology (DST), Government of India, Sir JC Bose National Fellowship Grant SR/S2/JCB-28/2010 (to T. K. K.), and an Integrated Ph.D. program scholarship of Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR) (to S. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3, Files S1–S9, and Tables S1–S6.
The RNA sequencing data reported in this paper have been submitted to the Gene Expression Omnibus (GEO) database under GEO accession no. GSE119654.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- DBD
- DNA-binding domain
- OSCC
- oral squamous cell carcinoma
- EMSA
- electrophoretic mobility shift assay
- GuHCl
- guanidine hydrochloride
- TSG
- tumor suppressor gene
- DEG
- differentially expressed genes
- MMP1
- matrix metalloproteinase 1
- P4H
- prolyl 4-hydroxylase
- qPCR
- quantitative PCR
- β-gal
- β-galactosidase
- EGFR
- epidermal growth factor receptor
- GOF
- gain of function.
References
- 1. Beckerman R., and Prives C. (2010) Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2, a000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friedman P. N., Chen X., Bargonetti J., and Prives C. (1993) The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 3319–3323 10.1073/pnas.90.8.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McLure K. G., and Lee P. W. (1998) How p53 binds DNA as a tetramer. EMBO J. 17, 3342–3350 10.1093/emboj/17.12.3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weinberg R. L., Veprintsev D. B., and Fersht A. R. (2004) Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 341, 1145–1159 10.1016/j.jmb.2004.06.071 [DOI] [PubMed] [Google Scholar]
- 5. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., and Vogelstein B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- 6. Agarwal M. L., Agarwal A., Taylor W. R., and Stark G. R. (1995) p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 92, 8493–8497 10.1073/pnas.92.18.8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuler M., Bossy-Wetzel E., Goldstein J. C., Fitzgerald P., and Green D. R. (2000) p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 275, 7337–7342 10.1074/jbc.275.10.7337 [DOI] [PubMed] [Google Scholar]
- 8. Vousden K. H., and Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- 9. Olivier M., Hollstein M., and Hainaut P. (2010) TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2, a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soussi T., Ishioka C., Claustres M., and Béroud C. (2006) Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat. Rev. Cancer 6, 83–90 10.1038/nrc1783 [DOI] [PubMed] [Google Scholar]
- 11. Hollstein M., Sidransky D., Vogelstein B., and Harris C. C. (1991) p53 mutations in human cancers. Science 253, 49–53 10.1126/science.1905840 [DOI] [PubMed] [Google Scholar]
- 12. Caron de Fromentel C., and Soussi T. (1992) TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer 4, 1–15 10.1002/gcc.2870040102 [DOI] [PubMed] [Google Scholar]
- 13. Pavletich N. P., Chambers K. A., and Pabo C. O. (1993) The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 7, 2556–2564 10.1101/gad.7.12b.2556 [DOI] [PubMed] [Google Scholar]
- 14. Hainaut P., and Hollstein M. (2000) p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 77, 81–137 [DOI] [PubMed] [Google Scholar]
- 15. Freed-Pastor W. A., and Prives C. (2012) Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong K. B., DeDecker B. S., Freund S. M., Proctor M. R., Bycroft M., and Fersht A. R. (1999) Hot-spot mutants of p53 core domain evince characteristic local structural changes. Proc. Natl. Acad. Sci. U.S.A. 96, 8438–8442 10.1073/pnas.96.15.8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bullock A. N., Henckel J., and Fersht A. R. (2000) Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene 19, 1245–1256 10.1038/sj.onc.1203434 [DOI] [PubMed] [Google Scholar]
- 18. Joerger A. C., and Fersht A. R. (2008) Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 10.1146/annurev.biochem.77.060806.091238 [DOI] [PubMed] [Google Scholar]
- 19. Willis A., Jung E. J., Wakefield T., and Chen X. (2004) Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 23, 2330–2338 10.1038/sj.onc.1207396 [DOI] [PubMed] [Google Scholar]
- 20. van Oijen M. G., and Slootweg P. J. (2000) Gain-of-function mutations in the tumor suppressor gene p53. Clin. Cancer Res. 6, 2138–2145 [PubMed] [Google Scholar]
- 21. Bossi G., Lapi E., Strano S., Rinaldo C., Blandino G., and Sacchi A. (2006) Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 25, 304–309 10.1038/sj.onc.1209026 [DOI] [PubMed] [Google Scholar]
- 22. Oren M., and Rotter V. (2010) Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2, a001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strano S., Dell'Orso S., Di Agostino S., Fontemaggi G., Sacchi A., and Blandino G. (2007) Mutant p53: an oncogenic transcription factor. Oncogene 26, 2212–2219 10.1038/sj.onc.1210296 [DOI] [PubMed] [Google Scholar]
- 24. Poeta M. L., Manola J., Goldwasser M. A., Forastiere A., Benoit N., Califano J. A., Ridge J. A., Goodwin J., Kenady D., Saunders J., Westra W., Sidransky D., and Koch W. M. (2007) TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 357, 2552–2561 10.1056/NEJMoa073770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lazarus P., Stern J., Zwiebel N., Fair A., Richie J. P. Jr., and Schantz S. (1996) Relationship between p53 mutation incidence in oral cavity squamous cell carcinomas and patient tobacco use. Carcinogenesis 17, 733–739 10.1093/carcin/17.4.733 [DOI] [PubMed] [Google Scholar]
- 26. Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S. V., Hainaut P., and Olivier M. (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629 10.1002/humu.20495 [DOI] [PubMed] [Google Scholar]
- 27. Brachmann R. K. (2004) p53 mutants: the Achilles' heel of human cancers? Cell Cycle 3, 1030–1034 [PubMed] [Google Scholar]
- 28. Hsieh L. L., Wang P. F., Chen I. H., Liao C. T., Wang H. M., Chen M. C., Chang J. T., and Cheng A. J. (2001) Characteristics of mutations in the p53 gene in oral squamous cell carcinoma associated with betel quid chewing and cigarette smoking in Taiwanese. Carcinogenesis 22, 1497–1503 10.1093/carcin/22.9.1497 [DOI] [PubMed] [Google Scholar]
- 29. Casson A. G., Mukhopadhyay T., Cleary K. R., Ro J. Y., Levin B., and Roth J. A. (1991) p53 gene mutations in Barrett's epithelium and esophageal cancer. Cancer Res. 51, 4495–4499 [PubMed] [Google Scholar]
- 30. Wagner J., Portwine C., Rabin K., Leclerc J. M., Narod S. A., and Malkin D. (1994) High frequency of germline p53 mutations in childhood adrenocortical cancer. J. Natl. Cancer Inst. 86, 1707–1710 10.1093/jnci/86.22.1707 [DOI] [PubMed] [Google Scholar]
- 31. Xie T. X., Zhou G., Zhao M., Sano D., Jasser S. A., Brennan R. G., and Myers J. N. (2013) Serine substitution of proline at codon 151 of TP53 confers gain of function activity leading to anoikis resistance and tumor progression of head and neck cancer cells. Laryngoscope 123, 1416–1423 10.1002/lary.23846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wasserman J. D., Novokmet A., Eichler-Jonsson C., Ribeiro R. C., Rodriguez-Galindo C., Zambetti G. P., and Malkin D. (2015) Prevalence and functional consequence of TP53 mutations in pediatric adrenocortical carcinoma: a children's oncology group study. J. Clin. Oncol. 33, 602–609 10.1200/JCO.2013.52.6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stenger J. E., Mayr G. A., Mann K., and Tegtmeyer P. (1992) Formation of stable p53 homotetramers and multiples of tetramers. Mol. Carcinog. 5, 102–106 10.1002/mc.2940050204 [DOI] [PubMed] [Google Scholar]
- 34. Mandal A. K., Samaddar S., Banerjee R., Lahiri S., Bhattacharyya A., and Roy S. (2003) Glutamate counteracts the denaturing effect of urea through its effect on the denatured state. J. Biol. Chem. 278, 36077–36084 10.1074/jbc.M211207200 [DOI] [PubMed] [Google Scholar]
- 35. Shen L., Yang M., Lin Q., Zhang Z., Zhu B., and Miao C. (2016) COL11A1 is overexpressed in recurrent non-small cell lung cancer and promotes cell proliferation, migration, invasion and drug resistance. Oncol. Rep. 36, 877–885 10.3892/or.2016.4869 [DOI] [PubMed] [Google Scholar]
- 36. Pardo A., and Selman M. (2005) MMP-1: the elder of the family. Int. J. Biochem. Cell Biol. 37, 283–288 10.1016/j.biocel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 37. Gilkes D. M., Chaturvedi P., Bajpai S., Wong C. C., Wei H., Pitcairn S., Hubbi M. E., Wirtz D., and Semenza G. L. (2013) Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 73, 3285–3296 10.1158/0008-5472.CAN-12-3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou F., Drabsch Y., Dekker T. J., de Vinuesa A. G., Li Y., Hawinkels L. J., Sheppard K. A., Goumans M. J., Luwor R. B., de Vries C. J., Mesker W. E., Tollenaar R. A., Devilee P., Lu C. X., Zhu H., Zhang L., and Dijke P. T. (2014) Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat. Commun. 5, 3388 10.1038/ncomms4388 [DOI] [PubMed] [Google Scholar]
- 39. Pollak M. (2008) Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 8, 915–928 10.1038/nrc2536 [DOI] [PubMed] [Google Scholar]
- 40. Minard M. E., Kim L. S., Price J. E., and Gallick G. E. (2004) The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res. Treat 84, 21–32 10.1023/B:BREA.0000018421.31632.e6 [DOI] [PubMed] [Google Scholar]
- 41. Yoshikawa K., Hamada J., Tada M., Kameyama T., Nakagawa K., Suzuki Y., Ikawa M., Hassan N. M., Kitagawa Y., and Moriuchi T. (2010) Mutant p53 R248Q but not R248W enhances in vitro invasiveness of human lung cancer NCI-H1299 cells. Biomed. Res. 31, 401–411 10.2220/biomedres.31.401 [DOI] [PubMed] [Google Scholar]
- 42. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 43. Larsson A. (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaypee S., Sahadevan S. A., Patil S., Ghosh P., Roy N. S., Roy S., and Kundu T. K. (2018) Mutant and wild-type tumor suppressor p53 induces p300 autoacetylation. iScience 4, 260–272 10.1016/j.isci.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hartley S. W., and Mullikin J. C. (2015) QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics 16, 224 10.1186/s12859-015-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kumar S., Gomez E. C., Chalabi-Dchar M., Rong C., Das S., Ugrinova I., Gaume X., Monier K., Mongelard F., and Bouvet P. (2017) Integrated analysis of mRNA and miRNA expression in HeLa cells expressing low levels of Nucleolin. Sci. Rep. 7, 9017 10.1038/s41598-017-09353-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Merabet A., Houlleberghs H., Maclagan K., Akanho E., Bui T. T., Pagano B., Drake A. F., Fraternali F., and Nikolova P. V. (2010) Mutants of the tumour suppressor p53 L1 loop as second-site suppressors for restoring DNA binding to oncogenic p53 mutations: structural and biochemical insights. Biochem. J. 427, 225–236 10.1042/BJ20091888 [DOI] [PubMed] [Google Scholar]
- 51. Zambetti G. P., and Levine A. J. (1993) A comparison of the biological activities of wild-type and mutant p53. FASEB J. 7, 855–865 10.1096/fasebj.7.10.8344485 [DOI] [PubMed] [Google Scholar]
- 52. Abarzúa P., LoSardo J. E., Gubler M. L., Spathis R., Lu Y. A., Felix A., and Neri A. (1996) Restoration of the transcription activation function to mutant p53 in human cancer cells. Oncogene 13, 2477–2482 [PubMed] [Google Scholar]
- 53. Ahn J., Poyurovsky M. V., Baptiste N., Beckerman R., Cain C., Mattia M., McKinney K., Zhou J., Zupnick A., Gottifredi V., and Prives C. (2009) Dissection of the sequence-specific DNA binding and exonuclease activities reveals a superactive yet apoptotically impaired mutant p53 protein. Cell Cycle 8, 1603–1615 10.4161/cc.8.10.8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Friedlander P., Legros Y., Soussi T., and Prives C. (1996) Regulation of mutant p53 temperature-sensitive DNA binding. J. Biol. Chem. 271, 25468–25478 10.1074/jbc.271.41.25468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.