LONG ABSTRACT:
M (microfold) cells of the intestine function to transport antigen from the apical lumen to the underlying Peyer’s patches and lamina propria where immune cells reside and therefore contribute to mucosal immunity in the intestine. A complete understanding of how M cells differentiate in the intestine as well as the molecular mechanisms of antigen uptake by M cells is lacking. This is because M cells are a rare population of cells in the intestine and because in vitro models for M cells are not robust. The discovery of a self-renewing stem cell culture system of the intestine, termed enteroids, has provided new possibilities for culturing M cells. Enteroids are advantageous over standard cultured cell lines because they can be differentiated into several major cell types found in the intestine, including goblet cells, Paneth cells, enteroendocrine cells and enterocytes. The cytokine RANKL is essential in M cell development, and addition of RANKL and TNF-α to culture media promotes a subset of cells from ileal enteroids to differentiate into M cells. The following protocol describes a method for the differentiation of M cells in a transwell epithelial polarized monolayer system of the intestine using human ileal enteroids. This method can be applied to the study of M cell development and function.
Keywords: ileal enteroids, stem cells, polarized epithelial monolayers, Transwells, M cells, immunofluorescence, goblet cells, RANKL, GP2, SpiB, lgA, Lgr5
SHORT ABSTRACT:
This protocol describes how to induce the differentiation of M cells in human stem cell-derived ileal monolayers and methods to assess their development.
INTRODUCTION:
M (microfold) cells are specialized intestinal epithelial cells found primarily in the follicle associated epithelium (FAE) of the intestine overlying small lymphoid regions termed Peyer’s patches1. M cells have short irregular apical microvilli and are deeply invaginated on their basolateral side, which allows immune cells to reside closely to their cell body2. This unique morphology enables M cells to sample antigen from the apical lumen of the intestine and deliver it directly to the underlying immune cells2. In this way, M cells are important for immune surveillance in the intestine but can also be exploited by pathogens for entry into the lamina propria1–7.
The study of M cells has been hindered by several factors. First, M cells are found at a low frequency in the mouse and human intestine8. In cultured cells systems, M cell-like cells have been induced to differentiate by co-culturing a polarized adenocarcinoma cell line, Caco-2, with either B lymphocytes from mouse Peyer’s patches or the B cell lymphoma cell line, Raji B9,10. This results in a subset of Caco-2 cells that express the M cell markers Sialyl Lewis A antigen and UEA-1 in the polarized epithelium9,10. (These markers are also expressed on goblet cells in intestinal tissues, so nowadays are less frequently used as definitive M cell markers11,12.) This Caco-2-M cell system has been used to study particle uptake and bacteria translocation13,14. However, Caco-2 cells are an established cell line from a large intestinal adenocarcinoma with the confounding factor that different sources of Caco-2 cells display different phenotypes among labs15. Further, they may not fully recapitulate the transcription levels of true M cells, as they lack expression of currently known M cell markers GP2 and SpiB16. Therefore, additional and more physiologically relevant culture models are needed to be able to study M cell development and functions.
Within the past ten years, the field of enteroid-derived model systems of the intestine has rapidly been progressing forward from the initial discovery that intestinal stem cells derived from human intestinal biopsy could self-propagate and self-renew in culture17,18. Importantly, removal of stem cell promoting factors from the growth media allows these stem cell cultures to differentiate into the many cell types found in the intestine18. Furthermore, recent work suggests the importance of RANKL-RANK signaling in M cell development in the intestine19,20. The RANK receptor is a member of the TNF family of receptors that is expressed on epithelial precursor cells in the intestine19 while RANKL (the RANK receptor ligand) is released by stromal cells of the Peyer’s patches20. Since the epithelial cell types present in ileal enteroids do not produce RANKL, M cell differentiation in ileal enteroid cultures can be induced by the addition of RANKL to the culture media21,22. Inclusion of TNFα in the culture media helps support M cell development in ileal enteroids23. Here, we describe the methods for inducing differentiation of M cells in intestinal monolayers derived from human ileal enteroids. Our methods are based in part on modifications from the following protocols21–23.
PROTOCOL:
All methods described here have been approved by the Tufts University IBC and IRB.
-
Inducing M cell differentiation in human ileal enteroid-derived monolayers
NOTE: This protocol uses ileal enteroids derived from human tissue biopsy. Please refer to published protocols for methods on how to grow and passage these cells18,24. The following methods for developing monolayers were adapted from Zou et al24. Methods for inducing M cells in ileal enteroid-derived cultures were adapted from previous reports21–23. All work is carried out in a sterile tissue culture hood and incubations are in hood or tissue culture incubator as indicated. See Table of Materials needed to prepare ileal enteroid monolayers and various medias.-
1.1Grow ileal enteroids for 4–10 days in extracellular matrix (ECM) (see Table of Materials) (Figure 1), depending on their intrinsic growth rates, before seeding onto transwells.
-
1.2Coating transwell membranes
-
1.2.1Place desired number of transwells in a 24-well plate creating a two-chamber system.
-
1.2.2Dilute ECM 25-fold in cold sterile phosphate-buffered saline (PBS) and add 100 μL of cold diluted solution into each upper chamber onto the membrane.NOTE: ECM and diluted ECM solution must be kept on ice until immediately before addition.
-
1.2.3Cover the 24-well plate with lid and place the plate into a tissue culture incubator at 37 °C for 2 h to permit ECM solidification on the membrane.
-
1.2.4After 2 h, remove the plate from the incubator and place in a tissue culture hood. Using sterile tweezers, invert each transwell to gently remove remaining solution. Allow the membranes to airdry in hood with the lid open while cells are being collected (Steps 1.3.1–1.3.11).
-
1.2.1
-
1.3Dissociating the ileal enteroids into single cells
-
1.3.1Remove the plate of ileal enteroids from the incubator and gently remove the culture media from each well by vacuum aspiration or with a pipette.NOTE: One well of ileal enteroids containing approximately 100 healthy cysts is sufficient to seed 1.5–2 wells.
-
1.3.2Add 500 μL of ice cold 0.5 mM ethylenediaminetetraacetic acid (EDTA) to each well containing ileal enteroids suspended in ECM to break up the ECM. Pipette up and down vigorously with a P1000 pipettor set at 500 μL to break up ECM thereby releasing ileal enteroids into the solution. To improve dissolution of ECM, after pipetting, shake the plate vigorously at 4 °C for 30 min.
-
1.3.3Collect the solution from each well into 15 mL conical tubes.NOTE: Collect up to 10 wells per 15 mL conical tube for optimal single cell collection.
-
1.3.4Pellet the cells in a centrifuge at 140 × g and 4 °C for 5 min. Pellet should be visible but can easily be dislodged, so slowly remove the supernatant by vacuum aspiration or with a pipette.NOTE: If concerned about loss of pellet and cells, use a pipette and save the supernatant in a separate tube.
-
1.3.5To digest tight junction linkages and break up the ileal enteroids into single cells, resuspend the pellet in 500 μL of room temperature trypsin per every 5 wells collected in step 1.3.3. Using a P1000, pipette up and down to disaggregate the clumps and incubate the tubes in a 37 °C water bath for 5 min or less.NOTE: Optimization is needed to determine the appropriate amount of time required to incubate the tubes so that the cells are broken up but not over-trypsinized to the point that they die. Use Trypan blue in step 1.3.9 to ensure that the cells are viable after trypsin treatment.
-
1.3.6Add 1 mL of Advanced DMEM/F12 with 10% Fetal Bovine Serum (FBS) per 500 μL of trypsin to inactivate the trypsin.
-
1.3.7Pipette up and down with a P1000 set at 500 μL at least 50 times against the side of the conical tube to further disaggregate remaining clumps into single cells.
-
1.3.8Place a 40 μm cell strainer over a 50 mL conical and add 1 mL of Advanced DMEM/F12 with 10% FBS to wet the cell strainer. Pipette the single cell suspension from the 15 mL conical onto the strainer. Wash the strainer with 1 mL of Advanced DMEM/F12 with 10% FBS.
-
1.3.9Transfer the cells that went through the cell strainer from the 50 mL conical into a new 15 mL conical tube. During the centrifugation step 1.3.10, the cellular pellet will be more easily seen in a 15 mL conical tube. Count the cells using a hemocytometer. Use Trypan blue to verify that cells are still alive. Typically, >95% viability is observed.
-
1.3.10While counting the cells, centrifuge the cells in the new 15 mL tube at 400 × g and room temperature for 5 min. Cell pellet should be visible. Carefully remove the supernatant with a pipette, again saving the supernatant in case the pellet becomes dislodged.
-
1.3.11Prepare modified complete growth media25 (MCMGF+ media) supplemented with 10 μM Y-27632. Resuspend pelleted cells at 2.5 × 105 cells/200 μL in MCMGF+. See remarks in discussion about optimizing cell seeding number.NOTE: MCMGF+ media is Advanced DMEM/F12 with 75% L-Wnt3a conditioned media, 10% R-spondin conditioned media, 5% Noggin conditioned media, 1x B27 Supplement, 1x N2 Supplement, 1 mM N-acetylcysteine, 50 ng/mL mouse recombinant EGF, 500 nM A-8301, 10 nM [Leu15]-Gastrin I, 10 mM HEPES, 2 mM GlutaMAX, and 1x Penicillin/Streptomycin (optional).
-
1.3.12Ensure that the ECM-coated membranes prepared in step 1.2 have fully dried, as assessed by eye. Wash the upper chamber with 200 μL of MCMGF+. Add 200 μL of cell solution into each upper chamber.
-
1.3.13Add 700 μL of MCMGF+ with 10 μM Y-27632 to each lower chamber. Place the plate in a 37 °C tissue culture incubator with 5% CO2.
-
1.3.14After 1 day of growth, remove the media from the upper chamber and replace with 200 μL of fresh MCMGF+, to prevent growth of multiple cell layers.
-
1.3.1
-
1.4Replacing medium
-
1.4.1Once monolayers are ~80% confluent, usually between days 1–3 post-seeding, replace basolateral media with differentiation media (DM) for control wells (see step 1.4.2 for more detail) or with M cell media for M cell induction wells (see step 1.4.3 for more detail). Replace the media in upper chamber with DM for both conditions.NOTE: DM is Advanced DMEM/F12 with 5% Noggin conditioned media, 1x B27 Supplement, 1x N2 Supplement, 1 mM N-acetylcysteine, 50 ng/mL mouse recombinant EGF, 500 nM A-8301, 10 nM [Leu15]-Gastrin I, 10 mM HEPES Buffer, 2 mM GlutaMAX, and 1x Penicillin/Streptomycin (optional). M cell media is DM supplemented with 200 ng/mL RANKL and 50 ng/mL TNFα.
-
1.4.2For control wells that should not contain M cells, add 200 μL of DM to the upper chamber and 700 μL DM to the bottom chamber.
-
1.4.3To induce M cells, add 200 μL of DM to the upper chamber and 700 μL of M cell media to the bottom chamber.
-
1.4.4Replace the media every 2 days. For control wells, replace DM in the upper and lower chambers. For M cell wells, replace DM in the upper chamber and M cell media in the lower chamber.NOTE: By day 7 post cell-seeding, M cells are fully induced in the monolayers.
-
1.4.1
-
1.1
-
Verifying M cell differentiation by qRT-PCR
NOTE: Perform the following work at a sterile RNAse-free bench space. See Table of Materials for a list of preferred materials for qRT-PCR.-
2.1Remove the media from upper and bottom chambers and wash the upper chamber 2x gently with 300 μL of room temperature PBS.
-
2.2Add 300 μL of Trizol to each upper chamber. Incubate at room temperature for 5 min.CAUTION: Wear gloves and eye protection when using Trizol to avoid contact with skin as indicated in manufacturer’s instructions.
-
2.3Meanwhile, label microcentrifuge tubes for each well and add 700 μL of Trizol to each tube.
-
2.4Collect cell homogenate by pipetting up and down 3x gently with a P1000 and transfer the contents into corresponding microcentrifuge tube. Vortex for 5 s to mix.
-
2.5Keep the samples at room temperature for an additional 3 min. Then store at −80 °C for up to one month.
-
2.6Follow standard qRT-PCR methodology for RNA isolation, DNase treatment, Reverse Transcription and qRT-PCR reactions. Refer to primer list in Table of Materials.
-
2.1
-
Verifying M cell differentiation by immunofluorescence
NOTE: Always keep the lower chamber of the plate filled with PBS so that the membranes remain wet. This procedure is performed on the bench. See Table of Materials for a list of preferred materials for immunofluorescence.-
3.1Remove the media from the upper chamber and wash 2x gently with 300 μL of room temperature PBS. Add 100 μL of room temperature 4% PFA in PBS to the upper chamber. Cover the plate with foil and let stand for 25 min at room temperature. Remove 4% PFA.CAUTION: 4% PFA should be properly disposed of as hazardous chemical waste.
-
3.2Wash the upper chamber 3x with 300 μL of room temperature PBS. At this point, samples can remain at 4 °C for up to a month prior to staining. Once stained, samples should be visualized within a week for best quality images.
-
3.3Incubate the monolayers with 100 μL of 5% Bovine Serum Albumin (BSA) dissolved in PBS for 30 min in dark at room temperature to block the monolayers.
-
3.4Prepare GP2 primary antibody solution in 1% BSA in PBS at a dilution of 1:100. Add 100 μL per well. Stain for 1 h at room temperature in the dark. Remove the solution.NOTE: Do not permeabilize the monolayers before primary stain for GP2 occurs because optimal primary GP2 surface staining of M cells is achieved without permeabilization.
-
3.5Wash the upper chamber 3x times with 300 μL of room temperature PBS.
-
3.6Prepare secondary stain solution of fluorescently tagged goat anti-mouse IgG at 1:200, phalloidin at 1:100 and DAPI in 1% BSA+ 0.1% triton in PBS. Add 100 μL per well. Stain for 30 min at room temperature in the dark.NOTE: Triton is added to the secondary stain solution to permeabilize the cells during this step for proper phalloidin stain.
-
3.7Wash 3x with 300 μL PBS.
-
3.8Place a 5 μL drop of mounting solution (Table of Materials) on a glass slide. Remove the well from the 24 well plate and invert. Carefully cut the membrane from the well using a scalpel. Place the membrane with the cells facing up onto the droplet of mounting solution on the glass slide. Add 10 μL of mounting solution onto the top and center of the membrane and place a coverslip on top to seal the membrane between the glass slide and coverslip.
-
3.9Dry the slides at room temperature in the dark for 24 h. Stained slides should be visualized on confocal microscope within 1-week post-staining.
-
3.1
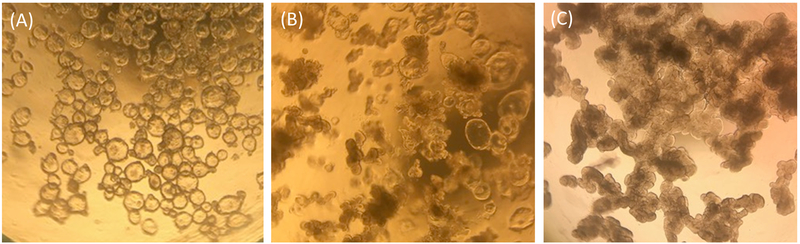
Figure 1. Representative morphology of human ileal enteroids in ECM one-week post-splitting.
(A) Clear and cystic undifferentiated ileal enteroids. (B) Intermediate phenotype with some cystic ileal enteroids and some opaque lobular ileal enteroids. (C) Darkened and lobular differentiated ileal enteroids. Images taken through the lens of an optical light microscope at 4X magnification using an iPhone7 camera.
REPRESENTATIVE RESULTS:
Ileal enteroids grown in ECM are analyzed visually and by qRT-PCR for their relative health status and differentiation states as a means of quality control for ileal enteroid cultures and for use in monolayers. Undifferentiated ileal enteroids grown in ECM appear clear and cystic in morphology, indicating the presence of many stem cells (Figure 1A). Over time, undifferentiated ileal enteroids grown in growth media may take on an intermediate phenotype where some will appear cystic and some appear opaque (Figure 1B). Frequently, our undifferentiated samples resemble those shown in Figure 1B rather than Figure 1A. These intermediate cultures contain more terminally differentiated enterocytes as measured by expression of the enterocyte marker, sucrase isomaltase (SI), and presumably extruded dead enterocytes in the lumen contribute to their dense appearance. Ileal enteroids can be used in this intermediate state for monolayer development, but it must be kept in mind that the quantity of intestinal stem cells present in the cultures may be low, and some differentiated cell types may be present (for example, see qRT-PCR levels in undifferentiated samples grown in ECM resembling Figure 1B in Figure 2). For comparison, ileal enteroids cultured with differentiation media in ECM for 5+ days will appear uniformly darkened and lobular and cultures with this morphology are not good candidates for seeding monolayers (Figure 1C).
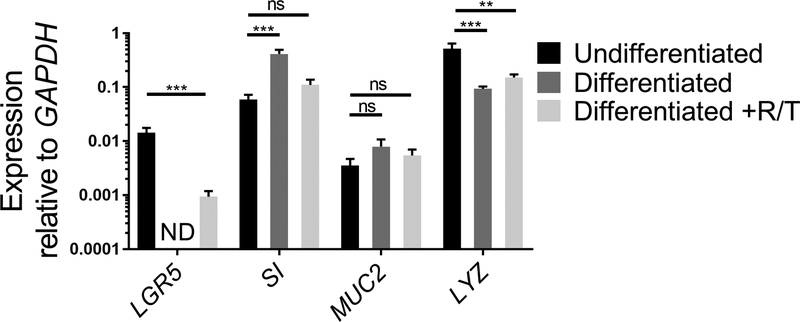
Figure 2. Relative expression of stem cell and differentiation markers of human ileal enteroids grown in ECM or differentiated as monolayers.
Ileal enteroids were grown for 7 days in ECM (Undifferentiated) or grown and differentiated as monolayers without (Differentiated) or with RANKL and TNFα (Differentiated +R/T). Ileal enteroid cultures or monolayers were harvested in Trizol for RNA extraction. Gene expression was determined by qRT-PCR and is expressed relative to GAPDH. Data is average of 3 independent wells of ileal enteroids or monolayers per condition. Error bars indicate SEM. ND is not detected. Statistical significance was determined on log-transformed values using one-way ANOVA with Dunnett’s multiple comparisons test comparing to the Undifferentiated. ** p<0.01, *** p<0.001
Expression of stem cell genes and genes of intestinal cell differentiation can be analyzed by qRT-PCR as another means to assess the health status of ileal enteroids grown in ECM and their differentiation capabilities once seeded as monolayers on transwells. The expression of a stem cell gene, LGR5, an enterocyte gene, SI, a goblet cell gene, MUC2, and a Paneth cell gene, LYZ, is compared between undifferentiated ileal enteroid cultures grown in ECM and differentiated ileal enteroid monolayers in the presence or absence of RANKL/TNFα (Figure 2). While the values may differ between experiments, expression of LGR5 should decrease after differentiation of monolayers18,26. LGR5 expression is usually not detected in the differentiated ileal monolayers without RANKL and TNFα by day 7. Conversely, expression of markers of differentiation of specific cell types, such SI and MUC2, increase after differentiation18. Expression of LYZ generally decreases after differentiation in our cultures. If the ileal enteroid cultures used for making monolayers look more like Figure 1B than Figure 1A, increases in intestinal differentiation markers may be modest after differentiation because these initial cultures are heterogeneous in intestinal cell types and have a higher basal level of SI and MUC2. However, differentiation in monolayers still occurs as assessed by loss of LGR5 expression and microscopy (see below). Furthermore, addition of RANKL and TNFα to the differentiation media reduces the loss of LGR5 expression (Figure 2). In parallel, the expression of SI and MUC2 are slightly lower than in the differentiated condition lacking RANKL and TNFα although their levels increase above the undifferentiated condition.
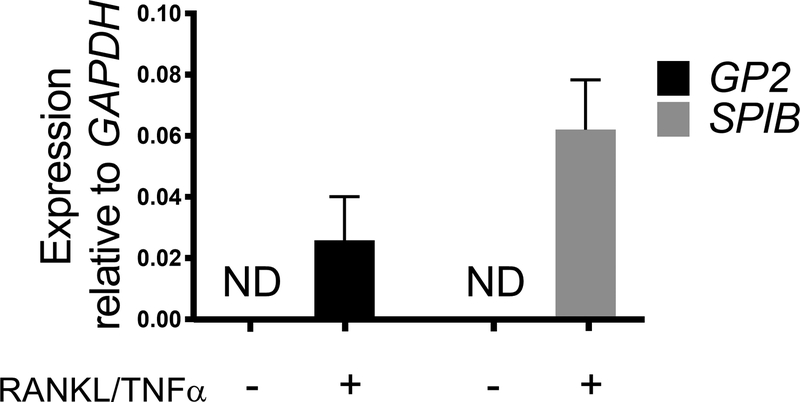
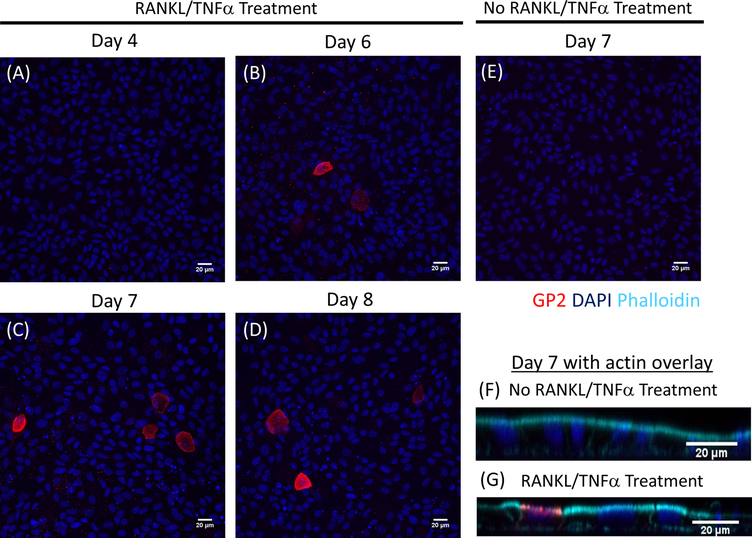
M cell differentiation in monolayers is determined both by qRT-PCR and immunofluorescence using two M cell specific markers including cell surface glycoprotein 2 (GP2) and transcription factor SpiB21. Expression of GP2 and SPIB is upregulated in the ileal enteroid-derived monolayers in the presence of RANKL and TNFα and is not detected in non-RANKL and TNFα treated samples (Figure 3). Expression of these markers can also be normalized to a piece of small bowel tissue22, if available. This permits the fold change of these M cell markers to be compared to tissue that has M cells rather than to control monolayers that have no expression of these markers and allows standardization between experiments in one lab. M cells are also detected by surface expression of GP2 by immunofluorescence (Figure 4). Typically, in a confluent monolayer, 1 to 5 M cells are observed in a given microscope field at 40X magnification by days 6 through 8 post-seeding in samples treated with RANKL and TNFα (Figure 4A–D). No GP2 expression is seen in the untreated samples (Figure 4E). The orthogonal view of the XZ plane overlaid with a phalloidin probe shows actin structures surrounding each cell and GP2 expression on the apical surface of M cells (Figure 4F–G). This model recapitulates the low frequency of M cells found in the human intestine1,2,8. To purify and isolate M cells for further study, M cells can be stained using GP2 surface expression and sorted using FACS for GP2+ cells.
Figure 3. Relative expression of M cell specific markers GP2 and SPIB from human ileal enteroid-derived monolayers.
RANKL/TNFα treated and non-treated human ileal enteroid-derived monolayers were harvested in Trizol for RNA extraction after 7 days post-seeding. Gene expression was determined by qRT-PCR and is expressed relative to GAPDH. Data is average of 6 independent monolayers per condition. Error bars indicate SEM. ND is not detected.
Figure 4. Immunofluorescence of surface GP2 expression on M cells in human ileal enteroid-derived monolayers over time.
RANKL/TNFα treated and non-treated human ileal enteroid-derived monolayers were fixed in 4% PFA and stained for immunofluorescence on various indicated days post-seeding. Images were analyzed using ImageJ software. DAPI = Blue; Glycoprotein 2 (GP2) = Red. (A-D) RANKL/TNFα treated monolayers at various days post-seeding. (E) Non-treated monolayer harvested at day 7 post-seeding. (F-G) Orthogonal XZ plane of monolayers at day 7 post-seeding overlaid with phalloidin probe for F-actin. Phalloidin = Cyan.
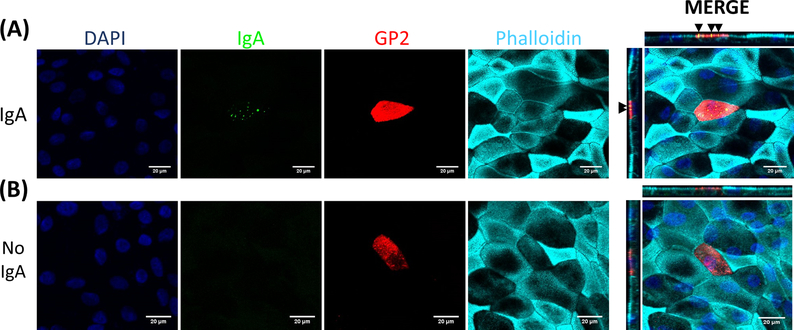
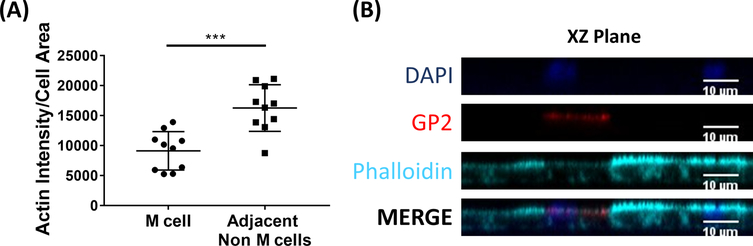
M cells bind to and transport antigen from the intestinal lumen to the immune cells residing beneath the epithelium2. Secretory IgA produced in the intestine binds to bacteria and can bind to the apical surface of M cells to facilitate transport of the microbes27,28. To determine if the M cells developed in this model are able to bind to IgA, human serum IgA is added to the upper chamber, allowed to bind for 1 h, and then the monolayers are prepared for immunofluorescence analysis. The presence of IgA on M cells is visualized using a fluor-conjugated secondary antibody that recognizes the heavy chain of human serum IgA. M cells treated with IgA for 1 h have IgA bound to the apical surface (Figure 5A), whereas M cells in control wells that were only treated with the secondary antibody to IgA have no detectable signal (Figure 5B). Further, IgA specifically binds to the apical surface of M cells and is not found bound to any cells lacking GP2 surface stain. In addition, M cells have characteristically shorter dense actin on their apical surface2. To analyze M cell morphology in this model, ileal enteroid-derived monolayers are grown for 7 days and harvested for immunofluorescence analysis of F-actin using phalloidin. Measurements of actin pixel intensity are calculated for M cells and for non-M cells that are directly adjacent to each M cell using ImageJ software (Figure 6A). Actin intensity is reduced on GP2+ M cells in this model and a representative image is shown in Figure 6B. Overall, the M cells developed in this ileal enteroid-derived monolayer model have characteristic gene expression, morphology and some M cell functions of human intestinal M cells, such as binding to IgA.
Figure 5. IgA binds specifically to the apical surface of M cells.
RANKL/TNFα-treated human ileal enteroid-derived monolayers were grown for 7 days and then (A) treated with 10 μg of human serum IgA for 1 h or (B) mock-treated with PBS only (No IgA control). After 1 h, monolayers were washed 2x in PBS, were fixed in 4% PFA, permeabilized with 0.1% TritonX-100, and stained for immunofluorescence. Images were analyzed using ImageJ software and are representative of 3 independent experiments. DAPI = Blue; Glycoprotein 2 (GP2) = Red; Antibody to human serum IgA = Green; Phalloidin = Cyan. Black arrows denote IgA bound to apical surface of M cell.
Figure 6. M cells have reduced actin intensity compared to adjacent non-M cells.
RANKL/TNFα-treated human ileal enteroid-derived monolayers were grown for 7 days and then fixed in 4% PFA and were stained for immunofluorescence. (A) Using ImageJ, GP2+ M cells were outlined using the Freehand Selection Tool and measurements of Area and Integrated Density were taken in the Phalloidin channel. The same analysis was then completed for each adjacent non-M cell that neighbors the M cell. The Raw Integrated Density was divided by the Area of each individual cell for normalization. The average Integrated Density/Area was calculated for each adjacent non-M cells for each M cell. Images were analyzed from 3 independent experiments; each dot is an M cell or average of neighboring cells. Error bars indicate SD. Statistical significance was determined on log-transformed values using a Paired t test. *** p=0.0001 (B) Representative image of XZ Plane from plot in A. Images were analyzed using ImageJ software. DAPI = Blue; Glycoprotein 2 (GP2) = Red; Phalloidin = Cyan.
DISCUSSION:
To develop monolayers that differentiate properly into the major intestinal cell types and M cells, it is critical to be aware of several factors. Ileal enteroids must be harvested from ECM cultures that are undifferentiated and have a high proportion of Lgr5+ stem cells. Visually, the majority of the ileal enteroids in the ECM cultures should not be darkened and multilobular, and LGR5 expression should be detected in these cultures by qRT-PCR analysis. Quality control of conditioned media is essential for the propagation of undifferentiated cultures over time and must be completed for each batch of conditioned media that is produced. Quality control can be completed by testing a new batch of media on some ECM cultures and comparing the morphology of the ileal enteroids to a previous batch of media over the course of a week. LGR5 expression should remain relatively similar in the ileal enteroid cultures grown in the new batch of media compared to the previous batch.
During preparation of the ileal enteroids for seeding as monolayers, it is important to vigorously pipette the cell solution after incubation with trypsin to break up the ileal enteroids into single cells. Cell clumps can lead to multi-layer formation when seeded for monolayers. In addition, it is essential to empirically determine the number of cells required to form a monolayer for each individual ileal enteroid line that is obtained. Typically, this value can range from 2.5 × 105 – 5.0 × 105 cells/well but depends on the degree of cystic to non-cystic ileal enteroids in cultures and varies for each individual ileal enteroid line. From experience, ileal enteroids grown in ECM that appear less cystic require higher cell seeding density to achieve monolayers. It is advisable to wash the upper chamber after 1 day of growth by gently pipetting the media up and down 2–3 times and replacing with fresh growth media. This process dislodges cells that have landed on top of other cells reducing the likelihood of multi-layer formation. Switching the media in the upper chamber from growth media to M cell media when the monolayers are ~80% confluent, which usually occurs at day 2 post-seeding, helps achieve good M cell differentiation. Addition of RANKL/TNFα to the upper chamber during M cell induction does not lead to the development of a greater number of M cells per monolayer and therefore can be left out of the upper chamber media. Transwells of varying pore sizes can be used in this protocol without affecting M cell development; however, cell seeding density must be optimized for those with larger pore sizes. Collagen IV can be substituted for ECM as a basement membrane protein coating for transwells or well plates which may be better suited for certain applications.
Ileal enteroid-derived monolayers on transwells provide a two-chamber system that allows for the creation of defined apical and basolateral surfaces such that the 4–5 different types of epithelial intestinal cells can polarize to express surface markers on each side relative to that found in the intestine. Additional factors can be added to either side such as particles, infectious agents, or other cell types. However, to date some limitations remain. As described, this system is a static system that lacks physiological flow, intestinal contractions, and intestinal contents. In addition, the villus-crypt architecture is lost by the formation of a flat monolayer. These systems lack Peyer’s patch regions, immune cells, and stromal cells. Whether the lack of immune and stromal cells residing closely underneath M cells affects the invaginations which are not observed in this system and other physiological functioning is an important future area of investigation. This protocol can be adapted to a 96-well plate or a multi-well plate format. The procedure for coating the 96-well plate with ECM and seeding with single cells from ileal enteroids remains the same as for transwells. Titration of the cell seeding density required to obtain monolayers must be done, but typically ranges from 1.0 × 105 – 3.0 ×105 cells/well in a 96-well plate format. M cells are induced by replacing the growth media with M cell media when the monolayers are 80% confluent typically by days 1–3 depending on initial cell seeding density.
This method of differentiating M cells from ileal enteroids in vitro provides significant improvements over the Caco-2 method. The ileal enteroids are primary cells and at least 4–5 epithelial cells types are present in the system. In addition, ileal enteroid lines derived from different people can be studied to investigate how genetics or disease state influence M cell development and behavior. Additional manipulation of the ileal enteroids during M cell differentiation will allow a better understanding of M cell development including characterizing M cell precursor cells. Finally, since the molecular mechanisms of M cell phagocytosis and transcytosis are still not completely understood3,29, this model provides the opportunity to study and visualize antigen and particle uptake by M cells.
Supplementary Material
ACKNOWLEDGMENTS:
This work was supported by NIAID U19AI131126 to Dr. Isberg (Tufts University School of Medicine) and Dr. Kaplan (Tufts University); (JM is Project 2 leader) and NIAID R21AI128093 to JM. ACF was supported in part by NIAID T32AI007077. SEB and MKE were supported by NIAID U19AI116497–05. We thank members of the Mecsas lab, the Ng lab, and Dr. Isberg at Tufts University School of Medicine for useful discussions. The confocal imaging was performed at the Tufts Center for Neuroscience Research, P30 NS047243.
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES:
- 1.Kraehenbuhl JP & Neutra MR Epithelial M cells: differentiation and function. Annual Review of Cell and Developmental Biology. 16 301–332, (2000). [DOI] [PubMed] [Google Scholar]
- 2.Neutra MR, Frey A & Kraehenbuhl JP Epithelial M cells: gateways for mucosal infection and immunization. Cell. 86 (3), 345–348, (1996). [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y, Kimura S & Hase K M cell-dependent antigen uptake on follicle-associated epithelium for mucosal immune surveillance. Inflammation and Regeneration. 38 15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark MA, Hirst BH & Jepson MA M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer’s patch M cells. Infection and Immunity. 66 (3), 1237–1243, (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen VB, Harty JT & Jones BD Interactions of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer’s patches. Infection and Immunity. 66 (8), 3758–3766, (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BD, Ghori N & Falkow S Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. Journal of Experimental Medicine. 180 (1), 15–23, (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marra A & Isberg RR Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer’s patch intestinal epithelium. Infection and Immunity. 65 (8), 3412–3421, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno H Intestinal M cells. Journal of Biochemistry. 159 (2), 151–160, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerneis S, Bogdanova A, Kraehenbuhl JP & Pringault E Conversion by Peyer’s patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 277 (5328), 949–952, (1997). [DOI] [PubMed] [Google Scholar]
- 10.Gullberg E et al. Expression of specific markers and particle transport in a new human intestinal M-cell model. Biochemical and Biophysical Research Communications. 279 (3), 808–813, (2000). [DOI] [PubMed] [Google Scholar]
- 11.Giannasca PJ, Giannasca KT, Leichtner AM & Neutra MR Human intestinal M cells display the sialyl Lewis A antigen. Infection and Immunity. 67 (2), 946–953, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang MH et al. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proceedings of the National Academy of Sciences of the United States of America. 101 (16), 6110–6115, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beloqui A, Brayden DJ, Artursson P, Preat V & des Rieux A A human intestinal M-cell-like model for investigating particle, antigen and microorganism translocation. Nature Protocols. 12 (7), 1387–1399, (2017). [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Argudo I & Jepson MA Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology. 154 (Pt 12), 3887–3894, (2008). [DOI] [PubMed] [Google Scholar]
- 15.Lee JB et al. Quantitative analysis of lab-to-lab variability in Caco-2 permeability assays. European Journal of Pharmaceutics and Biopharmaceutics. 114 38–42, (2017). [DOI] [PubMed] [Google Scholar]
- 16.Mabbott NA, Donaldson DS, Ohno H, Williams IR & Mahajan A Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunology. 6 (4), 666–677, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262–265, (2009). [DOI] [PubMed] [Google Scholar]
- 18.Sato T et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141 (5), 1762–1772, (2011). [DOI] [PubMed] [Google Scholar]
- 19.Knoop KA et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. Journal of Immunology. 183 (9), 5738–5747, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor RT et al. Lymphotoxin-independent expression of TNF-related activation-induced cytokine by stromal cells in cryptopatches, isolated lymphoid follicles, and Peyer’s patches. Journal of Immunology. 178 (9), 5659–5667, (2007). [DOI] [PubMed] [Google Scholar]
- 21.de Lau W et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Molecular and Cellular Biology. 32 (18), 3639–3647, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouch JD et al. Development of Functional Microfold (M) Cells from Intestinal Stem Cells in Primary Human Enteroids. PloS One. 11 (1), e0148216, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood MB, Rios D & Williams IR TNF-alpha augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. American Journal of Physiology: Cell Physiology. 311 (3), C498–507, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou WY et al. Human Intestinal Enteroids: New Models to Study Gastrointestinal Virus Infections. Methods in Molecular Biology. 10.1007/7651_2017_1, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozuka K et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Reports. 9 (6), 1976–1990, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker N et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449 (7165), 1003–1007, (2007). [DOI] [PubMed] [Google Scholar]
- 27.Mantis NJ et al. Selective adherence of IgA to murine Peyer’s patch M cells: evidence for a novel IgA receptor. Journal of Immunology. 169 (4), 1844–1851, (2002). [DOI] [PubMed] [Google Scholar]
- 28.Rios D et al. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunology. 9 (4), 907–916, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller H, Zhang J, Kuolee R, Patel GB & Chen W Intestinal M cells: the fallible sentinels? World Journal of Gastroenterology. 13 (10), 1477–1486, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heijmans J et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Reports. 3 (4), 1128–1139, (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.