Abstract
Lipid phosphate phosphatase 3 (LPP3), encoded by the PLPP3 gene, is an integral membrane enzyme that dephosphorylates phosphate esters of glycero- and sphingophospholipids. Cell surface LPP3 can terminate the signaling actions of bioactive lysophosphatidic acid (LPA) and sphingosine 1 phosphate, which likely explains its role in developmental angiogenesis, vascular injury responses, and cell migration. Heritable variants in the final intron PLPP3 associate with interindividual variability in coronary artery disease risk that may result from disruption of enhancer sequences that normally act in cis to increase expression of the gene. However, the mechanisms regulating PLPP3 expression are not well understood. We show that the human PLPP3 promoter contains three functional NF-κB response elements. All of these are required for maximal induction of PLPP3 promoter activity in reporter assays. The identified sequences recruit RelA and RelB components of the NF-κB transcription complex to chromatin, and these transcription factors bind to the identified target sequences in two different cell types. LPA promotes binding of Rel family transcription factors to the PLPP3 promoter and increases PLPP3 gene expression through mechanisms that are attenuated by an NF-κB inhibitor, LPA receptor antagonists, and inhibitors of phosphoinositide 3 kinase. These findings indicate that up-regulation of PLPP3 during inflammation and atherosclerosis results from canonical activation of the NF-κB signaling cascade to increase PLPP3 expression through nuclear import and binding of RelA and RelB transcription factors to the PLPP3 promoter and suggest a mechanism by which the LPP3 substrate, LPA, can regulate PLPP3 expression.
Keywords: glycerosphingolipid, lysophospholipid, glycerophospholipid, phosphatase, transcription promoter
Introduction
Lipid phosphate phosphatases (LPPs)2 are members of a larger family of integral membrane proteins with a common core transmembrane topology that comprises six α helices linked by extramembrane sequences (1). The catalytic residues of these proteins are related to bacterial haloacid dehydrogenases and are oriented close to the membrane surface (2). LPPs are localized to both intracellular membrane compartments and to the plasma membrane. The transmembrane topology places the active site facing the luminal side of intracellular membrane organelles or the extracellular face of the plasma membrane. This enables LPPs to function as “ecto” phosphatases that can dephosphorylate extracellularly presented substrates (3, 4). In in vitro assays using detergent solubilized substrates, LPPs have broad specificity for phosphate esters of isoprenols, glycerol, and sphingophospholipids (4, 5). Although less information is available about the physiological substrates of these enzymes, one well-documented function is to dephosphorylate and inactivate the signaling functions of lysophosphatidic acid (LPA) and sphingosine 1 phosphate (S1P) (1). These are bioactive lipids that signal through G protein–coupled receptors to promote cellular responses that include migration, proliferation, and survival (6, 7). Studies using genetic loss-of-function and overexpression approaches support the idea that LPPs can function as negative regulators of signaling responses to these bioactive lipids (8–10).
The mammalian genome contains three PLPP genes encoding structurally related LPP enzymes, termed LPP1–LPP3 (2). Although these enzymes exhibit broad partially overlapping expression patterns and have essentially identical substrate specificities and enzymatic activities in vitro, they are not functionally redundant. Homozygous inactivation of one of these genes, PLPP3, encoding LPP3, results in early embryonic lethality because of defects in embryonic patterning and vascular development (11). This embryonic lethality is phenocopied by inactivation of PLPP3 in vascular endothelial cells and likely results from dysregulated migration and survival of these cells, which normally drive development of the embryonic vasculature (12). Studies of mice with tissue-specific and/or inducible post-natal inactivation of PLPP3 have revealed essential functions for LPP3 in maintenance of vascular integrity, vascular responses to injury, immune cell trafficking between the lymphatic system and the circulation, and experimentally induced atherosclerosis (9, 12–14). Although other mechanisms involving intracellular lipid metabolism and signaling cannot be discounted, the most parsimonious explanation for these effects is that LPP3 normally inactivates extracellular bioactive lipids, including LPA and S1P, and that loss of LPP3 leads to exaggerated or spatially dysregulated signaling responses to these lipid mediators.
Although the above observations are clearly biologically important, the need to understand the regulation and function of LPP3 is further underscored by the observation that heritable variants in the final intron of the PLPP3 gene associate with significant interindividual variability in human risk of coronary artery disease (15, 16). This effect is independent of traditional risk factors, including elevations in low-density lipoprotein–associated cholesterol, suggesting that PLPP3 may be a component of a previously unappreciated coronary artery disease risk mechanism that, if defined, might be of benefit for diagnosis and treatment of the disease.
PLPP3 expression is strongly up-regulated by inflammatory stimuli in multiple cell types (9, 17). The risk-associated PLPP3 genotype is associated with lower levels of PLPP3 mRNA (18). Differentiated monocytes from subjects that are heterozygous for the risk-associated alleles exhibit lower PLPP3 expression in response to oxidized low-density lipoprotein, whereas these increases in PLPP3 expression are lost in cells from individuals that are homozygous for the risk allele (19). Although more work is needed to understand the significance of these observations, the risk-associated PLPP3 locus contains binding sites for the CCAAT enhancer–binding protein transcriptional enhancer (C/EBP) that are disrupted in the risk-associated allele, which decreases their activity in reporter gene assays (19). However, this locus contains binding sites for other transcriptional enhancers that could also contribute to variant-associated cis effects on PLPP3 gene expression. Effects of these heritable intronic PLPP3 variants on C/EBP function were demonstrated using minimal promoter-based reporter constructs and therefore need to be extrapolated to studies using the endogenous PLPP3 gene promoter.
The work summarized above clearly establishes the importance of understanding mechanisms regulating PLPP3 expression during development and in settings of inflammation and cardiovascular disease. To investigate this issue, we identified and cloned the PLPP3 promoter and used bioinformatic approaches, reporter assays, and mutagenesis and functional genomic experiments to identify mechanisms linking activation of the promoter to cell surface receptor–initiated inflammatory signaling. Our studies identify an important role of the NF-κB transcription factor complex in regulation of PLPP3 promoter activity and expression of functional LPP3.
Results
Transcription factor binding sites in the human PLPP3 promoter
We used online databases and data visualization tools (20–22), as detailed in the legend of Fig. S1, to examine predicted and experimentally validated functional elements in the vicinity of the human PLPP3 gene transcriptional initiation codon and 5′ UTR. We identified predicted binding sites for multiple transcription factors in active chromatin, as identified by association with histone methylation and acetylation marks. Several of these transcription factor–binding sites appeared to be functional based on publicly accessible ChIP sequencing data from multiple cell types (Fig. S1, A and C). Given the strong up-regulation of PLPP3 expression observed by others and ourselves in settings of inflammation, vascular injury, and atherosclerosis, we were particularly interested in the presence of three distinct binding sites for cRel components of the NF-κB transcription factor complex in the human PLPP3 promoter. These are identified as Rel-responsive elements (ReREs) (Fig. S1D). Many of the abovementioned observations of increased LPP3 expression in inflammation have been made in mouse models (17). Two of these Rel-responsive element sequences (denoted sites 2 and 3) were highly conserved in multiple vertebrate species, including mice. Site 1 was only present in human and rhesus monkey gene sequences (Fig. S1, B and D).
Identification of functional NF-κB response elements in the human PLPP3 promoter
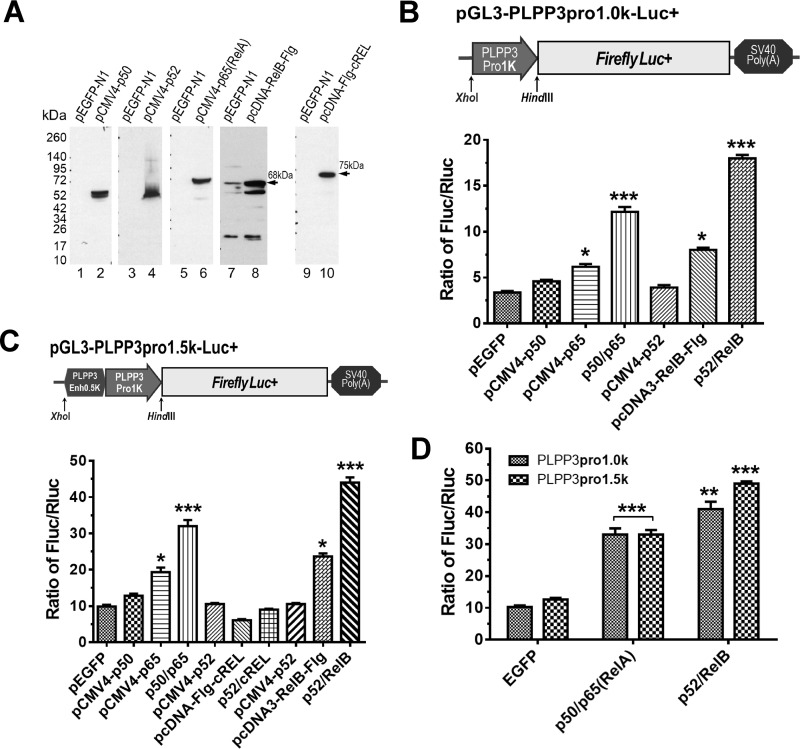
We cloned the human PLPP3 promoter and used this in reporter gene expression assays to test the hypothesis that transcriptional activity of the promoter is regulated by NF-κB. We generated two reporter constructs, one containing the core ∼1-kb PLPP3 promoter and another containing this promoter with an additional ∼0.5 kb upstream sequence. We did this because the bioinformatic analysis discussed above led us to postulate that this additional sequence might function as a transcriptional enhancer because of the presence of a third Rel-responsive sequence element. This third sequence element is uniquely present in this region of the human PLPP3 promoter but absent from the PLPP3 promoter in other vertebrate species, including mice (Figs. S1 and S2). We also generated constructs for ectopic expression of epitope-tagged NF-κB complex subunits: p50, p52, p65, RelB, and c-Rel/RelA. These epitope-tagged proteins were strongly expressed after transient transfection of the corresponding constructs into HEK293T cells and detection by Western blotting (Fig. 1A). Overexpression of some of these proteins, particularly p50/p65RelA and p52/RelB, significantly increased transcriptional activity of the PLPP3 promoter. In separate experiments, these effects appeared to be more pronounced with the ∼1.5-kb PLPP3 promoter/enhancer construct (Fig. 1, B and C), and this difference was confirmed in a separate experiment, suggesting that the human PLPP3 promoter–specific Rel-responsive element present in this promoter construct is active. (Fig. 1D). In these experiments overexpression of c-Rel/RelA and RelB had the strongest transcription-enhancing effect, and these subunits were used for subsequent experiments.
Figure 1.
Identification and functional activity of NF-κB response elements in the human PLPP3 promoter. A, cDNAs encoding epitope-tagged forms of five NF-κB protein subunits were overexpressed by transfection of HEK293T cells using pcDNA vectors. Cells were grown for 24 h, and recombinant proteins were detected by immunoblotting using relevant antibodies. B, top panel, diagram of PLPP3 promoter reporter constructs containing the ∼1000-bp upstream sequence of the translational start site including the 569 bp 5′ UTR sequence. Bottom panel, Luciferase activity in HEK293T cells that were cotransfected with the PLPP3 promoter reporter vector and with constructs containing a or combinations of NF-κB transcription factors, as indicated in the legend. C, top panel, the experiment shown in B was repeated using a PLPP3 reporter constructs containing an additional ∼500-bp sequence upstream of the ∼1-kb promoter/5′ UTR which, as our bioinformatic analysis suggested, might function as a transcriptional enhancer. Bottom panel, luciferase activity after cotransfection of this ∼1500-bp PLPP3 promoter/enhancer reporter vector with the indicated NF-κB transcription factors. D, the apparent functional differences between the PLPP3 promoter (∼1000 bp) and the promoter/enhancer (∼1500 bp) in their responses to RelA or RelB were compared directly in a single experiment. The data shown are representative of experiments that were repeated at least three times. Luciferase activity is presented as the ratio of Firefly to Renilla luciferase activity (Fluc/Rluc). Summarized data are means ± S.D. The p values for D were calculated using two-tailed paired t tests and for B and C using one-way ANOVA with Greenhouse–Geisser correction and Tukey's multiple comparisons tests. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Synergistic effects of Rel response elements on human PLPP3 promoter activity
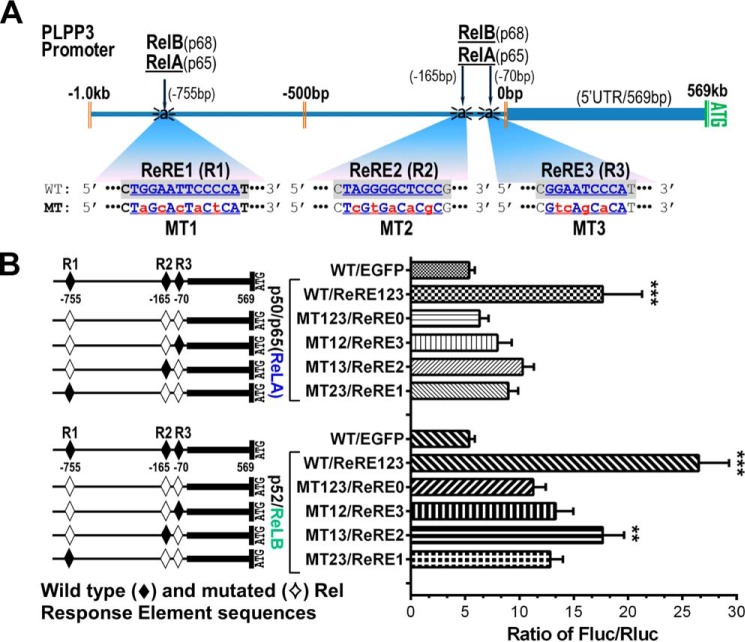
The human PLPP3 promoter contains three separate predicted Rel-responsive elements (Fig. S2). To examine the relative contributions of these to activation of the promoter by ectopic overexpression of NF-κB components, we generated a series of variants of our 1.5-kb promoter/reporter construct in which these sites were inactivated by mutagenesis, either separately or in all possible combinations (Fig. 2A, see also Table S1). Transfection of these reporter constructs with RelA or RelB revealed that all three of these Rel-responsive elements are functional, with the second of these (ReRE2) having the greatest transcription-enhancing activity. These experiments also clearly showed that integrity of all three of these Rel-responsive elements was necessary for maximal activation of the ∼1500-bp PLPP3 promoter by RelA and RelB. Importantly, the human PLPP3 promoter construct in which Rel-responsive element 1 was inactivated by mutation to mimic the PLPP3 promoter of multiple vertebrate species, including mice, was still strongly activated by RelA or RelB (Fig. 2B). Taken together with the results shown in Fig. 1, this suggests that activation of transcription by NF-κB family transcription factors is a common property of the PLPP3 promoter in many vertebrate species.
Figure 2.
Identification and functional validation of ReREs in the human PLPP3 promoter. A, schematic of the localization of the three RelA/p50 and RelB/p52 dimer binding sites in the PLPP3 promoter. The relative locations of binding sites are identified by their distance (in base pairs) from the transcriptional start site. The core consensus NF-κB (RelA/RelB) binding sites (ReRE1–ReRE3) are indicated in blue bold (WT sequence), and changes made to generate mutant (MT1–MT3) variants of these sites are highlighted in lowercase red bold. B, normalized luciferase activity in HEK293T cells transfected with constructs containing mutated ReREs (either single site mutants or combination site mutants, as denoted in the right panel; WT sites are denoted by solid diamonds, and mutant sites are denoted by open diamonds) compared with the activity of the WT promoter reporter vector. Summarized data (mean ± S.D.) are shown from three independent transfection experiments. The p values were calculated using one-way ANOVA with Greenhouse–Geisser correction and Tukey's multiple comparisons tests. **p ≤ 0.01, ***p ≤ 0.001.
Recruitment of RelB to the human PLPP3 promoter
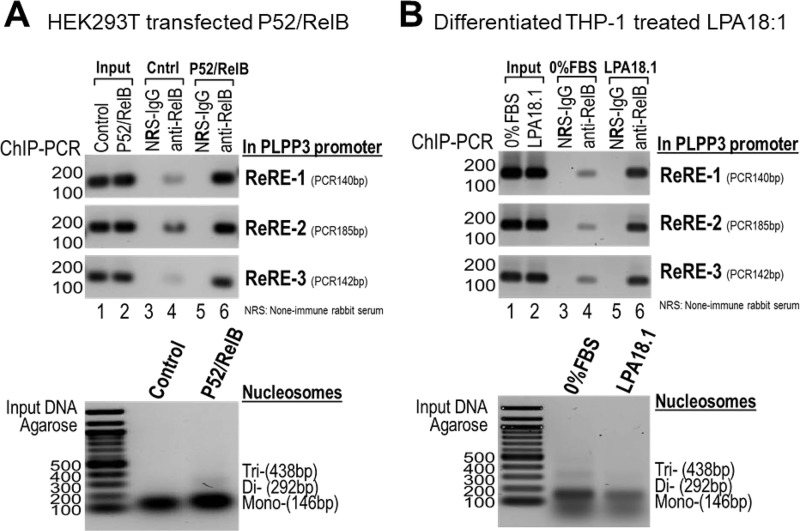
The most parsimonious explanation for the data discussed above is that NF-κB transcription factors bind directly to the Rel-responsive elements in the PLPP3 promoter to increase transcription. We used ChIP assays to demonstrate this directly. In ectopic expression assays, sequences corresponding to all three Rel-responsive elements in the human PLPP3 promoter could be detected in RelB immunoprecipitates. These sequences could also be detected at lower levels without overexpression of RelB, presumably reflecting recruitment of endogenous RelB to the PLPP3 promoter, but, as might be expected, these interactions were more pronounced with ectopic overexpression of RelB (Fig. 3A). To further explore the association of endogenous NF-κB with the PLPP3 promoter, we conducted similar experiments using differentiated THP-1 cells, which resemble human monocytes/macrophages. As explained above, PLPP3 expression is strongly up-regulated by treatment of differentiated human monocytes with oxidized low-density lipoprotein, which may underlie or contribute to the association of PLPP3 with coronary artery disease risk (19). As with HEK293 cells, we observed physical association of all three Rel-responsive element target sequences with RelB in these studies using differentiated THP-1 cells. Interestingly these associations were moderately strengthened by treatment of the cells with the PLPP3 substrate 18:1 LPA, suggesting that NF-κB–dependent transcriptional activation of the PLPP3 promoter might be controlled by activation of cell-surface LPA receptors (Fig. 3B). Although we did not test RelA in these assays because RelA and Rel B bind to the same target sequence, we presume that similar results would be obtained with RelA.
Figure 3.
Binding of NF-κB components to chromatin in proximity to the three Rel response element sites in the human PLPP3 promoter. A and B, ChIP assays demonstrating interactions between the p52/RelB complex and the endogenous PLPP3 promoter in HEK293T cells after ectopic expression of p52/RelB (A) and with endogenous p52/RelB in differentiated THP-1 cells treated with LPA18.1 (B). Bottom panels, the integrity of the nucleosome preparations used for these experiments. Both experiments were repeated three times with similar results.
Overexpression of NF-κB and activation of LPA signaling increases PLPP3 expression
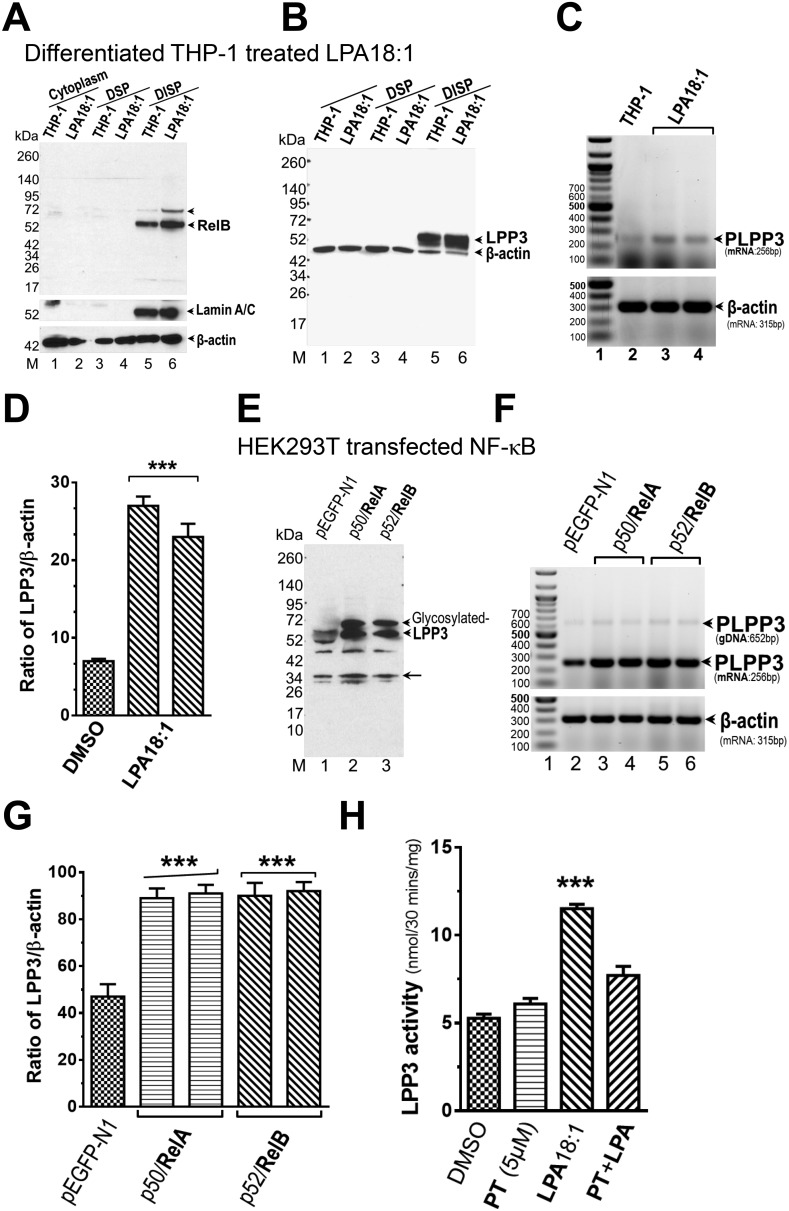
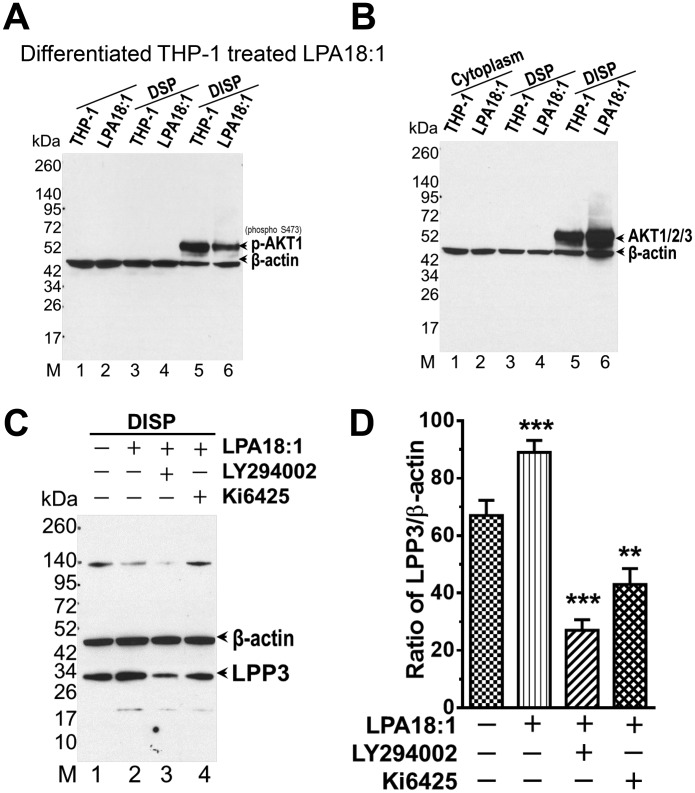
The studies above largely involved ectopic expression of reporter constructs and RelA/B and do not demonstrate functional effects of NF-κB signaling on transcriptional activity of the endogenous PLPP3 promoter. We used the differentiated THP-1 cell model to investigate the effects of NF-κB signaling on LPP3 mRNA and protein expression and on LPP enzymatic activity. For these studies, cells were fractionated into cytoplasmic (i.e. post-nuclear supernatant) and detergent-soluble and -insoluble total particulate fractions. As expected, RelB protein was readily detected in these cells, predominantly present in the detergent-insoluble total particulate fraction, and the levels were moderately increased by 18:1 LPA treatment. LPP3 protein was also detected in this detergent-insoluble particulate fraction. The levels of LPP3 protein and mRNA were both moderately increased (∼2-fold) by 18:1 LPA treatment of the cells (Fig. 4, A–D). These results support the idea that endogenously expressed NF-κB functions as a regulator of PLPP3 promoter activity to increase LPP3 mRNA and protein levels. As might be expected, when we repeated these experiments using ectopic overexpression of RelA or RelB in HEK293T cells, we observed similar increases in LPP3 mRNA levels and protein (Fig. 4, E–G). To further demonstrate the functional relevance of these observations and investigate the role of NF-κB in increases in PLPP3 expression, we measured LPP activity in detergent-extracted membrane proteins from differentiated THP-1 cells that were stimulated with 18:1 LPA. LPA treatment resulted in a 1.9-fold increase in LPA or S1P phosphatase activity, measured under LPP-specific assay conditions (i.e. using Triton X-100–solubilized substrates in the presence of EDTA) (5). This LPA-stimulated increase in LPP activity was blocked by preincubation of the cells with the NF-κB inhibitor parthenolide, supporting a role of NF-κB in the mechanism by which LPA increases LPP3 expression in these cells (Fig. 4H). Increases in LPP3 expression likely account for the increase in total LPP activity in these cells because LPA stimulated an ∼2-fold increase in LPP activity recovered in immunoprecipitates from these detergent extracts generated using our LPP3-specific antibody but not an irrelevant control antibody (data not shown).
Figure 4.
Ectopic overexpression of NF-κB component proteins and stimulation of cells with 18:1 LPA increases LPP3 protein, enzymatic activity, and mRNA levels. A–H, LPA18:1 treatment increases the levels of RelB (A) and LPP3 protein (B) in a DISP fraction and LPP3 mRNA levels (C and D) in PMA-differentiated THP-1 cells. Increases in LPP3 protein (D and E) and PLPP3 mRNA levels (F and G) in HEK293 cells were also observed after ectopic overexpression of P50/RelA or P52/RelB. Quantification of mRNA expression (D and G) depicts mean normalized ratio values ± S.D. from three independent experiments. LPA 18:1 treatment increases LPP activity in the detergent-extractable membrane protein fraction from PMA-differentiated THP-1 cells, and this increase is blocked by pretreatment of the cells with the NF-κB inhibitor parthenolide (H). The p values were calculated using an ordinary one-way ANOVA with Tukey's multiple comparisons test where necessary. ***, p ≤ 0.001. DSP, detergent-soluble particulate.
LPA increases PLPP3 expression through an LPA receptor–activated PI3K pathway
Well-characterized, highly selective pharmacological tools were used to further explore the mechanisms and functional significance of these effects of LPA on PLPP3 expression. PI3K is well-established as an important upstream regulator of the NF-κB pathway (23). Activation of PI3K, in turn, activates the lipid-responsive protein kinase PDK1, which then phosphorylates AKT family protein kinases. This phosphorylation, coupled with membrane recruitment, initiates downstream signaling responses (24). These cells expressed both AKT1 and AKT2 and/or AKT3, as detected by Western blotting. AKT phosphorylation and association of AKT with the detergent-insoluble particulate fraction was increased by treatment of the cells with 18:1 LPA. Concurrently, 18:1 LPA increased LPP3 protein levels in this fraction (Fig. 5, A–C). This is consistent with previous reports that LPP3 protein is largely localized to a detergent-insoluble membrane compartment (25). These effects of 18:1 LPA on LPP3 protein expression were attenuated by a selective antagonist of LPA1/3 receptors, Ki6425 (26), and by treatment of the cells with a selective PI3K inhibitor, LY294002 (27) (Fig. 5D).
Figure 5.
LPA increases LPP3 expression in THP-1 cells through an LPA receptor–activated PI3K pathway. A and B, AKT1/2/3 association with the DISP was increased in PMA-differentiated THP-1 cells after treatment with LPA18.1. C, LPA 18:1–stimulated increases in LPP3 expression in the DISP isolated from THP-1 cells were attenuated after treatment with the LPA receptor (LPA1/3) antagonist Ki16425 or the PI3K inhibitor LY294002. D, quantification of LPP3 protein levels after blocking LPA1/3 receptors and inhibiting the PI3K pathway. Bars show summarized data (mean ± S.D.) of normalized ratio values from three independent experiments. The p values were calculated using an ordinary one-way ANOVA with Tukey's multiple comparisons test. **, p ≤ 0.01; ***, p ≤ 0.001.
NF-κB–dependent mechanisms regulating PLPP3 expression
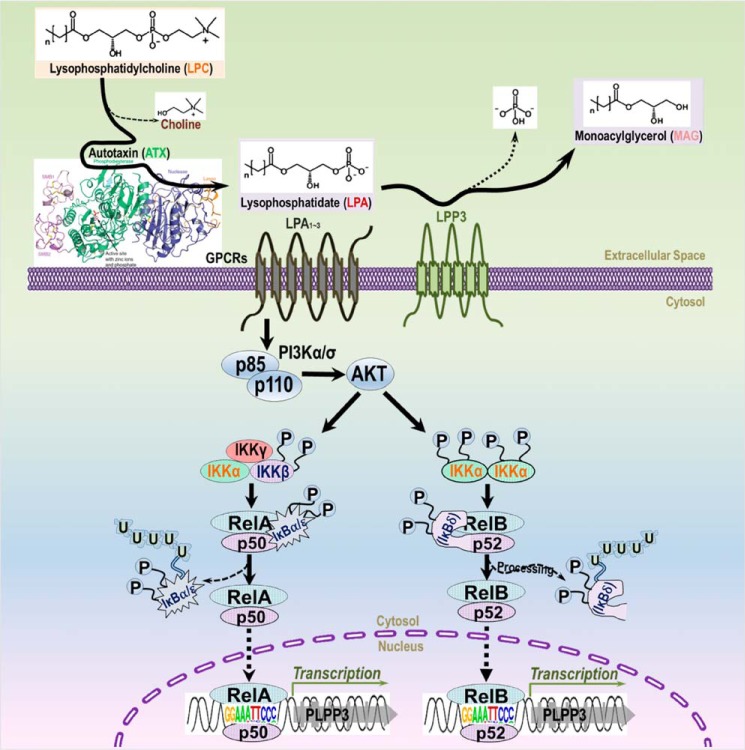
The data presented above support a model in which transcriptional activity of the PLPP3 promoter is regulated by NF-κB transcription factors to increase LPP3 mRNA and protein levels. LPA1/2 receptors can function as activators of this pathway by initiating activation of PI3K-dependent signaling pathways that likely impact the well-described canonical activation of NF-κB signaling through dissociation and degradation of the IκB inhibitor (28) (Fig. 6).
Figure 6.
Schematic of the pathway by which LPA increases LPP3 expression. Our data suggest that LPA activates the PI3K–AKT pathway through LPA receptors (LPA1/3) to increase PI3K–AKT activity, which then activates NF-κB through the canonical pathway involving dissociation and ubiquitin-dependent degradation of inhibitory IκB protein, resulting in nuclear translocation of RelA/P50 or RelB/P52 transcriptional activator complexes. These activated NF-κB complexes then bind to the Rel response elements in the PLPP3 promoter that were identified in this study to initiate PLPP3 gene transcription. GPCR, G protein–coupled receptors.
Discussion
PLPP3 is an essential gene for murine development (11). Studies in mice and humans indicate that PLPP3 expression is tightly regulated, with dramatic increases observed in response to broadly pro-inflammatory signals, both in vitro and in vivo (17, 19). Despite these observations, very little is known about mechanisms regulating PLPP3 expression. Here we show that the human PLPP3 promoter is highly responsive to activation by the NF-κB pathway. The promoter contains three distinct Rel-responsive sequence elements. Two of these sequence elements are conserved in the PLPP3 gene in multiple vertebrate species. All three of these sequence elements are functional in reporter gene assays, and all three sequences interact with NF-κB transcription factor complex components in ChIP assays. Although these observations were made using ectopic overexpression assays, they clearly reveal a physiologically important mechanism for regulation of PLPP3 transcription because we were able to demonstrate interactions of endogenously expressed NF-κB components with these target sequences in two different cell types. Moreover, activation of the NF-κB pathway increased both LPP3 mRNA and protein and LPP enzymatic activity in these cells. These effects were blocked by a small-molecule NF-κB inhibitor. Finally, although more would need to be done to investigate receptor-dependent signaling pathways controlling PLPP3 transcription, we found that activation of LPA1/3 receptors acting through a PI3K-dependent pathway increased PLPP3 expression and levels of LPP3 protein and mRNA in a model cell type. These findings also raise the interesting possibility that LPA, a known LPP3 substrate, could activate expression of PLPP3 to promote LPA degradation and termination of LPA signaling through increased expression of LPP3. This is an interesting possibility because desensitization of LPA-initiated receptor signaling pathways has been observed in a number of systems, but the mechanisms responsible have not been completely delineated (29). NF-κB–mediated increases in the expression of cell-surface LPP3 in response to LPA might be a way to terminate LPA signaling and down-regulate LPA signaling responses in inflammation or other settings where LPP3 has been shown to have a protective role. LPP3 is not specific for LPA and can dephosphorylate other lipid phosphate substrates, including S1P. Increased degradation of S1P in inflammation, where PLPP3 gene expression is increased, might down-regulate S1P signaling responses, leading to decreases in endothelial barrier function and attenuation of lymphocyte egress from the lymphatic system (7, 17, 30).
Common heritable variants in the final intron of PLPP3 associate with interindividual variation in coronary artery disease risk (15, 16). The risk-associated variants predict lower PLPP3 expression in vascular endothelial cells and differentiated peripheral blood monocytes (18, 19). Two studies indicate that sequences within the coronary artery risk–associated haplotype block can function as transcriptional enhancers and that this function is disrupted or attenuated by the risk-associated PLPP3 alleles (19, 31). Evidence of this function comes from reporter assays using minimal promoter constructs. Clearly, it will be very important to demonstrate that these putative cis-acting enhancer functions are enabled by functional interactions between the intronic enhancer and the authentic PLPP3 promoter under control of physiologically relevant transcription factors.
As noted in the introduction, PLPP3 expression is dramatically increased in vitro and in vivo in some settings of inflammation and injury; for example, in mouse models of mechanically induced vascular injury and experimentally induced atherosclerosis (9, 12). The PLPP3 gene product LPP3 is also prominently expressed in the core of human atheromas (19). Activation of NF-κB is strongly associated with inflammation and vascular injury, so our observation that PLPP3 is an NF-κB target gene very likely accounts for these powerful increases in PLPP3 expression (32). PLPP3 expression is necessary for mouse development. Although NF-κB is best known as a regulator of immune function and inflammation, loss of NF-κB family members in mouse models also leads to developmental defects during angiogenesis, including patterning defects that, in some respects, phenocopy PLPP3 deficiency (33). However, NF-κB deficiency does not appear to result in defects in developmental angiogenesis that characterize whole-body and vascular endothelial cell–specific PLPP3 deficiency (11), suggesting additional mechanisms, perhaps involving chromatin remodeling, promoter–enhancer interactions, or some of the other transcription factors that exhibit ChIP signals at the PLPP3 promoter, identified in Fig. S1. In particular, LPP3 expression in cancer likely involves additional mechanisms because levels of LPP3 have been reported to be reduced in some cancers, even in settings where cytokine-driven inflammation is high (1, 34). Given the necessary requirement for LPP3 expression during development and the powerful role of this gene in responses to inflammation, at least in blood and vascular cells, it is not surprising that PLPP3 expression appears to be tightly regulated.
Experimental procedures
Antibodies and other reagents
Specific antibodies against the following antigens were used: Lamin A/C (sc-7293, mouse), β-actin (sc-47778, mouse), c-Rel (C, sc-71, rabbit), NF-κB p50 (C-19, sc-1190, goat), NF-κB p65 (A, sc-109, rabbit), and RelB (C-19, sc-226, rabbit), all from Santa Cruz Biotechnology; NF-κB p52 (05-361, mouse) from EMD Millipore Corp.; and AKT1 (phospho-Ser-473, ab81283, rabbit) and AKT3+AKT2+AKT1 (ab32505, rabbit), both from Abcam. Affinity-purified rabbit polyclonal antibody raised against residues 2–17 (QNYKYDKAIVPESKNG) of the sequence of human LPP3 was used for the detection of LPP3 by Western blot analysis (35). Complete protease inhibitor mixture without EDTA was purchased from Roche Applied Science. Phorbol 12-myristate 13-acetate (PMA), the PI3K inhibitor LY294002 hydrochloride (L9908), and the LPA1/3 antagonist Ki6425 (SML0971) were obtained from Sigma. LPA (C18:1, 857230) was obtained from Avanti Polar Lipids (Alabaster, AL). Parthenolide and fatty acid–free BSA were from Sigma-Aldrich.
Cell culture and transfections
The THP-1 monocyte cell line was obtained from the ATCC (Manassas, VA). Cells were cultured at 0.5–7 × 105 cells/ml in RPMI 1640 medium containing 10% heat-inactivated FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained at 37 °C under 5% CO2. THP-1 monocytes (1 × 106 cells) were differentiated into THP-1 macrophages in 10-cm dishes by directly adding 5 ng/ml PMA into complete RPMI 1640 medium over 48 h. The differentiated THP-1 macrophages were then used immediately. For these experiments, THP-1 macrophages were pretreated with 10 μm LY294002, 1 μm Ki16425, or 5 μm parthenolide for 6 h before addition of 5 μm LPA 18:1 complexed with 0.1% (w/v) fatty acid–free BSA in FBS-free RPMI 1640 medium (36). HEK293T cells were cultured in DMEM following ATCC recommendations, supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. HEK293T cells were used for all transfection experiments, including NF-κB ectopic expression and luciferase assays, using FuGENE® HD transfection reagent (Roche Diagnostics) according to the manufacturer's instructions, with cells cultured in serum-free medium for the indicated times or until the cells reached 95% confluence.
Expression constructs/vectors
The mammalian expression vector containing NF-κB subunits, RelB cFLAG pcDNA3, was a gift from Stephen Smale (Addgene plasmid 20017). pcDNA-FLAG-REL was a gift from Thomas Gilmore (Addgene plasmid 27253). pCMV4 p50, pCMV4p52, and pCMV4 p65 were gifts from Warner Greene (Addgene plasmids 21965, 23289, and 21966, respectively).
The human PLPP3 gene promoter identified by the UCSC Genome Browser in the GRCh37/hg19 database is around 1000 bp upstream of the ATG codon (translation start site), which corresponds with layered trimethyl lysine 4 histone H3 H3K4me3 and enhanced histone 3 lysine 3 and 27 acetylation K3K27AC signals. The putative upstream enhancer was identified as 1000∼1500 bp on the basis of the presence of multiple H3K4me1 active enhancer marks (Fig. S1, A and C). The PLPP3 promoter sequence ∼1000 bp and ∼1500 bp with this enhancer were PCR-amplified from normal human vascular smooth muscle cell genomic DNA and cloned into pGL3-Basic (Promega) at the XhoI and HindIII sites to construct the pGL3-PLPP3pro1.0k-Luc+ and pGL3-PLPP3pro1.5k-Luc+ report vectors. The three putative NF-κB (RelA or RelB) response elements present in the PLPP3 promoter/enhancer (Fig. S2A) were mutated using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The PCR primers used for cloning or mutagenesis are listed in Table S1. All constructs were confirmed by sequencing.
Luciferase reporter assays
HEK293T cell lines were used for luciferase reporter assays. The cells were seeded into 96-well plates and transfected with the luciferase reporter DNA construct after 24 h. 1 μg of pGL3-PLPP3pro1.0k-Luc+ or pGL3-PLPP3pro1.5k-Luc+ firefly luciferase vector and 50 ng of pGL4.74[hRluc/TK] (Promega) Renilla luciferase vectors were cotransfected into each well. In some experiments, 500 ng of the NF-κB subunit expression vectors (p50/RelA, p52/RelB) were cotransfected with the reporter constructs. Transfections used FuGENE® HD transfection reagent (Roche Diagnostics). Forty-eight hours after transfection, luciferase activity was measured using the Dual-Luciferase reporter system (Promega) and detected using the GloMax®-Multi+ Detection System with Instinct® software (Promega), according to the manufacturer's instructions using a BioTek Synergy plate reader and analyzed using BioTek Gen5 software.
Subcellular fractionation
Cytoplasm, nucleoplasm, and detergent-soluble and -insoluble total particulate fractions were isolated as described previously with slight modifications (37, 38). In brief, HEK293T or THP-1 cells were washed with cold PBS and resuspended in buffer A containing 10 mm Hepes (pH 7.8), 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, 5 mm sodium butyrate, 1 mm Na3VO4, 0.2 mm PMSF, and 1× Roche protease inhibitor mixture. Cells were incubated on ice for 15 min. NP-40 was added (0.75%), and cells were vortexed for 10 s. The detergent extracts were centrifuged at 3000 rpm for 3 min at 4 °C in a microcentrifuge. The supernatant (cytoplasm) was collected, and the pellet (nuclei) was resuspended in buffer A containing 25% (w/v) sucrose and underlaid with half a volume of 50% (w/v) sucrose in the same buffer A. The sample was centrifuged at 10,000 rpm for 10 min at 4 °C in a microcentrifuge, and the pellet was resuspended in buffer A containing 1 mm CaCl2 and 0.05 units/μl micrococcal nuclease and incubated for 5 to 30 min in a 37 °C water bath. After addition of 0.2 mm EDTA, the samples were centrifuged at 10,000 rpm for 10 min at 4 °C in a microcentrifuge, and the supernatant (detergent-soluble particulate) and pellet (detergent-insoluble particulate (DISP)) fractions were isolated. The sedimented pellet was solubilized in buffer A containing 10 mm EDTA and 500 mm NaCl and incubated for between 2 and 12 h at 4 °C on a tube shaker/rotator. After centrifugation at 10,000 rpm for 10 min at 4 °C in a microcentrifuge, the supernatant was collected and analyzed by Western blotting with specific antibodies as indicated in the figure legends.
Western blotting
HEK293T or THP-1 cells were collected at 48 h after NF-κB expression vector transfection or LPA stimulation with or without inhibitor treatment and suspended in 100 μl (for a 6-well plate) of RPA buffer (50 mm Tris-HCl (pH 7.5), 1 mm EDTA (pH 8.0), 150 mm NaCl, 0.5% Na-deoxycholate, and 1% Nonidet P-40) containing 1 unit/μl benzonase nuclease (Novagen) and 1× Complete Protease Inhibitor Mixture (Roche Diagnostics). Protein extracts were agitated at 4 °C for 30 min and then centrifuged at 14,000 × g for 30 min at 4 °C. 50 μg of protein-containing supernatants was analyzed on 10% SDS-polyacrylamide gels and transferred to Hybond-C Extra membranes (GE Lifesciences). The primary antibodies used in the study and their sources are detailed above. Protein bands were visualized by enhanced chemiluminescence using Super Signal (Pierce).
ChIP assays
ChIP assays were performed with a ChIP assay kit (Upstate Biotechnology) according to the manufacturer's instructions. In brief, HEK293T or THP-1 cells were cross-linked with 1% formaldehyde for 10 min before quenching with glycine (0.1375 m) for 5 min at room temperature. The cross-linked cells were rinsed with ice-cold PBS twice, suspended in cell lysis buffer (50 mm Tris (pH 8), 10 mm EDTA, and 1% SDS), sonicated by 10-s pulses on ice with 2-min intervals a total of nine times, and then centrifuged. Sonicated lysate supernatants were precleared for 1 h at 4 °C, adding Pierce ChIP-grade protein A/G magnetic beads (Thermo Scientific) that were preabsorbed with sonicated single-stranded herring salmon sperm DNA and precoated with BSA by the manufacturer. Soluble chromatin was immunoprecipitated with 5 μg of rabbit polyclonal RelB antibody or normal rabbit IgG. DNA–protein complexes were isolated with Pierce ChIP-grade protein A/G magnetic beads for 2 h at 4 °C. The immunoprecipitated complexes were sequentially washed with low-salt buffer (20 mm Tris (pH 8.1), 10 mm EDTA, 150 mm NaCl, 1.0% NP-40, and 0.1% SDS,), high-salt buffer (20 mm Tris (pH 8.1), 10 mm EDTA, 500 mm NaCl, 1% NP-40, and 0.1% SDS), LiCl buffer (10 mm Tris (pH 8.1), 0.25 m LiCl, 1 mm EDTA, and 0.5% NP40), and 20 mm Tris, pH8.1, 10 mm EDTA before eluting in 1% SDS in 0.1 m NaHCO3. Cross-links were reversed by heating at 65 °C overnight in 0.3 m NaCl, and then samples were treated with proteinase K for 2 h at 55 °C. Input samples were also similarly treated with proteinase K. DNA was extracted with phenol extraction and ethanol precipitation. PCR was performed using specific primers designed for three RelB and/or RelA response elements (ReRE1, ReRE2, and ReRE3) (Fig. 2A) in the human PLPP3 promoter/enhancer. Primer sequences are listed in Table S1.
RT-PCR
Total RNA from HEK293 or THP-1 cells was extracted using Qiagen the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Before reverse transcription, the total RNA samples were treated with RQ1 RNase-Free DNase (Promega) One microgram of RQ1 DNase-treated total RNA was added to each synthesis reaction (100-μl volume containing TaqMan reverse transcription reagent random primers (Applied Biosystems). Quantitative real-time PCR reactions were performed in a 7300 Real Time PCR System (Applied Biosystems), with SYBR® Green PCR Master Mix (2×) kit (Applied Biosystems). The above experiments were done by following the company protocol (Power SYBR® Green PCR Master Mix and RT-PCR, Applied Biosystems). Human β-actin mRNA was measured as an internal control. All reactions were carried out in quadruplicate with reference dye normalization, and the median cycle threshold value was used for subsequent analysis. Each real-time PCR quantification experiment was conducted in triplicate. The results are presented as the relative ratios to the β-actin control, which was arbitrarily assigned a value of 1. The hPLPP3 complementary DNA-specific primer was designed to span two adjacent exons, exon 3 and exon 4, of hPLPP3 genomic DNA, containing a 396-bp intron 3 sequence for the DNA template, which can distinguish PCR products from genomic DNA contamination. Primer sequences are listed in Table S1.
Measurement of LPP activity
Cells were cultured in 6-well plates and grown to ∼50% confluence, and the culture medium was replaced with RPMI 1640 medium not containing FBS. Cells were cultured for 48 h in medium containing vehicle or 5 μm parthenolamide for 12 h and then stimulated with vehicle or 5 μm 18:1 LPA in a final concentration of 0.1% (w/v) fatty acid–free BSA (Millipore-Sigma) for a further 48 h, during which time the cells reached ∼90% confluence. The medium was removed, the cells were washed once in PBS, and then whole-cell lysates were prepared as described above. Membrane proteins were extracted in lysis buffer containing 1% Triton X-100 with mixing for 1 h at 4 °C, and then insoluble material was removed by centrifugation. The detergent extracted was used for measurements of LPP activity using Triton X-100–solubilized 16:0 LPA or S1P as substrate, as described previously (4). The remaining substrates and reaction products (16:0 monoacylglycerol and sphingosine) were quantitated by HPLC-coupled electrospray ionization tandem MS as described previously (39). LPP3 was immunoaffinity-isolated from these detergent extracts by incubation with 1 μg of the LPP3-specific antibody described above or an irrelevant antibody control for 1 h. Immune complexes were captured with protein A/G–Sepharose, washed once in lysis buffer, and then resuspended in lysis buffer for measurements of LPP activity using the method detailed above for detergent-extracted proteins.
Statistical analysis
All statistical assays, Student's t test, and one-way analysis of variance (ANOVA) were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.). Statistical significance (p values) are reported in the figure legends and in the text. Where appropriate, corrections for multiple comparisons were performed as indicated in the figure legends.
Author contributions
G. M., S. S. S., and A. J. M. conceptualization; G. M. and A. J. M. data curation; G. M., S. S. S., and A. J. M. investigation; G. M. methodology; G. M., S. S. S., and A. J. M. writing-original draft; S. S. S. and A. J. M. supervision; S. S. S. and A. J. M. funding acquisition; S. S. S. and A. J. M. project administration; S. S. S. and A. J. M. writing-review and editing.
Supplementary Material
Acknowledgment
This work benefitted from resources at the Lexington Veterans Affairs Medical Center.
This work was supported by grants from NHLBI, National Institutes of Health Grant R01 HL120507 and Veterans Health Administration Grants I01CX001550 and 1I01BX002769 (to A. J. M. and S. S. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Table S1.
- LPP
- Lipid phosphate phosphatase
- LPA
- lysophosphatidic acid
- ReRE
- Rel-responsive element
- PMA
- phorbol 12-myristate 13-acetate
- DISP
- detergent-insoluble particulate
- ANOVA
- analysis of variance.
References
- 1. Tang X., Benesch M. G., and Brindley D. N. (2015) Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res. 56, 2048–2060 10.1194/jlr.R058362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sigal Y. J., McDermott M. I., and Morris A. J. (2005) Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem. J. 387, 281–293 10.1042/BJ20041771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pilquil C., Ling Z. C., Singh I., Buri K., Zhang Q. X., and Brindley D. N. (2001) Co-ordinate regulation of growth factor receptors and lipid phosphate phosphatase-1 controls cell activation by exogenous lysophosphatidate. Biochem. Soc. Trans. 29, 825–830 10.1042/bst0290825 [DOI] [PubMed] [Google Scholar]
- 4. Roberts R., Sciorra V. A., and Morris A. J. (1998) Human type 2 phosphatidic acid phosphohydrolases: substrate specificity of the type 2a, 2b, and 2c enzymes and cell surface activity of the 2a isoform. J. Biol. Chem. 273, 22059–22067 10.1074/jbc.273.34.22059 [DOI] [PubMed] [Google Scholar]
- 5. Waggoner D. W., Gómez-Muñoz A., Dewald J., and Brindley D. N. (1996) Phosphatidate phosphohydrolase catalyzes the hydrolysis of ceramide 1-phosphate, lysophosphatidate, and sphingosine 1-phosphate. J. Biol. Chem. 271, 16506–16509 10.1074/jbc.271.28.16506 [DOI] [PubMed] [Google Scholar]
- 6. Yung Y. C., Stoddard N. C., and Chun J. (2014) LPA receptor signaling: pharmacology, physiology, and pathophysiology. J. Lipid Res. 55, 1192–1214 10.1194/jlr.R046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Proia R. L., and Hla T. (2015) Emerging biology of sphingosine-1-phosphate: its role in pathogenesis and therapy. J. Clin. Invest. 125, 1379–1387 10.1172/JCI76369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng H. Y., Dong A., Panchatcharam M., Mueller P., Yang F., Li Z., Mills G., Chun J., Morris A. J., and Smyth S. S. (2012) Lysophosphatidic acid signaling protects pulmonary vasculature from hypoxia-induced remodeling. Arterioscler. Thromb. Vasc. Biol. 32, 24–32 10.1161/ATVBAHA.111.234708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panchatcharam M., Miriyala S., Salous A., Wheeler J., Dong A., Mueller P., Sunkara M., Escalante-Alcalde D., Morris A. J., and Smyth S. S. (2013) Lipid phosphate phosphatase 3 negatively regulates smooth muscle cell phenotypic modulation to limit intimal hyperplasia. Arterioscler. Thromb. Vasc. Biol. 33, 52–59 10.1161/ATVBAHA.112.300527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ren H., Panchatcharam M., Mueller P., Escalante-Alcalde D., Morris A. J., and Smyth S. S. (2013) Lipid phosphate phosphatase (LPP3) and vascular development. Biochim. Biophys. Acta 1831, 126–132 10.1016/j.bbalip.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Escalante-Alcalde D., Hernandez L., Le Stunff H., Maeda R., Lee H. S. Jr., Gang C., Sciorra V. A., Daar I., Spiegel S., Morris A. J., and Stewart C. L. (2003) The lipid phosphatase LPP3 regulates extra-embryonic vasculogenesis and axis patterning. Development 130, 4623–4637 10.1242/dev.00635 [DOI] [PubMed] [Google Scholar]
- 12. Panchatcharam M., Salous A. K., Brandon J., Miriyala S., Wheeler J., Patil P., Sunkara M., Morris A. J., Escalante-Alcalde D., and Smyth S. S. (2014) Mice with targeted inactivation of ppap2b in endothelial and hematopoietic cells display enhanced vascular inflammation and permeability. Arterioscler. Thromb. Vasc. Biol. 34, 837–845 10.1161/ATVBAHA.113.302335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bréart B., Ramos-Perez W. D., Mendoza A., Salous A. K., Gobert M., Huang Y., Adams R. H., Lafaille J. J., Escalante-Alcalde D., Morris A. J., and Schwab S. R. (2011) Lipid phosphate phosphatase 3 enables efficient thymic egress. J. Exp. Med. 208, 1267–1278 10.1084/jem.20102551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Busnelli M., Manzini S., Hilvo M., Parolini C., Ganzetti G. S., Dellera F., Ekroos K., Jänis M., Escalante-Alcalde D., Sirtori C. R., Laaksonen R., and Chiesa G. (2017) Liver-specific deletion of the Plpp3 gene alters plasma lipid composition and worsens atherosclerosis in apoE−/− mice. Sci. Rep. 7, 44503 10.1038/srep44503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. CARDIoGRAMplusC4D Consortium, Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T. L., Thompson J. R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B. A., Stirrups K., König I. R., Cazier J. B., Johansson A., et al. (2013) Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33 10.1038/ng.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schunkert H., König I. R., Kathiresan S., Reilly M. P., Assimes T. L., Holm H., Preuss M., Stewart A. F., Barbalic M., Gieger C., Absher D., Aherrahrou Z., Allayee H., Altshuler D., Anand S. S., et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Busnelli M., Manzini S., Parolini C., Escalante-Alcalde D., and Chiesa G. (2018) Lipid phosphate phosphatase 3 in vascular pathophysiology. Atherosclerosis 271, 156–165 10.1016/j.atherosclerosis.2018.02.025 [DOI] [PubMed] [Google Scholar]
- 18. Erbilgin A., Civelek M., Romanoski C. E., Pan C., Hagopian R., Berliner J. A., and Lusis A. J. (2013) Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J. Lipid Res. 54, 1894–1905 10.1194/jlr.M037085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reschen M. E., Gaulton K. J., Lin D., Soilleux E. J., Morris A. J., Smyth S. S., and O'Callaghan C. A. (2015) Lipid-induced epigenomic changes in human macrophages identify a coronary artery disease-associated variant that regulates PPAP2B expression through altered C/EBP-β binding. PLoS Genet. 11, e1005061 10.1371/journal.pgen.1005061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diehl A. G., and Boyle A. P. (2016) Deciphering ENCODE. Trends Genet. 32, 238–249 10.1016/j.tig.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 21. Li S., Wan C., Zheng R., Fan J., Dong X., Meyer C. A., and Liu X. S. (2019) Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks. Nucleic Acids Res. 47, W206–W211 10.1093/nar/gkz332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jegga A. G., Chen J., Gowrisankar S., Deshmukh M. A., Gudivada R., Kong S., Kaimal V., and Aronow B. J. (2007) GenomeTrafac: a whole genome resource for the detection of transcription factor binding site clusters associated with conventional and microRNA encoding genes conserved between mouse and human gene orthologs. Nucleic Acids Res. 35, D116–D121 10.1093/nar/gkl1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoesel B., and Schmid J. A. (2013) The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12, 86 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan T. O., Rittenhouse S. E., and Tsichlis P. N. (1999) AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu. Rev. Biochem. 68, 965–1014 10.1146/annurev.biochem.68.1.965 [DOI] [PubMed] [Google Scholar]
- 25. Sciorra V. A., and Morris A. J. (1999) Sequential actions of phospholipase D and phosphatidic acid phosphohydrolase 2b generate diglyceride in mammalian cells. Mol. Biol. Cell 10, 3863–3876 10.1091/mbc.10.11.3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohta H., Sato K., Murata N., Damirin A., Malchinkhuu E., Kon J., Kimura T., Tobo M., Yamazaki Y., Watanabe T., Yagi M., Sato M., Suzuki R., Murooka H., Sakai T., et al. (2003) Ki16425, a subtype-selective antagonist for EDG-family lysophosphatidic acid receptors. Mol. Pharmacol. 64, 994–1005 10.1124/mol.64.4.994 [DOI] [PubMed] [Google Scholar]
- 27. Vlahos C. J., Matter W. F., Hui K. Y., and Brown R. F. (1994) A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269, 5241–5248 [PubMed] [Google Scholar]
- 28. Gilmore T. D. (2006) Introduction to NF-κB: players, pathways, perspectives. Oncogene 25, 6680–6684 10.1038/sj.onc.1209954 [DOI] [PubMed] [Google Scholar]
- 29. Alcántara-Hernández R., Hernández-Méndez A., Campos-Martínez G. A., Meizoso-Huesca A., and García-Sáinz J. A. (2015) Phosphorylation and internalization of lysophosphatidic acid receptors LPA1, LPA2, and LPA3. PLoS ONE 10, e0140583 10.1371/journal.pone.0140583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hisano Y., and Hla T. (2019) Bioactive lysolipids in cancer and angiogenesis. Pharmacol. Ther. 193, 91–98 10.1016/j.pharmthera.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu C., Huang R. T., Kuo C. H., Kumar S., Kim C. W., Lin Y. C., Chen Y. J., Birukova A., Birukov K. G., Dulin N. O., Civelek M., Lusis A. J., Loyer X., Tedgui A., Dai G., et al. (2015) Mechanosensitive PPAP2B regulates endothelial responses to atherorelevant hemodynamic forces. Circ. Res. 117, e41–e53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Winther M. P., Kanters E., Kraal G., and Hofker M. H. (2005) Nuclear factor κB signaling in atherogenesis. Arterioscler. Thromb. Vasc. Biol. 25, 904–914 10.1161/01.ATV.0000160340.72641.87 [DOI] [PubMed] [Google Scholar]
- 33. Espín-Palazón R., and Traver D. (2016) The NF-κB family: key players during embryonic development and HSC emergence. Exp. Hematol. 44, 519–527 10.1016/j.exphem.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 34. Tanyi J. L., Morris A. J., Wolf J. K., Fang X., Hasegawa Y., Lapushin R., Auersperg N., Sigal Y. J., Newman R. A., Felix E. A., Atkinson E. N., and Mills G. B. (2003) The human lipid phosphate phosphatase-3 decreases the growth, survival, and tumorigenesis of ovarian cancer cells: validation of the lysophosphatidic acid signaling cascade as a target for therapy in ovarian cancer. Cancer Res. 63, 1073–1082 [PubMed] [Google Scholar]
- 35. Sciorra V. A., Rudge S. A., Wang J., McLaughlin S., Engebrecht J., and Morris A. J. (2002) Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 159, 1039–1049 10.1083/jcb.200205056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park E. K., Jung H. S., Yang H. I., Yoo M. C., Kim C., and Kim K. S. (2007) Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56, 45–50 10.1007/s00011-007-6115-5 [DOI] [PubMed] [Google Scholar]
- 37. Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., and Spiegel S. (2009) Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325, 1254–1257 10.1126/science.1176709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu X., and Fagotto F. (2011) A method to separate nuclear, cytosolic, and membrane-associated signaling molecules in cultured cells. Sci. Signal. 4, pl2 [DOI] [PubMed] [Google Scholar]
- 39. Selim S., Sunkara M., Salous A. K., Leung S. W., Berdyshev E. V., Bailey A., Campbell C. L., Charnigo R., Morris A. J., and Smyth S. S. (2011) Plasma levels of sphingosine 1-phosphate are strongly correlated with haematocrit, but variably restored by red blood cell transfusions. Clin. Sci. 121, 565–572 10.1042/CS20110236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.