Abstract
The constitutively nuclear histone deacetylases (HDACs) 1, 2, and 3 erase acetyl marks on acetyllysine residues, alter the landscape of histone modifications, and modulate chromatin structure and dynamics and thereby crucially regulate gene transcription in higher eukaryotes. Nuclear HDACs exist as at least six giant multiprotein complexes whose nonenzymatic subunits confer genome targeting specificity for these enzymes. The deacetylase activity of HDACs has been shown previously to be enhanced by inositol phosphates, which also bridge the catalytic domain in protein–protein interactions with SANT (Swi3, Ada2, N-Cor, and TFIIIB) domains in all HDAC complexes except those that contain the Sin3 transcriptional corepressors. Here, using purified recombinant proteins, coimmunoprecipitation and HDAC assays, and pulldown and NMR experiments, we show that HDAC1/2 deacetylase activity in one of the most ancient and evolutionarily conserved Sin3L/Rpd3L complexes is inducibly up-regulated by inositol phosphates but involves interactions with a zinc finger motif in the Sin3-associated protein 30 (SAP30) subunit that is structurally unrelated to SANT domains, indicating convergent evolution at the functional level. This implies that this mode of regulation has evolved independently multiple times and provides an evolutionary advantage. We also found that constitutive association with another core subunit, Rb-binding protein 4 chromatin-binding factor (RBBP4), further enhances deacetylase activity, implying both inducible and constitutive regulatory mechanisms within the same HDAC complex. Our results indicate that inositol phosphates stimulate HDAC activity and that the SAP30 zinc finger motif performs roles similar to that of the unrelated SANT domain in promoting the SAP30–HDAC1 interaction and enhancing HDAC activity.
Keywords: transcription regulation, chromatin modification, histone deacetylase (HDAC), protein–protein interaction, allosteric regulation, inositol phosphate, signaling, gene expression, epigenetics, cofactor-mediated protein–protein interaction, convergent evolution
Introduction
Lysine acetylation is an abundant posttranslational histone modification found in euchromatin that is correlated with increased chromatin dynamics and access to the underlying DNA template to regulatory factors, effectors, and molecular machines (1). Histone deacetylation reverses these effects and the chemical transformation is mediated in large part by the constitutively nuclear class I Zn2+-dependent HDACs3 1, 2, and 3 (2, 3). In mammals, these enzymes function in the context of at least six giant multiprotein complexes, including the Sin3L/Rpd3L, Sin3S/Rpd3S, NuRD, CoREST, MiDAC, and SMRT–NCoR complexes, which, in turn, are recruited via protein–protein interactions by sequence-specific DNA-binding transcription factors and/or posttranslational modifications in the tail regions of histones (4–7). Although some progress has been made regarding how these complexes are assembled, including the recruitment and integration of HDACs in these complexes, the precise molecular role(s) of individual subunits has been, with few exceptions, enigmatic.
The mammalian 1.2- to 2-MDa Sin3L/Rpd3L HDAC complex regulates the expression of a large variety of genes with roles in cell cycle control, differentiation, metabolism, and stem cell maintenance (4, 7–9). Five subunits of the complex, including Sin3A/B, SAP30/SAP30L, Sds3/BRMS1/BRMS1L, HDAC1/2, and RBBP4/7 (paralogous subunits are distinguished by a slash) are found in a broad range of eukaryotes from yeast to human and are thought to comprise the core complex (10–14). Among these five subunits, HDAC1/2 and RBBP4/7 are shared with multiple chromatin-modifying complexes, Sin3A and Sin3B assort into the Sin3L/Rpd3L and Sin3S–Rpd3S complexes, respectively, and the SAP30 and Sds3 subunits (and their paralogs) are unique to the Sin3L/Rpd3L complex.
We and others have shown previously, through biochemical and/or structural studies, that the Sin3A subunit, besides being directly recruited by transcription factors via interactions with the PAH1 and PAH2 domains, performs a scaffolding function by engaging in direct interactions with HDAC1, Sds3, and SAP30 (4, 7). Although the interactions with HDAC1 and Sds3 are mediated by the HDAC interaction domain (HID) (15), the interaction with SAP30 occurs via the PAH3 domain, located immediately N-terminal to the HID (16). In these studies, we also showed that Sds3 provides dimerization and putative nucleic acid binding functions for the complex. However, the role of SAP30 has been less obvious because, besides a Sin3 interaction domain (SID), the polypeptide contains only one other domain that harbors a unique zinc finger (ZnF) motif that, as we and others proposed, could bind to negatively charged molecules through a highly conserved, positively charged surface (17, 18). Indeed, elucidation of the precise molecular roles of the Sin3, Sds3, and SAP30 polypeptides has been hampered in large part because of the lack of broadly distributed and well-characterized protein–protein interaction domains within these proteins.
Recent studies have unexpectedly suggested that the class I HDACs 1, 2, and 3 are regulated by inositol phosphates derived from membrane lipids (19–21). Besides promoting interactions between HDAC1/3 and SANT domain–bearing proteins via a conserved surface located near the enzyme active site, certain inositol phosphates have been shown in these studies to significantly enhance the activity of the enzyme. However, unlike in the NuRD and SMRT HDAC complexes, where these inositol phosphate–dependent interactions have been described, none of the subunits in the Sin3L/Rpd3L complex harbor SANT domains. Here we show that the SAP30 ZnF motif, which is structurally unrelated to SANT domains, performs similar roles in promoting the interaction between SAP30 and HDAC1 while concomitantly enhancing enzyme activity. We further show that the histone H3/H4-interacting subunit RBBP4 also enhances the basal deacetylase activity of HDAC1 through constitutive interactions with the catalytic domain.
Results
SAP30 engages with HDAC1 in the Sin3L/Rpd3L complex
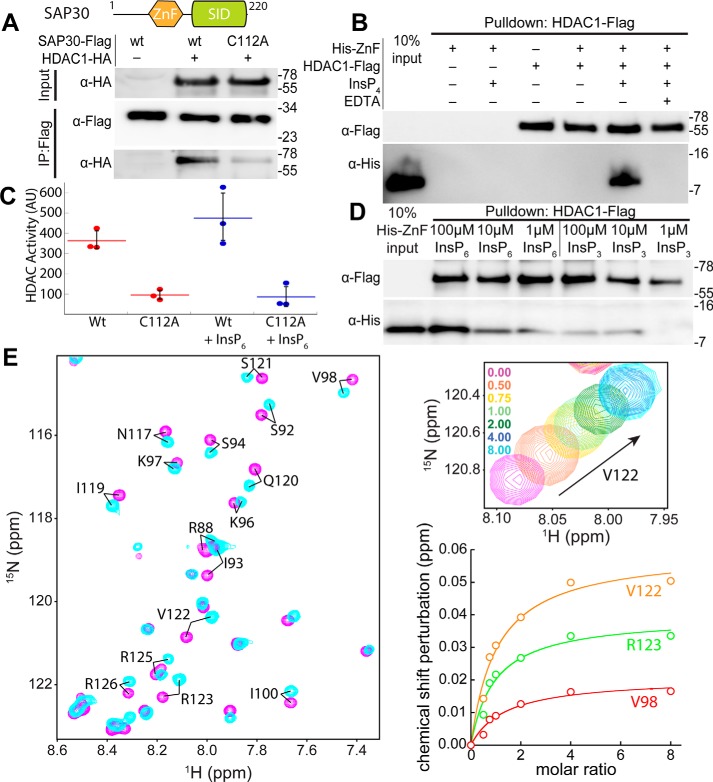
Because the SAP30 ZnF motif has been shown previously to bind to molecules enriched in negatively charged moieties, we surmised that the SAP30 subunit might have a role akin to the SANT domains of MTA1 and SMRT corepressors in the NuRD and SMRT complexes (19–21). Specifically, we asked whether SAP30, and the ZnF motif in particular, could directly interact with HDAC1. In coimmunoprecipitation (coIP) experiments with HA-tagged HDAC1 and FLAG-tagged SAP30, the WT proteins exhibited a robust interaction (Fig. 1A). However, mutation of C112, a key Zn2+-coordinating residue, to alanine in SAP30, which, as we showed previously, precluded proper folding of the ZnF (17), failed to efficiently immunoprecipitate HDAC1, implying that a properly folded ZnF motif is necessary for a robust interaction with HDAC1. We then asked whether full-length HDAC1 or a construct lacking a large section of the unstructured C-terminal tail but retaining the catalytic domain, was sufficient for the interaction. Both constructs were efficiently immunoprecipitated by SAP30 (Fig. S1A), implying that the catalytic domain of HDAC1 serves as the primary site of interaction.
Figure 1.
SAP30 physically associates with the catalytic domain of HDAC1 via the ZnF domain. A, domain map of SAP30 and coIP of HA-tagged HDAC1 by FLAG-tagged, WT, and the C112A mutant of SAP30. B, pulldown assays conducted with immobilized FLAG-tagged HDAC1 and His6-tagged SAP30 ZnF in the presence or absence of InsP4 and EDTA. C, endpoint activity assays with WT and C112A SAP30 immunoprecipitates with and without addition of 100 μm InsP6. Error bars represent standard deviations (n = 3). D, pulldown assays conducted as in C except in the presence or absence of varying amounts of InsP3 or InsP6. E, 1H-15N–correlated spectrum of a 250 μm sample of [15N]SAP30 ZnF in the absence (magenta) and presence (cyan) of eight equivalents of InsP6 (left panel). The expanded plot shows changes in peak position for Val-122 in the same titration (top right) and binding isotherms deduced from changes in chemical shifts for selected residues (bottom right). In A, B, and D, molecular mass (in kilodaltons) is shown on the right of each blot. AU, arbitrary units.
To test whether the SAP30 ZnF motif was sufficient to engage HDAC1 in direct interactions, we expressed and purified the zinc finger and conducted pulldown assays with immobilized FLAG-tagged HDAC1. Although no binding was detected for the His6-tagged SAP30 ZnF when presented alone (Fig. 1B), inclusion of inositol 1,4,5,6-tetrakisphosphate (InsP4) resulted in a robust interaction between the two proteins. However, the interaction was abrogated when HDAC1 was presented with SAP30 ZnF pretreated with EDTA, even though the binding reaction was supplemented with InsP4. This again confirmed that the interaction was specific and that a properly folded zinc finger was critical for interactions with HDAC1. Previous studies involving HDAC1/3 and SANT domains indicated that a minimum of four phosphate groups at strategic positions in the inositol ring are necessary for efficient interactions (21). We tested whether inositol 1,4,5-triphosphate (InsP3) and inositol 1,2,3,4,5,6-hexakisphosphate (InsP6) could similarly promote interactions between the SAP30 ZnF and HDAC1 over a range of inositol phosphate concentrations. Although InsP6 promoted a robust interaction at high micromolar concentrations, in contrast, InsP3 did so poorly, even at 100 μm concentration (Fig. 1D). This implies that the same trends noted previously involving inositol phosphate–mediated protein–HDAC interactions (21) also apply to the SAP30–HDAC1 interaction.
We then asked whether SAP30 immunoprecipitates harbored deacetylase activity in cells. FLAG-tagged WT SAP30 or the C112A mutant was coexpressed with HDAC1 and, following immunoprecipitation, eluted from the anti-FLAG resin (Fig. S1B). Deacetylase activity assays demonstrated ∼4-fold increased activity in the WT immunoprecipitates compared with the C112A mutant (Fig. 1C). However, addition of InsP6 did not lead to any significant increase in deacetylase activity, likely because of the presence of endogenous InsPs in the immunoprecipitates. Collectively, these results demonstrate that SAP30 can directly and independently recruit deacetylase activity via interactions with HDAC1 in a manner that depends on an intact ZnF in mammalian cells.
To gain insights into the SAP30 ZnF interaction with inositol phosphates at the molecular level, we performed titrations with 15N-labeled protein and InsP6. The addition of increasing amounts of InsP6 induced relatively small but readily discernible changes in the NMR spectrum, with only a subset of resonances showing significant perturbations (Fig. 1E). The resonances “shifted” as a function of added ligand, indicative of fast dissociation kinetics for the complex. Indeed, the modest stability of the complex was confirmed by an equilibrium dissociation constant of 250 ± 11 μm, obtained through nonlinear least squares fitting of the binding isotherms (Fig. 1E). Mapping the chemical shift changes on to the structure of the SAP30 ZnF motif reveals a contiguous surface with the most significant perturbations (Fig. S2). This surface overlaps with the conserved, basic surface formed by residues in the α1 and α2 helices that was anticipated previously to be functionally relevant (17). However, NMR titration experiments to probe whether the same surface is also involved in HDAC1 interactions was precluded by the limited solubility of the HDAC.
The SAP30 ZnF and HDAC1 interact via conserved surfaces
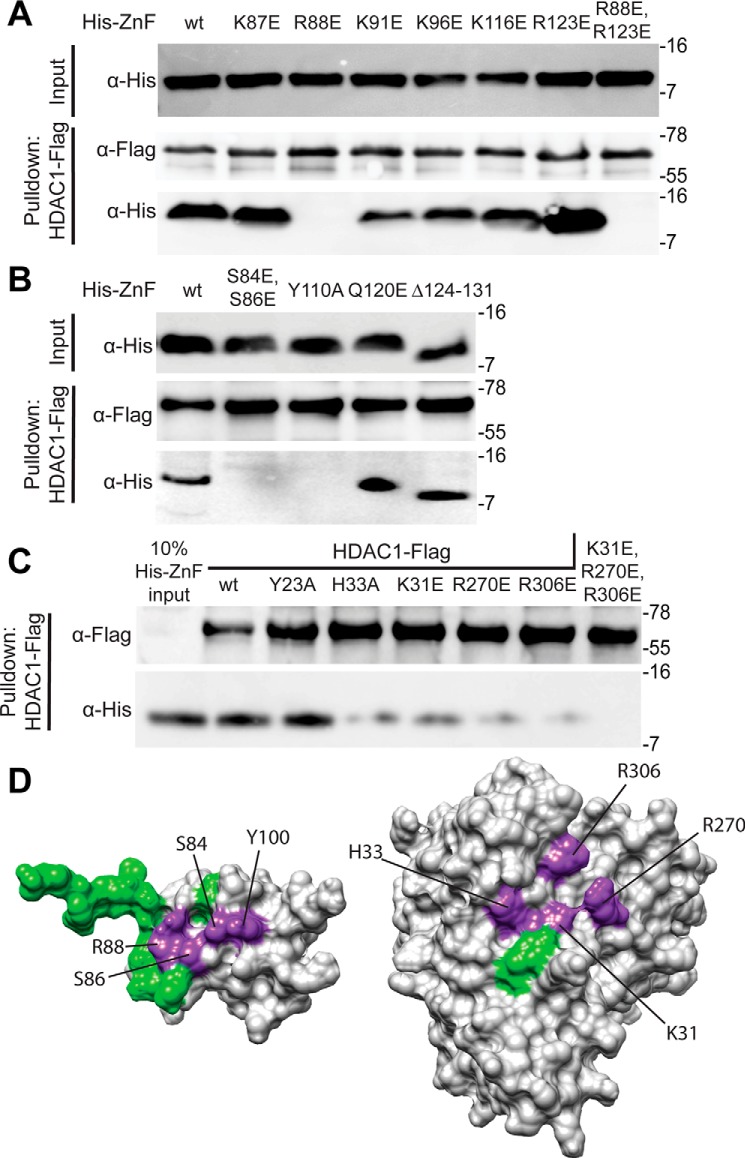
To determine the role of individual residues in promoting the formation of the SAP30 ZnF–HDAC1 complex, we mutated several basic residues located in the α1 and α2 helices that were implicated in inositol phosphate binding by our NMR studies and evaluated them by pulldown assays. To maximize disruption, the residues were mutated to glutamic acid either in isolation or in combination with another residue. Rather unexpectedly, most of the mutations that were screened produced only modest or no effects on binding to HDAC1 (Fig. 2A). The sole exception was R88E, which, either in isolation or in combination with R123E, completely abrogated the interaction, implying that the Arg-88 side chain is most likely involved in engaging with phosphate moieties in inositol phosphates.
Figure 2.
Mapping residues at the interfaces of the SAP30 ZnF–InsP6–HDAC1 complex via mutagenesis and interaction assays. A and B, pulldown assays conducted with immobilized FLAG-tagged HDAC1 and His6-tagged SAP30 ZnF single-site mutants of candidate basic residues involved in engaging with inositol phosphates (A) or other solvent-exposed conserved residues (B). The protein labeled Δ124–131 corresponds to SAP30 ZnF, which has a C-terminal deletion; the construct spans residues 64–123. C, pulldown assays with immobilized FLAG-tagged WT or mutant HDAC1 and His6-tagged SAP30 ZnF. All pulldown experiments were conducted in the presence of InsP6. D, mutations mapped onto the surface of SAP30 ZnF (right) and HDAC1 (left). Purple indicates mutations that lead to loss of binding in pulldown experiments, and green indicates that no change was observed. In A–C, molecular mass (in kilodaltons) is shown on the right of each blot. AU, arbitrary units.
We then asked whether other residues in the general vicinity of Arg-88 in the three-dimensional structure were also important for the interaction. In this regard, we mutated Gln-120 and two serines (Ser-84 and Ser-86) in the loop segment preceding helix α1 along with Tyr-110 located in another loop segment preceding α2 in these interactions. Although the Q120E mutant bound to HDAC1 just like the WT protein, the S84E,S86E double mutant as well as the Y110A mutant showed complete loss of binding to HDAC1 (Fig. 2B). Consistent with the other results, deletion of the SAP30 ZnF C-terminal unstructured tail segment (spanning residues 124–131), which is located distally from Arg-88 and harbors multiple basic residues that showed chemical shift perturbations in InsP6 titrations, had little or no effect on HDAC1-binding activity.
To define the complementary protein surface involved in formation of the ternary complex, we mutated basic residues in HDAC1 that have been shown previously to bind to inositol phosphates, including Lys-31, Arg-270, and Arg-306, to glutamic acid both individually and in combination (21). The individual mutations significantly reduced SAP30 ZnF binding activity, whereas the triple mutant completely abrogated binding (Fig. 2C). We also mutated residues near these residues, including Tyr-23 and His-33, which comprise the binding site in the MTA1–InsP6–HDAC1 complex. Although the Y23A mutant bound the SAP30 ZnF comparably as the WT protein, binding by the H33A mutant was significantly diminished, implying that residues beyond those involved in binding inositol phosphates are important for the stability of the ternary complex.
Collectively, our mutagenesis studies reveal that Arg-88, Ser-84, Ser-86, and Tyr-110 side chains in the SAP30 ZnF and His-33, Lys-31, Arg-270, and Arg-306 in HDAC1 are critical for the stability of the ternary complex. In both proteins, the residues define contiguous surfaces for interactions with inositol phosphates and HDAC1 (Fig. 2D). To gain more detailed insights into the mode of interaction involving the two proteins and InsP6, we performed multibody molecular docking of all three components using HADDOCK, with residues found to be critical for the interaction designated as “active” residues for docking (22). The resulting docked structures were clustered based on the fraction of common intermolecular contacts (>0.75). Twenty-eight structures from the largest cluster (of 100 structures used for the final refinement in explicit solvent) with no NOE violations of more than 0.3 Å and favorable energies were selected for analysis.
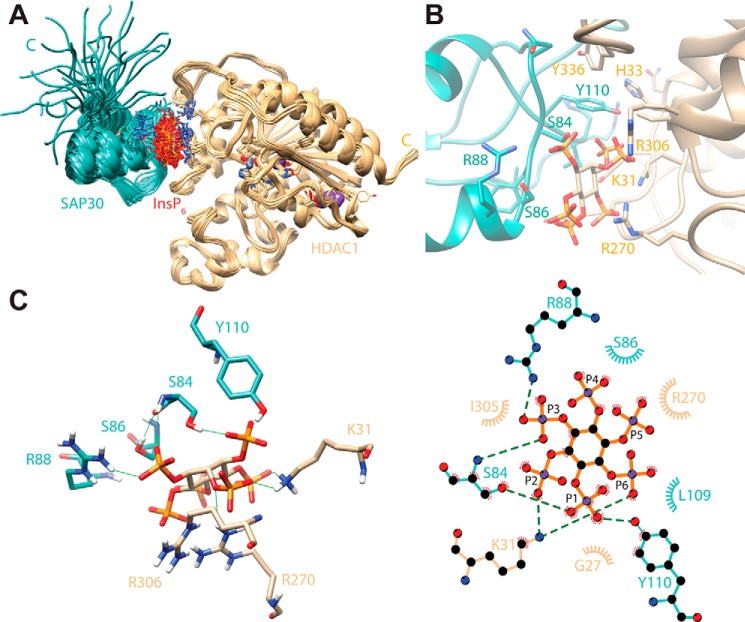
Although the backbone level precision of the ensemble for ordered regions is reasonable (0.71 Å from the average structure), the precision at the side-chain level for interfacial residues is significantly lower because the restraints were sparse, and side chains at the interface were allowed to move during the docking (Fig. 3A). Nevertheless, certain features of the complex could be readily and consistently detected. InsP6 adopts the chair-like conformation in previously reported structures and is sandwiched between the two proteins. One face of the “ring” of phosphates in InsP6 engages in electrostatic and/or hydrogen bonding interactions with Lys-33, Arg-270, and Arg-306 of HDAC1, as noted previously in the case of the NuRD complex (21) (Fig. 3, B and C). Arg-88 is the sole side chain in the SAP30 ZnF, engaging in salt bridging interactions with the other face of the phosphate ring. The hydroxyl groups of Ser-84, Ser-86, and Tyr-110 all engage different phosphates in hydrogen bonding interactions. The short side chain of the serine residues brings the polypeptide backbone in close proximity to the phosphates, allowing it to engage in hydrogen bonding interactions. The side chain of Tyr-110 inserts into a hydrophobic pocket formed by the side chains of His-33, Ile-305, and Tyr-336. Additional hydrophobic interactions are observed, albeit less consistently, whereas other types of favorable interactions involving interfacial residues are plausible even though they are less common in the ensemble. Notwithstanding these issues, the size of the protein–protein interface is small, amounting to a little under 300 Å2 on average, implying that the primary affinity determinants stem from interactions with the inositol phosphate moiety. Indeed, every residue in both proteins involved in these interactions is invariant over a broad range of species (Fig. S3), suggesting strong evolutionary pressure to preserve these interactions.
Figure 3.
A structural model of the SAP30 ZnF-InsP6–HDAC1 complex obtained via molecular docking. SAP30 ZnF backbone and side-chain carbon atoms are colored in cyan, whereas HDAC1 backbone and side-chain carbon and InsP6 carbon atoms are colored in tan; noncarbon atoms are shown in Corey–Pauling–Koltun colors. A, the ensemble of 28 docked structures deemed to be consistent with the functional studies described in Fig. 2. B, close-up view of the interface, showing the side chains of interacting residues in the representative structure of the ensemble. C, stabilizing intermolecular hydrogen bonding interactions (green lines) involving the side chains of essential residues in the two proteins and InsP6 in the representative model in 3D (left panel) and 2D (right panel). Dashed lines in the latter representation indicate hydrogen bonds, whereas semicircles with radiating spokes identify residues involved in intermolecular hydrophobic contacts. Carbon atoms in the 2D representation are colored in black, whereas the bond colors are in cyan for SAP30 ZnF, tan for HDAC1, and orange for InsP6.
Inositol phosphates, SAP30, and RBBP4 enhance HDAC1 deacetylase activity
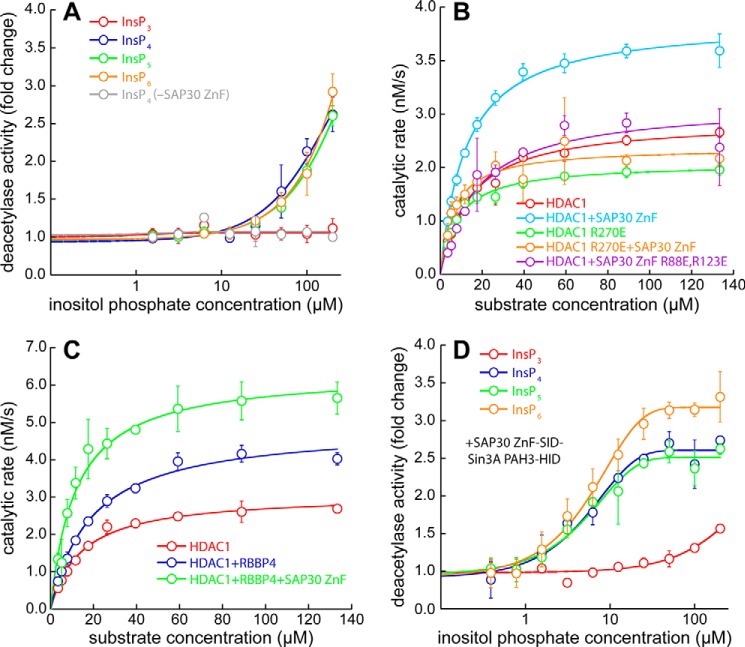
Because certain inositol phosphates enhance deacetylase activity in other HDAC-containing multiprotein complexes, we asked whether this was also the case for the Sin3L/Rpd3L complex. To answer this question, HDAC1 deacetylase activity assays using a model acetylated peptide substrate in endpoint format were conducted in the presence of excess SAP30 ZnF (23). Although InsP4, InsP5, and InsP6 enhanced deacetylase activity 2- to 3-fold, albeit at high inositol phosphate concentrations (>100 μm), InsP3 showed no effect (Fig. 4A). The deacetylase activity was unaffected by InsP4 in the absence of SAP30 ZnF. These results thus imply that a minimum of four phosphates in inositol phosphates was critical for the enhancement of HDAC1 deacetylase activity.
Figure 4.
Enhancement in the catalytic activity of HDAC1 by SAP30 and RBBP4. A, changes in the level of HDAC1 deacetylase activity measured in an endpoint assay format as a function of inositol phosphate concentration in the presence of 30 μm SAP30 ZnF. Changes in activity are reported relative to the level of activity measured in the absence of inositol phosphates. The data shown in gray and denoted InsP4 (−SAP30 ZnF) measured deacetylase activity in the presence of InsP4 but in the absence of SAP30 ZnF. B, deacetylase assays conducted using WT or mutant SAP30 ZnF and/or WT or mutant HDAC1 in the presence of 100 μm InsP6. C, changes in the kinetic parameters in deacetylase assays of HDAC1 conducted in the absence or presence of RBBP4, SAP30 ZnF, and 100 μm InsP6. D, changes in the level of deacetylase activity measured in an endpoint assay format as a function of inositol phosphate concentration in the presence of stoichiometric amounts of HDAC1 and SAP30–Sin3A fusion protein. Changes in activity are reported relative to the level of activity measured in the absence of inositol phosphates.
To characterize the effects of HDAC1 and SAP30 ZnF mutations on deacetylase activity, we measured the kinetic parameters of the enzyme using the same model substrate as above. These assays were conducted in the presence of 100 μm InsP6. Although the inclusion of SAP30 ZnF only had little effect on the Km, the kcat was enhanced almost 2-fold (Fig. 4B; Table 1). The HDAC1 R270E mutant exhibited a diminished kcat, although the catalytic efficiency (kcat/Km ratio) was unchanged from the WT enzyme. The inclusion of SAP30 ZnF led to only a slight enhancement in both kcat and kcat/Km for this HDAC1 mutant. Similarly, the SAP30 ZnF R88E,R123E mutant produced little or no increase in these parameters relative to that measured for the enzyme alone. Collectively, these results indicate that the enhancement of HDAC1 deacetylase activity emanates from stable formation of the SAP30 ZnF–InsP6–HDAC1 ternary complex.
Table 1.
Kinetic parameters for wild-type and mutant HDAC1 enzyme activity
Uncertainties correspond to 95% confidence intervals.
| Protein(s) | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1 s−1 | |
| HDAC1 | 13.98 ± 1.08 | 2.88 ± 0.07 | 2.06 × 105 |
| HDAC1+SAP30 ZnFa | 12.37 ± 0.68 | 4.75 ± 0.08 | 3.84 × 105 |
| HDAC1 R270E | 10.24 ± 0.70 | 2.12 ± 0.05 | 2.07 × 105 |
| HDAC1 R270E+SAP30 ZnFa | 9.65 ± 1.25 | 2.46 ± 0.11 | 2.55 × 105 |
| HDAC1+SAP30 ZnF R88E,R123Ea | 17.36 ± 3.93 | 3.19 ± 0.24 | 1.84 × 105 |
| HDAC1+RBBP4 | 18.92 ± 1.63 | 4.89 ± 0.14 | 2.58 × 105 |
| HDAC1+RBBP4+SAP30 ZnFa | 11.78 ± 1.62 | 6.35 ± 0.25 | 5.39 × 105 |
a Experiments with SAP30 ZnF proteins conducted in the presence of 30 μm ZnF and 100 μm InsP6.
Because RBBP4 is known to constitutively interact with HDAC1, we asked whether it might affect HDAC1 deacetylase activity. Compared with HDAC1 alone, the inclusion of RBBP4 enhanced the kcat almost 2-fold, although the Km also increased significantly to only modestly increase the catalytic efficiency (Fig. 4C; Table 1). Addition of SAP30 ZnF further enhanced the kcat while diminishing the Km to produce a robust surge in kcat/Km. However, the effects of RBBP4 and SAP30 ZnF on HDAC1 deacetylase activity appear to be additive rather than synergistic. In contrast to the enhancement in deacetylase activity produced by RBBP4 and SAP30 ZnF individually and in combination, inclusion of the Sin3A HID, which can also associate constitutively with HDAC1, produced no such effect (Fig. S4).
Because enhancements in deacetylase activity occurred at somewhat high concentrations of inositol phosphates, presumably reflecting a modest affinity interaction between SAP30 ZnF and HDAC1, we asked whether the enhancements might be more physiologically relevant in the context of the Sin3L–Rpd3L complex. Because the interaction between Sin3 and SAP30 is of nanomolar affinity, and the structure of this complex is known (16), we coexpressed and copurified a fusion protein of SAP30 and Sin3A spanning the ZnF domain and SID in the former and the HID of the latter with HDAC1. Because Sin3A HID and HDAC1 can associate constitutively, we conducted activity assays as a function of inositol phosphate concentration. Whereas InsP3 caused a modest increase in deacetylase activity at high concentrations (>100 μm), InsP4, InsP5, and InsP6 produced severalfold enhancements even at low micromolar concentrations (Fig. 4D; Table 2), which is well within the intracellular concentrations measured for these inositol phosphates (24), implying that these cofactors most likely have a role in regulating the deacetylase activity of HDAC1.
Table 2.
EC50 parameters for HDAC1 activation by inositol phosphates
Uncertainties correspond to standard deviations from three independent measurements.
| Inositol phosphate | EC50 |
|---|---|
| μm | |
| InsP3 | NDa |
| InsP4 | 5.18 ± 1.03 |
| InsP5 | 5.19 ± 0.88 |
| InsP6 | 6.07 ± 0.57 |
a ND, not determined.
Discussion
The Sin3L/Rpd3L complex is ancient and one of the most broadly distributed HDAC complexes in eukaryotes, with half a dozen subunits conserved from yeast to human (14, 25). In yeast, Rpd3, the ortholog of mammalian HDAC1/2, is found in two complexes, Rpd3L and Rpd3S (26). Although Rpd3 has been reported to be regulated by inositol phosphates and shares a conserved inositol phosphate–binding surface with its orthologs from a variety of species (27), the identity of the subunit in these HDAC complexes involved in inositol phosphate–based regulation has remained elusive until now. Our study establishes that this role is performed by the evolutionarily conserved SAP30 subunit in the mammalian Sin3L/Rpd3L complex. The involvement of the SAP30 zinc finger motif in these interactions is unexpected because it shares no overt sequence or structural similarity with SANT domains, which have been implicated previously in this role (19–21). Interestingly, the ZnF motif is narrowly distributed and found in only one other protein in higher eukaryotes, that of the SAP30 paralog SAP30L (28), which is also found in a subset of Sin3L/Rpd3L complexes, targeting the complex to the nucleolus (29). The human paralogs share 79% identity and 89% similarity in this region, with all residues deemed to be critical for engaging with inositol phosphates or HDAC1 by our studies invariant in the two proteins. Thus, SAP30L is expected to play similar roles as SAP30 in regulating the deacetylase activity of HDACs 1 and 2 in these complexes.
Because inositol phosphate–based activation of HDAC activity was first described for the human HDAC3-containing SMRT complex (20), this paradigm has been extended for the human HDAC1/2–containing NuRD (19, 21) and now to the mammalian Sin3L–Rpd3L complexes (Fig. 5). Despite differences in the identities of the proteins involved and the structural motifs used for engagement with inositol phosphates, each one of these complexes relies on a minimum of four phosphate groups in these cofactors for efficient engagement and activation. Interestingly, although InsP3 has minimal impact on the deacetylase activity in each of these complexes, InsP4, InsP5, and InsP6 potentiate the deacetylase activity to similar extents. The same three basic residues in the catalytic domain are involved in engaging with the cofactors in all three complexes, although the protein–protein interaction surface extends beyond these residues to varying extents. In all three cases, engagement with the cofactor and the catalytic domain results in tangible enhancement in deacetylase activity. Although a structural basis for the enhancement is not readily apparent from the crystal structures, molecular dynamics simulations have suggested that the effect could be due to allosteric modulation of protein motions within the loops comprising the active site (30).
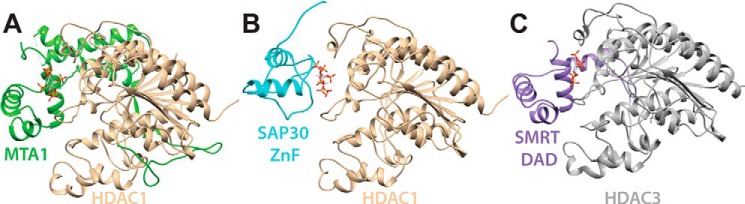
Figure 5.
Structural comparison of various HDAC complexes. A, crystal structure of the MTA1–InsP6–HDAC1 complex (PDB code 5ICN). B, structural model of the SAP30 ZnF–InsP6–HDAC1 complex from molecular docking. C, crystal structure of the SMRT DAD–InsP4–HDAC3 complex (PDB code 4A69). Note the parallels in InsP and HDAC engagement by the MTA1 and SMRT subunits featuring structurally similar SANT domains, whereas the mode of InsP and HDAC engagement by the structurally unrelated SAP30 ZnF domain is unique, implying convergent evolution at the functional level.
Interestingly, the level of enhancement of deacetylase activity differs for each complex but is most critically dependent on the identity of the HDAC. Although the deacetylase activity of HDAC3 in the SMRT complex is potentiated substantially by inositol phosphates (∼100-fold) (21), that of HDAC1 in the NuRD and Sin3L/Rpd3L complexes is comparably modest (2- to 3-fold) (19). Although the baseline catalytic rate and efficiency for HDAC1 is twice that of HDAC3, the cofactor-mediated activation for the latter is much more substantial (23) (note that the baseline activity of HDAC3 corresponds to when SMRT is present because the enzyme is otherwise inactive). Despite the seemingly modest inositol phosphate–mediated enhancement in deacetylase activity for HDAC1, mutations in the inositol phosphate–binding site causes a substantial decrease in cell viability (31), highlighting a critical role of these small-molecule cofactors and HDAC-associating subunits in multiprotein complexes.
In all three complexes characterized so far, HDACs are recruited and integrated into these complexes through constitutive associations (by SMRT, MTA1/2, and Sin3A/B in the SMRT, NuRD and Sin3L/Rpd3L complexes, respectively); therefore, even in the absence of inositol phosphates, all three complexes retain significant deacetylase activity. The role of inositol phosphates thus appears to be in turbocharging the activity of these enzymes. Because these HDAC complexes have well-documented roles in regulating the cell cycle (4–7), and because InsP4, InsP5, and InsP6 levels fluctuate, peaking in G1 (24), this would suggest that these cofactors have evolved to function as facile molecular signals to tune HDAC activity.
A less appreciated role of inositol phosphates is that they add another point of contact between the HDAC and the interacting subunit, contributing to the tight association and integration of the HDAC subunits in these complexes. Unlike the MTA1 subunit in the NuRD complex, which engages with HDAC1 both constitutively and inducibly via structurally distinct domains, association with HDAC1 in the Sin3L/Rpd3L complex is more distributed, with specific domains of separate polypeptides, Sin3A and SAP30, performing these roles. Interestingly, RBBP4, which is shared by both of these complexes and engages in constitutive interactions with HDAC1, also enhances deacetylase activity. Structural studies of this interaction are needed to understand the basis for this activation.
Its evolutionary conservation notwithstanding, SAP30 is a subunit unique to the Sin3L/Rpd3L complex, which raises questions regarding the identity of the corresponding subunit in the Sin3S–Rpd3S complex. Because the latter complex has a different role in transcription elongation in actively transcribed genes, it is possible that a need for turbocharging deacetylase activity does not exist for this complex. Alternatively, because SAP30 ZnF and the SANT domains have evolved independently to enhance HDAC activity, it is plausible that additional motifs might exist. Indeed, although SAP30 is evolutionarily conserved, the SAP30 ortholog in yeast lacks the highly conserved ZnF motif found in a broad range of organisms from fly to human (17). Because the inositol phosphate–binding site is conserved in yeast Rpd3, we surmise that another structural motif, either within SAP30 or in a different subunit of the Rpd3L complex, likely targets this site, suggesting that this mechanism of HDAC regulation has evolved independently multiple times and provides an evolutionary advantage to the organism.
Experimental procedures
Protein expression and purification
Escherichia coli BL21(DE3) was transformed with the pMCSG7 vector encoding SAP30 ZnF (aa 64–131) and grown at 37 °C until it reached an A600 nm of 0.6. Cultures were transferred to 16 °C and induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 16 h prior to harvesting. Cells were suspended in 50 mm Tris-HCl buffer (pH 8.0) containing 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine (TCEP) hydrochloride, 1 mm PMSF, 1 μm leupeptin, 1 mm pepstatin, and 0.1% Triton X-100 and lysed by sonication. After centrifugation, the lysate was loaded onto a HiTrap SP cation exchange column (GE Healthcare). The protein was eluted using a gradient from 0.05 to 1 m NaCl over 5 column volumes. Fractions were screened by SDS-PAGE, and those containing SAP30 ZnF were combined and incubated with tobacco etch virus (TEV) protease overnight to remove the His6 tag at 4 °C; this step was skipped in cases where His6-tagged protein was required. Further purification was achieved by reverse-phase HPLC using a linear gradient of 0.1% TFA and 0.1% TFA with 80% acetonitrile. Fractions were screened by MALDI and lyophilized. 15N-labeled samples were purified using the same method except that cells were grown in 15N ammonium sulfate containing M9 minimal medium. Point mutations in the SAP30 ZnF expression construct were introduced using the QuikChange mutagenesis protocol (Agilent), and the mutants were produced using the same procedure as for the WT protein. All mutations were confirmed by DNA sequencing.
Full-length human HDAC1 was cloned into a pcDNA3.1 vector with a C-terminal FLAG tag and TEV protease cleavage site. FLAG-tagged HDAC1 was expressed in nonadherent HEK293F cells following a protocol established previously (32). Cells were harvested after 48 h of expression, and the cell pellet was resuspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 50 mm potassium acetate, 1 mm EDTA, 1 mm PMSF, 1 μm leupeptin, 1 mm pepstatin, 0.2% Triton X-100, and 5% glycerol) and sonicated. After centrifugation, lysates were precleared by incubation with Sepharose 4B resin (Sigma-Aldrich). Clarified lysates were incubated with anti-FLAG M2 affinity resin (Sigma-Aldrich) for 1 h at 4 °C. The resin was then washed with lysis buffer, high-salt buffer (50 mm Tris-HCl (pH 7.5), 300 mm NaCl, 50 mm potassium acetate, 1 mm EDTA, and 5% glycerol), and rinse buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 50 mm potassium acetate, and 1 mm EDTA) and incubated overnight at 4 °C with TEV protease, and the flow-through containing untagged HDAC1 was collected. HDAC1 elutions were combined and dialyzed against 50 mm Tris (pH 7.5), 100 mm NaCl, 50 mm KCl, and 1 mm TCEP and then 50 mm Tris (pH 7.5), 100 mm NaCl, 50 mm KCl, and 1 mm TCEP with 10 μm ZnCl2. HDAC1 was coexpressed and copurified with RBBP4 using the same protocol as for HDAC1 alone.
HA-tagged HDAC1 was coexpressed with a C-terminally FLAG-tagged SAP30 (aa 64–131)–Sin3A (aa 532–766) fusion construct subcloned into the pcDNA3.1 vector. The complex was purified by suspending the cell pellet in lysis buffer, followed by sonication. Lysates were centrifuged and precleared with Sepharose 4B resin and incubated with anti-FLAG M2 affinity resin (Sigma-Aldrich) for 1 h at 4 °C. The resin was washed with lysis, high salt, and rinse buffers, and then it was incubated with 10 μm Sds3 SID (201–234) for 1 h at 4 °C. The resin was washed with rinse buffer, and the complex was eluted using 150 μg/ml FLAG peptide. The sample was immediately dialyzed in the same manner as HDAC1 alone.
Coimmunoprecipitation experiments
HEK293T cells were transfected using calcium phosphate with the indicated expression plasmids and collected 48 h post-transfection. Cells were lysed in 20 mm HEPES (pH 7.9), 150 mm KCl, 5% glycerol, 10 μm zinc acetate, 0.2% NP-40, 1 mm PMSF, 1 μm leupeptin, and 1 mm pepstatin. Cell extracts were incubated for 1 h at 4 °C with anti-FLAG M2 affinity resin (Sigma-Aldrich). The resin was washed five times with high-salt buffer (20 mm HEPES (pH 7.9), 300 mm KCl, 5% glycerol, 10 μm zinc acetate, and 0.2% NP-40), and samples were boiled with SDS-PAGE loading buffer. Proteins were then resolved using SDS-PAGE, transferred to nitrocellulose, and probed with primary antibodies, either anti-FLAG (Sigma-Aldrich, F3165, 1:500 dilution) or anti-HA (Sigma-Aldrich, H3663, 1:500 dilution). Thereafter, anti-mouse HRP-conjugated secondary antibody (Thermo Fisher Scientific, OB617005, 1:1000 dilution) was used. The blot was imaged using West Pico chemiluminescent substrate (Thermo Scientific, 34080) and a Syngene Pxi chemiluminescence imager. Mutations in the FLAG-tagged, full-length human SAP30 and HA-tagged human HDAC1 constructs were introduced using the QuikChange protocol (Agilent); all mutations were confirmed by DNA sequencing. WT and mutant SAP30–HDAC1 complexes were immunoprecipitated and eluted using excess FLAG peptide (ApexBio) for deacetylase activity assays.
Pulldown experiments
HDAC1 was expressed and purified as described above, except that it was left bound to the anti-FLAG M2 affinity resin. SAP30 ZnF WT and mutants were expressed as described previously, and pellets were resuspended in pulldown buffer (20 mm HEPES (pH 7.9), 100 mm NaCl, 50 mm potassium acetate, 5% glycerol, 0.1% NP-40, 1 mm PMSF, 1 μm leupeptin, and 1 mm pepstatin) and lysed by sonication. Cell lysates were incubated with HDAC1-bound anti-FLAG resin in the presence of 100 μm InsP4 or InsP6 for 30 min. The resin was washed five times with pulldown buffer and boiled in SDS-PAGE loading buffer. Samples were resolved by SDS-PAGE, transferred to nitrocellulose, and stained with either anti-FLAG or anti-His (Thermo Fisher, MA121315, 1:1000 dilution) antibody. Blots were visualized in the same manner as described above for the coIP experiments.
NMR experiments
NMR data were acquired on a 600-MHz Agilent DD2 NMR spectrometer at 25 °C. Dry, lyophilized [15N]SAP30 ZnF was dissolved in 50 mm Tris-d11 acetate-d4 buffer (pH 6.0) and 0.2% NaN3 with equimolar ZnCl2 added. The inositol phosphates InsP4 (Cayman Chemicals) and InsP6 (Sigma-Aldrich) used for the titrations were dissolved in the same buffer and used without additional purification. Data processing and analysis were performed using Felix (Felix NMR) and NMRFAM-Sparky (33).
HDAC1 deacetylase activity assays
A model peptide substrate containing an aminocoumarin (AMC) moiety was used for the deacetylase assays. The Ac-Gly-Ala-Lys(Ac)-AMC peptide was purchased from Bachem Americas and used in a slightly modified protocol as described previously (23). Reactions were performed in a 384-well plate at room temperature with 1 nm HDAC1 in 50 mm HEPES (pH 7.4), 100 mm KCl, 0.001% Tween 20, 5% DMSO, and 200 nm trypsin. Fluorescence data were acquired at room temperature for 1 h on a Biotek Synergy 4 microplate reader with excitation and emission wavelengths set to 360 and 440 nm, respectively. Data were fitted with R using nonlinear least squares fitting. Endpoint assays were conducted in a similar manner as above but using a fixed concentration of 60 μm AMC peptide. Steady-state fluorescence in these assays was measured after 1 h of equilibration.
Molecular modeling
The previously determined NMR structure of SAP30 ZnF (PDB code 2KDP) (17) was docked with the crystal structure of HDAC1 bound to InsP6 (PDB code 5ICN) (21) using HADDOCK version 2.2 (22). Active residues for docking were assigned based on the results of pulldown experiments with mutant proteins. These included Ser-84, Ser-86, Arg-88, and Tyr-110 in SAP30 ZnF and Lys-31, His-33, Arg-270, and Arg-306 in HDAC1 besides the InsP6 moiety. To sample a broader range of conformations, the first 10 conformers of SAP30 ZnF in the NMR ensemble were used in the calculations. Force field parameters for InsP6 were generated for CNS using the PRODRG server (34); these were supplemented with additional chirality restraints. One thousand structures were calculated during the rigid body docking phase; the 200 best structures were used for semiflexible docking, of which the 100 best structures were refined in explicit solvent. Default parameters were used, except the distance restraints were weighted more significantly (0.1, 0.5, and 1.0 for the three phases of the docking calculation). Distance restraints between specific InsP6 phosphate groups and Lys-31, Arg-270, and Arg-306 to target distances observed in the crystal structure were enforced throughout the calculation to ensure that the InsP6 moiety was anchored to HDAC1 (21). Analyses of the resulting structures were conducted using PLIP (35), LIGPLOT (36), and MONSTER (37).
Author contributions
R. D. M. and I. R. conceptualization; R. D. M. investigation; R. D. M. and I. R. methodology; R. D. M. writing-original draft; I. R. supervision; I. R. funding acquisition; I. R. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Gregory David (New York University) for critical reading of this manuscript and the Robert Lurie Comprehensive Cancer Center at Northwestern for supporting structural biology research.
This work was supported by American Heart Association Grants 14GRNT20170003 and 17GRNT33680167 (to I. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- HDAC
- histone deacetylase
- HID
- HDAC interaction domain
- SID
- Sin3 interaction domain
- ZnF
- zinc finger
- IP
- immunoprecipitation
- InsP4
- inositol 1,4,5,6-tetrakisphosphate
- InsP3
- inositol 1,4,5-triphosphate
- InsP6
- inositol 1,2,3,4,5,6-hexakisphosphate
- aa
- amino acids
- TCEP
- tris(2-carboxyethyl)phosphine
- TEV
- tobacco etch virus
- AMC
- aminocoumarin
- SANT
- Swi3, Ada2, N-Cor, and TFIIIB
- NuRD
- nucleosome remodeling and deacetylase
- CoREST
- corepressor with RE1 silencing and transcription factor
- MiDAC
- mitotic deacetylase complex
- SMRT
- silencing mediator of retinoic acid and thyroid hormone
- NCoR
- nuclear receptor corepressor
- CNS
- crystallography and NMR system
- DAD
- deacetylase activation domain.
References
- 1. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seto E., and Yoshida M. (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang X. J., and Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams G. E., Chandru A., and Cowley S. M. (2018) Co-repressor, co-activator and general transcription factor: the many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 475, 3921–3932 10.1042/BCJ20170314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayakawa T., and Nakayama J. (2011) Physiological roles of class I HDAC complex and histone demethylase. J. Biomed. Biotechnol. 2011, 129383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson P. J., Fairall L., and Schwabe J. W. (2012) Nuclear hormone receptor co-repressors: structure and function. Mol. Cell Endocrinol. 348, 440–449 10.1016/j.mce.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grzenda A., Lomberk G., Zhang J. S., and Urrutia R. (2009) Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450 10.1016/j.bbagrm.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silverstein R. A., and Ekwall K. (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 10.1007/s00294-004-0541-5 [DOI] [PubMed] [Google Scholar]
- 9. Chaubal A., and Pile L. A. (2018) Same agent, different messages: insight into transcriptional regulation by SIN3 isoforms. Epigenetics Chromatin 11, 17 10.1186/s13072-018-0188-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. David G., Turner G. M., Yao Y., Protopopov A., and DePinho R. A. (2003) mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 17, 2396–2405 10.1101/gad.1109403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleischer T. C., Yun U. J., and Ayer D. E. (2003) Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell Biol. 23, 3456–3467 10.1128/MCB.23.10.3456-3467.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McDonel P., Costello I., and Hendrich B. (2009) Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 41, 108–116 10.1016/j.biocel.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowman C. J., Ayer D. E., and Dynlacht B. D. (2014) Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat. Cell Biol. 16, 1202–1214 10.1038/ncb3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banks C. A. S., Thornton J. L., Eubanks C. G., Adams M. K., Miah S., Boanca G., Liu X., Katt M. L., Parmely T. J., Florens L., and Washburn M. P. (2018) A structured workflow for mapping human Sin3 histone deacetylase complex interactions using Halo-MudPIT affinity-purification mass spectrometry. Mol. Cell Proteomics 17, 1432–1447 10.1074/mcp.TIR118.000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark M. D., Marcum R., Graveline R., Chan C. W., Xie T., Chen Z., Ding Y., Zhang Y., Mondragón A., David G., and Radhakrishnan I. (2015) Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc. Natl. Acad. Sci. U.S.A. 112, E3669–E3678 10.1073/pnas.1504021112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie T., He Y., Korkeamaki H., Zhang Y., Imhoff R., Lohi O., and Radhakrishnan I. (2011) Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding. J. Biol. Chem. 286, 27814–27824 10.1074/jbc.M111.252494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y., Imhoff R., Sahu A., and Radhakrishnan I. (2009) Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition. Nucleic Acids Res. 37, 2142–2152 10.1093/nar/gkp051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Viiri K. M., Jänis J., Siggers T., Heinonen T. Y., Valjakka J., Bulyk M. L., Mäki M., and Lohi O. (2009) DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell Biol. 29, 342–356 10.1128/MCB.01213-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Millard C. J., Watson P. J., Celardo I., Gordiyenko Y., Cowley S. M., Robinson C. V., Fairall L., and Schwabe J. W. (2013) Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol. Cell 51, 57–67 10.1016/j.molcel.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watson P. J., Fairall L., Santos G. M., and Schwabe J. W. (2012) Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340 10.1038/nature10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watson P. J., Millard C. J., Riley A. M., Robertson N. S., Wright L. C., Godage H. Y., Cowley S. M., Jamieson A. G., Potter B. V., and Schwabe J. W. (2016) Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 7, 11262 10.1038/ncomms11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dominguez C., Boelens R., and Bonvin A. M. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- 23. Schultz B. E., Misialek S., Wu J., Tang J., Conn M. T., Tahilramani R., and Wong L. (2004) Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry 43, 11083–11091 10.1021/bi0494471 [DOI] [PubMed] [Google Scholar]
- 24. Barker C. J., Wright J., Hughes P. J., Kirk C. J., and Michell R. H. (2004) Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem. J. 380, 465–473 10.1042/bj20031872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., Yates J., Washburn M. P., and Workman J. L. (2005) Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731, 77–87; discussion 75–76 10.1016/j.bbaexp.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 26. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., and Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- 27. Worley J., Luo X., and Capaldi A. P. (2013) Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 3, 1476–1482 10.1016/j.celrep.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laitaoja M., Tossavainen H., Pihlajamaa T., Valjakka J., Viiri K., Lohi O., Permi P., and Jänis J. (2016) Redox-dependent disulfide bond formation in SAP30L corepressor protein: implications for structure and function. Protein Sci. 25, 572–586 10.1002/pro.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Viiri K. M., Korkeamäki H., Kukkonen M. K., Nieminen L. K., Lindfors K., Peterson P., Mäki M., Kainulainen H., and Lohi O. (2006) SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus. Nucleic Acids Res. 34, 3288–3298 10.1093/nar/gkl401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arrar M., Turnham R., Pierce L., de Oliveira C. A., and McCammon J. A. (2013) Structural insight into the separate roles of inositol tetraphosphate and deacetylase-activating domain in activation of histone deacetylase 3. Protein Sci. 22, 83–92 10.1002/pro.2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jamaladdin S., Kelly R. D., O'Regan L., Dovey O. M., Hodson G. E., Millard C. J., Portolano N., Fry A. M., Schwabe J. W., and Cowley S. M. (2014) Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 9840–9845 10.1073/pnas.1321330111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Portolano N., Watson P. J., Fairall L., Millard C. J., Milano C. P., Song Y., Cowley S. M., and Schwabe J. W. (2014) Recombinant protein expression for structural biology in HEK 293F suspension cells: a novel and accessible approach. J. Vis. Exp. e51897 10.3791/51897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schüttelkopf A. W., and van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- 35. Salentin S., Schreiber S., Haupt V. J., Adasme M. F., and Schroeder M. (2015) PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43, W443–W447 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- 37. Salerno W. J., Seaver S. M., Armstrong B. R., and Radhakrishnan I. (2004) MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 32, W566–W568 10.1093/nar/gkh434 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.