Abstract
Soft-tissue sarcomas are increasingly characterized and subclassified by genetic abnormalities that represent underlying drivers of their pathology. Hallmark tumor suppressor gene mutations and pathognomonic gene fusions collectively account for approximately one-third of all sarcomas. These genetic abnormalities most often result in global transcriptional misregulation via disruption of protein regulatory complexes which govern chromatin architecture. Specifically, alterations to mammalian SWI/SNF (mSWI/SNF or BAF) ATP-dependent chromatin remodeling complexes and polycomb repressive complexes cause disease-specific changes in chromatin architecture and gene expression across a number of sarcoma subtypes. Understanding the functions of chromatin regulatory complexes and the mechanisms underpinning their roles in oncogenesis will be required for the design and development of new therapeutic strategies in sarcomas.
Keywords: soft-tissue sarcoma; epigenetic regulation; chromatin remodeling; mSWI/SNF complexes; BAF complexes; polycomb repressive complexes; fusion oncoproteins, oncogenesis
Introduction
Sarcomas are rare, highly aggressive malignancies marked by the formation of solid tumors in soft tissues or bone that are derived from cells of mesenchymal origin. Sarcoma subtypes possess diverse genetic profiles ranging from a single pathognomonic genetic event, such as chromosomal translocations in synovial sarcoma and Ewing sarcoma, to extensive mutational burden and complex genomes [1–7]. Sarcomas arise in all age groups, but are disproportionally present in children, accounting for a significant number of cancer-related mortalities in both pediatric and young adult patients [8]. In the absence of molecularly targeted therapies, treatment of sarcomas most often relies on cytotoxic chemotherapy in combination with radiation and surgery [9–11]. Upon treatment with these traditional approaches, patients with cases of localized sarcomas achieve 5-year event-free survival rates of 60–70% [12,13]. However, these aggressive malignancies are prone to metastasize, particularly to the lungs, and survival rates for patients with metastatic disease are 20–25% at 2 years [14,15]. Improved patient outcomes rely on the development of targeted therapies tailored to counter the transformative oncogenic activity of the underlying genetic alterations driving each sarcoma subtype. The development of such therapies requires a detailed molecular understanding of the mechanism by which certain genetic perturbations are capable of co-opting specific tissue environments to activate cellular proliferation programs that facilitate solid tumor growth. This review explores the current understanding of recurrent genetic alterations to epigenetic transcriptional regulators across a set of molecularly well-characterized sarcoma subtypes and proposes clinical strategies for reversing their oncogenic capabilities (Table 1).
Table 1.
An overview of the chromatin regulator-associated genetic abnormalities in sarcomas
| Sarcoma subtype | Gene (protein) involved | Associated genetic abnormality (% of cases) | References |
|---|---|---|---|
| Synovial sarcoma (SS) | SS18, SSX1/2/4 | Chromosomal translocation forming SS18–SSX1/2/4 fusion (~100%) |
[1,2,70] |
| Ewing sarcoma (ES) | EWSR1, FLI1, ERG, ETV1, E1AF, FEV | Chromosomal translocation forming EWS–FLI1/ERG/ETV1/E1AF/FEV fusion (~100%) |
[3,87–90] |
| Malignant rhabdoid tumor (MRT)/atypical teratoid/rhabdoid tumor (AT/RT)/epithelioid sarcoma (EpS) | SMARCB1 (BAF47) | Homozygous loss (98%, 98%, > 90%) |
[47,103–105] |
| Malignant peripheral nerve sheath tumors (MPNSTs) | SUZ12, EED | Heterozygous or homozygous loss (70–92%) |
[128–130] |
Epigenetic gene regulation
Genetic events that perturb the protein machinery responsible for epigenetic regulation can drive widespread changes in transcriptional programs. The human genome consists of approximately 3 billion base pairs of DNA compacted in the nucleus of each cell through the organized wrapping of 146 DNA base pairs around histone protein octamers which form nucleosomes [16–18]. Nucleosomes pack tightly against one another to facilitate the storage of genetic information and in doing so form compacted chromatin. This compaction provides structural integrity and serves as a physical barrier to safeguard the DNA content from genetic alterations (i.e. mutations). This mechanism of DNA compaction sterically inhibits protein machineries involved in transcription, replication, recombination, and DNA repair from accessing and binding to DNA. Consequently, the tight compaction of DNA must be relaxed in a highly controlled manner to facilitate access to specific gene elements, such as gene promoters by transcription factors and RNA Pol II, to permit appropriate gene transcription and other essential cellular processes to occur.
The regulation of chromatin architecture, required for proper lineage-specific gene expression, occurs by controlling the DNA–histone interactions through a number of mechanisms that include DNA methylation, covalent modification of histones, and ATP-dependent chromatin remodeling. Control of DNA accessibility to maintain the precise balance between transcriptional activation and repression is achieved through the enzymatic activities of large multi-subunit protein complexes. Two such classes of complexes form the basis of this review, and include the mammalian switch/sucrose non-fermentable (SWI/SNF) complexes and the polycomb group (PcG) protein complexes. SWI/SNF complexes are trithorax group (TrxG) protein members that utilize the power of ATP hydrolysis to carry out the energetically unfavorable process of remodeling nucleosome–DNA interactions to facilitate DNA accessibility [19,20]. This activity is antagonistic to forms of PcG protein complexes that covalently modify the N-terminal tails of histone proteins post-translation to increase chromatin compaction associated with transcriptional repression [20–24]. The precise balance between SWI/SNF and PcG function is tailored to the specific transcriptional needs of different cells through differentiation and development. In embryonic stem (ES) cells, PcG proteins bind cell lineage-specific genes to repress transcription and prevent untimely activation of differentiation pathways [25,26]. SWI/SNF complex activity is critically important for activating transcription of lineage-specific genes to promote cell differentiation [27,28]. In terminally differentiated cells, PcG proteins continue to repress genes of other tissue lineages to maintain the cellular identity [29]. This careful balance between the activity of SWI/SNF and PcG protein complexes facilitates the accessibility and expression of genes required for differentiation of a progenitor cell into a cell of committed linage, while maintaining repression of genes involved in alternative cell fates.
Composition and function of mSWI/SNF (BAF) complexes
The TrxG proteins were first discovered and characterized in Drosophila, in which they are required for the activation of Hox genes and others for proper body segmentation [30–35]. In mammals, TrxG proteins are a diverse and heterogeneous group involved in the transcriptional activation of genes through a number of chromatin modifying activities. TrxG protein members include the SWI/SNF family of chromatin remodeling complexes that possess an ATPase to harness the power of ATP to modify the nucleosome landscape in a variety of manners.
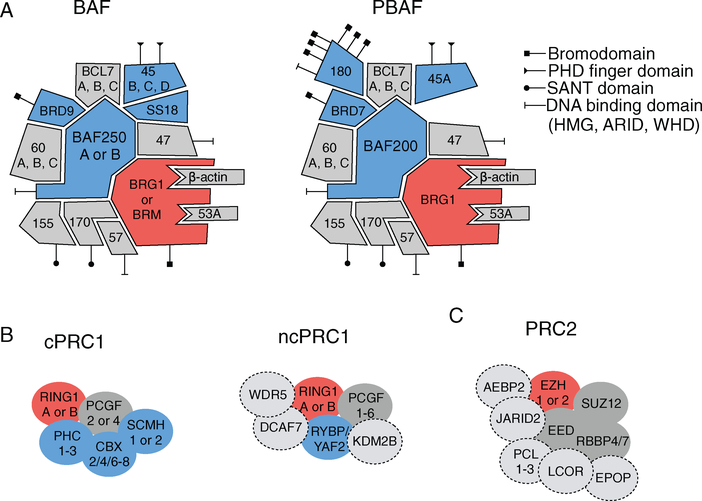
SWI/SNF complexes were initially discovered via the identification of redundant phenotypes upon mutation of genes involved in mating type switching (SWI) and nutrient switching (SNF) regulation in yeast [36,37]. Mutation of histone coding genes suppressed these phenotypes, indicating that these SWI/SNF proteins were involved in altering chromatin structure to regulate transcription [38]. Subsequently, purification in yeast produced an ~1MDa complex consisting of 9–12 subunits that was capable of disrupting mono-nucleosomes in vitro [39–41]. The SWI/SNF family of proteins is evolutionarily conserved with homologous members in flies, plants, and mammals [19]. The mammalian SWI/SNF (mSWI/SNF) complexes have a greater number of subunits, up to 15 subunits in a given complex assembly, to scale with the greater demand for transcriptional regulation and enable specialized cellular functions in multicellular organisms. Furthermore, there are at least two distinct populations of mSWI/SNF complexes, named BAF (BRG1 or BRM-associated factors) and PBAF (polybromo-associated BAF), characterized by differential subunit compositions (Figure 1A) [42]. Both complexes possess a catalytic ATPase subunit of either BRG1 or BRM, along with other core subunits (BAF155, BAF170, BAF47, BAF57), but differ in their ARID domain-containing subunit (BAF250A/B in BAF and BAF200 in PBAF) and the incorporation of the BAF180, BRD7, and BAF45A subunits specifically in PBAF complexes. To allow for further specialization of transcriptional regulatory capacity of mSWI/SNF complexes, a number of subunit members possess multiple paralogs with tissue-specific expression that play a role in directing ES cells toward terminal differentiation, such as in neuronal and cardiac differentiation [43–46]. The diversity of subunit members provides mSWI/SNF complexes with various capabilities including ATP hydrolysis, maintenance of structural integrity, and binding of DNA and histones. Mutations in SMARCB1 (BAF47) represented the first link between SWI/SNF complexes and cancer, upon the discovery by Delattre and co-workers that 98% of malignant rhabdoid tumors exhibit biallelic inactivation of SMARCB1 [47]. Exome sequencing studies have further revealed that members of these complexes are mutated in over 20% of cancers and that certain subunits are recurrently mutated in specific cancer types [48,49]. For example, ARID1A (BAF250A) is mutated in 46% of ovarian clear cell carcinoma cases; SMARCA4 (BRG1) is mutated in 10–35% of non-small cell lung cancer cases; and PBRM1 (BAF180) is mutated in 41% of clear cell renal cell carcinoma cases [50–53]. These findings underscore that each subunit has a specific role as a member of these complexes and that loss of specific subunit functions results in oncogenic transformation only in the context of certain cellular lineages.
Figure 1.
Composition of mammalian SWI/SNF and polycomb group complexes PRC1 and PRC2. (A) mSWI/SNF chromatin remodeling complexes assemble into two distinct complexes, each containing an ATPase subunit (depicted in red), subunits specific to one of the two assembly forms (depicted in blue), and core subunits present in both complex forms (gray). DNA-binding and histone recognition domains are marked according to the legend. (B) Polycomb repressive complex 1 assembles into canonical PRC1 (cPRC1) and non-canonical PRC1 (ncPRC1) complexes possessing a ubiquitination subunit (depicted in red), subunits unique to one complex form (depicted in blue), and accessory subunits that are incorporated into different ncPRC1 forms (depicted by a dashed line). (C) Polycomb repressive complex 2 (PRC2) assembly includes a methyltransferase subunit (depicted in red) which places the H3K27me3 repressive mark, other core subunits (shown in gray), and accessory subunits that are incorporated into different PRC2 forms (depicted by a dashed line).
Composition and function of PcG complexes
Genes that produce the PcG proteins were first discovered in Drosophila as repressors of Hox gene expression in development and differentiation [20]. Homozygous loss of PcG gene members resulted in dramatic homeotic transformation in the body segmentation of flies [54–58]. Protein purifications from Drosophila revealed that individual PcG members formed a protein complex, called polycomb repressive complex 1 (PRC1), which was capable of compacting nucleosomes [23]. Further purifications revealed a second complex with distinct members, called polycomb repressive complex 2 (PRC2), possessing a methyltransferase specific for lysine 27 on the H3 tail (H3K27) [59,60]. Homologous PRC1 and PRC2 complexes exist in mammals and similar to SWI/SNF complexes, subunits of each complex have multiple paralogs to facilitate greater compositional and thus functional diversity for their wide-ranging repressive roles in both embryonic and tissue development [61]. Canonical PRC1 complexes (cPRC1) consist of CBX, PHC, PCGF, RING, and SCMH proteins, with non-canonical PRC1 complexes (ncPRC1) replacing CBX with RYBP or YAF2 along with a number of additional proteins specific to different ncPRC1 forms (Figure 1B) [22]. The primary chromatin modifying role of PRC1 comes from the RING subunit, which is an E3 ligase which catalyzes the ubiquitination of lysine 119 of histone H2A (H2AK119). The core members of PRC2 complexes are SUZ12, EED, RBBP4/7, and either EZH1 or EZH2, with a variety of accessory proteins involved in chromatin binding (Figure 1C). The EZH1/2 proteins are SET methyltransferases that produce mono-, di-, and tri-methylated H3K27 in a SAM-dependent manner. The core member EED binds H3K27me3 to further propagate methylation of H3K27me3 by PRC2, and it has been shown that the methyltransferase activity of EZH1/2 requires a minimal complex that includes SUZ12 and EED [62–64]. While the potential role of cooperation between PRC1 and PRC2 complexes in transcriptional repression is still being fully elucidated, these two complexes modify chromatin through the placement of the H2AK119 ubiquitin and H3K27me3 marks to compact chromatin and inhibit transcription.
Gain-of-function oncogenic mechanisms of mSWI/SNF complexes within sarcomas
Synovial sarcoma: the gained SS18–SSX subunit of mSWI/SNF complexes results in loss of PRC2-induced repression
Synovial sarcoma comprises 8–10% of soft-tissue malignancies and forms solid tumors that in over 80% of cases arise in the extremities [65,66]. Synovial sarcoma occurs in both adult and pediatric patients, with a median age of onset within the third decade of life [67]. Tumors are high-grade and prone to metastasize to the lungs, with a higher frequency of metastasis observed in adult patients [68]. Synovial sarcomas are divided into three histological subtypes consisting of monophasic (spindle cells only), biphasic (both spindle and epithelial cells that form glandular patterns), and a third class termed poorly differentiated synovial sarcoma (PDSS) characterized by an undifferentiated/round cell appearance [69]. The hallmark genetic event, initially identified in 1994 and present in nearly 100% of cases, is a recurrent chromosomal translocation, t(X;18)(p11.2;q11.2), that fuses an SSX gene family member (SSX1, SSX2, and rarely SSX4) to the SS18 gene [1,70,71]. The resulting product is the SS18–SSX fusion protein, in which the last eight C-terminal amino acids of SS18 are replaced by 78 C-terminal amino acids of SSX. Clinical diagnosis for this disease is confirmed through detection of SS18–SSX by next-generation sequencing (NGS), RT-PCR or FISH [72–74]. The SSX gene family consists of nine members with high (73–92%) homology that are restricted in gene expression to the testes [75]. The SSX fusion partner is SSX1 in two-thirds of cases and SSX2 in the remaining third of cases, with a few documented cases of SSX4 [70,76,77]. As the SS18 gene is ubiquitously expressed, following translocation, the resulting SS18–SSX fusion protein is expressed in all tumor cells. The presence of this fusion is the hallmark genetic feature of these tumors and as there are no other recurrent cytogenetic abnormalities, it has been determined that expression of this fusion is necessary and sufficient for the initiation and progression of synovial sarcoma [78,79].
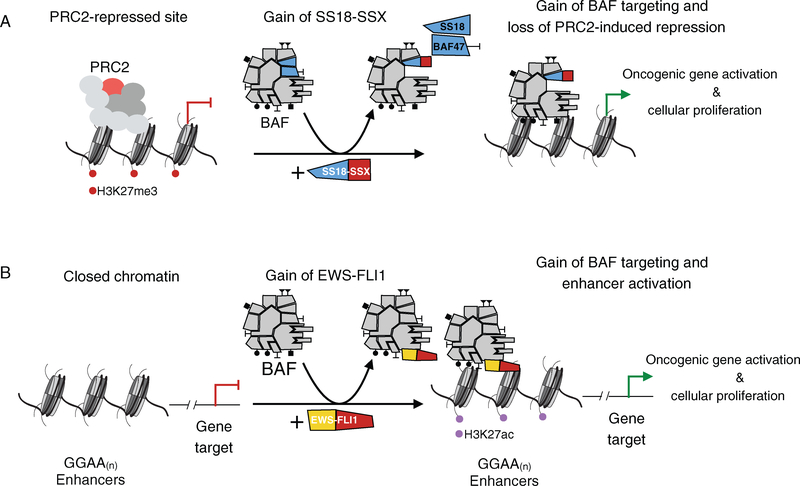
Historically, genes encoding mSWI/SNF complex members have been considered as tumor suppressors, in that protein-level loss of a subunit member results in oncogenesis in a particular cellular context [80]. Synovial sarcoma presents an example in which mSWI/SNF complexes gain oncogenic activity through aberrant fusion of the SS18 subunit gene to SSX to form the SS18–SSX oncogene. The SS18–SSX fusion protein is stably and dominantly incorporated into BAF complexes, replacing wild-type SS18, and furthermore, results in eviction of the BAF47 (SMARCB1/SNF5/INI1) subunit, a known tumor suppressor [81] (Figure 2A). Consequently, upon expression of SS18–SSX, the majority (>90%) of BAF complexes become altered to form a population of oncogenic SS18–SSX-containing complexes. The eviction of BAF47 from these complexes results in its proteasome-mediated degradation resulting in decreased BAF47 protein levels in these tumors, as determined by immunohistochemical staining of paraffin-embedded primary tumor samples [81–84]. This gain of SS18–SSX and loss of BAF47 represent co-occurring alterations to BAF complexes; their respective contributions to the oncogenic mechanism in this malignancy have not, to date, been elucidated. SS18–SSX has been demonstrated to provide BAF complexes with novel targeting capabilities on chromatin to activate aberrant transcription. For example, SS18–SSX-containing BAF complexes localize to the SOX2 locus, at which levels of the PRC2-placed H3K27me3 repressive mark are then reduced, resulting in expression of the SOX2 mRNA transcript [81]. This exemplifies aberrantly gained genomic targeting of BAF complexes to a site of PRC2 repression. While the cell of origin of synovial sarcoma is unknown, there likely exists a highly specific cellular context that is permissive of SS18–SSX-mediated BAF complex recruitment and gene activation that leads to oncogenic transformation, as expression of SS18–SSX in most primary fibroblast cultures (as well as other cell lines) does not yield transformation. Some studies have suggested that the cell of origin of these tumors may be of a myogenic lineage; however, the specific features and/or stage of myogenic differentiation remain incompletely defined [85].
Figure 2.
Fusion oncoprotein-bound mSWI/SNF complexes activate oncogenic transcription via cancer-specific genomic targeting. (A) In synovial sarcoma, the SS18–SSX fusion is incorporated into BAF complexes, resulting in the eviction of wild-type SS18 and BAF47 subunits. These SS18–SSX-containing complexes are targeted to sites of PRC2-mediated repression, such as SOX2, leading to a reduction in H3K27me3 levels and activation of transcription. (B) In Ewing sarcoma, the EWS–FLI1 fusion transiently interacts with BAF complexes and results in cancer-specific targeting to GGAA microsatellites. This de novo targeting of BAF complexes leads to increased H3K27ac levels, enhancer activation, and oncogenic transcription.
Ewing sarcoma: gained functionality of mSWI/SNF complexes through interaction with the FET family protein EWSR1
Ewing sarcoma represents the second most common pediatric bone cancer, with tumors that arise in bones or surrounding soft tissues [86]. Tumors most often present in teenagers and young adults, and these cancers, similar to cases of synovial sarcoma, share a pathognomonic chromosomal translocation [86]. In 90% of cases, this translocation results in the EWSR1 gene becoming fused to the FLI1 gene, which encodes for an E-twenty-six (ETS) transcription factor [3]. Less common but related translocations in the remaining 10% of Ewing sarcoma cases form EWSR1 gene fusions with other members of the ETS family of transcription factors, including ERG, ETV1, E1AF, and FEV [87–90]. Recent exome sequencing efforts indicate that these resulting oncogenic gene fusions are the only reoccurring genetic alterations in this malignancy as few other mutations are detected [91]. Studies have demonstrated that expression of the fusion protein is sufficient for transformation of mesenchymal stem cells [92–97].
While exome sequencing studies have identified extensive mutations in dedicated subunits of mSWI/SNF complexes, the contribution of mSWI/SNF complexes in oncogenesis likely extends well beyond perturbation to mSWI/SNF complex members themselves [49]. Indeed, mSWI/SNF complex mistargeting due to genetic alterations to transient protein interaction partners, such as transcription factors, represents one route to altered, cancer-specific gene activation. Ewing sarcoma is an example of such a malignancy where mSWI/SNF complexes acquire gained oncogenic function due to genetic perturbation to a protein interaction partner. The ETS DNA binding domain of FLI1 enables the EWS–FLI1 fusion to aberrantly bind DNA at GGAA repeats [3,98–101]. However, binding of FLI1 alone at GGAA repeats is not sufficient for inducing oncogenic gene activation and thus this oncogenic fusion possesses a gained function [102]. Mass spectrometry-based proteomics coupled with biochemical validation experiments demonstrate that the EWSR1 protein binds BAF complexes and that the EWS–FLI1 fusion retains this binding capability [102]. This interaction between EWS–FLI1 fusion and BAF complexes represents a weak, transient interaction, in comparison to the highly stable interactions of integrated BAF complex subunits. Importantly, the prion-like domain of the EWSR1 N-terminus confers unique phase transition properties, providing BAF complexes with gain-of-function capability to be recruited to GGAA repeats and to remodel closed chromatin for enhancer activation (gained H3K27ac levels) that leads to oncogenic transcription [102] (Figure 2B). This is a novel example of a perturbation to mSWI/SNF complexes that drives cancer resulting from a gained interaction with an oncogenic fusion protein in the absence of any genetic perturbations to individual subunit members. It highlights that identifying malignancies driven fully or in part by mSWI/SNF complexes will not be comprehensively flagged through exome sequencing studies, but instead requires rigorous biochemical interrogation into both alterations in complex composition and transient interactions with transcription factors or other protein partners.
Oncogenic loss-of-function mechanisms of mSWI/SNF and PcG complexes in sarcomas
SMARCB1-deficient sarcomas: loss of mSWI/SNF complex function results in gain of PRC2-induced repression
Biallelic inactivation of SMARCB1 (BAF47/hSNF5/ INI1) is the defining genetic alteration in a specific set of malignancies. Protein loss of BAF47 occurs in ~98% of malignant rhabdoid tumor (MRT) and atypical teratoid/rhabdoid tumor (AT/RT) cases, and over 90% of epithelioid sarcoma (EpS) cases [47,103–105]. These sarcomas are all aggressive tumors arising in the kidney and soft tissues (MRT) and central nervous system (AT/RT) of pediatric patients in the infant and toddler age groups, and distal and proximal sites of extremities (EpS) in young adults. The genetic perturbation of BAF47 has also been documented and implicated in a number of other neoplasms including poorly differentiated chordoma, renal medullary carcinoma, and meningiomas and schwannomas in patients with schwannomatosis [106–109]. Germline mutation resulting in loss of a single allele of SMARCB1 predisposes the development of MRT or another BAF47-deficient cancer owing to loss of SMARCB1 via a second genetic event within certain cell types. These sarcomas possess stable genomes with low mutational burdens, suggesting that BAF47 loss is responsible for tumor initiation and progression [91,110]. BAF47 has been well established as a tumor suppressor as conditional biallelic inactivation of Smarcb1 (Baf47) in a mouse model resulted in tumor formation in 100% of cases with a median onset of 11 weeks, representing the most rapid inception of tumorigenesis by deletion of a single gene currently documented [111].
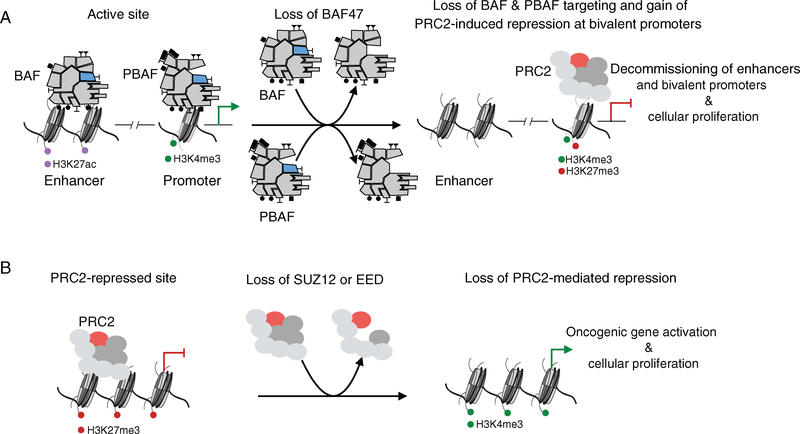
BAF47 is a stable subunit in both BAF and PBAF assembly forms of mSWI/SNF complexes, and the discovery of BAF47 loss in MRT was the first evidence that alterations to the composition of mSWI/SNF complexes perturbed their tumor suppressive functions [47]. This is an example of a genetic event which results in loss of protein expression of a single subunit member, causing altered stability, targeting capability, or enzymatic activity of mSWI/SNF complexes. Studies examining the role of BAF47 in complex stability have been somewhat contradicting, with some studies suggesting that complexes lose the ability to assemble in the absence of BAF47, while others demonstrate that complexes lacking BAF47 form stable complexes that have diminished chromatin-binding capabilities [24,112–116]. Providing further support that BAF47 is not required for the structural integrity and assembly of mSWI/SNF complexes, a study in yeast examining SWI/SNF complexes lacking the Snf5 subunit determined that aberrant complexes were formed that exhibited dramatically attenuated ATPase function [117]. In the context of MRT, mSWI/SNF complexes lacking BAF47 demonstrated decreased affinity for binding chromatin, which resulted in significantly decreased genome-wide targeting [118]. Reintroduction of BAF47 expression and subsequent incorporation into BAF and PBAF complexes resulted in gained occupancy of BAF complexes on enhancers and PBAF complexes on gene promoters, leading to enhancer (increased H3K27ac levels) and gene activation [118] (Figure 3A). In the absence of recruitment by BAF complexes due to loss of BAF47 assembly, a number of bivalent promoters of neural and kidney development-associated genes are silenced owing to the co-presence of the activating H3K4me3 and repressive H3K27me3 marks, and thus are transcriptionally repressed [118]. Reincorporation of BAF47 into mSWI/SNF complexes displaces PRC2 complexes at these bivalent genes, resulting in reduction of the H3K27me3 mark and resolution of these bivalent genes toward activation to permit cell development toward terminal differentiation [118]. The ability of mSWI/SNF complexes to oppose PcG occupancy and induce repression is reliant on the BAF47 subunit. This is supported by previous evidence that mSWI/SNF complexes lose the ability to oppose PcG complexes in the absence of BAF47 at the p16INK4A locus and now it is clear that this opposition is lost at a number of bivalent genes genome-wide [119,120].
Figure 3.
Genome-wide loss of mSWI/SNF and PRC2 complex activity and targeting results in oncogenic transcription. (A) In BAF47-deficient sarcomas, such as malignant rhabdoid tumor and epithelioid sarcoma, the loss of BAF47 incorporation into BAF and PBAF complexes results in residual complexes with decreased chromatin affinity. BAF complex occupancy and H3K27ac levels are reduced genome-wide at enhancers and PBAF complexes are attenuated at gene promoters, while PRC2 occupancy increases at active gene promoters, leading to deposition of H3K27me3 and the creation of transcriptionally-silent bivalent gene promoters. (B) In cases of malignant peripheral nerve sheath tumors with SUZ12 or EED subunit loss, PRC2 occupancy and H3K27me3 levels are reduced, contributing to increased activation of oncogenic transcription.
Malignant peripheral nerve sheath tumors (MPNSTs): loss of PRC2 complex function results in transcriptional activation
MPNSTs are highly aggressive, soft-tissue tumors that arise in the cellular components associated with peripheral nerves [121]. These tumors can arise in pediatric patients, but are more prominent in adult patients [121]. They have a high propensity to metastasize and are often resistant to the standard treatment of radiation and chemotherapy, and patients with this disease have a 5-year survival rate of 20% [122]. Cases of MPNSTs account for 5–10% of all soft-tissue sarcoma cases and are composed of three types: sporadic (45% of cases), associated with neurofibromatosis type 1 or NF1-associated (45% of cases), and associated with previous radiotherapy (10% of cases) [123–125]. NF1 is an autosomal-dominant hereditary cancer syndrome that affects 1 in 3000 individuals due to a germline mutation that inactivates one allele of the NF1 gene [126]. Acquisition of a second somatic mutation that silences the NF1 gene of the allele inherited from the unaffected parent is the key driver in developing neurofibroma, which forms benign tumors and increases the risk of developing the more aggressive MPNSTs [127]. Whole-exome sequencing studies and transcriptional analysis of MPNST cases of all three types have uncovered recurrent inactivating genetic alterations to NF1 (72% of non-NF1-associated cases), CDKN2A (81% of all cases), and multiple subunit members of the PRC2 complex (70–92% of all cases) [128–130].
While MRT and synovial sarcomas discussed above contain genetic alterations to mSWI/SNF members that result in gained and lost H3K27me3-mediated repression, respectively, MPNST-associated mutations in PRC2 complex components directly cause loss of the H3K27me3 mark. The genetic alterations to PRC2 complex members consist of loss-of-function mutations to EED or SUZ12 that result in decreased levels of the H3K27me3 mark detected by immunohistochemical staining of paraffin-embedded tumor samples from patients (Figure 3B) [128]. Decreased H3K27me3 levels in MPNSTs due to loss of function of PRC2 complexes are associated with poor survival [131]. PRC2 loss has not been observed in cases of neurofibroma alone, the precursor to a subset of MPNST cases. Recent transcriptional analyses in MPNSTs have revealed that this loss of PRC2 function results in increased expression of master regulator genes, known to be repressed by PRC2, as would be expected given the well-documented opposition between PRC2 and TrxG complexes [128]. However, to date, specific changes to the activity or localization of mSWI/SNF complexes have not been reported in this disease setting. As evidence that this loss of PRC2-mediated transcriptional repression contributes to tumorigenesis, reintroduction of SUZ12 expression in a cell line established from MPNSTs with a SUZ12 silencing mutation results in substantially decreased cell growth accompanied by decreased expression of developmental genes, increased recruitment of PRC2 complexes to gene promoters, increased levels of H3K27me3, and subsequent loss of the H3K4me3 and H3K27ac activation marks [128,130]. Cases of MPNSTs display clinical and histological diversity due to divergent differentiations, yet possess and share a convergent set of genetically perturbed molecular pathways (stemming from NF1, CDKN2A, and PRC2 complex gene mutations). The wide range of genes silenced by PRC2 complexes to prevent aberrant differentiation toward an alternative cell lineage suggests a mechanism by which perturbation of PRC2 complexes can result in disruption of gene repression, allowing for the formation of tumors through activation of one of a number of differentiation pathways. Due to the contribution of loss-of-function genetic alterations to PRC2 complexes in driving tumorigenesis, MPNST is a unique setting where genes encoding PRC2 complex members act as tumor suppressors rather than oncogenes [132–134].
Therapeutic strategies and conclusions
The discovery of actionable synthetic lethal dependencies resulting from an imbalance of mSWI/SNF and PcG complex activities in sarcomas presents an important foundation for the identification of targeted therapies. Large, genome-scale fitness dropout screens using shRNA-mediated gene knockdown and CRISPR-Cas9-mediated gene knockout have been carried out across hundreds of cancer cell lines, including some derived from sarcomas [135–137]. These screens are designed to identify genes and gene pathways that are essential for proliferation selectively within the context of specific sarcoma subtypes and their underlying genetic abnormalities. Due to the diverse nature of mSWI/SNF–PcG opposition genome-wide in sarcomas and the resulting heterogeneity in oncogenic transcription, it remains to be determined whether there are unique downstream dependencies that are shared across any individual sarcoma subtype. Consequently, as the mechanistic underpinnings of epigenetic dysregulation are further uncovered, altered mSWI/SNF or PcG activity driving oncogenic transformation may be targeted directly.
A range of small molecules that inhibit PRC2-mediated deposition of H3K27me3 by targeting the EZH2 and EED subunits have been reported, and are actively being evaluated across a range of malignant contexts [138–140]. Depletion of EZH2 protein levels has been shown to induce apoptosis in some MRT cell lines, which is consistent with the observed gain of PRC2-induced repression in BAF47-deficient sarcomas [119]. Potent and specific EZH2 inhibition has been shown to attenuate BAF47-deficient solid tumor growth in preclinical models [141,142]. A phase 2 clinical trial () investigated the anti-tumor activity of EZH2 inhibition in BAF47-deficient tumors and reported potentially promising anti-tumor response within the patient cohort [143]. This trial included synovial sarcoma patients due to the low, residual levels of BAF47 protein expression present in these tumors, yet results to date indicate a lack of objective response in synovial sarcoma patients following treatment with EZH2 inhibitor [144]. This is likely due to the fact that BAF47 loss in this disease is a concurrent event along with gain of the SS18–SSX fusion. Rather than loss of BAF47 resulting in gain of PRC2-mediated repression, as is the case in other BAF47-deficient sarcomas, the SS18–SSX-containing BAF complexes appear to lead to gene activation at loci such as the SOX2 locus. While further studies are ongoing, this provides preliminary evidence that oncogenesis is dependent on gained mSWI/SNF complex-mediated transcription rather than overactivity of PRC2-induced gene repression. Consequently, targeted therapies for synovial sarcoma patients will likely need to target mSWI/SNF complex activity or other regulators required for their activity, as opposed to PRC2 function. Furthermore, while it is clear that loss of PRC2 function in MPNSTs results in gene activation, it remains to be determined whether a transcriptional activator, such as a SWI/SNF family chromatin remodeler, contributes to this activation. The characterization of such an activator and validation of its essentiality in cell proliferation of MPNSTs would suggest a promising therapeutic route in this disease.
Reversing the oncogenic activity of fusion proteins could be achieved through chemical biology-centered strategies that inhibit a certain function of the protein or promote protein-specific degradation. The SS18–SSX fusion results in novel mSWI/SNF targeting which suggests that this fusion interacts directly and uniquely with chromatin or a chromatin-associated protein [81]. Identification of a specific interaction partner would provide the foundational knowledge required for the design of small molecule screens to discover chemical ligands capable of disrupting SS18–SSX chromatin-targeting activity. Similarly, the EWS–FLI1 fusion interacts with mSWI/SNF complexes to activate enhancers through the EWSR1 member of the fusion protein [102]. Discovery of a chemical ligand that inhibits this interaction with mSWI/SNF complexes could be used to prevent the mistargeting of complexes driving this disease. Modern chemical biology techniques have laid the groundwork for using a small molecule chemical binder of a protein as a targeting module for protein-specific proteasome degradation. The selective chemical binder is tethered to a module that recruits an E3 ubiquitin ligase, rendering the targeted protein marked for degradation [145–148]. Discovery of a high-affinity, specific chemical binder to either the SS18–SSX or the EWS–FLI1 fusion could be used to target these fusions for protein degradation using this approach. While these binders do not currently exist and would require a great deal of biochemical and preclinical validation prior to their use in a clinical setting, this is a promising strategy for the protein-level removal of oncogenic fusions in sarcomas.
In summary, there remains a major need to mechanistically understand the functional ramifications of recurrent genetic abnormalities that drive sarcomagenesis via gene regulatory changes, specifically due to alteration to chromatin regulatory complexes. This knowledge will be requisite for the design and development of cancer-specific therapies tailored to address the underlying molecular events driving tumorigenesis within each sarcoma subtype.
Acknowledgements
We thank Zachary McKenzie, John Pulice, Alfredo Valencia, and other members of the Kadoch laboratory for insightful discussions, guidance, and feedback on the manuscript. MJM is supported by a research fellowship from Harvard University Graduate School of Arts and Science (GSAS) and NIH Grant Number 5 T32 GM095450–04. CK is supported in part by the NIH DP2 New Innovator Award 1DP2CA195762–01, the American Cancer Society Research Scholar Award RSG-14–051-01-DMC, the Pew-Stewart Scholars for Cancer Research Grant, and the Alex’s Lemonade Stand Foundation Young Investigator Award.
Footnotes
Conflict of interest statement: CK is a Scientific Founder and consultant of Foghorn Therapeutics, Inc (Cambridge, MA).
References
- 1.Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994; 7: 502–508. [DOI] [PubMed] [Google Scholar]
- 2.Crew AJ, Clark J, Fisher C, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995; 14: 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992; 359: 162–165. [DOI] [PubMed] [Google Scholar]
- 4.Turc-Carel C, Aurias A, Mugneret F, et al. Chromosomes in Ewing’s sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12). Cancer Genet Cytogenet 1988; 32: 229–238. [DOI] [PubMed] [Google Scholar]
- 5.Zucman J, Delattre O, Desmaze C, et al. Cloning and characterization of the Ewing’s sarcoma and peripheral neuroepithelioma t(11;22) translocation breakpoints. Genes Chromosomes Cancer 1992; 5: 271–277. [DOI] [PubMed] [Google Scholar]
- 6.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer 2003; 3: 685–694. [DOI] [PubMed] [Google Scholar]
- 7.Borden EC, Baker LH, Bell RS, et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res 2003; 9: 1941–1956. [PubMed] [Google Scholar]
- 8.Bleloch JS, Ballim RD, Kimani S, et al. Managing sarcoma: where have we come from and where are we going? Ther Adv Med Oncol 2017; 9: 637–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Graaf WT, Gelderblom H. New systemic therapy options for advanced sarcomas. Curr Treat Options Oncol 2012; 13: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frezza AM, Stacchiotti S, Gronchi A. Systemic treatment in advanced soft tissue sarcoma: what is standard, what is new. BMC Med 2017; 15: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark MA, Fisher C, Judson I, et al. Soft-tissue sarcomas in adults. N Engl J Med 2005; 353: 701–711. [DOI] [PubMed] [Google Scholar]
- 12.Gorlick R, Janeway K, Lessnick S, et al. Children’s Oncology Group’s 2013 blueprint for research: bone tumors. Pediatr Blood Cancer 2013; 60: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins DS, Spunt SL, Skapek SX. Children’s Oncology Group’s 2013 blueprint for research: soft tissue sarcomas. Pediatr Blood Cancer 2013; 60: 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol 2013; 5: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss A, Gill J, Goldberg J, et al. Advances in therapy for pediatric sarcomas. Curr Oncol Rep 2014; 16: 395. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science 1974; 184: 868–871. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science 1974; 184: 865–868. [DOI] [PubMed] [Google Scholar]
- 18.Olins AL, Olins DE. Spheroid chromatin units (v bodies). Science 1974; 183: 330–332. [DOI] [PubMed] [Google Scholar]
- 19.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poynter ST, Kadoch C. Polycomb and trithorax opposition in development and disease. Wiley Interdiscip Rev Dev Biol 2016; 5: 659–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuettengruber B, Bourbon HM, Di Croce L, et al. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 2017; 171: 34–57. [DOI] [PubMed] [Google Scholar]
- 23.Shao Z, Raible F, Mollaaghababa R, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999; 98: 37–46. [DOI] [PubMed] [Google Scholar]
- 24.Kadoch C, Williams RT, Calarco JP, et al. Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 2017; 49: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441: 349–353. [DOI] [PubMed] [Google Scholar]
- 26.Pasini D, Bracken AP, Hansen JB, et al. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 2007; 27: 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011; 476: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo AS, Staahl BT, Chen L, et al. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009; 460: 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jadhav U, Nalapareddy K, Saxena M, et al. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell 2016; 165: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingham PW. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 1983; 306: 591–593. [DOI] [PubMed] [Google Scholar]
- 31.Ingham PW. Genetic control of the spatial pattern of selector gene expression in Drosophila. Cold Spring Harb Symp Quant Biol 1985; 50: 201–208. [DOI] [PubMed] [Google Scholar]
- 32.Sato T Genetic interaction between homoeotic Sex combs reduced and Regulator of bithorax (or trithorax) genes of Drosophila melanogaster. Roux Arch Dev Biol 1988; 197: 435–440. [DOI] [PubMed] [Google Scholar]
- 33.Tamkun JW, Deuring R, Scott MP, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 1992; 68: 561–572. [DOI] [PubMed] [Google Scholar]
- 34.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A 1988; 85: 8136–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 1995; 29: 289–303. [DOI] [PubMed] [Google Scholar]
- 36.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 1984; 108: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 1992; 68: 573–583. [DOI] [PubMed] [Google Scholar]
- 38.Hirschhorn JN, Brown SA, Clark CD, et al. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 1992; 6: 2288–2298. [DOI] [PubMed] [Google Scholar]
- 39.Cairns BR, Kim YJ, Sayre MH, et al. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci U S A 1994; 91: 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cote J, Quinn J, Workman JL, et al. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 1994; 265: 53–60. [DOI] [PubMed] [Google Scholar]
- 41.Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci U S A 1991; 88: 2687–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulice JL, Kadoch C. Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb Symp Quant Biol 2016; 81: 53–60. [DOI] [PubMed] [Google Scholar]
- 43.Ho L, Crabtree GR. Chromatin remodelling during development. Nature 2010; 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A 2009; 106: 5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lessard J, Wu JI, Ranish JA, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007; 55: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singhal N, Graumann J, Wu G, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 2010; 141: 943–955. [DOI] [PubMed] [Google Scholar]
- 47.Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998; 394: 203–206. [DOI] [PubMed] [Google Scholar]
- 48.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 2015; 1: e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010; 363: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011; 469: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson BG, Helming KC, Wang X, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol 2014; 34: 1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oike T, Ogiwara H, Tominaga Y, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res 2013; 73: 5508–5518. [DOI] [PubMed] [Google Scholar]
- 54.Carroll SB, Laymon RA, McCutcheon MA, et al. The localization and regulation of Antennapedia protein expression in Drosophila embryos. Cell 1986; 47: 113–122. [DOI] [PubMed] [Google Scholar]
- 55.Dura JM, Ingham P. Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 1988; 103: 733–741. [DOI] [PubMed] [Google Scholar]
- 56.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276: 565–570. [DOI] [PubMed] [Google Scholar]
- 57.Riley PD, Carroll SB, Scott MP. The expression and regulation of Sex combs reduced protein in Drosophila embryos. Genes Dev 1987; 1: 716–730. [DOI] [PubMed] [Google Scholar]
- 58.Smouse D, Goodman C, Mahowald A, et al. polyhomeotic: a gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes Dev 1988; 2: 830–842. [DOI] [PubMed] [Google Scholar]
- 59.Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111: 185–196. [DOI] [PubMed] [Google Scholar]
- 60.Muller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002; 111: 197–208. [DOI] [PubMed] [Google Scholar]
- 61.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 2015; 16: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell 2004; 15: 57–67. [DOI] [PubMed] [Google Scholar]
- 63.Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009; 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasini D, Bracken AP, Jensen MR, et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaganathan S, Goyal A, Gadodia A, et al. Spectrum of synovial pathologies: a pictorial assay. Curr Probl Diagn Radiol 2012; 41: 30–42. [DOI] [PubMed] [Google Scholar]
- 66.Spillane AJ, A’Hern R, Judson IR, et al. Synovial sarcoma: a clinicopathologic, staging, and prognostic assessment. J Clin Oncol 2000; 18: 3794–3803. [DOI] [PubMed] [Google Scholar]
- 67.Sultan I, Rodriguez-Galindo C, Saab R, et al. Comparing children and adults with synovial sarcoma in the Surveillance, Epidemiology, and End Results program, 1983 to 2005: an analysis of 1268 patients. Cancer 2009; 115: 3537–3547. [DOI] [PubMed] [Google Scholar]
- 68.Lagarde P, Przybyl J, Brulard C, et al. Chromosome instability accounts for reverse metastatic outcomes of pediatric and adult synovial sarcomas. J Clin Oncol 2013; 31: 608–615. [DOI] [PubMed] [Google Scholar]
- 69.van de Rijn M, Barr FG, Xiong QB, et al. Poorly differentiated synovial sarcoma: an analysis of clinical, pathologic, and molecular genetic features. Am J Surg Pathol 1999; 23: 106–112. [DOI] [PubMed] [Google Scholar]
- 70.Skytting B, Nilsson G, Brodin B, et al. A novel fusion gene, SYT–SSX4, in synovial sarcoma. J Natl Cancer Inst 1999; 91: 974–975. [DOI] [PubMed] [Google Scholar]
- 71.de Leeuw B, Balemans M, Olde Weghuis D, et al. Identification of two alternative fusion genes, SYT–SSX1 and SYT–SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet 1995; 4: 1097–1099. [DOI] [PubMed] [Google Scholar]
- 72.Amary MF, Berisha F, Bernardi FDC, et al. Detection of SS18–SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol 2007; 20: 482–496. [DOI] [PubMed] [Google Scholar]
- 73.Hiraga H, Nojima T, Abe S, et al. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998; 7: 102–110. [DOI] [PubMed] [Google Scholar]
- 74.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Synovial sarcoma. Cancer Genet Cytogenet 2002; 133: 1–23. [DOI] [PubMed] [Google Scholar]
- 75.Smith HA, McNeel DG. The SSX family of cancer-testis antigens as target proteins for tumor therapy. Clin Dev Immunol 2010; 2010: 150591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guillou L, Benhattar J, Bonichon F, et al. Histologic grade, but not SYT–SSX fusion type, is an important prognostic factor in patients with synovial sarcoma: a multicenter, retrospective analysis. J Clin Oncol 2004; 22: 4040–4050. [DOI] [PubMed] [Google Scholar]
- 77.Ladanyi M, Antonescu CR, Leung DH, et al. Impact of SYT–SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res 2002; 62: 135–140. [PubMed] [Google Scholar]
- 78.Limon J, Mrozek K, Mandahl N, et al. Cytogenetics of synovial sarcoma: presentation of ten new cases and review of the literature. Genes Chromosomes Cancer 1991; 3: 338–345. [DOI] [PubMed] [Google Scholar]
- 79.dos Santos NR, de Bruijn DR, Balemans M, et al. Nuclear localization of SYT, SSX and the synovial sarcoma-associated SYT–SSX fusion proteins. Hum Mol Genet 1997; 6: 1549–1558. [DOI] [PubMed] [Google Scholar]
- 80.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481–492. [DOI] [PubMed] [Google Scholar]
- 81.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18–SSX oncogenic fusion in synovial sarcoma. Cell 2013; 153: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kohashi K, Oda Y, Yamamoto H, et al. Reduced expression of SMARCB1/INI1 protein in synovial sarcoma. Mod Pathol 2010; 23: 981–990. [DOI] [PubMed] [Google Scholar]
- 83.Ito J, Asano N, Kawai A, et al. The diagnostic utility of reduced immunohistochemical expression of SMARCB1 in synovial sarcomas: a validation study. Hum Pathol 2016; 47: 32–37. [DOI] [PubMed] [Google Scholar]
- 84.Rekhi B, Vogel U. Utility of characteristic ‘Weak to Absent’ INI1/SMARCB1/BAF47 expression in diagnosis of synovial sarcomas. APMIS 2015; 123: 618–628. [DOI] [PubMed] [Google Scholar]
- 85.Haldar M, Hancock JD, Coffin CM, et al. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell 2007; 11: 375–388. [DOI] [PubMed] [Google Scholar]
- 86.Grier HE. The Ewing family of tumors. Ewing’s sarcoma and primitive neuroectodermal tumors. Pediatr Clin North Am 1997; 44: 991–1004. [DOI] [PubMed] [Google Scholar]
- 87.Ginsberg JP, de Alava E, Ladanyi M, et al. EWS–FLI1 and EWS–ERG gene fusions are associated with similar clinical phenotypes in Ewing’s sarcoma. J Clin Oncol 1999; 17: 1809–1814. [DOI] [PubMed] [Google Scholar]
- 88.Kaneko Y, Yoshida K, Handa M, et al. Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer 1996; 15: 115–121. [DOI] [PubMed] [Google Scholar]
- 89.Peter M, Couturier J, Pacquement H, et al. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 1997; 14: 1159–1164. [DOI] [PubMed] [Google Scholar]
- 90.Jeon IS, Davis JN, Braun BS, et al. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 1995; 10: 1229–1234. [PubMed] [Google Scholar]
- 91.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Riggi N, Cironi L, Provero P, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 2005; 65: 11459–11468. [DOI] [PubMed] [Google Scholar]
- 93.Riggi N, Suva ML, Suva D, et al. EWS–FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 2008; 68: 2176–2185. [DOI] [PubMed] [Google Scholar]
- 94.Herrero-Martin D, Fourtouna A, Niedan S, et al. Factors affecting EWS–FLI1 activity in Ewing’s sarcoma. Sarcoma 2011; 2011: 352580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tirode F, Surdez D, Ma X, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014; 4: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crompton BD, Stewart C, Taylor-Weiner A, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014; 4: 1326–1341. [DOI] [PubMed] [Google Scholar]
- 97.Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014; 10: e1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Riggi N, Knoechel B, Gillespie SM, et al. EWS–FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014; 26: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A 2008; 105: 10149–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guillon N, Tirode F, Boeva V, et al. The oncogenic EWS–FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One 2009; 4: e4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patel M, Simon JM, Iglesia MD, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res 2012; 22: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boulay G, Sandoval GJ, Riggi N, et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 2017; 171: 163–178.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chun HJ, Lim EL, Heravi-Moussavi A, et al. Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell 2016; 29: 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Biegel JA, Zhou JY, Rorke LB, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res 1999; 59: 74–79. [PubMed] [Google Scholar]
- 105.Modena P, Lualdi E, Facchinetti F, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res 2005; 65: 4012–4019. [DOI] [PubMed] [Google Scholar]
- 106.Hulsebos TJ, Plomp AS, Wolterman RA, et al. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet 2007; 80: 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Christiaans I, Kenter SB, Brink HC, et al. Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet 2011; 48: 93–97. [DOI] [PubMed] [Google Scholar]
- 108.Mobley BC, McKenney JK, Bangs CD, et al. Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathol 2010; 120: 745–753. [DOI] [PubMed] [Google Scholar]
- 109.Sigauke E, Rakheja D, Maddox DL, et al. Absence of expression of SMARCB1/INI1 in malignant rhabdoid tumors of the central nervous system, kidneys and soft tissue: an immunohistochemical study with implications for diagnosis. Mod Pathol 2006; 19: 717–725. [DOI] [PubMed] [Google Scholar]
- 110.McKenna ES, Sansam CG, Cho YJ, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol 2008; 28: 6223–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roberts CW, Leroux MM, Fleming MD, et al. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2002; 2: 415–425. [DOI] [PubMed] [Google Scholar]
- 112.Jamshidi F, Bashashati A, Shumansky K, et al. The genomic landscape of epithelioid sarcoma cell lines and tumours. J Pathol 2016; 238: 63–73. [DOI] [PubMed] [Google Scholar]
- 113.Doan DN, Veal TM, Yan Z, et al. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 2004; 23: 3462–3473. [DOI] [PubMed] [Google Scholar]
- 114.Wang X, Sansam CG, Thom CS, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res 2009; 69: 8094–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Lee RS, Alver BH, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 2017; 49: 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei D, Goldfarb D, Song S, et al. SNF5/INI1 deficiency redefines chromatin remodeling complex composition during tumor development. Mol Cancer Res 2014; 12: 1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sen P, Luo J, Hada A, et al. Loss of Snf5 induces formation of an aberrant SWI/SNF complex. Cell Rep 2017; 18: 2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakayama RT, Pulice JL, Valencia AM, et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 2017; 49: 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010; 18: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kia SK, Gorski MM, Giannakopoulos S, et al. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b–ARF–INK4a locus. Mol Cell Biol 2008; 28: 3457–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thway K, Fisher C. Malignant peripheral nerve sheath tumor: pathology and genetics. Ann Diagn Pathol 2014; 18: 109–116. [DOI] [PubMed] [Google Scholar]
- 122.Zou C, Smith KD, Liu J, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Ann Surg 2009; 249: 1014–1022. [DOI] [PubMed] [Google Scholar]
- 123.LaFemina J, Qin LX, Moraco NH, et al. Oncologic outcomes of sporadic, neurofibromatosis-associated, and radiation-induced malignant peripheral nerve sheath tumors. Ann Surg Oncol 2013; 20: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodriguez FJ. Peripheral nerve sheath tumors: the elegant chapter in surgical neuropathology. Acta Neuropathol 2012; 123: 293–294. [DOI] [PubMed] [Google Scholar]
- 125.Eilber FC, Brennan MF, Eilber FR, et al. Validation of the postoperative nomogram for 12-year sarcoma-specific mortality. Cancer 2004; 101: 2270–2275. [DOI] [PubMed] [Google Scholar]
- 126.Carey JC, Baty BJ, Johnson JP, et al. The genetic aspects of neurofibromatosis. Ann N Y Acad Sci 1986; 486: 45–56. [DOI] [PubMed] [Google Scholar]
- 127.Liu C, Shi X, Wang L, et al. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol 2014; 35: 6073–6082. [DOI] [PubMed] [Google Scholar]
- 128.Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 2014; 46: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang M, Wang Y, Jones S, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet 2014; 46: 1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.De Raedt T, Beert E, Pasmant E, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014; 514: 247–251. [DOI] [PubMed] [Google Scholar]
- 131.Cleven AH, Al Sannaa GA, Briaire-de Bruijn I, et al. Loss of H3K27 tri-methylation is a diagnostic marker for malignant peripheral nerve sheath tumors and an indicator for an inferior survival. Mod Pathol 2016; 29: 1113. [DOI] [PubMed] [Google Scholar]
- 132.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629. [DOI] [PubMed] [Google Scholar]
- 133.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008; 647: 21–29. [DOI] [PubMed] [Google Scholar]
- 134.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010; 42: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meyers RM, Bryan JG, McFarland JM, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 2017; 49: 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsherniak A, Vazquez F, Montgomery PG, et al. Defining a cancer dependency map. Cell 2017; 170: 564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McDonald ER 3rd, de Weck A, Schlabach MR, et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017; 170: 577–592.e10. [DOI] [PubMed] [Google Scholar]
- 138.Qi W, Zhao K, Gu J, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol 2017; 13: 381–388. [DOI] [PubMed] [Google Scholar]
- 139.Kim W, Bird GH, Neff T, et al. Targeted disruption of the EZH2–EED complex inhibits EZH2-dependent cancer. Nat Chem Biol 2013; 9: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med 2016; 22: 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Knutson SK, Warholic NM, Wigle TJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 2013; 110: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alimova I, Birks DK, Harris PS, et al. Inhibition of EZH2 suppresses self-renewal and induces radiation sensitivity in atypical rhabdoid teratoid tumor cells. Neuro Oncol 2013; 15: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gounder MM, Stacchiotti S, Schöffski P, et al. Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with INI1 negative epithelioid sarcoma (). J Clin Oncol 2017; 35: 11058–11058. [Google Scholar]
- 144.Schoffski P, Agulnik M, Stacchiotti S, et al. Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with synovial sarcoma (). J Clin Oncol 2017; 35: 11057–11057. [Google Scholar]
- 145.Sakamoto KM, Kim KB, Verma R, et al. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol Cell Proteomics 2003; 2: 1350–1358. [DOI] [PubMed] [Google Scholar]
- 146.Winter GE, Buckley DL, Paulk J, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015; 348: 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Raina K, Lu J, Qian Y, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 2016; 113: 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sakamoto KM, Kim KB, Kumagai A, et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A 2001; 98: 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]