Abstract
Research over the past several decades has unmasked a major contribution of disrupted chromatin regulatory processes to human disease, particularly cancer. Advances in genome-wide technologies have highlighted frequent mutations in genes encoding chromatin-associated proteins, identified unexpected synthetic lethal opportunities and enabled increasingly comprehensive structural and functional dissection. Here, we review recent progress in our understanding of oncogenic mechanisms at each level of chromatin organization and regulation, and discuss new strategies towards therapeutic intervention.
The dynamic control of genomic architecture is required for virtually every cellular function. Nearly 2 m of DNA is organized in each cell nucleus by interacting with histones to form chromatin, a structure that enables its packaging into a less- than 10-μm diameter space1. Chromatin can be regulated by several processes, including modifications of DNA2, modifications of histones3 and protein complexes that remodel its architecture4. These mechanisms function individually and in concert to modulate genome-wide topology and gene expression, thereby regulating cell differentiation, cell division and tissue and organismic development.
Disruptions in chromatin regulation can have profoundly detrimental effects. The role for chromatin regulatory processes in development and disease has been studied in depth and has recently been brought to the forefront of attention by exome-wide and genome-wide studies, which have identified mutations in genes involved in chromatin organization and regulation in over 50% of cancers5–7. In a subset of cancers, such mutations represent the sole genetic abnormalities, providing strong support for their initiating, causative functions, rather than roles as permissive passenger mutations.
The impetus to mechanistically understand chromatin regulatory machinery in diseases such as cancer stems from several important features, perhaps the most compelling of which is the fact that epigenetic changes are, in principle, reversible. However, considering the diverse functions of each class of chromatin regulators in normal tissues and disease contexts, safe and efficacious therapeutic targeting remains a major yet promising challenge. In this Review, we discuss the state of the field investigating epigenetic dysregulation in cancer and highlight the range of current and emerging opportunities for clinical development.
Mechanisms governing chromatin structure
The primary functional unit of chromatin is the nucleosome core particle, which consists of ~146 base pairs of DNA wrapped around a histone octamer assembled by two molecules of histones H2A, H2B, H3 and H4 (ref. 8) (Fig. 1a). Linker DNA connects nucleosome core particles to create the classical ‘beads on string’ analogy that is commonly used to describe primary chromatin structure. The linker histone protein H1 binds to the nucleosome core particle at the DNA entry and exit sites to impart nucleosome stability and facilitate higher-order chromatin structure9,10 (Fig. 1a). Chromatin can be either densely packed, in the form of heterochromatin, which is largely inaccessible to transcriptional machinery and hence encompasses inactive genes, or as open and accessible euchromatin, which contains greater numbers of active genes. The dynamic, tightly controlled regulation of these chromatin configurations is essential for timely, coordinated and appropriately scaled gene expression.
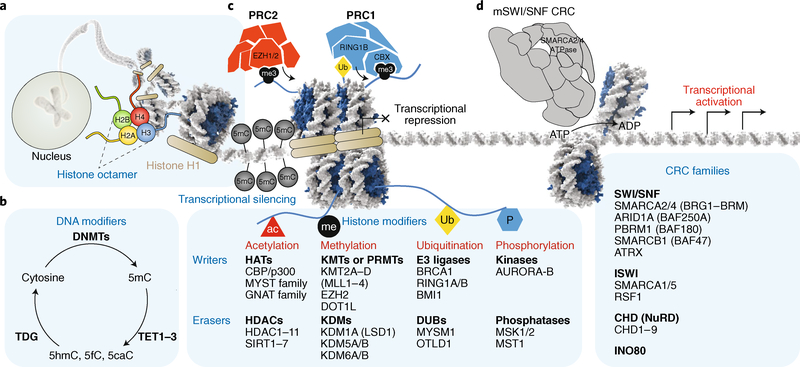
Fig. 1 |. Chromatin regulatory processes in mammalian cells.
a, DNA is wrapped around a histone octamer containing two copies each of histones H2A, H2B, H3 and H4, forming the primary functional unit of chromatin: the nucleosome. Histone H1 binds to DNA at the entry and exit site of the nucleosome. b, DNA methylation is achieved by DNMTs, which are responsible for creating the 5mC mark, associated with transcriptional repression, and TET enzymes, which oxidize 5mC to create 5hmC, 5fC and 5caC. c, Histone modifications, such as acetylation (ac), methylation (me), ubiquitination (Ub) and phosphorylation (P), serve as instructive marks for both gene activation and gene repression. PRC2 and PRC1 deposit the H3K27me3 and H2AK119Ub marks, respectively, both of which correlate with transcriptional repression. d, Four families of ATP-dependent CRCs alter chromatin architecture by mobilizing, depositing or evicting nucleosomes. AURORA-B, Aurora kinase B; BRCA1, breast cancer type 1 susceptibility protein; CBP, CREB-binding protein; DUBs, deubiquitinating enzymes; GNAT, Gcn5-related N-acetyltransferases; HATs, histone acetyltransferases; ISWI, imitation SWI; KDMs, methyl demethylases; KMTs, methyl transferases; MSK1/2, mitogen- and stress-activated protein kinase 1/2; MST1, mammalian STE20-like protein kinase 1 (also known as STK4); PBRM1, protein polybromo-1; PRMTs, protein arginine N-methyltransferases; RSF1, remodelling and spacing factor 1; SIRT, sirtuin; TDG, thymine DNA glycosylase.
DNA- and histone-modifying proteins, and ATP-dependent chromatin remodelling complexes (CRCs) are the three groups of proteins that facilitate changes in chromatin topology and regulation. DNA-modifying proteins place covalent modifications, such as cytosine methylation (5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC)), on DNA itself2,11 (Fig. 1b), rendering genes controlled by such sequences as inactive or active, respectively. Histone-modifying proteins mediate >200 distinct covalent post-translational modifications on histone globular domains or on histone tails to alter local chromatin compaction, nucleosome dynamics, recruitment of other chromatin-bound proteins, and hence, transcription3,12 (Fig. 1c). Finally, a diverse group of CRCs comprising > 100 different protein subunits utilize ATP hydrolysis to mobilize nucleosomes, thereby modulating chromatin structure and regulation4 (Fig. 1d). CRC activity is believed to locally increase DNA accessibility through nucleosome sliding or ejection. However, recent studies also suggest ATPase-dependent functions that affect the targeting and activity of other chromatin regulatory proteins13,14. Taken together, genomic architecture and gene expression are governed by a diverse collection of proteins and modifications, acting both globally and focally at specific sites to orchestrate dynamic processes, such as cell division, differentiation and development, as well as basal maintenance of cell homeostasis.
History and mechanisms of covalent DNA modifications
DNA methylation regulates gene silencing including X-chromosome inactivation, genomic imprinting and tissue-specific transcriptional repression15,16. In vertebrates, 5mC is predominantly found in CpG dinucleotides and localizes to transcriptionally silenced centromeres, telomeres and repetitive transposable elements (short interspersed nuclear elements and long interspersed nuclear elements). Additionally, regions with highly methylated CpG density have been shown to be associated with high nucleosome occupancy in facultative and constitutive heterochromatin17. 5mC is present throughout the vertebrate genome with distinct localizations required for cell differentiation during development and in somatic cells2. Although most CpG dinucleotides are methylated, dense regions called CpG islands, which overlap with over half of mammalian promoters, are largely unmethylated and therefore active. Early studies have established global levels of hypomethylation with focal hypermethylation of promoters and enhancers as a common feature among several cancers18 (Fig. 2a).
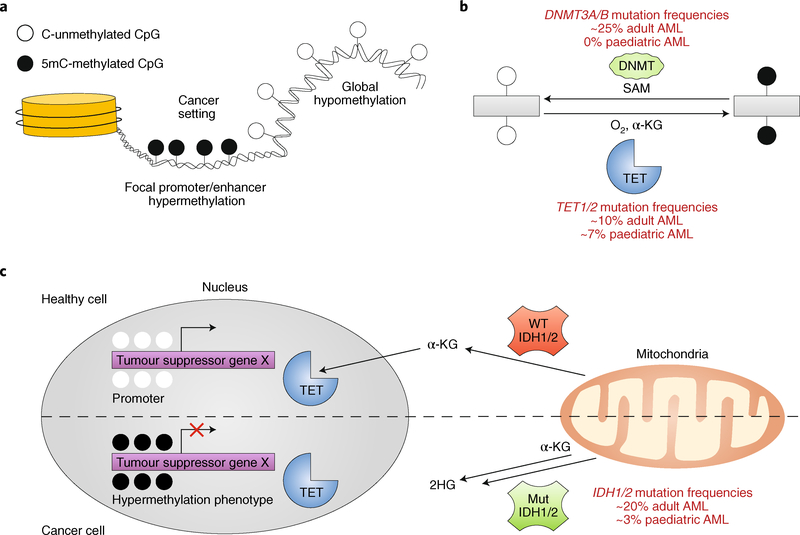
Fig. 2 |. DNMT and TET enzymes and related perturbations in AML.
a, Global hypomethylation and focal promoter/enhancer hypermethylation phenotypes are commonly detected in cancer. b, DNMT and TET enzymes are commonly mutated in adult AML and counteract one another via deposition or removal of the 5mC mark, respectively. DNMTs deposit a methyl group on to the carbon-5 position of cytosine using S-adenosyl-methionine (SAM) as a substrate, and TET enzymes rely on ɑ-ketoglutarate (ɑ-KG) and oxygen to oxidize 5mC and promote cytosine demethylation. c, IDH1/2 (encoding isocitrate dehydrogenase 1) mutations, which are common in AML, inhibit TET activity by converting the TET substrate ɑ-KG to 2-hydroxyglutarate (2HG), resulting in a hypermethylation phenotype. WT, wild type.
DNA methyltransferases
DNA methylation is catalysed by DNA methyltransferases (DNMTs), which deposit methyl groups on the carbon-5 position of cytosine via the S-adenosyl-methionine methyl donor19 (Fig. 2b). Among the five members of the DNMT family, DNMT3A and DNMT3B are canonically considered de novo methyltransferases that localize to pericentromeric heterochromatin to silence gene expression20,21. DNMT1 serves more general maintenance-centred roles owing to relaxed substrate specificity and preference for methylation of hemimethylated CpG dinucleotides, particularly in proliferating cells22. Collectively, DNMTs are involved in appropriate haematopoietic stem cell differentiation, tissue development, adult tissue integrity23 and immune function24.
DNA methylation patterns are disrupted in various malignancies25–30, but aberrations in the genes encoding DNA methylation and demethylation machinery have only been recently identified. DNMT3A mutations, including those found in the hotspot catalytic domain31, are present in ~25% of adult acute myeloid leukaemia (AML) cases32–34, pointing to DNMT3A as an important tumour suppressor31 (Fig. 2b).
Although genome-wide distributions of DNMTs, particularly those of DNMT3A/B, have been extensively studied35,36, the mechanisms responsible for their chromatin deposition and activity in steady state or in cancer have not been fully elucidated. The crystal structure of the DNMT3A–DNMT3L–DNA complex has revealed the mechanisms governing DNMT3 substrate recognition and enzymatic specificity37, by demonstrating that DNMT3A monomers attack two CpGs through a target recognition domain. Intriguingly, DNMT3A cancer-associated somatic missense mutations of the substrate-binding domain decrease in vitro methyltransferase activity, thereby inducing CpG hypomethylation37. Similar studies of DNMT1 indicate that histone post-translational modifications recruit and activate DNMT1 at specific DNA methylation sites38. Cytidine analogues, such as 5-azacytidine and 5-aza-2′ -deoxycytidine (also known as decitabine) are potent DNMT inhibitors that have shown modest efficacy in the treatment of AML, chronic myelomonocytic leukaemia and myelodysplastic syndromes39, and are potentiated in combination with other epigenetic and/or chemotherapeutic agents40.
TET enzymes
The identification and biochemical characterization of ten-eleven translocation (TET) enzymes over the past decade has been a particularly important advance in the field of epigenetics. The fact that ~7–10% of all patients with AML harbour deletion or truncating mutations in TET genes highlights the importance of characterizing bidirectional implications of epigenetic modifications in cancer. The TET1–3 enzymes directly oppose the activity of DNMTs by erasing DNA methylation through the iterative oxidation of 5mC to both relatively stable (5hmC) and transient (5-formyl-methylcytosine (5fC) and 5-carboxyl-methylcytosine (5caC)) derivatives, in an Fe(II) and α-ketoglutarate-dependent mechanism41 (Fig. 2b). The 5fC and 5caC derivatives are then thought to be processed by thymine DNA glycolase and DNA base excision repair machinery11,42, resulting in unmethylated cytosines. Structural investigations suggest that catalytic domains of TET1–3 preferentially bind to CpG dinucleotides43. TET1 and TET3 each harbour an additional CXXC domain, which favours binding of 5mC-, 5hmC- and 5caC-modified CpGs44,45. Excitingly, recent results using novel TET inhibitors in AML cell lines and mouse models suggest potential therapeutic benefits46. As this class of inhibitors advances towards the clinic, the development of readily available, paired genomics-based diagnostic approaches to determine DNMT versus TET mutational status in patient tumours will be required.
Histone modifiers and their implications in cancer
A large collection of histone tail modifications and the proteins that control them represent critical components of the chromatin regulatory system as they contribute to the positioning and function of chromatin regulatory proteins and protein complexes genome wide47–49. Histone-modifying enzymes are grouped into histone deacetylases (HDACs) and histone acetyltransferases, which control lysine acetylation; methyl transferases and demethylases, which regulate lysine methylation; arginine methyltransferases, which facilitate arginine methylation; and various kinases and phosphatases (Fig. 3a). Additional histone modifications include citrullination, SUMOylation, ADP ribosylation, deamination and crotonylation (reviewed elsewhere12,49). Over 150 histone-modifying proteins have been identified and their dysregulation can result in the inappropriate activation of oncogenes or, conversely, the inactivation of tumour suppressors50–52.
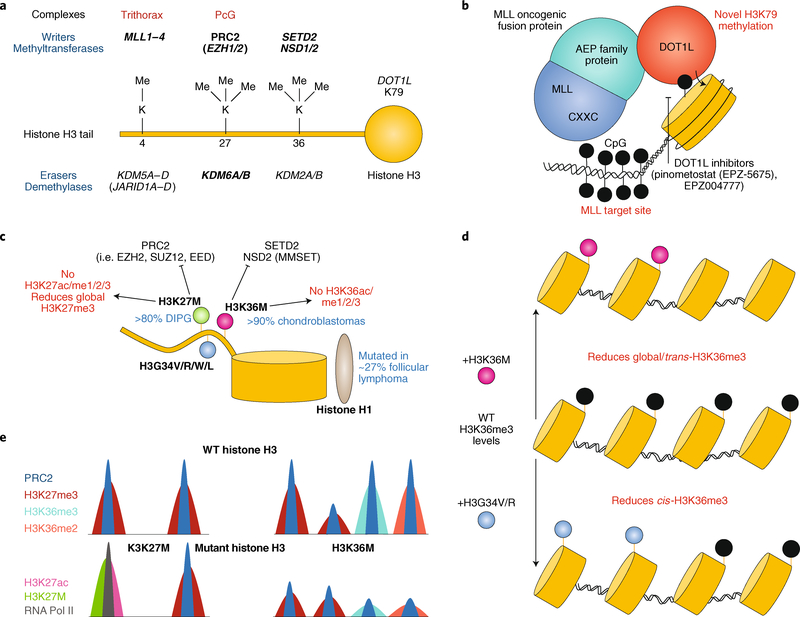
Fig. 3 |. Histone H3 methylation modifications and disruption in cancer.
a, Histone H3 methyltransferases and demethylases. Mutations to genes in bold are implicated in cancer. b, Depiction of MLL–ENL-rearranged leukaemia. The MLL CXXC domain targets the fusion protein to MLL target sites and the ENL domain recruits DOT1L methyltransferase activity, resulting in aberrant methylation. c, Schematic of oncohistone mutations in cancer and their antagonism with methyltransferases. d, The H3K36M mutation results in the global reduction of H3K36me3 levels, whereas the H3K34 mutation diminishes only cis-H3K36me3 levels. e, Schematic of H3K27M and H3K36M oncohistone chromatin occupancy compared to wild-type (WT) histone H3. H3K27M inhibits H3K27me3, resulting in RNA polymerase II (RNA Pol II) recruitment and activation, as assessed by H3K27ac levels. The H3K36M mutation reduces genome-wide H3K36me2/3 and H3K27me3 levels. DIPG, diffuse intrapontine glioma.
Changes to global histone modification signatures are common in cancer (reviewed elsewhere12,53,54), therapeutic interventions for which have recently been reviewed55. Here, we highlight key examples of perturbed histone modification machinery in cancer, which have catalysed the development of several new targeted therapies. In particular, we examine HDACs, Polycomb group (PcG) repressive complexes 1 and 2 (PRC1 and PRC2), and mixed-lineage leukaemia (MLL; also known as KMT2A).
HDACs
HDACs and their associated deacetylation functions play important roles in cell-cycle regulation, apoptosis, DNA-damage repair and other cellular processes56, and their dysregulation is commonly observed in cancer. For example, upregulation of classes I, II and IV HDACs is observed in breast and colorectal cancers, as well as haematological malignancies, whereas their downregulation is observed at lower frequencies57. Oncogenic fusions also deregulate HDAC activity, such as the AML–ETO (eight-twenty-one oncoprotein) and promyelocytic leukaemia protein–retinoic acid receptor-ɑ fusions, which aberrantly retarget HDACs to repress AML and retinoic acid receptor-ɑ target genes, respectively, resulting in cellular transformation58–60. Several HDAC inhibitors have been US FDA approved for cancer treatment or are currently being evaluated in clinical trials for both solid and haematological malignancies61. Given that HDAC inhibitor monotherapy has been largely ineffective in solid tumours62, careful evaluation of various combination regimens is currently ongoing in clinical trials. As with other chromatin regulators, effective therapeutic responses may necessitate the development of potent small molecules that target specific rather than global HDAC activities.
PRC2 and EZH2
PcG proteins form multiprotein complexes that bind to chromatin and repress transcription through methylation and ubiquitination of histones. PRC2 catalyses the monomethylation, dimethylation and trimethylation of histone H3 (that is, H3K27me, H3K27me2 and H3K27me3, respectively) and is canonically associated with long-term transcriptional silencing through deposition of the H3K27me3 mark63,64. Enhancer of zeste homologues 1 and 2 (EZH1/2) are the mutually exclusive catalytic subunits of PRC2, which function in a multiprotein complex with EED, SUZ12 and additional subunits65. EZH2 has garnered substantial attention as both an oncogene and a tumour suppressor, even in the same cancer type (reviewed elsewhere66). Given that Polycomb can repress both oncogenes67 and tumour suppressors68,69, misregulation of either gene class could promote oncogenesis in a context-specific manner. Loss-of-function mutations of genes encoding PRC2 subunits have been identified in leukaemia, myeloid disorders and malignant peripheral nerve sheath tumours70–73. Conversely, EZH2 upregulation has also been implicated in various cancers, including melanoma and breast cancers74, and gain-of-function mutations in the SET domain of EZH2 have been identified in diffuse large B cell lymphoma75–77.
The EED subunit of PRC2 forms an aromatic cage around the H3K27me3 (or Jumonji/ARID domain-containing protein 2 (JARID2)-K116me3) mark, which allosterically activates EZH2. This is reliant on the ordered activity between the SRM and SET domains of EZH2 (refs. 78–80). Mutations in this recognition site are implicated in cancer and Weaver’s syndrome81 and have been shown to impart deficient allosteric activation profiles. Remarkably, although mutants were deficient in activating genome-wide H2K27me3 deposition, no significant changes were observed in their genomic localization profiles81, and allosteric activation of the hyperactive Y646N mutation present in diffuse large B cell lymphoma could be selectively inhibited. Such studies decouple chromatin binding and activity and suggest that allosteric inhibition of hyperactive EZH2 is a potential therapeutic avenue, consistent with newly developed allosteric inhibitors of PRC2, which target the EED subunit78,82,83. These results indicate that decoupling chromatin binding, enzymatic activity and allosteric modulation of other non-enzymatic subunits may afford additional opportunities for complex targeting, for PRC2 as well as a concept for other chromatin regulatory protein complexes. EZH2 inhibitors show acceptable safety profiles and some efficacy in treating various cancers, including multiple myeloma84, B cell non-Hodgkin’s lymphoma and epithelioid sarcoma85.
PRC1 and BMI1
PRC1 complexes contain a RING1 E3 ubiquitin ligase (RING1A/B), which catalyses the monoubiquitylation of histone H2A (that is, H2AK119Ub) and PcG RING finger proteins (PCGF1–6), which dictates downstream PRC1 subunit associations and thereby differential genome-wide localization of complexes86. Additional subcomplex-specific subunits include chromobox proteins, which bind to methylated histones, including PRC2-deposited H3K27 methylation, to promote gene silencing87,88. BMI1 (also known as PCGF4) is a PRC1 complex member that can form homodimers and heterodimers with RING1 (ref. 89) or PHC subunits90 that are important for chromatin compaction91. BMI1 is frequently upregulated in in AML33,34,92 and it is necessary for selfrenewal and maintenance of healthy and leukaemic stem cells93–96. Depletion of BMI1 reduces proliferation and results in apoptosis of epithelial97 and leukaemic cell lines, and in murine colorectal cancer xenograft models98.
Therapeutic PRC1 targeting has only recently transitioned into preclinical and clinical settings. The first BMI1 inhibitor, PTC209 (ref. 98), results in dose-dependent decreases of BMI1 protein levels, associated with global decreases of H2AK119Ub levels. Despite consistent anticancer activities in cell99,100, murine xenograft98,101 and preclinical102 studies, the poor pharmacokinetic properties of PTC209 have stifled progress towards clinical trials92. The development of a more potent, orally available BMI1 inhibitor, PTC596 (ref. 103), which also downregulates the anti-apoptotic factor MCL1, has progressed through phase I clinical trials for patients with advanced solid tumours104 (ClinicalTrials.gov identifier: ), and an additional trial for patients with ovarian cancer is ongoing (ClinicalTrials.gov identifier: ).
MLL
The MLL gene encodes a histone methyltransferase, translocations of which are implicated in AML and acute lymphoid leukaemia with poor prognosis. MLL maintains the expression of HOX genes during development105, and thus, its rearrangement often upregulates this gene cluster106,107, including HOXA9, which is necessary for leukaemic cell proliferation108. Over 100 translocations and 60 fusion partners have been documented, with over two-thirds of these fusions involving members of the AEP (AF4–ENL (eleven nineteen leukaemia)–P-TEFb (positive transcription elongation factor b)) family109–113. Typically the amino-terminal MLL fusion breakpoint maintains CXXC, AT hooks and menininteracting regions, with loss of the carboxy-terminal SET, PHD and other domains114. The retained CXXC domain involved in CpG recognition is essential for transactivation and myeloid differentiation of MLL fusions115. Furthermore, the AEP subunit fusion partner recruits the DOT1-like protein (DOT1L) histone methyltransferase (Fig. 3b), resulting in aberrant H3K79 methylation at MLL target genes, such as HOXA9 and MEIS1. A clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9)-based screen in MLL–AF4 AML cells identified the ENL gene as a target of AML proliferation116. Targeted degradation further demonstrated that ENL regulates global transcription, with HOXA10 and MEIS1 exhibiting significant downregulation in expression.
Mutations targeting other histone-modifying methyltransferases also suggest new therapeutic potential. For instance, the DOT1L inhibitor pinometostat (also known as EPZ-5676) exhibited a favourable safety profile during phase I clinical trials, but requires further investigation, probably in combination with other therapies or in different cancer contexts, to maximally exploit its potential for efficacy117 (ClinicalTrials.gov identifier: ).
Oncohistones in cancer
Mutations in histone genes represent some of the most recently identified gene classes and have emerged through exome-wide sequencing of highly rare tumour types. Exome sequencing uncovered mutations in the genes encoding histone H3 variants (H3.1–H3.3) that convert lysine 27 to methionine (H3K27M) or glycine 34 to arginine or valine (H3G34R/V) in aggressive paediatric brain tumours (such as diffuse intrapontine glioma)118,119 (Fig. 3c). These mutations specifically occur in the H3 tail, which prevents post-translational modification of H3 residues, and exhibit dominant functional changes. Moreover, specific mutations are found in tumours that arise in distinct brain regions, correlating with their distinct molecular characteristics120. Tumours harbouring the H3K27M mutation show dramatic reduction of H3K27me3 levels121, suggesting that such mutants may dominantly inhibit normal lysine methylation pathways, such as those catalysed by PRC2 complexes122. Additional studies in Drosophila demonstrated that H3K27M mutant expression phenotypically mimics PRC2 loss123,124 and mirrors the replacement of all histone H3s with a H3K27R mutant125. H3G34R/V mutations were also shown to block methylation of H3K36 in cis rather than inhibit bulk H3K27me3 or H3K36me3 in trans113 (Fig. 3d). The precise, dominant mechanism(s) by which H3K27M affects the chromatin landscape and the activities of chromatin regulatory machineries remains to be identified. Some studies have suggested that H3K27M interacts with EZH2 (ref. 122); however, others have found interaction with bromodomain-containing protein 4 (BRD4), consistent with increased histone acetylation levels observed in H3K27M mutant cells126 (Fig. 3e).
Sequencing of chondroblastoma and giant cell tumours of the bone have identified additional oncohistone mutations127. Over 95% of chondroblastomas possess the histone H3.3 lysine 36 to methionine (H3.3K36M) mutation in the H3F3B gene, and about 92% of giant cell tumours of the bone harbour mutations of histone H3.3 glycine 34 to tryptophan or leucine (H3.3G34W/L)127 (Fig. 3c). Similar to H3K27M, H3K36M reduces methylation of H3K36 by inhibiting the SETD2 and NSD2 methyltransferases128,129 (Fig. 3e). These findings extend to mutations in genes encoding histone H1 in follicular lymphoma130–132, which are predominantly single amino acid missense mutations scattered throughout the globular H1 domain involved in chromatin compaction, and have been shown to result in reduced association of histone H1 with chromatin131 and DNMT3B133.
Taken together, recurrent driver mutations in histone-encoding genes have added a new layer to the mechanisms of chromatin disruption in human cancer. Moreover, results such as those in paediatric brain tumours have underscored the specific cellular, developmental, anatomical and chromatin architecture contexts required for the high penetrance of such mutations.
Emerging mechanisms of ATP-dependent CRCs
Chromatin remodellers are multi-subunit complexes that use the energy of ATP hydrolysis to reposition, eject, slide or alter the composition of nucleosomes, enabling access of DNA-binding proteins and transcriptional machinery to DNA in order to facilitate gene expression4 (Fig. 4a). Chromatin remodelling proteins play critical functions in cellular differentiation, division and DNA replication4. The four classes of chromatin remodellers include the mammalian SWI/SNF (mSWI/SNF (BAF)), imitation SWI, INO80, and nucleosome remodelling and deacetylation chromodomain helicase DNA-binding complexes4 (Fig. 4b). Chromatin remodeller genes are evolutionarily conserved from yeast to humans, although higherorder organisms have evolved paralogous as well as new subunits in response to evolutionary pressures. The catalytic activity of each complex relies on the activity of a SWI/SNF2-like core ATPase/helicase, with accessory subunits harbouring DNA and histone-binding motifs. Across these families, combinatorial subunit assembly provides extensive complex diversity. Thus, most mammalian chromatin remodellers are typically further subclassified into multiple subcomplexes per family134.
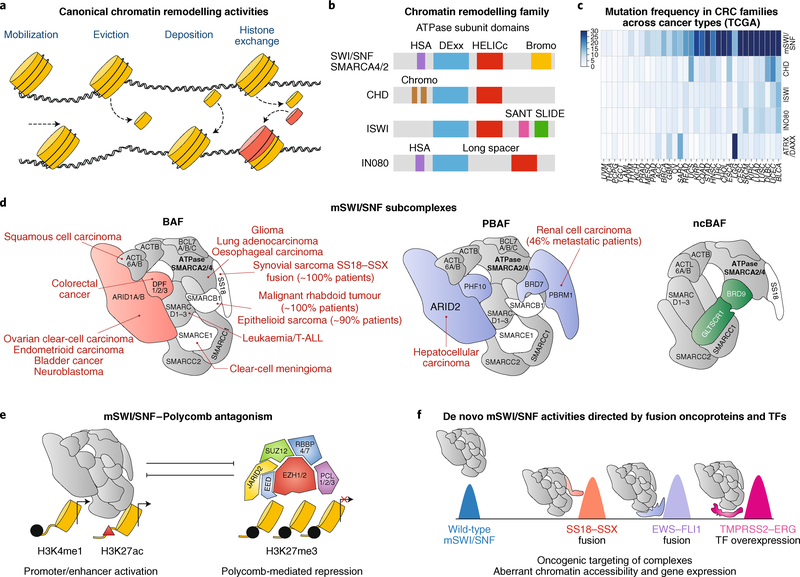
Fig. 4 |. CRCs in cancer: a focus on mSWI/SNF (BAF) complexes.
a, Cartoon depiction of the chromatin remodelling activities: nucleosome sliding, ejection and placement, and histone variant exchange. b, Domain organization within the ATPase subunit of each class of CRCs. c, Pan-cancer mutation frequency across chromatin remodelling families. Mutation frequencies for all genes encoding members of each family were summed and represented as a heatmap. Analysis of public The Cancer Genome Atlas (TCGA) data for 33 available cancer types showing mutation frequency rates across 4 CRC families and SWI/SNF-like ATRX/DAXX. d, mSWI/SNF subcomplex protein associations overlayed with subunit-specific mutations identified across cancer types. The mSWI/SNF subcomplex-defining subunits are coloured in red (BAF), purple (PBAF) and green (ncBAF). e, mSWI/SNF complexes are typically associated with active chromatin landscapes and directly oppose Polycomb-mediated repression. f, Gain-of-function perturbations to mSWI/SNF complexes include fusion oncoproteins and transcription factors (TFs) that tether to mSWI/SNF complex surfaces. The SS18–SSX fusion oncoprotein replaces the SS18 subunit to hijack complexes genome wide, the EWS–FLI1 (friend leukaemia integration 1 transcription factor) fusion directs complexes to GGAA repeat sites in Ewing sarcoma and the ERG transcription factor targets BAF complexes to ETS DNA sequence motifs genome wide, each of which results in aberrant, cancer-specific transcriptional regulation. ACTB, actin, cytoplasmic 1; ACTL6, actin-like protein 6; BCL7, B-cell CLL/lymphoma 7 protein family member; CHD, chromodomain helicase DNA-binding; DPF, zinc-finger protein neuro-d4; GLTSCR1, BRD4-interacting CRC-associated protein; HELICc, helicase superfamily C-terminal; HSA, helicase/SANT associated; PCL, Polycomb-like protein; PHF10, PHD finger protein 10; SLIDE, SANT-like but with several insertions; T-ALL, T cell acute lymphoblastic leukaemia; TMPRSS2, transmembrane protease serine 2.
Among the CRC families, the most extensively mutated class is the mSWI/SNF complex (Fig. 4c). These complexes were originally characterized in yeast135 in screens for mating-type switching and sucrose fermentation (hence the name SWI/SNF), and were later characterized in Drosophila136 and mammals137. Over the course of evolution, SWI/SNF complexes have gained, lost and altered subunits to accommodate increasing genomic complexity and size. Recently, the modular organization and order of assembly of mSWI/ SNF family complexes, including canonical BRG1/BRM-associated factor (BAF) complexes, polybromo-associated BAF (PBAF) complexes and newly discovered non-canonical BAF (ncBAF) complexes, were extensively characterized138,139. Exome-wide sequencing studies have revealed that > 20% of all cancers harbour mutations in mSWI/SNF-encoding genes140,141, several of which are considered to be the key drivers of oncogenesis142 (Fig. 4d). In particular, rare cancers, such as synovial sarcoma, malignant rhabdoid tumour (MRT), clear-cell meningioma and others, are known to be uniformly or near-uniformly caused by perturbations to mSWI/SNF complex subunit genes. In Drosophila and humans13,143, SWI/SNF complexes oppose PRCs to activate gene expression134 (Fig. 4e), suggesting that they may exert specific, ATP-dependent functions other than direct nucleosome remodelling. However, the biochemical and structural basis of such mechanisms remains unknown.
mSWI/SNF complexes were first linked to cancer through the identification of biallelic inactivation of the SMARCB1 gene, which encodes the BAF47 subunit, in ~98% of MRT144,145. SMARCB1 loss has been shown to result in decreased chromatin affinity, largely over distal enhancers, and an inability to oppose Polycomb-mediated repression at bivalent promoters146,147. These data provide a mechanistic explanation for early cell-based and murine model-based findings, indicating that MRT may be uniquely sensitive to PRC2 inhibition; however, clinical trials using EZH2 inhibitors in MRT are still ongoing (ClinicalTrials.gov identifier: ).
Recent systems biology-centred studies indicate that loss of other mSWI/SNF subunits, such as AT-rich interactive domain-containing protein 1A (ARID1A), SMARCE1 or the SMARCA4 ATPase, may exhibit mechanistic convergence with SMARCB1 loss148, particularly in generating and maintaining accessibility over enhancer regions. Coupled with the recent comprehensive architectural characterization of mSWI/SNF complexes138, these findings provide additional evidence for the mutational patterns observed in human disease and suggest the utility of such studies for other chromatin regulatory complexes. These studies also obviate the need for 3D-structure-based interrogation of disease-associated mutations in these specific components.
Gain-of-function perturbations of mSWI/SNF subunits have also been recently discovered (Fig. 4f). For example, the SS18 (also known as SSXT)–SSX fusion oncoprotein is observed in nearly 100% of cases of synovial sarcoma and integrates as a stable mSWI/ SNF complex subunit149. Despite the gain-of-function nature of this event, the SMARCB1 core BAF complex subunit was found to be concurrently displaced from BAF complexes. Recent genome-wide studies defined the targeting profiles of the SS18–SSX-containing complexes, and showed that the SSX tail redirects BAF complexes to new genomic loci and, hence, target genes, at which antagonism of PRC2 facilitates transcriptional activation150. These studies support the opposing functions of BAF and PRC2 complexes151,152 and provide mechanistic evidence for the lack of observed efficacy of EZH2 inhibitors in patients with synovial sarcoma to date153.
Nevertheless, the promising results from cell line and mouse models have prompted the evaluation of EZH2 inhibitors in clinical trials for mSWI/SNF-perturbed MRT and epithelioid sarcoma, with preliminary positive results85,154,155. Inhibitors and ligand-based degrader compounds targeting the bromodomains of the BRD9 and BRD7 mSWI/SNF subunits have been recently developed156, with BRD9 degradation (and hence ncBAF complex inhibition) showing promise in canonical BAF-perturbed cancers such as synovial sarcoma and malignant rhabdoid tumours139,157. The potential for therapeutic efficacy of subunit protein degradation approaches in mSWI/SNF-perturbed cancers has yet to be tested extensively in vivo in disease model systems.
Conclusions and future outlook
Over the past decade, the field of chromatin regulation has made tremendous progress, ranging from understanding the basic chromatin-associated hallmarks of cancer to identifying the underlying genetic changes driving distinct, oncogenic gene expression programmes and promoting tumour development. The challenges and opportunities associated with translating these findings into actionable, mechanism-specific therapeutic strategies have captured the attention of academic research groups and the pharmaceutical industry.
This surge in impactful discoveries continues to be potentiated by the genomic sequencing of human tumours, but also by techniques such as chromatin immunoprecipitation followed by sequencing (ChIP-seq), RNA sequencing (RNA-seq), assay for transposase-accessible chromatin using sequencing (ATAC-seq), Hi-ChIP, Hi-Seq and others, that permit the investigation of chromatin-bound features and topology, modifications, and protein interactions at a genome-wide level. These methodologies are increasingly being paired with advanced computational and bioinformatic approaches. In parallel, state-of-the-art protein identification and mapping approaches based on quantitative mass spectrometry are further elucidating the satellite protein interactome of the chromatin regulatory system described herein. Genetic manipulation strategies, such as CRISPR–Cas9-mediated gene disruption and base editing, as well as major improvements in structural biology approaches, such as cryo-electron microscopy, continue to uncover clinically relevant functional and structural properties of chromatin regulatory proteins and protein complexes. The identification of high-resolution 3D structures of uncharacterized epigenome modifiers and regulators will vastly potentiate both rational design and discovery screens of inhibitors, small molecule or otherwise. Finally, genome-scale RNA interference-based and CRISPR–Cas9-based synthetic lethal studies in hundreds of cancer cell lines and cancer model systems have identified dependencies specific to chromatin regulatory system perturbations, continuing to prompt the development of a wide range of new therapeutic discovery efforts. The systematic, integrative pursuit of such approaches promises a bright future for the further study of the chromatin regulatory system and the elucidation of new therapeutic avenues in cancer.
Acknowledgements
We thank members of the Kadoch Laboratory for helpful discussions during preparation of this Review, and Andrew R. D’Avino for computational analysis presented in Fig. 4c. A.M.V. is supported by the Howard Hughes Medical Institute Gilliam Fellowship Program, NIH 5 T32 GM095450–04, and the Ford Foundation Fellowship. C.K. is supported by awards from the NIH DP2 New Innovator Award 1DP2CA195762–01, the American Cancer Society Research Scholar Award RSG-14–051-01-DMC, the Pew-Stewart Scholars in Cancer Research Grant and the Alex’s Lemonade Stand Foundation ‘A’ Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Competing interests
C.K. is a Scientific Founder, Board of Directors member, Scientific Advisory Board member, shareholder and consultant of Foghorn Therapeutics, Inc. (Cambridge, MA, USA).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Annunziato A DNA packaging: nucleosomes and chromatin. Nat. Educ 1, 26 (2008). [Google Scholar]
- 2.Schübeler D Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Tessarz P & Kouzarides T Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol 15, 703–708 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Clapier CR & Cairns BR The biology of chromatin remodeling complexes. Annu. Rev. Biochem 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Beck S et al. A blueprint for an International Cancer Epigenome Consortium. A report from the AACR Cancer Epigenome Task Force. Cancer Res. 72, 6319–6324 (2012). [DOI] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. et al. The Cancer Genome Atlas pan-cancer analysis project. Nature Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polak P et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesh S & Workman JL Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Fyodorov DV, Zhou B-R, Skoultchi AI & Bai Y Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol 19, 192–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hergeth SP & Schneider R The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16, 1439–1453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohli RM & Zhang Y TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Audia JE & Campbell RM Histone modifications and cancer. Cold Spring Harb. Perspect. Biol 8, a019521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadoch C et al. Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet 49, 213–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanton BZ et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet 49, 282–288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riising EM et al. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347–360 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Jones PA Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Collings CK & Anderson JN Links between DNA methylation and nucleosome occupancy in the human genome. Epigenet. Chromatin 10, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulis M & Esteller M DNA methylation and cancer. Adv. Genet 70, 27–56 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Lyko F The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet 19, 81–92 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Okano M, Bell DW, Haber DA & Li E DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Okano M, Xie S & Li E Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet 19, 219–220 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Sen GL, Reuter JA, Webster DE, Zhu L & Khavari PA DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap DB et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PP et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Peters SL et al. An essential role for Dnmt1 in the prevention and maintenance of MYC-induced T-cell lymphomas. Mol. Cell Biol 33, 4321–4233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laird PW et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81, 197–205 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Bröske A-M et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet 41, 1207–1215 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Kim MS, Kim YR, Yoo NJ & Lee SH Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. APMIS 121, 85–94 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Kandoth C et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Rau R & Goodell MA DNMT3A in haematological malignancies. Nat. Rev. Cancer 15, 152–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao J et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet 47, 469–478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baubec T et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z-M et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishiyama S et al. Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell 68, 350–360.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Shapiro RM & Lazo-Langner A Systematic review of azacitidine regimens in myelodysplastic syndrome and acute myeloid leukemia. BMC Hematol. 18, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dombret H et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with>30 % blasts. Blood 126, 291–299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen KD & Helin K Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H & Zhang Y Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu L et al. Crystal structure of TET2–DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Xu Y et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin S-G et al. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 14, 493–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X et al. Targeted inhibition of STAT/TET1 axis as a therapeutic strategy for acute myeloid leukemia. Nat. Commun 8, 2099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenuwein T & Allis CD Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Schreiber SL & Bernstein BE Signaling network model of chromatin. Cell 111, 771–778 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y & Garcia BA Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol. 7, a025064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khare SP et al. HIstome—a relational knowledgebase of human histone proteins and histone modifying enzymes. Nucleic Acids Res. 40, D337–D342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dawson MA & Kouzarides T Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Bannister AJ & Kouzarides T Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennett RL & Licht JD Targeting epigenetics in cancer. Annu. Rev. Pharmacol. Toxicol 58, 187–207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seto E & Yoshida M Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol 6, a018713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West AC & Johnstone RW New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest 124, 30–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Licht JD AML1 and the AML1–ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene 20, 5660–5679 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Liu Y et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell 9, 249–260 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Di Croce L et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079–1082 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Suraweera A, O’Byrne KJ & Richard DJ Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol 8, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y & Seto E HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med 6, a026831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laugesen A, Højfeldt JW & Helin K Role of the Polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med 6, a026575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chittock EC, Latwiel S, Miller TCR & Müller CW Molecular architecture of Polycomb repressive complexes. Biochem. Soc. Trans 45, 193–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Margueron R & Reinberg D The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Comet I, Riising EM, Leblanc B & Helin K Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803–810 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Vo BT et al. Inactivation of Ezh2 upregulates Gfi1 and drives aggressive Myc-driven group 3 medulloblastoma. Cell Rep. 18, 2907–2917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bracken AP, Dietrich N, Pasini D, Hansen KH & Helin K Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao S-B et al. EZH2 represses target genes through H3K27-dependent and H3K27-independent mechanisms in hepatocellular carcinoma. Mol. Cancer Res 12, 1388–1397 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Puda A et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol 87, 245–250 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Ntziachristos P et al. Genetic inactivation of the Polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med 18, 298–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Score J et al. Inactivation of Polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 119, 1208–1213 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Lee W et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet 46, 1227–1232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bachmann IM et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol 24, 268–273 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Bödör C et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 122, 3165–3168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morin RD et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet 42, 181–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yap DB et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Justin N et al. Structural basis of oncogenic histone H3K27M inhibition of human Polycomb repressive complex 2. Nat. Commun 7, 11316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Margueron R et al. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanulli S et al. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 57, 769–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee C-H et al. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol. Cell 70, 422–434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brooun A et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun 7, 11384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiao L & Liu X Structural basis of histone H3K27 trimethylation by an active Polycomb repressive complex 2. Science 350, aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arora S et al. EZH2 inhibitors are broadly efficacious in multiple myeloma as single agent and in combination with standard of care therapeutics. Blood 128, 5672 (2016). [Google Scholar]
- 85.Italiano A et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Gao Z et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernstein E et al. Mouse Polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol 26, 2560–2569 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morey L et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62 (2012). [DOI] [PubMed] [Google Scholar]
- 89.McGinty RK, Henrici RC & Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gray F et al. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat. Commun 7, 13343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdouh M, Hanna R, El Hajjar J, Flamier A & Bernier G The Polycomb repressive complex 1 protein BMI1 is required for constitutive heterochromatin formation and silencing in mammalian somatic cells. J. Biol. Chem 291, 182–197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nishida Y et al. The novel BMI-1 inhibitor PTC596 downregulates MCL-1 and induces p53-independent mitochondrial apoptosis in acute myeloid leukemia progenitor cells. Blood Cancer J. 7, e527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lessard J & Sauvageau G Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 (2003). [DOI] [PubMed] [Google Scholar]
- 94.Yuan J et al. Bmi1 is essential for leukemic reprogramming of myeloid progenitor cells. Leukemia 25, 1335–1343 (2011). [DOI] [PubMed] [Google Scholar]
- 95.Park I-K et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Rizo A, Dontje B, Vellenga E, de Haan G & Schuringa JJ Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood 111, 2621–2630 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Liang W et al. Knockdown BMI1 expression inhibits proliferation and invasion in human bladder cancer T24 cells. Mol. Cell Biochem 382, 283–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kreso A et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med 20, 29–36 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Dimri M, Kang M & Dimri GP A miR-200c/141–BMI1 autoregulatory loop regulates oncogenic activity of BMI1 in cancer cells. Oncotarget 7, 36220–36234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mourgues L et al. The BMI1 Polycomb protein represses cyclin G2-induced autophagy to support proliferation in chronic myeloid leukemia cells. Leukemia 29, 1993–2002 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Bansal N et al. BMI-1 targeting interferes with patient-derived tumor-initiating cell survival and tumor growth in prostate cancer. Clin. Cancer Res 22, 6176–6191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nishida Y et al. Preclinical activity of the novel B-cell-specific Moloney murine leukemia virus integration site 1 inhibitor PTC-209 in acute myeloid leukemia: implications for leukemia therapy. Cancer Sci. 106, 1705–1713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim MJ et al. Abstract 5517: PTC596-induced Bmi1 hyperphosphorylation via Cdk1/2 activation resulting in tumor stem cell depletion. Cancer Res. 74, 5517 (2014). [Google Scholar]
- 104.Infante JR et al. Phase 1 results of PTC596, a novel small molecule targeting cancer stem cells (CSCs) by reducing levels of BMI1 protein. J. Clin. Oncol 35, 2574 (2017). [Google Scholar]
- 105.Yu BD, Hess JL, Horning SE, Brown GAJ & Korsmeyer SJ Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995). [DOI] [PubMed] [Google Scholar]
- 106.Armstrong SA et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet 30, 41–47 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Yeoh E-J et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1, 133–143 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Faber J et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 113, 2375–2385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meyer C et al. The MLL recombinome of acute leukemias in 2013. Leukemia 27, 2165–2176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meyer C et al. The MLL recombinome of acute leukemias in 2017. Leukemia 32, 273–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yokoyama A, Lin M, Naresh A, Kitabayashi I & Cleary ML A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meeks JJ & Shilatifard A Multiple roles for the MLL/COMPASS family in the epigenetic regulation of gene expression and in cancer. Annu. Rev. Cancer Biol 1, 425–446 (2017). [Google Scholar]
- 113.Krivtsov AV & Armstrong SA MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Thiel AT et al. MLL–AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 17, 148–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ayton PM, Chen EH & Cleary ML Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol. Cell Biol 24, 10470–10478 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Erb MA et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nat. Genet 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stein EM et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131, 2661–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cohen KJ, Jabado N & Grill J Diffuse intrinsic pontine gliomas—current management and new biologic insights. Is there a glimmer of hope? Neuro-Oncology 19, 1025–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 120.Sturm D et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012). [DOI] [PubMed] [Google Scholar]
- 121.Venneti S et al. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 23, 558–564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan KM et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pengelly AR, Copur Ö, Jäckle H, Herzig A & Müller J A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Herz H-M et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lewis PW et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Piunti A et al. Therapeutic targeting of Polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Genet 23, 493–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Behjati S et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet 45, 1479–1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu C et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fang D et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lohr JG et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA 109, 3879–3884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Morin RD et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole genome sequencing. Blood 122, 1256–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morin RD et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li H et al. Mutations in linker histone genes HIST1H1 B, C, D and E, OCT2 (POU2F2), IRF8 and ARID1A underlying the pathogenesis of follicular lymphoma. Blood 123, 1487–1498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poynter ST & Kadoch C Polycomb and trithorax opposition in development and disease. Wiley Interdiscip. Rev. Dev. Biol. 5, 659–688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peterson CL & Herskowitz I Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68, 573–583 (1992). [DOI] [PubMed] [Google Scholar]
- 136.Tamkun JW et al. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2SWI2. Cell 68, 561–572 (1992). [DOI] [PubMed] [Google Scholar]
- 137.Kwon H, Imbalzano AN, Khavari PA, Kingston RE & Green MR Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370, 477–481 (1994). [DOI] [PubMed] [Google Scholar]
- 138.Mashtalir N et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Michel BC et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol 20, 1410–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kadoch C & Crabtree GR Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv 1, e1500447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kadoch C et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bailey MH et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson BG et al. Epigenetic antagonism between Polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chun H-JE et al. Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell 29, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Versteege I et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 146.Wang X et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet 49, 289–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakayama RT et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet 49, 1613–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pan J et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst. 6, 555–568.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kadoch C & Crabtree GR Reversible disruption of mSWI/SNF (BAF) complexes by the SS18–SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McBride MJ et al. The SS18–SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 33, 1128–1141.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kawano S et al. Preclinical evidence of anti-tumor activity induced by EZH2 inhibition in human models of synovial sarcoma. PLoS ONE 11, e0158888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Su L et al. Deconstruction of the SS18–SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell 21, 333–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schoffski P et al. Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with synovial sarcoma (). J. Clin. Oncol 35, 11057 (2017). [Google Scholar]
- 154.Chi SN et al. A phase I study of the EZH2 inhibitor tazemetostat in pediatric subjects with relapsed or refractory INI1-negative tumors or synovial sarcoma. J. Clin. Oncol 34, TPS10587 (2017). [Google Scholar]
- 155.Gounder MM et al. Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with INI1 negative epithelioid sarcoma (). J. Clin. Oncol 35, 11058 (2017). [Google Scholar]
- 156.Remillard D et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brien GL et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. eLife 15, e41305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]