Abstract
Background
Cardiac arrest (CA) has been a leading cause of death for many decades. Despite years of research, we still do not understand how each organ responds to the reintroduction of blood flow after prolonged CA. Following changes in metabolites of individual organs after CA and resuscitation gives context to the efficiency and limitations of current resuscitation protocols.
Methods and Results
Adult male Sprague–Dawley rats were arbitrarily assigned into 3 groups: control, 20 minutes of CA, or 20 minutes of CA followed by 30 minutes of cardiopulmonary bypass resuscitation. The rats were euthanized by decapitation to harvest brain, heart, kidney, and liver tissues. The obtained tissue samples were analyzed by ultra‐high‐performance liquid chromatography–high‐accuracy mass spectrometry for comprehensive metabolomics evaluation. After resuscitation, the brain showed decreased glycolysis metabolites and fatty acids and increased amino acids compared with control. Similarly, the heart displayed alterations mostly in amino acids. The kidney showed decreased amino acid and fatty acid pools with severely increased tricarboxylic acid cycle metabolites following resuscitation, while the liver showed minimal alterations with slight changes in the lipid pool. Each tissue has a distinct pattern of metabolite changes after ischemia/reperfusion. Furthermore, resuscitation worsens the metabolic dysregulation in the brain and kidney, while it normalizes metabolism in the heart.
Conclusions
Developing metabolic profiles using a global metabolome analysis identifies the variable nature of metabolites in individual organs after CA and reperfusion, establishing a stark contrast between the normalized heart and liver and the exacerbated brain and kidney, only after the reestablishment of blood circulation.
Keywords: asphyxia, cardiopulmonary bypass, ischemia reperfusion injury, mass spectrometry
Subject Categories: Sudden Cardiac Death, Animal Models of Human Disease, Metabolism, Basic Science Research
Clinical Perspective
What Is New?
Metabolic dysregulations observed after cardiac arrest and subsequent resuscitation demonstrate a unique, individualized profile for each organ.
Metabolic alterations after 20 minutes of cardiac arrest are found to be more severe in the heart and kidney than in the brain and liver.
Metabolic alterations after 30 minutes of cardiopulmonary bypass resuscitation are rescued in the heart and liver but become more severe in the brain and kidney.
What Are the Clinical Implications?
More severe metabolic dysregulation of the brain indicates that brain damage may directly be caused by resuscitation rather than ischemia.
This result shows a critical limitation of current resuscitation protocols, requiring the development of more targeted interventions for individual organs with a particular emphasis on the brain.
Introduction
Cardiac arrest (CA) is a leading cause of death in the United States, with an overall survival rate of approximately 10%. While many patients are temporarily resuscitated, poor neurological outcome is a major cause for the high mortality rate.1, 2, 3 Because CA is a time‐dependent pathology, it is imperative to quickly initiate resuscitation protocols, which can successfully treat many patients who experience CA; however, the longer a patient resides in the ischemic phase, the likelihood of his/her survival decreases drastically.4 As such, survival is significantly lower with patients who experience prolonged CA. Despite improved resuscitation guidelines and new therapeutic interventions, restoring blood flow to ischemic organs remains the primary constituent of current resuscitation medicine5, 6, 7, 8; furthermore, it is commonly known that reperfusion may also produce elements of “reperfusion injury.”9 Although reperfusion is imperative for survival, we do not fully understand the metabolic alterations that occur after restoring blood flow to each organ, particularly after a prolonged duration of cardiac arrest.
Under ischemic conditions, numerous cellular pathways are altered and these alterations in turn cause changes in the content and/or the composition of metabolites in tissues; metabolites are important indicators of physiological or pathological states of organs.10 Therefore, following metabolic alterations caused by the full body ischemia of CA and alterations that remain or newly arise after reperfusion of these ischemic tissues will help assess the effects of the restoration of blood flow on individual organs.
Current metabolomics approach allows the simultaneous measurement of numerous metabolites in various pathways, explicating many fundamental questions about cellular physiology/metabolism.11 Not only is this approach becoming more essential in discovering new molecular biomarkers, but it has been useful in understanding the overall cellular metabolism in organs.12, 13, 14 This is of great advantage in studying CA pathology, where whole‐body ischemia and subsequent resuscitation affect multiple pathways in multiple organs simultaneously.15, 16, 17, 18, 19, 20, 21, 22 These prior studies have mainly used isolated organ ischemia/reperfusion injury models, which do not accommodate for whole‐body ischemia, which is a distinctive feature of cardiac arrest, thus limiting the applicability of their metabolic findings in understanding CA pathology.
A lack of a severe injury model that mimics the human condition of prolonged CA has also been an obstacle in CA research. A majority of patients with CA die of prolonged CA within several hours, even with advanced life support. However, most preclinical studies are established on the basis of CA models with relatively short CA times (up to 10 minutes), after which animals can survive for several days without any treatment.23, 24 Therefore, models with short CA time represent only a minor portion of patients who experience CA with mild injury, and alterations observed in these models have limited clinical applicability. The lack of a severe injury model is partly attributable to the limitation of conventional chest compression to resuscitate animals up to 10 minutes after CA.
To overcome this limitation, we previously established a customized rat model of prolonged CA followed by resuscitation using cardiopulmonary bypass (CPB). The rodent CPB model mimics elements of human extracorporeal membrane oxygenation resuscitation,25, 26, 27 which provides better survival and neurological outcomes compared with conventional cardiopulmonary resuscitation (CPR) in human patients with CA.28, 29, 30, 31 In animal experiments, CPB resuscitation can restore native heart contractions and full reperfusion after 30 minutes of CA, allowing the examination of postresuscitation metabolic status after sustained CA. Our model shows that even if rats are able to achieve return of spontaneous circulation (ROSC), their survival time is ≈4 hours after resuscitation, which is translatable to most human patients, who also die within a few hours despite achieving ROSC, validating that our model mimics human disease.
Using this model, we applied a global metabolomics strategy to evaluate how each tissue responds to 20 minutes of prolonged CA and then 30 minutes of reperfusion, and whether the metabolic alterations are seen during ischemia or rather develop during reperfusion. An ultra‐high‐performance liquid chromatography (LC)–high‐accuracy mass spectrometry (MS) system was used to follow the changes in lipids, amino acids, carbohydrates, and other metabolites in the brain, heart, kidney, and liver. This information about tissue‐specific metabolite alterations can be used to further understand the pathophysiological changes that occur following ischemia/reperfusion as a function of cardiac arrest, especially in the brain. Furthermore, the nuances of metabolite abnormalities in different tissues can be used to develop novel, more targeted therapeutic strategies to treat the metabolic injury in patients with CA.
Materials and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Chemicals and Materials
Reagents used for metabolomics analyses include methanol, acetonitrile, and ultra‐pure water (LC‐MS grade; EMD Millipore, Gibbstown, NJ); formic acid, acetic acid, ammonium hydroxide (Optima LC‐MS Grade; Fisher Chemical, Pittsburgh, PA); and ammonium carbonate (Sigma Aldrich, St. Louis MO).
Animals and Surgical Procedures
The experimental protocol was approved by the Institutional Animal Care and Use Committee of the Feinstein Institute of Medical Research. All procedures were conducted following the approved protocol. Adult male Sprague–Dawley rats (3–4 months of age, weight 410–450 g; Charles River Production, Wilmington, MA) were used for this study. Rats were maintained under a 12‐hour light/dark cycle with free access to food and water and used without starvation. Rats were arbitrarily assigned into 3 groups: control, in which no CA is performed, 20 minutes of CA, and 20 minutes of CA followed by 30 minutes of resuscitation via CPB. This study used an established asphyxia‐induced CA rat model, of which the detailed surgical procedures have been reported previously.25, 26 Briefly, rats were anesthetized with 4% isoflurane and ventilated with an intubated catheter with 2% isoflurane. After the injection of heparin (150 U) and vecuronium (1 mg), asphyxia was induced by stopping the ventilator and isoflurane discontinued. Animals in the CA group were euthanized 20 minutes after the initiation of asphyxia. In the resuscitation group, resuscitation was started following 20 minutes of asphyxia with the initiation of CPB flow and resumption of ventilation. All rats used in this study achieved ROSC. After 30 minutes of CPB resuscitation, rats were euthanized by decapitation to harvest brain, heart, kidney, and liver tissue samples or maintained on the ventilator to measure survival time. Time to reach CA is defined as mean arterial pressure dropping below 20 mm Hg.26, 32 Time to achieve ROSC was monitored to ensure consistency in our animal model. Control group animals were euthanized by decapitation 7 minutes after administration of isoflurane to ensure deep anesthesia before harvesting tissues. We chose 20‐minute CA for this study because it produces a severe injury to the animal model that mimics most human patients in critical conditions, but also retains the potential to be treatable with interventions. The harvested tissues from all 3 groups were immediately flash‐frozen in liquid nitrogen, pulverized, and stored at −80°C until analysis. Six animals were included in each group, and each animal was used to generate one data point except for liver in the CPB group, which has only 5 samples because of failure in MS analysis. Data are represented as mean±SEM.
Sample Extraction
The pulverized tissues (50 mg, n=6 group) were transferred to 1.5‐mL Eppendorf tubes. Extraction solvents (500 μL, 50:50 v/v methanol:acetonitrile) were added into the tubes, and the mixture was homogenized with 0.5‐mm zirconium oxide beads (Next Advance, Inc., Averill Park, NY, product #ZrOB05) using a countertop bullet blender (Next Advance, Inc.) at 4°C for 4 minutes. Samples were incubated for 1 hour at −20°C, then centrifuged at 4°C at 15 000g for 10 minutes. Aliquots (400 μL) of the upper layer were transferred individually to new tubes and evaporated in a stream of nitrogen gas. The residue was reconstituted in solution (200 μL, 50:50 v/v acetonitrile:water), vortexed for 20 seconds, centrifuged at 4°C at 15 000g for 5 minutes, and then stored at −80°C until analysis via LC–tandem mass spectrometry (MS/MS). To ensure the stability and repeatability of the LC–MS system, quality control samples (n=4), which were generated by pooling 10‐μL aliquots from all tissues samples extracts, were analyzed along with the samples.
LC‐MS/MS for Metabolomics Analysis
Chromatography was performed with a Shimadzu Nexera system (Shimadzu, Columbia, MD) coupled to a high‐resolution hybrid quadrupole time‐of‐flight mass spectrometer (TripleTOF 5600; SCIEX, Framingham, MA). For each sample, 2 different analysis methods were performed using reverse‐phase or hydrophilic interaction LC columns.33 In reverse‐phase LC, chromatographic separations were carried out using a 2.1×100 mm Titan C18 column (1.9 μm, Supelco, Bellefonte, PA) for positive and negative ion analyses. The sample injection volume was 5 μL, and the flow rate was 0.2 mL/min. The mobile phases consisted of water (A) and methanol (B), both with 0.1% formic acid. The gradient was as follows: an initial hold at 3% B for 0.5 minutes, followed by a gradient of 3% to 20% B in 5 minutes, to 55% B in 13 minutes, to 95% B in 15.5 minutes, held until 18.5 minutes, then a shift to 5% B at 19 minutes until 25 minutes. The column temperature was held at 45°C. In metabolomics hydrophilic interaction LC analysis, separation was carried out using a 2.1×150 mm SeQuant ZIC‐pHILIC (5 μm, EMD Millipore, Billerica, MA).33 The flow rate was 0.2 mL/min and the injection volume was 5 μL. The 2 mobile phases consisted of 20 mmol/L ammonium carbonate, pH 9.2 with ammonium hydroxide in water (A) and acetonitrile (B). The gradient was as follows: an initial hold at 80% B for 2 minutes, followed by a gradient of 80% to 20% B in 17 minutes, then a shift to 80% B at 17.1 minutes until 22 minutes. The column temperature was 45°C.
Time‐of‐flight–MS was operated with an acquisition time of 0.25 seconds and a scan range of 70 to 1000 Da. MS/MS acquisition was performed with collision energy set at 35 V and collision energy spread of 15 V. Each MS/MS scan had an accumulation time of 0.12 seconds and a range of 40 to 1000 Da using information‐dependent acquisition (IDA). The source temperature was set at 500°C in reverse phase, 550°C in hydrophilic interaction LC and IonSpray voltage at 5.5 kV in positive ion mode and −4.5 kV negative ion mode, respectively.
Data Processing
Targeted metabolomics data were processed using Peak View 2.1 and MultiQuant software version 3.0.2 (SCIEX). Chromatographic peaks of targeted metabolites were annotated using the extract ion chromatogram lists on the basis of high‐accuracy MS, MS/MS fragmentation, isotopic distribution, and retention time compared with an in‐house library of 635 metabolite standards (IROA Technologies, Bolton, MA). In addition to the IROA database, the fragmentation spectra of all peaks were searched through Metlin and HMDB. Each identified metabolite was quantified by integrating peak area using MultiQuant software. The quantitative analysis was based on the total peak areas of extracted ion chromatograms of feature ions.
Multivariate and Statistical Analyses
Principal component analysis was used to examine the metabolic profile to reflect the overall changes following CA and following resuscitation compared with control in the brain, heart, kidney, and liver tissues. The principal component analysis plot was generated with MetaboAnalyst (v4.0) using normalized data (Pareto scaling feature, log transformation). Second, changes in individual metabolites were assessed and diagrammed in well‐recognized biochemical pathways. However, pathways may be carried out differently among the 4 tissues. Statistical analyses for each metabolite were performed with 1‐way ANOVA with Bonferroni's multiple comparison tests using GraphPad Prism 6.0 software (GraphPad, La Jolla, CA). Finally, Pathway Analysis using Metaboanalyst (v4.0) was used to evaluate the impact of individual metabolite alteration on different metabolic pathways. The levels of each metabolite were converted to relative changes compared with control values, which were set as 1, allowing 1 control column for all 4 tissues. To control for false discovery rates for these metabolomics data during follow‐up analyses, statistical significance was determined by q values (false discovery rate corrected P values) using an adaptive linear step‐up procedure; q<0.05 was considered to be significant.
Results
Physiological Outcome
The average time to reach CA was 186±42 seconds. After the initiation of CPB resuscitation, the average time to display heart electrical activity was 45±27 seconds and the time to achieve ROSC was 200±81 seconds. The average survival time of rats after 20 minutes of CA and 30 minutes of CPB resuscitation was 3 to 8±0.38 hours with limited brain function, seen as a lack of response to toe pinch and corneal agitation. This limited brain function and poor survival outcome is analogous to human patients with CA with severe injury.
Principal Component Analyses
Principal component analysis was performed to screen changes in the overall metabolic profile in each organ after 20 minutes of CA and 30 minutes of CPB resuscitation compared with control. The scores plot shown in Figure 1A contains PC1 and PC2 values of 30.4% and 17.7% of data variation, respectively, which represents a difference between CA and CPB in each tissue. In the brain, there is no direct cluster separation after 20 minutes of CA, but there is significant separation following 30 minutes of CPB resuscitation. In the heart, significant cluster separation from control exists after CA, but no further separation follows resuscitation. In the kidney, there is significant separation following CA and following resuscitation. The liver shows a significant separation following CA, which minimizes after resuscitation.
Figure 1.
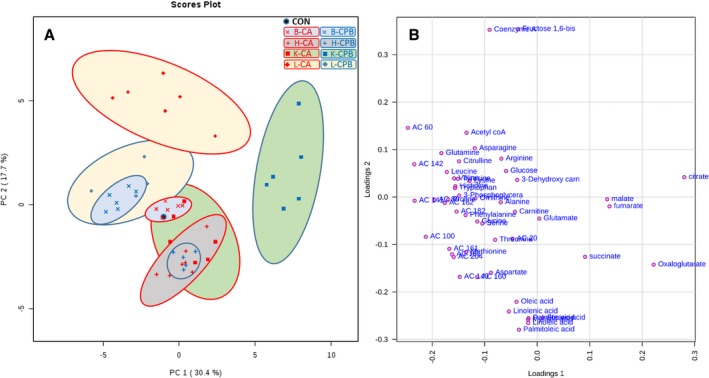
A, Principal components analysis of metabolites from tissues after cardiac arrest and resuscitation. B, Loading plot of metabolites. All 4 organs have distinct metabolic profiles after cardiac arrest and after resuscitation, with the brain being the one organ that exhibited drastic alterations only after resuscitation. We superimposed the controls of all 4 organs into 1 point. As such, the “control” serves as a control for all 4 organs. B indicates brain; CA, cardiac arrest; CON, control; CPB, cardiopulmonary bypass; H, heart; K, kidney; L, liver.
The loading plot (Figure 1B) shows the correlation of each metabolite in each tissue between 20 minutes of CA and 30 minutes of CPB resuscitation. The data show that acylcarnitines (ACs), for example, AC 6:0, AC 10:0, AC 14:1, AC 14:2, and AC 16:1, with negative loading 1 (about −0.2) and loading 2 (between ≥0.1 and 0.1) overlap in the brain and liver after resuscitation, as shown in the score plot. This correlation shows that changes in ACs are one of the main reasons for the distinct metabolic profiles of the liver and brain after CPB resuscitation. Similarly, citrate, α‐ketoglutarate (oxoglutarate), malate, fumarate, and succinate in the tricarboxylic acid (TCA) cycle are significantly changed in kidney tissue after CPB compared with control or CA. A greater degree of change in free fatty acid content is seen in the heart as compared with other organs after CA or CPB. In summary, 20 minutes of CA results in a distinct metabolic profile for individual organs. During 30 minutes of CPB resuscitation, the heart and liver have the most potential to be normalized; the brain and kidney show progressive abnormalities during this same reperfusion period.
We also provide a heatmap of targeted metabolites within pathways as a visual representation of the relative changes in each metabolite among control, 20‐minute CA, and 30‐minute CPB resuscitation groups for all 4 tissue types (Figure 2). The clustering tree was built on the basis of the distance metrics of Pearson correlation coefficient. All data are plotted in clusters showing relative values when compared with a control group, which is a superimposition of the control values for all 4 organs. For example, oxoglutarate is decreased in the brain and liver but increased in the heart and kidney after CPB resuscitation when compared with control.
Figure 2.
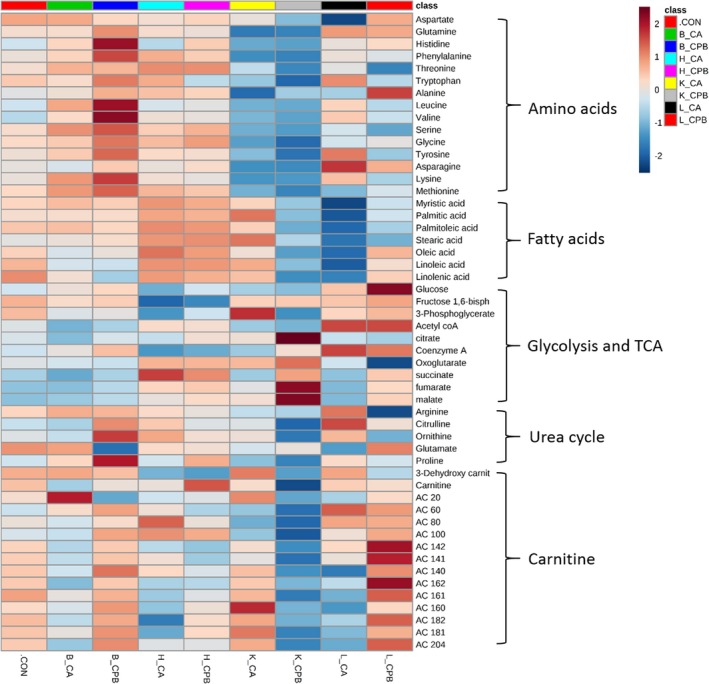
Heatmap. The heatmap provides intuitive visualization of a data table. Each colored cell on the map corresponds to peak area average value in the data table, with samples in columns and features/compounds in rows. Colors indicate Z score (standard deviation from the mean) with red representing increased and blue representing decreased with respect to control. The heatmap was generated with MetaboAnalyst 4.0 using normalized data (log transformation, Pareto scaling) using Euclidean distance measure. B indicates brain; CA, cardiac arrest; CON, control; CPB, cardiopulmonary bypass; H, heart; K, kidney; L, liver.
Changes in Individual Metabolites Following CA and CPB Resuscitation
Statistically significant changes in individual metabolites in each tissue after 20 minutes of CA followed by 30 minutes of CPB resuscitation are shown in Figures 3 and 4. These metabolic pathway maps provide TCA cycle intermediates, energy‐generating metabolites of glycolysis, amino acids, fatty acids, and urea cycle metabolites. This diagrammatic pathway map was constructed to facilitate visualization of post‐CA and post‐CPB resuscitation changes in metabolic constituents based on well‐accepted metabolic pathways.33 The color code represents a statistically significant increase (red) or decrease (green) with P<0.05 in metabolites after resuscitation compared with control, emphasizing the metabolic status in resuscitated rats.
Figure 3.
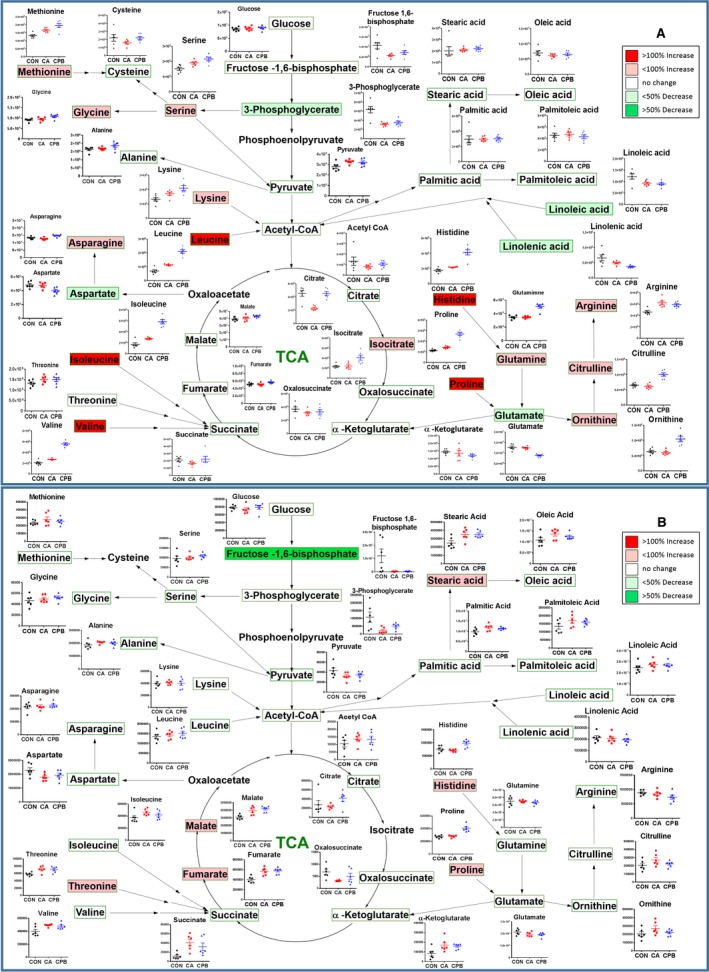
Assembly of the major metabolic pathways in the brain (A) and heart (B). Postresuscitation metabolites are shown, with the corresponding changes from control to cardiac arrest to resuscitation shown in graphs adjacent to respective metabolites. The colors correspond to the level of metabolites after resuscitation: red is increased levels, while green is decreased levels. TCA indicates tricarboxylic acid.
Figure 4.
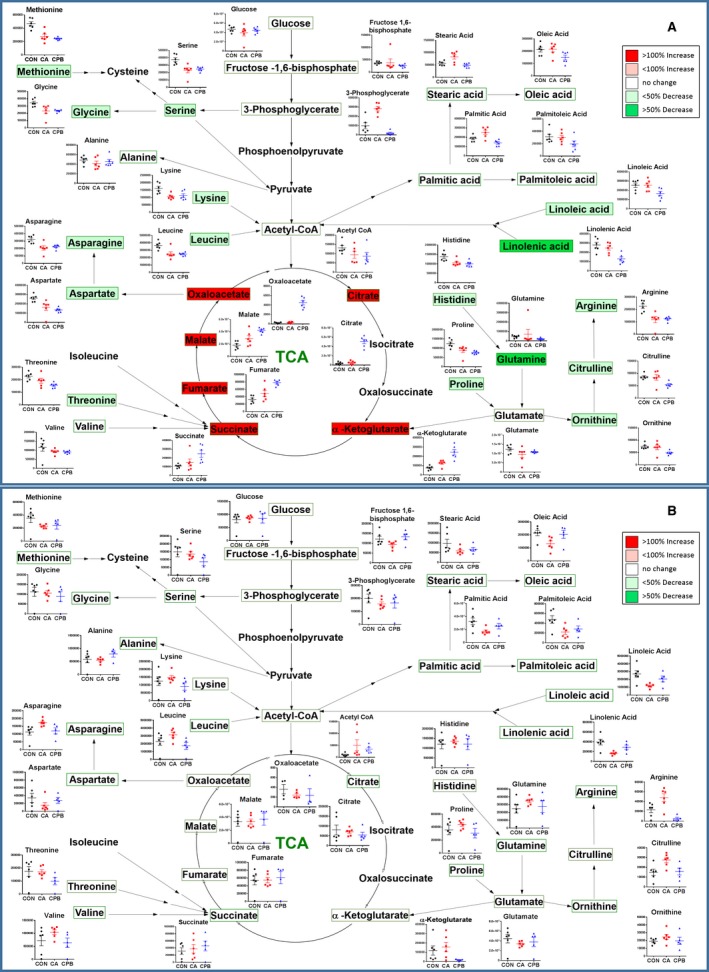
Assembly of the major metabolic pathways in the kidney (A) and liver (B). Postresuscitation metabolites are shown, with the corresponding changes from control to cardiac arrest to resuscitation shown in graphs adjacent to respective metabolites. The colors correspond to the level of metabolites after resuscitation: red is increased levels, while green is decrease levels. TCA indicates tricarboxylic acid.
In the brain tissue, 20 minutes of CA and subsequent 30‐minute CPB resuscitation results in significant changes in the amino acid pool; most of these changes occur mainly after resuscitation (Figure 3A). Particular attention is drawn to histidine, leucine, isoleucine, valine, serine, and methionine, which all increased above control levels after 20 minutes of CA. Moreover, these amino acids, along with glutamine, glycine, and lysine all significantly increased above control after 30 minutes of CPB resuscitation. The only amino acids that decreased after resuscitation compared with control were aspartate and glutamate, while the remaining measured amino acids remained unchanged throughout ischemia and reperfusion. Unlike the amino acid pool, the fatty acid pool in the brain remained relatively normal; after 20 minutes of CA, linoleic acid was significantly decreased. After 30 minutes of CPB resuscitation, linoleic and linolenic acids were significantly decreased. The brain tissue shows a decrease in glycolytic and TCA cycle metabolites, such as fructose‐1,6‐bisphosphate, 3‐phosphoglycerate, and citrate after 20 minutes of CA; however, after 30 minutes of CPB resuscitation, 3‐phosphoglycerate significantly decreased. Urea cycle metabolites, such as arginine and proline, displayed a significant elevation after 20 minutes of CA; however, 30 minutes of CPB resuscitation resulted in variations in all major urea cycle metabolites, such that arginine, citrulline, ornithine, and proline were all significantly increased, while glutamate was significantly decreased.
In the heart tissue, the amino acid pool remained relatively normal; 20 minutes of CA resulted in significant elevation in only threonine, valine, and isoleucine above control (Figure 3B). Resuscitation via 30 minutes of CPB resulted in threonine and histidine elevation, while isoleucine returned to control levels. The only amino acid that was significantly decreased after resuscitation was tryptophan. Heart tissue also displayed an increase in stearic acid as the only fatty acid that was altered after CA, which remained elevated after 30 minutes of CPB resuscitation. The heart tissue shows variations in glycolytic and TCA cycle metabolites, such as a decrease in 3‐phosphoglycerate and an increase in oxoglutarate, succinate, fumarate, and malate. However, 30 minutes of CPB resuscitation resulted in the normalization of 3‐phosphoglycerate and succinate, but lack of normalization of fumarate and malate. Urea cycle metabolites, such as proline significantly increased only after resuscitation. During CA, most of the urea cycle metabolites were not altered significantly.
The pool of measured amino acids in the kidney tissue displayed an intriguing pattern of dysregulation compared with the other tissues; the only amino acids that were unaffected by CA were glutamine, threonine, alanine, and valine, while the remaining measured amino acids were significantly decreased (Figure 4A). Furthermore, 30 minutes of CPB resuscitation resulted in even more dysregulation; all measured amino acids with the exception of glutamate, alanine, and valine were significantly decreased. Kidney tissue also displayed a significant increase in palmitic and stearic acids after 20 minutes of CA. However, 30 minutes of CPB resuscitation resulted in a significant decrease in linoleic and linolenic acids, while stearic acid returned to control levels. The general trend of lipids in the kidney is that they increase after ischemia, but either return to or fall below control levels after resuscitation. The kidney shows variations in glycolytic and TCA cycle metabolites, such as a significant elevation in 3‐phosphoglycerate and oxoglutarate after 20 minutes of CA. However, CPB resuscitation resulted in a significant increase in citrate, oxoglutarate, succinate, fumarate, malate, and oxaloacetate. The urea cycle metabolites were mostly altered after 30 minutes of CPB resuscitation in the kidney. Arginine and proline were significantly decreased after CA, but after CPB, arginine, citrulline, ornithine, and proline were all decreased.
The liver displayed the most resistance to amino acid alterations, such that 20 minutes of CA elevated asparagine levels significantly, which returned to control levels after resuscitation. CA resulted in a significant decrease in myristic, palmitic, palmitoleic, oleic, linoleic, and linolenic acids, which all normalized after 30 minutes of CPB resuscitation (Figure 4B). The liver tissue shows variations in glycolytic and TCA cycle metabolites, such as a significant decrease in fructose‐1,6‐bisphosphate and a significant increase in coenzyme A after CA. However, after resuscitation, fructose‐1,6‐bisphosphate was normalized. There was relative resistance to changes in the urea cycle metabolites in the liver, such that arginine and citrulline were both significantly elevated following CA, while after CPB, liver tissue normalized most of the metabolites.
Changes in Individual AC Species Following CA and CPB Resuscitation
AC species are fatty‐acyl chains connected to carnitine molecules via an ester bond to be oxidized via fatty acid oxidation.34 Figure 5 shows the comparison of many AC species between control, 20‐minute CA, and 30‐minute CPB resuscitation in all 4 tissues. In brain tissue, CA resulted in elevations in AC 2:0 and AC 6:0 but a decrease in AC 16:2 and AC 18:2 when compared with control. Resuscitation‐induced alterations of AC species when compared with post‐CA are as follows: decreased levels were observed in only AC 2:0, while increased levels were observed in AC 6:0, AC 10:0, AC 14:2, AC 14:0, AC 16:2, AC 16:0, AC 18:2, AC 18:1, and AC 20:4. Resuscitation resulted in an increase in AC 6:0, AC 10:0, and AC 16:0 as compared with control.
Figure 5.
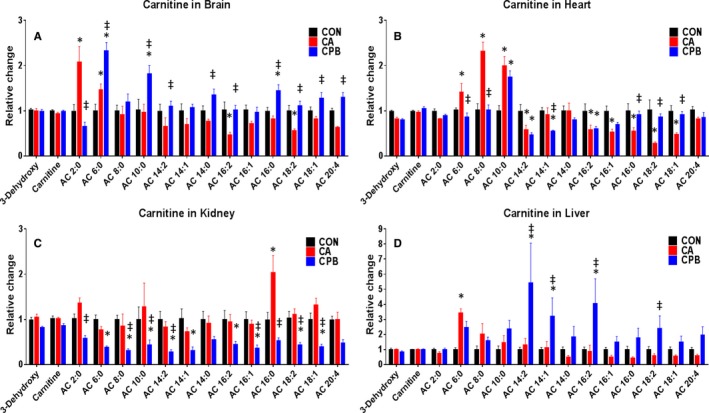
Acylcarnitine species with their relative alterations when compared between control, 20 minutes of cardiac arrest, and resuscitation shown for individual tissues. As compared with control, the brain shows a general decreasing trend of acylcarnitine species after cardiac arrest, which mostly increase following resuscitation without a distinct pattern (A); the heart shows an increase in short‐chain and a decrease in long‐chain acylcarninties after cardiac arrest, which mostly normalize following resuscitation (B); the kidney shows a generalized decrease in the levels of acylcarnitines after resuscitation (C); the liver has minimal changes after cardiac arrest and has a general increase in acylcarnitine levels following resuscitation (D). *P<0.05 between cardiac arrest or resuscitation and control; ‡ P<0.05 between resuscitation and cardiac arrest. AC indicates acylcarnitine; CA, cardiac arrest; CON, control; CPB, cardiopulmonary bypass.
In heart tissue, 20 minutes of CA resulted in elevations in AC 6:0, AC 8:0, and AC 10:0, while decreased levels were observed in AC 14:2, AC 16:2, AC 16:1, AC 16:0, AC 18:2, and AC 18:1 when compared with control. After 30 minutes of CPB resuscitation, increased levels of AC 16:0, AC 18:2, and AC 18:1 and decreased levels of AC 6:0, AC 8:0, and AC 14:1 were found when compared with post‐CA. After resuscitation, AC species that were decreased were AC 14:2, AC 14:1, and AC 16:2, while AC 10:0 was increased when compared with control. During ischemia, longer‐chain fatty acids decreased compared with control, while shorter‐chain fatty acids increased; however, resuscitation results in more normalization across the board.
In kidney tissue, CA resulted in elevation in only AC 16:0 when compared with control. Resuscitation resulted in a decrease from CA in AC 2:0, AC 8:0, AC 10:0, AC 14:2, AC 16:1, AC 16:0, AC 18:2, and AC 18:1. When compared with control, AC species that were decreased after resuscitation were AC 6:0, AC 8:0, AC 10:0, AC 14:2, AC 14:1, AC 16:2, AC 16:1, AC 18:2, and AC 18:1. The kidney was unable to normalize ACs after resuscitation, resulting in a generally diminished lipid reserve after 30 minutes of CPB resuscitation.
In liver tissue, CA resulted in elevation of one major AC species, AC 6:0. However, resuscitation resulted in an increase in AC 14:2, AC 14:1, and AC 16:2 when compared with control, and AC 14:2, AC 14:1, AC 16:2, and AC 18:2 when compared with post‐CA. The general trend observed following resuscitation in the liver is that there is an increased level of AC species.
Analysis of Specific Metabolite Pathway Deviations
To further assess the impact of the observed metabolic alterations, we performed a pathway analysis based on the Kyoto Encyclopedia of Genes and Genomes database using MetaboAnalyst (v4.0). This analysis uses a database of predefined metabolites in various individual or overlapping pathways to identify the most significantly affected pathways. Figure 6 highlights the altered pathways following CA and following resuscitation in the brain, heart, kidney, and liver. The detailed list of pathways that were changed following 20 minutes of CA and 30 minutes of CPB resuscitation are provided in Tables S1 through S8.
Figure 6.
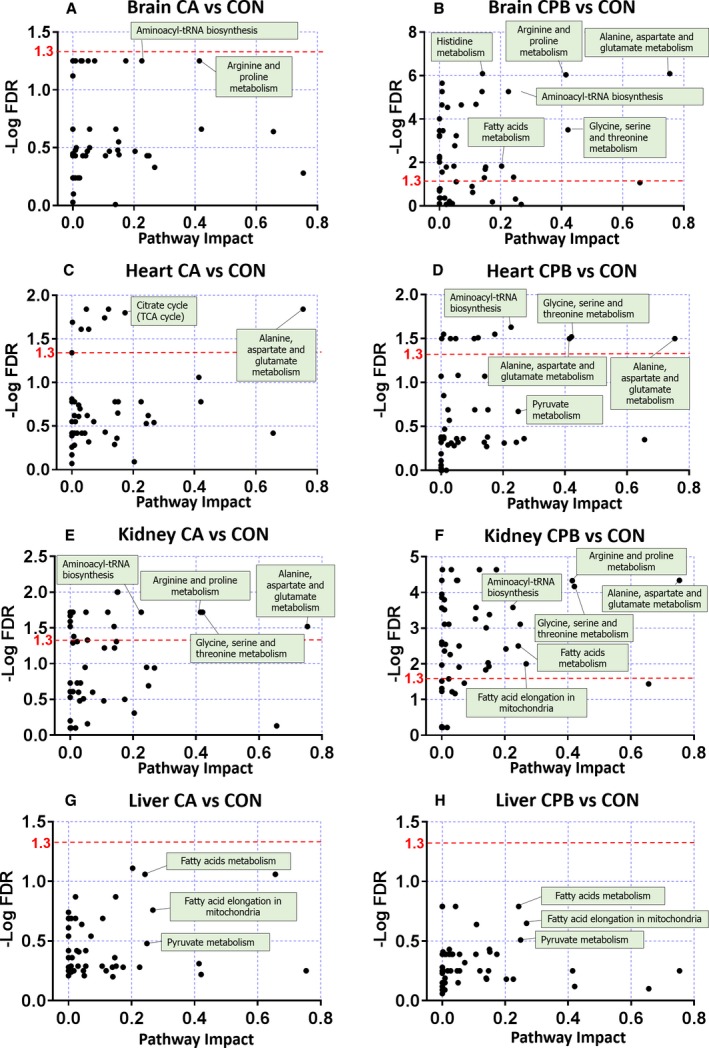
Pathway analysis. Analyses are based on all annotated metabolites (n=6; log transformation, Pareto Scaling) and pathway enrichment of significant metabolites in control vs cardiac arrest and control vs resuscitation using metabolic data sets, which were generated with MetaboAnalyst (4.0). Any data points above the red dotted line are statistically significant (–Log false discovery rate=1.3 is equal to q=0.05). The brain shows no significant changes in any pathway after cardiac arrest (A). However, resuscitation results in significant pathway alterations (B). Cardiac arrest alters some pathways in the heart (C), while resuscitation results in a decrease in the severity of pathway alterations (D). The kidney has many pathways that are affected by cardiac arrest (E), and similar to brain, the kidney, after resuscitation, demonstrates a greater number of pathways that are significantly altered (F). The liver does not demonstrate any pathway significantly altered after cardiac arrest or resuscitation (G and H). CA indicates cardiac arrest; CON, control; CPB, cardiopulmonary bypass.
In the brain, no pathways were found to be significantly changed upon 20 minutes of CA compared with control (q>0.05) (Figure 6A). However, 30 minutes of CPB resuscitation compared with post‐CA in brain tissue showed significant changes in 33 pathways (Figure 6B). Of these significantly altered pathways, the most notable metabolites were involved in aminoacyl‐tRNA biosynthesis; arginine and proline metabolism; and alanine, aspartate and glutamate metabolism, among others in CA compared with CPB in brain tissue.
In heart tissue, alanine, aspartate, and glutamate metabolism; linoleic acid metabolism; arginine and proline metabolism; and glycine, serine, and threonine metabolism showed significant alterations after 20 minutes of CA followed by 30 minutes of CPB resuscitation compared with control; 9 metabolic pathways were significantly altered after CA when compared with control (Figure 6C). Aminoacyl‐tRNA biosynthesis metabolism among the 13 pathways after CPB when compared with control showed the lowest q value (0.02), but the alanine, aspartate, and glutamate metabolism pathway showed the high‐impact value (0.75) (Figure 6D).
In kidney tissue, 24 pathways were significantly changed after CA compared with control, and lysine biosynthesis and purine metabolism were shown with the lowest q value (0.01) (Figure 6E). Following resuscitation, 46 pathways remained significantly altered compared with control, with nicotinate and nicotinamide metabolism, and phenylalanine metabolism having the lowest q values (Figure 6F). In liver tissue, although α‐linolenic acid metabolism, linoleic acid metabolism, and fatty acid metabolism showed the lowest q values, none of metabolic pathways were significantly altered after CA or after CPB resuscitation (Figure 6G and 6H).
Discussion
CA is a time‐sensitive pathology in which many patients succumb to the damage, especially in the brain, that ensues because of prolonged ischemia and the subsequent metabolic alterations.25, 26, 27, 35, 36 We applied a global metabolomics analysis on 4 major organs, the brain, heart, kidney, and liver, after 20 minutes of CA, or 20 minutes of CA followed by 30 minutes of CPB resuscitation in our rat model of asphyxia‐induced cardiac arrest. This severe injury model mimics the metabolic status and pathological injury of a majority of human CA patients.25, 36, 37 Our analysis demonstrates 3 novel findings: (1) the brain metabolome is more sensitive to alterations during resuscitation than during ischemia, illustrating the necessity of resuscitation protocols to focus on the brain; (2) each organ has a unique metabolic profile, which provides insight on organ‐specific targets for future resuscitation therapies; and (3) the strengths of a global metabolomics analysis supersedes that of isolated organs, which has been used by prior studies. Therefore, by improving current resuscitation methods with an emphasis on alleviating individual organ metabolome alterations, particularly in the brain, we can not only increase the survival of patients suffering from major brain damage after CA but also improve the survival of patients who are unable to immediately achieve ROSC.
People affected by longer durations of CA are greater in prevalence and have a worse survival rate than those suffering from a shorter duration. For this study, we have chosen 20 minutes of CA, which results in the death of rats within 18 hours, exemplifying the severity of this injury. This represents a majority of CA patients, who die within a day after achieving ROSC. Additionally, omega‐3 fatty acid treatment improved rat survival after 20 minutes of CA38 indicating that this degree of injury, although severe, still retains the potential of being a treatable injury. Traditionally, the longer durations of CA are not amenable to resuscitation or treatment. However, we have created this model of 20 minutes of CA that closely mimics a majority of human patients. This model is valuable for examining the key metabolic pathways that may be related to improving survival as key therapeutic targets.
The brain's metabolic pattern suggests an increased susceptibility during reperfusion, which is compounded to any damage inflicted during ischemia as seen in Figures 1 and 3A. The pathway analysis confirms minimal changes in the brain metabolome after ischemia and severe changes after resuscitation. This is consistent with our previous report that reperfusion results in only brain mitochondria experiencing cardiolipin alterations supplementing the injury already attained from ischemia.39 The other organs also demonstrated distinct metabolic profiles. The heart and liver normalized the ischemia‐induced alterations of their metabolome after reperfusion, which bolsters our previous data that these organs recover normal mitochondrial activity after reperfusion.40 One important caveat is that the liver displays marginally increased levels of AC species after reperfusion, which is expected because a key function of the liver is to regulate lipid metabolism (Figure 5).41 The liver may be accruing fatty acids after reperfusion for its own metabolic needs, and/or the liver is unable to effectively process and transport fatty acids to other tissues, resulting in an increased accumulation. The kidney displays severe metabolite alterations following 20 minutes of CA, which further worsen following 30 minutes of CPB resuscitation; this serves as evidence that poor recovery of its metabolites to homeostatic levels is consistent with our prior studies showing that the kidney has lower‐than‐normal mitochondrial function following resuscitation.40 The kidney was the only tissue observed to have a significant elevation in TCA metabolites, signifying diminished TCA cycle efficiency and/or turnover. Overall, our data suggest that reperfusion results in more normalized metabolites in the heart and liver but more dysregulated metabolic profiles in the brain and kidney.
Our global metabolomics analysis demonstrates that the brain, heart, kidney, and liver all have broad metabolic alterations responding to ischemia and reperfusion with a distinctive pattern, such that one organ displays a dramatic fluctuation in specific metabolite pools that may not be seen in other organs. This approach is valuable because several metabolites can be simultaneously screened and assessed for significant changes in each organ directly caused by this pathology.42 Many prior studies on the metabolic dysregulation attributable to ischemia/reperfusion have focused on individual tissues, such as inducing ischemia via direct ligation of the blood supply to the kidneys,18, 20, 43, 44, 45 brain,15, 17, 19, 46 heart,16, 47 or liver15, 48, 49, 50 without focusing on the metabolic interplay between organs during ischemia/reperfusion. Direct ligation experiments that induce ischemia to a particular organ are valuable in determining the effects of ischemia/reperfusion. In fact, it has been shown that injury to one organ can cause some alterations in other organs.51 An example of this phenomenon is that a few studies found succinate accumulation in either the plasma, brain, or heart tissues as a consequence of ischemia,19, 52, 53 while another study found that succinate increased universally when the brain, heart, kidney, or liver were individually subjected to ischemia via direct ligation.15 However, our study and one other study showed no such universality.21 Although direct ligation models may also obtain similar metabolite profiles for their individual diseased organs similar to our model, these models tend to have the other organs working in a near‐physiological manner that can serve in a compensatory role to help alleviate any pathological changes that are happening to that one targeted organ. In our global hypoperfusion model, all organs are being affected simultaneously in a closed system, an important facet of CA pathology, which suppresses the aforementioned compensatory role. This makes our study unique because it mimics the pathophysiological state of human cardiac arrest.54Therefore, our data obtained from a simultaneous assessment of metabolite alterations in various tissues in whole‐body ischemia provide vital information to assess the efficiency of current resuscitation methods on the basis of simply resuming blood circulation.
A limitation in this study is that we do not address any causative or correlative interactions between different metabolites, nor the role of each metabolite for CA pathology. Additionally, an altered metabolic pathway may have a significant functional impact in one organ but not in other organs. In fact, there may be regional specificity in individual organs for a particular pathway, as organs are not homogenous; for our analysis, we used whole organ samples. In this light, the extent of metabolic alterations may not directly correlate with the overall functional damage of each organ. Therefore, the important question about why the alterations in the brain and kidney become worse but normalize in the heart and liver cannot be answered simply by pattern analysis. The complex metabolic alterations may be caused by dysfunction of the mitochondria, which serve as the major site of energy generation. Energy generation is needed for normal physiological functions and is required for repair.55 Energy‐producing pathways are heavily affected following ischemia/reperfusion.56, 57 Furthermore, we have previously shown that decreased mitochondrial function correlated with poor organ function outcomes, particularly in the brain,40 which is also consistent with the metabolic alterations seen in Figures 3 and 4, which we organized on the basis of mitochondrial metabolism. As such, targeting the mitochondria may be a good first step in helping to identify and rescue the organ metabolic dysfunction after ischemia/reperfusion. Further studies that manipulate certain pathways using interventions such as hypothermia are required to understand the interactions between metabolites, the significance of alterations in each metabolite in individual organs, and how these alterations further affect other organs.
Despite these limitations, we found that resuscitation‐driven, not ischemia‐induced, alterations are the main cause for the overall brain metabolic alterations in these resuscitated rats. The worsening of brain metabolome after resuscitation implicates reperfusion as the major cause for brain damage and poor neurological function in these animals. This phenomenon may be extrapolated to human patients who clinically do not have neurological function when exposed to long CA times even after resuscitation, providing the impetus for modifying current resuscitation methodologies to alleviate reperfusion brain injury.57, 58 For this purpose, metabolic alterations observed in our study may provide potential target pathways for novel organ‐specific pharmacological therapies.59 Additionally, metabolic alterations found in the kidney may also be important for brain function. Kidney injury can result in changes in the blood‐brain barrier and neurotransmitter concentrations, possibly potentiating neural damage.60 Once brain injury is mitigated, the recovery of kidney function may be the driving factor in long‐term survival. This illustrates that the hypoperfusion from CA not only elicits damage in individual organs but also directly impacts the relationship between the organs, increasing the complexity of developing therapeutic strategies to treat CA.
In conclusion, determining global metabolic alterations in individual organs is valuable for understanding the metabolic status of patients with CA after resuscitation. Moreover, following the metabolic alterations that arise during CA and how these alterations further change responding to resuscitation is necessary to reevaluate the effects of current resuscitation protocols, which mainly depends on the restoration of blood circulation. A novel finding in our comprehensive metabolic analysis is that the brain, heart, kidney, and liver all undergo metabolic alterations following 20 minutes of CA, with the brain and liver less severe than the heart and kidney. However, 30 minutes of CPB resuscitation rescued metabolic alterations in the heart and liver but caused additional metabolic alterations in the brain and kidney. The more severe metabolome of the brain provides the basis for poor neurological outcome after prolonged CA. Overall, our results clearly show a critical limitation of current resuscitation protocols. We clearly show the requirement for additional interventions in future studies that are targeted to the individual organs to mitigate reperfusion injury and ultimately improve neurological function and overall survival.
Sources of Funding
This work was supported by the National Institutes of Health grant (S10RR027878).
Disclosures
None.
Supporting information
Table S1. Pathway Analysis of Brain Tissue Comparing Control With Cardiac Arrest
Table S2. Pathway Analysis of Brain Tissue Comparing Control With CPB Resuscitation
Table S3. Pathway Analysis of Heart Tissue Comparing Control With Cardiac Arrest
Table S4. Pathway Analysis of Heart Tissue Comparing Control With CPB Resuscitation
Table S5. Pathway Analysis of Kidney Tissue Comparing Control With Cardiac Arrest
Table S6. Pathway Analysis of Kidney Tissue Comparing Control With CPB Resuscitation
Table S7. Pathway Analysis of Liver Tissue Comparing Control With Cardiac Arrest
Table S8. Pathway Analysis of Liver Tissue Comparing Control With CPB Resuscitation
(J Am Heart Assoc. 2019;8:e012809 DOI: 10.1161/JAHA.119.012809.)
References
- 1. Stub D, Bernard S, Duffy SJ, Kaye DM. Post cardiac arrest syndrome: a review of therapeutic strategies. Circulation. 2011;123:1428–1435. [DOI] [PubMed] [Google Scholar]
- 2. Madl C, Holzer M. Brain function after resuscitation from cardiac arrest. Curr Opin Crit Care. 2004;10:213–217. [DOI] [PubMed] [Google Scholar]
- 3. Allen BS, Buckberg GD. Studies of isolated global brain ischaemia: I. Overview of irreversible brain injury and evolution of a new concept—redefining the time of brain death. Eur J Cardiothorac Surg. 2012;41:1132–1137. [DOI] [PubMed] [Google Scholar]
- 4. Cummins RO. From concept to standard‐of‐care? Review of the clinical experience with automated external defibrillators. Ann Emerg Med. 1989;18:1269–1275. [DOI] [PubMed] [Google Scholar]
- 5. Jentzer JC, Chonde MD, Dezfulian C. Myocardial dysfunction and shock after cardiac arrest. Biomed Res Int. 2015;2015:314796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callaway CW, Donnino MW, Fink EL, Geocadin RG, Golan E, Kern KB, Leary M, Meurer WJ, Peberdy MA, Thompson TM, Zimmerman JL. Part 8: post‐cardiac arrest care: 2015 American Heart Association guidelines update for cardiopulmonary resuscitation and emergency cardiovascular care. Circulation. 2015;132:S465–S482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang CH, Tsai MS, Chien KL, Chang WT, Wang TD, Chen SC, Ma MH, Hsu HY, Chen WJ. Predicting the outcomes for out‐of‐hospital cardiac arrest patients using multiple biomarkers and suspension microarray assays. Sci Rep. 2016;6:27187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nielsen N, Wetterslev J, Cronberg T, Erlinge D, Gasche Y, Hassager C, Horn J, Hovdenes J, Kjaergaard J, Kuiper M, Pellis T, Stammet P, Wanscher M, Wise MP, Aneman A, Al‐Subaie N, Boesgaard S, Bro‐Jeppesen J, Brunetti I, Bugge JF, Hingston CD, Juffermans NP, Koopmans M, Kober L, Langorgen J, Lilja G, Moller JE, Rundgren M, Rylander C, Smid O, Werer C, Winkel P, Friberg H; TTM Trial Investigators . Targeted temperature management at 33 degrees C versus 36 degrees C after cardiac arrest. N Engl J Med. 2013;369:2197–2206. [DOI] [PubMed] [Google Scholar]
- 9. Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1:a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newgard CB. Metabolomics and metabolic diseases: where do we stand? Cell Metab. 2017;25:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim SJ, Kim SH, Kim JH, Hwang S, Yoo HJ. Understanding metabolomics in biomedical research. Endocrinol Metab (Seoul). 2016;31:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trivedi DK, Hollywood KA, Goodacre R. Metabolomics for the masses: the future of metabolomics in a personalized world. New Horiz Transl Med. 2017;3:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodi V, Marrachelli VG, Husser O, Chorro FJ, Vina JR, Monleon D. Metabolomics in the diagnosis of acute myocardial ischemia. J Cardiovasc Transl Res. 2013;6:808–815. [DOI] [PubMed] [Google Scholar]
- 15. Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa ASH, Brookes PS, Davidson SM, Duchen MR, Saeb‐Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, Murphy MP. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heather LC, Pates KM, Atherton HJ, Cole MA, Ball DR, Evans RD, Glatz JF, Luiken JJ, Griffin JL, Clarke K. Differential translocation of the fatty acid transporter, FAT/CD36, and the glucose transporter, GLUT4, coordinates changes in cardiac substrate metabolism during ischemia and reperfusion. Circ Heart Fail. 2013;6:1058–1066. [DOI] [PubMed] [Google Scholar]
- 17. Irie M, Fujimura Y, Yamato M, Miura D, Wariishi H. Integrated MALDI‐MS imaging and LC‐MS techniques for visualizing spatiotemporal metabolomic dynamics in a rat stroke model. Metabolomics. 2014;10:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Q, Xiao X, Fogle P, Dong Z. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PLoS ONE. 2014;9:e106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang T, Wang W, Huang J, Liu X, Zhang H, Zhang N. Metabolomic investigation of regional brain tissue dysfunctions induced by global cerebral ischemia. BMC Neurosci. 2016;17:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jouret F, Leenders J, Poma L, Defraigne JO, Krzesinski JM, de Tullio P. Nuclear magnetic resonance metabolomic profiling of mouse kidney, urine and serum following renal ischemia/reperfusion injury. PLoS ONE. 2016;11:e0163021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wijermars LG, Schaapherder AF, Kostidis S, Wust RC, Lindeman JH. Succinate accumulation and ischemia‐reperfusion injury: of mice but not men, a study in renal ischemia‐reperfusion. Am J Transplant. 2016;16:2741–2746. [DOI] [PubMed] [Google Scholar]
- 22. Jiang H, Meng F, Li W, Tong L, Qiao H, Sun X. Splenectomy ameliorates acute multiple organ damage induced by liver warm ischemia reperfusion in rats. Surgery. 2007;141:32–40. [DOI] [PubMed] [Google Scholar]
- 23. Brucken A, Derwall M, Bleilevens C, Stoppe C, Gotzenich A, Gaisa NT, Weis J, Nolte KW, Rossaint R, Ichinose F, Fries M. Brief inhalation of nitric oxide increases resuscitation success and improves 7‐day‐survival after cardiac arrest in rats: a randomized controlled animal study. Crit Care. 2015;19:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hossmann KA, Oschlies U, Schwindt W, Krep H. Electron microscopic investigation of rat brain after brief cardiac arrest. Acta Neuropathol. 2001;101:101–113. [DOI] [PubMed] [Google Scholar]
- 25. Kim J, Lampe JW, Yin T, Shinozaki K, Becker LB. Phospholipid alterations in the brain and heart in a rat model of asphyxia‐induced cardiac arrest and cardiopulmonary bypass resuscitation. Mol Cell Biochem. 2015;408:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han F, Boller M, Guo W, Merchant RM, Lampe JW, Smith TM, Becker LB. A rodent model of emergency cardiopulmonary bypass resuscitation with different temperatures after asphyxial cardiac arrest. Resuscitation. 2010;81:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patil KD, Halperin HR, Becker LB. Cardiac arrest: resuscitation and reperfusion. Circ Res. 2015;116:2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nichol G, Karmy‐Jones R, Salerno C, Cantore L, Becker L. Systematic review of percutaneous cardiopulmonary bypass for cardiac arrest or cardiogenic shock states. Resuscitation. 2006;70:381–394. [DOI] [PubMed] [Google Scholar]
- 29. Morimura N, Sakamoto T, Nagao K, Asai Y, Yokota H, Tahara Y, Atsumi T, Nara S, Hase M. Extracorporeal cardiopulmonary resuscitation for out‐of‐hospital cardiac arrest: a review of the Japanese literature. Resuscitation. 2011;82:10–14. [DOI] [PubMed] [Google Scholar]
- 30. Chen YS, Yu HY, Huang SC, Lin JW, Chi NH, Wang CH, Wang SS, Lin FY, Ko WJ. Extracorporeal membrane oxygenation support can extend the duration of cardiopulmonary resuscitation. Crit Care Med. 2008;36:2529–2535. [DOI] [PubMed] [Google Scholar]
- 31. Shin TG, Choi JH, Jo IJ, Sim MS, Song HG, Jeong YK, Song YB, Hahn JY, Choi SH, Gwon HC, Jeon ES, Sung K, Kim WS, Lee YT. Extracorporeal cardiopulmonary resuscitation in patients with inhospital cardiac arrest: a comparison with conventional cardiopulmonary resuscitation. Crit Care Med. 2011;39:1–7. [DOI] [PubMed] [Google Scholar]
- 32. Kim J, Yin T, Yin M, Zhang W, Shinozaki K, Selak MA, Pappan KL, Lampe JW, Becker LB. Examination of physiological function and biochemical disorders in a rat model of prolonged asphyxia‐induced cardiac arrest followed by cardio pulmonary bypass resuscitation. PLoS ONE. 2014;9:e112012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDougall M, Choi J, Kim HK, Bobe G, Stevens JF, Cadenas E, Tanguay R, Traber MG. Lethal dysregulation of energy metabolism during embryonic vitamin E deficiency. Free Radic Biol Med. 2017;104:324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoin CS, Knotts TA, Adams SH. Acylcarnitines–old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11:617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gilmore CM, Rea TD, Becker LJ, Eisenberg MS. Three‐phase model of cardiac arrest: time‐dependent benefit of bystander cardiopulmonary resuscitation. Am J Cardiol. 2006;98:497–499. [DOI] [PubMed] [Google Scholar]
- 36. Cronberg T, Brizzi M, Liedholm LJ, Rosen I, Rubertsson S, Rylander C, Friberg H. Neurological prognostication after cardiac arrest–recommendations from the Swedish resuscitation council. Resuscitation. 2013;84:867–872. [DOI] [PubMed] [Google Scholar]
- 37. Katz L, Ebmeyer U, Safar P, Radovsky A, Neumar R. Outcome model of asphyxial cardiac arrest in rats. J Cereb Blood Flow Metab. 1995;15:1032–1039. [DOI] [PubMed] [Google Scholar]
- 38. Kim J, Yin T, Shinozaki K, Lampe JW, Becker LB. DHA‐supplemented diet increases the survival of rats following asphyxia‐induced cardiac arrest and cardiopulmonary bypass resuscitation. Sci Rep. 2016;6:36545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tam J, Hong A, Naranjo PM, Yin T, Shinozaki K, Lampe JW, Becker LB, Kim J. The role of decreased cardiolipin and impaired electron transport chain in brain damage due to cardiac arrest. Neurochem Int. 2018;120:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J, Villarroel JP, Zhang W, Yin T, Shinozaki K, Hong A, Lampe JW, Becker LB. The responses of tissues from the brain, heart, kidney, and liver to resuscitation following prolonged cardiac arrest by examining mitochondrial respiration in rats. Oxid Med Cell Longev. 2016;2016:7463407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ponziani FR, Pecere S, Gasbarrini A, Ojetti V. Physiology and pathophysiology of liver lipid metabolism. Expert Rev Gastroenterol Hepatol. 2015;9:1055–1067. [DOI] [PubMed] [Google Scholar]
- 42. McGarrah RW, Crown SB, Zhang GF, Shah SH, Newgard CB. Cardiovascular metabolomics. Circ Res. 2018;122:1238–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baranovicova E, Grendar M, Kalenska D, Tomascova A, Cierny D, Lehotsky J. Nmr metabolomic study of blood plasma in ischemic and ischemically preconditioned rats: an increased level of ketone bodies and decreased content of glycolytic products 24 h after global cerebral ischemia. J Physiol Biochem. 2018;74:417–429. [DOI] [PubMed] [Google Scholar]
- 44. Huang H, van Dullemen LFA, Akhtar MZ, Faro ML, Yu Z, Valli A, Dona A, Thezenas ML, Charles PD, Fischer R, Kaisar M, Leuvenink HGD, Ploeg RJ, Kessler BM. Proteo‐metabolomics reveals compensation between ischemic and non‐injured contralateral kidneys after reperfusion. Sci Rep. 2018;8:8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malagrino PA, Venturini G, Yogi PS, Dariolli R, Padilha K, Kiers B, Gois TC, Motta‐Leal‐Filho JM, Takimura CK, Girardi ACC, Carnevale FC, Canevarolo R, Malheiros D, de Mattos Zeri AC, Krieger JE, Pereira AC. Metabolomic characterization of renal ischemia and reperfusion in a swine model. Life Sci. 2016;156:57–67. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y, Wang YG, Ma TF, Li M, Gu SL. Dynamic metabolites profile of cerebral ischemia/reperfusion revealed by (1)h NMR‐based metabolomics contributes to potential biomarkers. Int J Clin Exp Pathol. 2014;7:4067–4075. [PMC free article] [PubMed] [Google Scholar]
- 47. Nadtochiy SM, Urciuoli W, Zhang J, Schafer X, Munger J, Brookes PS. Metabolomic profiling of the heart during acute ischemic preconditioning reveals a role for SIRT1 in rapid cardioprotective metabolic adaptation. J Mol Cell Cardiol. 2015;88:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruinsma BG, Sridharan GV, Weeder PD, Avruch JH, Saeidi N, Ozer S, Geerts S, Porte RJ, Heger M, van Gulik TM, Martins PN, Markmann JF, Yeh H, Uygun K. Metabolic profiling during ex vivo machine perfusion of the human liver. Sci Rep. 2016;6:22415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen L, Luo Z, Fu W, Liao X, Cui Z, Zhou J. Detection of urinary metabolomics before and after pringle maneuver‐induced liver ischemia and reperfusion injury in rats using gas chromatography‐mass spectrometry. Dis Markers. 2013;35:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hrydziuszko O, Perera MT, Laing R, Kirwan J, Silva MA, Richards DA, Murphy N, Mirza DF, Viant MR. Mass spectrometry based metabolomics comparison of liver grafts from donors after circulatory death (DCD) and donors after brain death (DBD) used in human orthotopic liver transplantation. PLoS ONE. 2016;11:e0165884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fox BM, Gil HW, Kirkbride‐Romeo L, Bagchi RA, Wennersten SA, Haefner KR, Skrypnyk NI, Brown CN, Soranno DE, Gist KM, Griffin BR, Jovanovich A, Reisz JA, Wither MJ, D'Alessandro A, Edelstein CL, Clendenen N, McKinsey TA, Altmann C, Faubel S. Metabolomics assessment reveals oxidative stress and altered energy production in the heart after ischemic acute kidney injury in mice. Kidney Int. 2019;95:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Varvarousis D, Xanthos T, Ferino G, Noto A, Iacovidou N, Mura M, Scano P, Chalkias A, Papalois A, De‐Giorgio F, Baldi A, Mura P, Staikou C, Stocchero M, Finco G, d'Aloja E, Locci E. Metabolomics profiling reveals different patterns in an animal model of asphyxial and dysrhythmic cardiac arrest. Sci Rep. 2017;7:16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang J, Wang YT, Miller JH, Day MM, Munger JC, Brookes PS. Accumulation of succinate in cardiac ischemia primarily occurs via canonical KREBS cycle activity. Cell Rep. 2018;23:2617–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Papadimitriou D, Xanthos T, Dontas I, Lelovas P, Perrea D. The use of mice and rats as animal models for cardiopulmonary resuscitation research. Lab Anim‐Uk. 2008;42:265–276. [DOI] [PubMed] [Google Scholar]
- 55. Molnar JA, Underdown MJ, Clark WA. Nutrition and chronic wounds. Adv Wound Care (New Rochelle). 2014;3:663–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elia M. Insights into energy requirements in disease. Public Health Nutr. 2005;8:1037–1052. [DOI] [PubMed] [Google Scholar]
- 57. Roberts BW, Kilgannon JH, Chansky ME, Mittal N, Wooden J, Parrillo JE, Trzeciak S. Multiple organ dysfunction after return of spontaneous circulation in postcardiac arrest syndrome. Crit Care Med. 2013;41:1492–1501. [DOI] [PubMed] [Google Scholar]
- 58. Geocadin RG, Koenig MA, Jia X, Stevens RD, Peberdy MA. Management of brain injury after resuscitation from cardiac arrest. Neurol Clin. 2008;26:487–506, ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Senn T, Hazen SL, Tang WH. Translating metabolomics to cardiovascular biomarkers. Prog Cardiovasc Dis. 2012;55:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nongnuch A, Panorchan K, Davenport A. Brain‐kidney crosstalk. Crit Care. 2014;18:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Pathway Analysis of Brain Tissue Comparing Control With Cardiac Arrest
Table S2. Pathway Analysis of Brain Tissue Comparing Control With CPB Resuscitation
Table S3. Pathway Analysis of Heart Tissue Comparing Control With Cardiac Arrest
Table S4. Pathway Analysis of Heart Tissue Comparing Control With CPB Resuscitation
Table S5. Pathway Analysis of Kidney Tissue Comparing Control With Cardiac Arrest
Table S6. Pathway Analysis of Kidney Tissue Comparing Control With CPB Resuscitation
Table S7. Pathway Analysis of Liver Tissue Comparing Control With Cardiac Arrest
Table S8. Pathway Analysis of Liver Tissue Comparing Control With CPB Resuscitation


