Abstract
To maximize a desired product, metabolic engineers typically express enzymes to high, constant levels. Yet permanent pathway activation can have undesirable consequences including competition with essential pathways and accumulation of toxic intermediates. Faced with similar challenges, natural metabolic systems compartmentalize enzymes into organelles or post-translationally induce activity under certain conditions. Here, we report that optogenetic control can be used to extend compartmentalization and dynamic control to engineered metabolisms in yeast. We describe a suite of optogenetic tools to trigger assembly and disassembly of metabolically-active enzyme clusters. Using the deoxyviolacein biosynthesis pathway as a model system, we find that light-switchable clustering can enhance product formation by six-fold and product specificity by 18-fold by decreasing the concentration of intermediate metabolites and reducing flux through competing pathways. Inducible compartmentalization of enzymes into synthetic organelles can thus be used to control engineered metabolic pathways, limit intermediates and favor the formation of desired products.
Introduction
Metabolic engineering aims to divert cellular metabolites to a desired biosynthetic pathway with the goal of maximizing a product of interest1. This goal is usually pursued by manipulating expression levels: overexpressing the enzymes required for product formation, and reducing enzyme expression in competing, endogenous pathways2,3. However, expressing an engineered pathway at constant, high levels can face numerous challenges. Diverting flux to engineered pathways may compete with essential natural pathways, necessitating the use of small-molecule inducers to turn enzyme expression on at a particular time or to an intermediate level4. Furthermore, the level of overexpression needed for efficient product generation can itself be deleterious due to the burden of synthesizing these high enzyme loads5. Considerable advantages could thus be gained by increasing flux through an engineered pathway on demand without altering enzyme expression levels, yet few examples exist of post-translational, dynamic regulation of metabolic flux in metabolic engineering6–9.
In contrast, dynamic, post-translational regulation is a hallmark property of natural metabolisms10–12. Cells can shift metabolic flux in response to diverse signals by redistributing existing enzymes into co-localized clusters13 or membrane-bound organelles14 without changing total enzyme quantities (Fig. 1a–c). Prior theoretical and experimental work also provides a mechanistic explanation for clustering-induced metabolic flux: when sequential enzymes in a pathway are co-localized, the product of the first enzyme can have a high probability of reaction through the second enzyme before diffusing away from the cluster15. An additional consequence of this mechanism is more efficient conversion of intermediate metabolites, lowering their steady-state concentrations. This effect can be crucial for limiting intermediates that are toxic at high concentrations16 or preventing loss to alternative, branched pathways8,9,15.
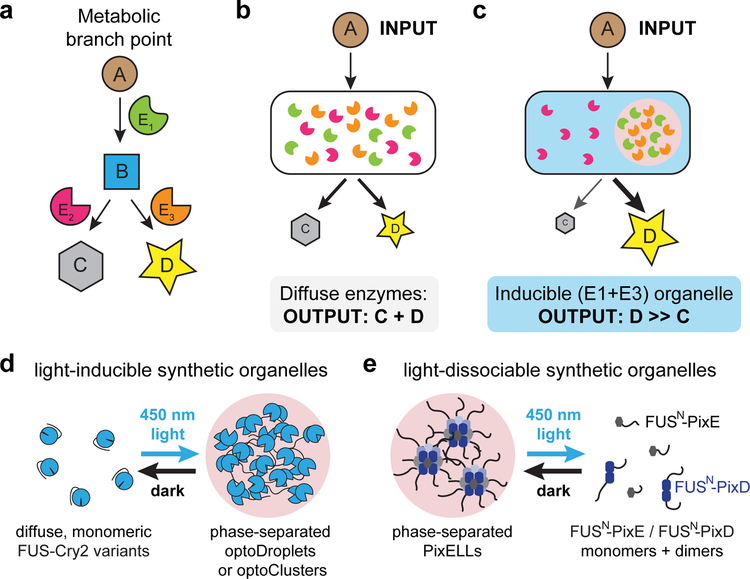
Figure 1. Light-switchable synthetic organelles for redirecting metabolic flux.
(a) A schematic of metabolic flux in a branched pathway. Metabolites A, B, C and D are converted through the action of enzymes E1, E2, and E3. (b) Under conditions where E3 and E2 are similarly efficient, metabolite B will be converted to mixture of products C and D. (c) Synthetic metabolic organelles can bias flux at branch points: by co-localizing enzymes E1 + E3, B will be shunted toward product D, thereby limiting production of the unwanted by-product C. (d, e) Two general strategies for optogenetic regulation of membraneless organelle assembly. FUSN fusions to variants of the Cry2 optogenetic system lead to light-induced protein phase separation in the optoDroplet and optoCluster systems (d), whereas FUSN fusions to the PixD/E proteins form the light-dissociable PixELL optogenetic system (e).
Due to these advantages, the idea of engineering synthetic organelles to co-localize the components of an engineered metabolism has gained considerable interest in recent years. Recent studies have achieved increased product yields by localizing metabolic enzymes to synthetic scaffolds17,18, encapsulins19, the host cell’s mitochondria20, peroxisomes21, and other organelles22, as well by expressing enzyme fusion proteins that aggregate into higher-order clusters15. However, no approaches yet exist to trigger the assembly or disassembly of these synthetic structures on demand to enable fast, reversible control over metabolic flux.
Optogenetics offers a potential solution to this challenge. Light can be applied and removed at will, enabling dynamic, reversible control over protein interactions23. Moreover, we and others have established a suite of tools for optogenetic control over protein clustering in live cells24–26. Here, we report that light-controlled clustering can be extended to the assembly of functional metabolic organelles. Starting from the recently-published optoDroplet and PixELL systems24,25, we developed methods for obtaining yeast strains in which protein clusters could be reliably assembled and dissolved. Using the deoxyviolacein pathway as a model metabolic pathway, we find that synthetic co-localization of sequential enzymes can enhance metabolic flux in a light-switchable manner. Light-regulated clustering leads to a 6.1 ± 0.9 fold-change in the level of the desired product, achieving the theoretical maximum expected fold-change for two-enzyme colocalization15, as well as an 18-fold increase in selectivity over an alternative, non-enzymatic product. The programmable assembly/disassembly of membraneless organelles thus offers a new strategy for rapid post-translational dynamic control of engineered metabolic pathways.
Results
Optogenetic clustering can be variable between cells
We set out to reversibly control metabolic flux in microbes using a set of recently developed optogenetic clustering tools24–27. Many light-induced clustering approaches are based on the A. thaliana Cry2 photolyase homology domain (henceforth termed Cry2) that oligomerizes upon blue light stimulation28. Cry2 oligomerization can be enhanced by a point mutation (in a variant termed Cry2olig)26 as well as by fusion to the N-terminal intrinsically disordered region (IDR) from the protein FUS (FUSN)24. Both FUSN-Cry2 and FUSN-Cry2olig fusion proteins exhibit rapid, reversible clustering in mammalian cells (Fig 1d) with different physical properties: FUSN-Cry2 forms liquid-like spherical droplets that rapidly exchange monomers in and out of clusters, whereas FUSN-Cry2olig forms rigid clusters that do not exchange subunits with the solution24. Throughout this study we will refer to these systems as optoDroplets and optoClusters, respectively (Fig. 1d).
We also tested an additional optogenetic clustering system with an inverted light response termed the PixELL system25. PixELLs are based on the PixD and PixE proteins from Synechocystis sp. PCC6803. PixD and PixE form a high-order complex in the dark that dissociates into PixE monomers and PixD dimers after blue light stimulation29. We previously found that IDR fusions of these two proteins (FUSN-PixD and FUSN-PixE) form liquid-like droplets when co-expressed in mammalian cells, and that these droplets are rapidly disassembled within seconds upon blue light illumination25 (Fig. 1e).
We began by adapting optoDroplets for expression in S. cerevisiae by constructing a 2μ plasmid containing a fusion of FUSN, the FusionRed fluorescent protein30, and Cry2 (Fig. 2a). We observed light-dependent changes in oligomerization of this fusion protein in some cells, suggesting that the optoDroplet system is indeed functional in yeast. However, the quality of light-induced droplet formation was variable from cell to cell. Many cells failed to assemble droplets upon light stimulation; in others, droplets remained assembled even in the dark (Fig. 2a; Supplementary Fig. 1a). We hypothesized that this heterogeneity was due to cell-to-cell variability in gene expression caused by differences in 2μ plasmid copy number. A strong dependence of clustering on expression level is to be expected from the principles of phase separation. If the protein concentration is too low, even maximum illumination will fail to induce clustering, whereas if it is too high, the intrinsic tendency for FUSN to aggregate will dominate even in the dark24. The kinetics of droplet assembly may also exhibit substantial concentration-dependent differences24,25,31. This cell-to-cell variability in droplet formation is a barrier to adopting light-regulated synthetic organelles for metabolic engineering.
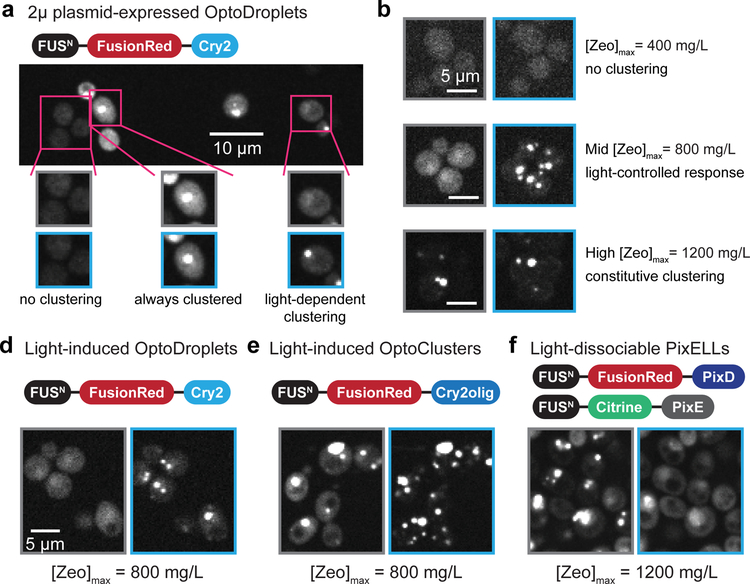
Figure 2. Light-regulated organelle formation depends on component concentration.
(a) A strain expressing optoDroplets from a 2μ plasmid (strain yEZ53) exhibits three different clustering responses in the same colony. Low-expressing cells fail to cluster in all illumination conditions, high-expressing cells remain clustered, and only a small portion of cells reversibly form clusters with light. Representative images of 4 independent experiments are shown. (b) Zeocin selection of multiply-integrated constructs enables titration of expression levels without increasing cell-to-cell variability in clustering (strain yNS47). Colonies show no organelle formation at [Zeo]max = 400mg/L (left), light-inducible organelle formation at [Zeo]max = 800mg/L (middle) and constitutive organelle formation at [Zeo]max = 1,200mg/L (right). (c-e) FusionRed fluorescence for three optogenetic systems in the presence or absence of blue light: optoDroplets (strain yNS47; c), optoClusters (strain yNS49; d) and PixELLs (strain YEZ232; e). Blue boxes indicate images acquired after cells were incubated in blue light; gray boxes indicate images acquired after dark incubation.
A selection strategy for obtaining homogenous clustering
To overcome cell-to-cell variability in droplet formation, we set out to find ideal protein levels for light-induced organelle assembly, while also obtaining homogeneous responses within the majority of cells in a colony. We turned to a genome integration and selection strategy using the antibiotic zeocin. A useful feature of zeocin resistance is that it is highly dose-dependent, so increasing zeocin levels selects for increasing ZeoR expression32. We thus hypothesized that by genomically integrating a variable number of zeocin-resistant optogenetic cassettes and replica-plating the transformed cells on plates containing different zeocin concentrations, we would be able to reproducibly obtain strains at optoDroplet expression levels that support light-switchable droplet formation in all cells within a colony (Supplementary Fig. 1b).
We built cassettes for expression of either the optoDroplet or optoCluster systems driven by the medium-strength PADH1 promoter as well as a zeocin resistance marker (plasmid pNS1 and pNS3; Table S1), and integrated variable numbers of copies of the construct into δ-sites of the yeast genome as previously described32. We plated transformed cells onto a nonselective plate for a short outgrowth (see Online Methods for details), followed by replica-plating at a series of zeocin concentrations. To estimate the expression levels of individual transformants, we define a quantity [Zeo]max as the maximum concentration of zeocin in which a colony can grow (Fig. 2b). Prior genomic analysis of similar constructs indicated that a [Zeo]max of 400 mg/L corresponds to 1–2 copies; a [Zeo]max of 800 mg/L corresponds to 3–4 copies; a [Zeo]max of 1,200 mg/L corresponds to 5–6 copies33. Indeed, we observed that [Zeo]max was predictive of optogenetic system expression and photoswitchable droplet formation (Fig. 2c). In colonies with [Zeo]max of 400 mg/L, we observed low expression and poor droplet formation (Fig. 2c, left). At the other extreme, colonies with [Zeo]max of 1,200 mg/L exhibited constitutive droplets and high expression (Fig. 2c, right). For colonies with intermediate [Zeo]max values, robust photoswitchable droplet formation was consistently observed (Fig. 2c, center).
To further characterize the dynamics and reversibility of photoswitchable organelle formation, we selected optoDroplet and optoCluster colonies with a [Zeo]max of 800 mg/L and imaged FusionRed localization by confocal microscopy in response to sequences of darkness and blue illumination (Fig. 2d–e; Supplementary Videos 1, 2)24,26. The inclusion of the IDR tag was crucial, as strains expressing FusionRed-Cry2 or FusionRed-Cry2olig without the FUSN tag failed to exhibit robust clustering at any [Zeo]max level (Supplementary Fig. 1c–d). We found that optoDroplets exhibited the cleanest change from diffuse to clustered localization upon light stimulation (Fig. 2d). In contrast, optoClusters exhibited some clusters in un-illuminated cells but also exhibited more overall redistribution into clusters upon illumination (Fig. 2e). This is consistent with previous observations that Cry2olig shows an increased propensity to cluster compared to Cry226.
We obtained similar results with the inverse PixELL system. Based on the observation that PixELL clusters contain PixD and PixE in a 2:1 stoichiometry34, we first integrated a single copy of FUSN-Citrine-PixE driven by the PPGK1 promoter into the HIS3 locus, and then integrated a variable number of copies of PADH1–driven FUSN-FusionRed-PixD into δ-sites (YEZ232, Supplementary Tables 1 and 2). Colonies having [Zeo]max of 1,200 mg/L exhibit robust PixD/PixE clustering that dissociate after blue light stimulation (Fig. 2f; Supplementary Video 3). As expected, both PixD and PixE constructs were required for clustering, as strains expressing only one or the other showed only diffuse localization (Supplementary Fig. 1e–f).
We further validated that optoDroplets, optoClusters and PixELLs are each functional in two yeast strains commonly used in cell biology and metabolic engineering studies, BY474135,36 and CEN.PK2–1C37 (for BY4741, see Supplementary Fig. 2; for CEN.PK2–1C, see Fig. 2d–f). For each optogenetic system, we quantified the number of clusters formed upon illumination and their assembly/disassembly kinetics after illumination changes (see Supplementary Fig. 3). Taken together, our results show that the assembly and disassembly of membraneless organelles can be robustly triggered with light across a colony of engineered budding yeast cells.
Light-triggered deoxyviolacein flux using optoClusters
The ability to induce the formation of synthetic membraneless organelles could have enormous potential for metabolic engineering, enabling the on-demand compartmentalization of metabolic enzymes and thus control of metabolic flux. To demonstrate this potential in a controlled model system, we set out to control the flux through the deoxyviolacein (DV) pathway.
The DV pathway produces two distinct end products depending on the level of activity of two enzymes: VioE and VioC (Supplementary Fig. 4). VioE catalyzes the formation of an intermediate, protodeoxyviolaceinate (PTDV), which is then converted by VioC to the pink-colored product deoxyviolacein (DV). Alternatively, PTDV can be spontaneously oxidized to a green product, prodeoxyviolacein (PDV). Both products, PDV and DV, can be detected by chromatographic methods (Supplementary Fig. 5)38. This ease of product quantification makes the DV pathway an ideal platform for assessing metabolic flux control by light-inducible enzyme clustering.
We hypothesized that by inducing the co-localization of VioE and VioC, we could shift flux from PDV to DV production. As a first test of this hypothesis we fused VioE and VioC to the components of our optoCluster system (Fig. 3a). We generated a yeast strain (yNS21) that constitutively expresses VioA and VioB (Supplementary Table 2). We then integrated several copies of a cassette containing VioE-optoCluster and VioC-optoCluster fusions, driven by PADH1, into δ-sites of yNS21 (see Online Methods and Supplementary Tables 1, 2). Both N- and C-terminal orientations were tested for the optoCluster/enzyme fusions, leading to a total of four yeast strains (yNS34, yNS34-cterm, yNS36, yNS36-cterm) (Supplementary Table 2). We then screened several colonies of each transformation with various [Zeo]max levels for light-dependent changes in PDV production.
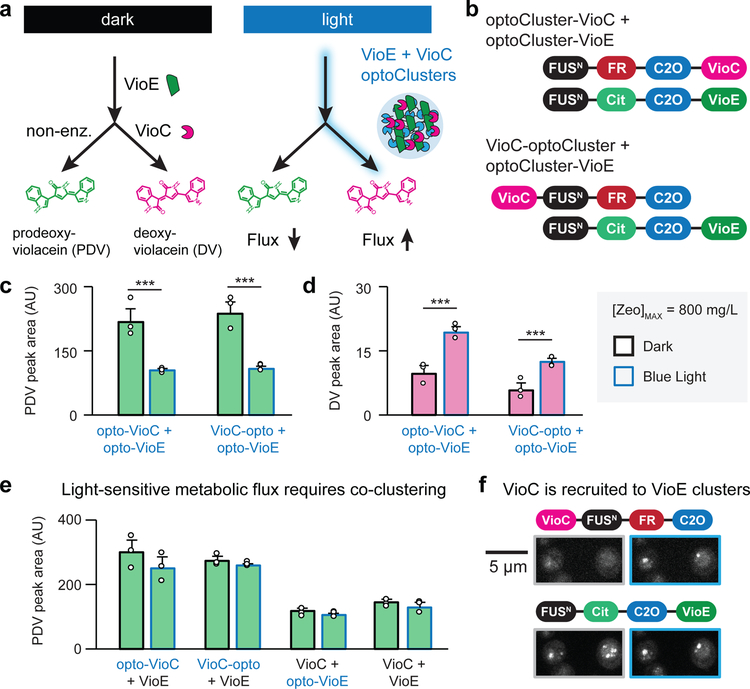
Figure 3. Redirecting flux in the deoxyviolacein pathway using light-inducible optoClusters.
(a) Light-induced co-clustering the VioE and VioC enzymes is expected to increase VioC-induced production of deoxyviolacein (DV) and limit production of prodeoxyviolacein (PDV). (b) optoCluster constructs tested for light-induced DV production. (c, d) HPLC quantification of PDV (c) and DV (d) from colonies with [Zeo]max = 800 mg/L (strains yNS34 and yNS36). Cultures were incubated in dark (black outlines) or light (blue outlines) for 4 days prior to harvesting. ***, p < 0.001. Statistics are derived using a one-sided t test. (e) HPLC quantification for PDV in strains lacking co-clustered VioC and VioE (strains yNS54, yNS55, yNS56, yNS57). (f) Microscopy images of strain yNS34 with [Zeo]max = 800 mg/L, under dark (gray outline) and blue light conditions (blue outline). Top images indicate FusionRed fluorescence; bottom images indicate Citrine fluorescence. Images are representative of four colonies picked at the conditions specified. For the bar graphs in c-e, error bars represent standard deviations of three 1 mL biological replicates (shown as individual points).
The two strains expressing VioE-optoCluster (yNS34-cterm, yNS36-cterm) failed to produce any detectable DV in either light or dark conditions, suggesting that VioE is non-functional with the optoCluster domains fused to its C-terminus. However, strains co-expressing optoCluster-VioE and either optoCluster-VioC (yNS34) or VioC-optoCluster (yNS36) exhibit approximately a two-fold increase in DV production and a corresponding decrease in PDV production when incubated under continuous blue light, relative to their production levels in the dark (Fig. 3b–d). The light-induced change in PDV/DV levels is exactly what would be expected in the current model of light-induced enzyme clustering15. Within a cluster, the PTDV intermediate produced by VioE would have an increased likelihood of encountering co-clustered VioC, leading to enhanced DV production. Moreover, this enhanced conversion would reduce steady-state PTDV levels, decreasing the production of the alternative PDV product.
We performed a series of control experiments to confirm that flux redirection was due to co-clustering of both enzymes rather than a clustering-induced change in the function of VioC or VioE alone. No light-dependent change in product formation was observed in strains expressing VioC-optoCluster and un-clustered VioE, un-clustered VioC and optoCluster-VioE, or VioE and VioC without clustering tags (Fig. 3e, Supplementary Fig. 6). We also verified that the total protein levels of VioC and VioE were not changed by light or dark incubation in either of two VioC/VioE optoCluster strains (yNS34 and yNS36; Supplementary Fig. 7). Finally, we used live-cell microscopy to verify that VioE-VioC clusters were indeed light-switchable (Fig. 3f). We found that VioE forms constitutive clusters even without light exposure, likely due to synergy between VioE’s innate tendency to oligomerize and the FUSN tag. In contrast, VioC’s clustering was light-inducible: VioC was diffuse in the dark, shifting to clusters that colocalized with VioE upon light stimulation (Fig 3f; right panels). Taken together, our results are consistent with a shift in metabolic flux from PDV to DV production driven by enhanced substrate conversion within light-induced VioE-VioC clusters.
We repeated colony screening, light stimulation and DV/PDV product analysis in analogous strains using the optoDroplet system (yNS34drop, yNS36drop) and with Cry2olig-VioC/VioE that lacked the FUSN tag (YEZ250), but did not observe an increase in DV production under blue light. As the FUSN tag and Cry2olig variant both serve to increase the extent of light-induced clustering, these data support a model where the strongest-clustering optogenetic variants are best-suited for shifting metabolic flux.
Light-suppressed deoxyviolacein flux using PixELLs
We have seen that light-induced clustering can shift flux towards a desired product upon illumination. We next sought to test whether light-dissociated synthetic organelles can invert this response. Light-dissociable enzyme clusters would have the benefit of enhancing flux towards a desired product upon a shift from light to darkness, which may be easier to achieve in high-cell-density fermentations and in large-scale bioreactors33. Furthermore, having both light-assembled and light-dissociated organelles in the same strain could enable bi-directional control, shifting cells from growth-promoting metabolism to an engineered pathway by changing light conditions33.
To generate light-dissociable metabolic organelles we turned to PixELL system (Fig. 4a). Starting from YEZ282 (with VioA/VioB in the LEU2 locus), we integrated one copy of FUSN-Citrine-PixE-VioE driven by the strong constitutive PPGK1 promoter into the HIS3 locus, and then integrated multiple copies of FUSN-FusionRed-PixD-VioC into δ-sites to create strain YEZ257 (Fig. 4b). We found that YEZ257 colonies with a [Zeo]max of 1,200 mg/L exhibited a pronounced metabolic shift between light and dark conditions (Fig. 4c–d), exceeding the fold change observed previously with the optoCluster system. The best colony showed a 6.1 ±0.9-fold change in DV production and a corresponding decrease in PDV titer (Fig. 4c,d), leading to an 18.4 ± 4.5-fold change in DV-to-PDV ratio from light to dark conditions (Fig. 4e). This effect was not observed for colonies where [Zeo]max was 400 mg/L, 800 mg/L or 1,600 mg/L, supporting the observation that the photoswitchable response is optimal at intermediate fusion protein expression levels, where light can efficiently assemble/disassemble clusters throughout the cell population.
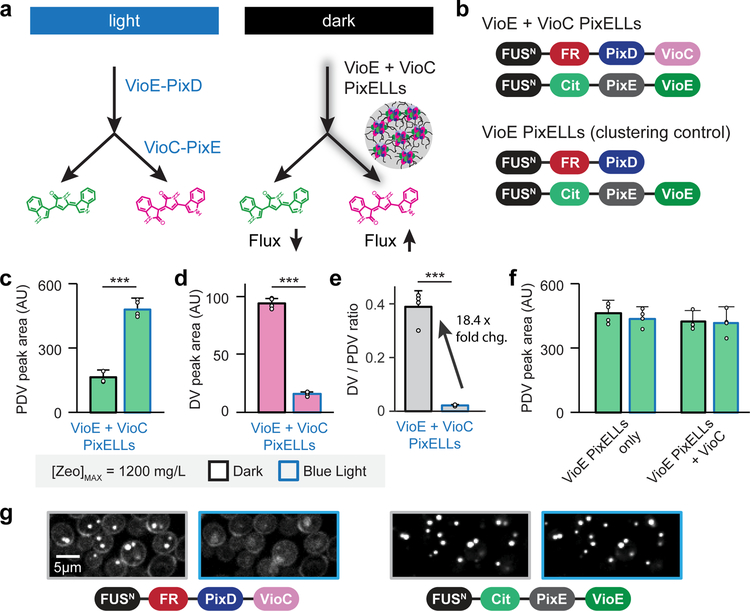
Figure 4. Redirecting flux in the prodeoxyviolacein pathway using light-dissociable PixELLs.
(a) Co-clustering the VioE and VioC enzymes is expected to increase VioC-induced production of deoxyviolacein (DV) and limit production of prodeoxyviolacein (PDV). Because PixELLs are dissociated in the light, this co-clustering enhancement should be lost upon light stimulation. (b) Constructs tested for darkness-induced DV production. (c-e) HPLC quantification of DV (c), of PDV (d), and of the DV/PDV ratio (e) from four-day fermentations of a YEZ257 colony with [Zeo]max = 1,200 mg/L. ***, p < 0.001. Statistics are derived using a one-sided t test. (f) HPLC quantification of PDV for strains YEZ281 and YEZ512 lacking co-clustered VioE and VioC. (g) Microscopy images of YEZ257 under different light conditions showing constitutively-clustered VioE but light-induced delocalization of VioC. Images are representative of four colonies picked at the conditions specified. For the bar graphs in c-f, error bars represent standard deviations of four 1 mL biological replicates (shown as individual points).
As in the case of DV-producing optoClusters, we confirmed that these light-dependent changes in metabolic flux reflect the assembly/disassembly of clusters containing VioC and VioE. Strains expressing VioE-only PixELLs in the presence or absence of un-clustered VioC (YEZ512 and YEZ281, respectively) did not exhibit light-dependent changes in PDV production (Fig. 4f, Supplementary Fig. 8, see Online Methods). The metabolic shift was also not due to light-induced changes in protein expression, as we observed no difference in VioC or VioE expression levels as a function of light stimulus (strains yEZ257 and yEZ281; Supplementary Fig. 7). We also used live-cell microscopy to confirm that VioE/VioC PixELLs were assembled in the dark and could be dissociated in blue light (Fig. 4g). Time-lapse imaging of strain YEZ257 revealed that blue light stimulation caused VioC to switch from a clustered to a diffuse subcellular distribution (Fig. 4g, left). In contrast, VioE remains clustered in both light and dark conditions (Fig. 4g, right), just as we had observed previously for the optoCluster system (Fig. 3f). Together, these data demonstrate that PixELL-enzyme fusions are a powerful platform for darkness-triggered metabolic flux, complementing the light-triggered flux of optoCluster-enzyme fusions.
One potential limitation of our optogenetic strategy lies in the size of the optogenetic tags, which incorporate a 200 amino acid disordered domain (FUSN), a fluorescent protein, and a light-sensitive domain (e.g. the FUSN-FusionRed-PixD tag is 483 amino acids). We thus tested whether shortened variants might still retain strong light-dependent clustering and metabolic flux enhancement. We found that the FUSN IDR could be shortened by over 50% to 93 amino acids (termed ‘short FUS’ or sFUS) while retaining potent light-regulated PixELL clustering (strain yEZ555; Supplementary Fig. 9a). By removing the fluorescent protein we generated a final sFUS-PixD tag that is approximately half the size of our original tag (247 vs 483 amino acids). The resulting PixELL-expressing strain (yEZ553) was still able to generate a strong light-induced flux shift (Supplementary Fig. 9b–d) and further increased the maximum overall DV yield by 3.2-fold (Supplementary Fig. 9d). These promising improvements suggest that additional gains may be possible through further optimization of the length, IDR sequence, or light-switchable clustering domains39. It may also be advantageous to more precisely control the subcellular localization of our optogenetic tools, which are expressed throughout the nucleus and cytosol and can cluster in either compartment (Supplementary Fig. 10). Adding subcellular localization tags (e.g. nuclear export sequences or mitochondrial localization tags) could increase yields by limiting clustering to subcellular compartments where the concentration of upstream metabolites is highest.
Light-controlled flux at an enzymatic branch point
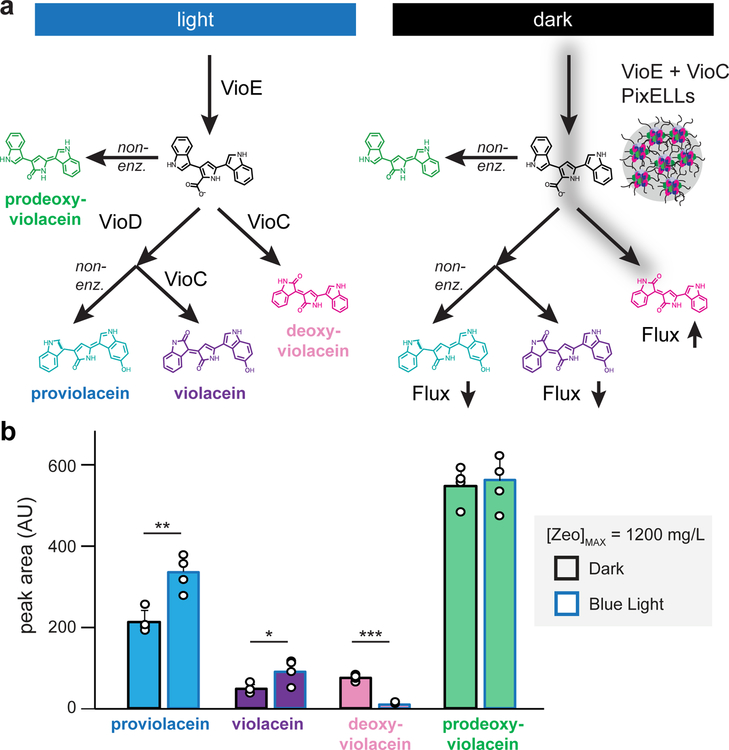
Finally, we sought to extend the use of light-controlled metabolic organelles to a more complex scenario. In the deoxyviolacein pathway used so far, enzyme-catalyzed deoxyviolacein production competes with a non-enzymatic side pathway. However, many metabolic pathways have branch points where two enzymes compete for access to a single intermediate (Fig. 1a–c), raising the question whether clustering would be effective at such a two-enzyme branch point. We reasoned that such a branch point could be created in our system by adding a single additional enzyme, VioD. VioD competes with VioC for the substrate PTDV, driving the formation of two other pigments: proviolacein (PV) and violacein (V) (Fig. 5a).
Figure 5. Light-switchable metabolic flux control at an enzymatic branch point.
(a) Co-clustering VioE and VioC is expected to enhance production of deoxyviolacein and suppress an alternate VioD-catalyzed branch that produces proviolacein and violacein. Because PixELLs are dissociated in the light, this co-clustering enhancement should be present in the dark and lost upon blue light stimulation. (b) HPLC quantification of proviolacein, violacein, deoxyviolacein, and prodeoxyviolacein from four-day fermentations of strain yEZ511. Error bars represent standard deviations of four 1 mL biological replicates (shown as individual points). *, p < 0.05; **, p < 0.01; ***, p < 0.001. Statistics are derived using a one-sided t test.
We expressed VioD driven by the PPGK1 promoter from a 2μ plasmid into strain YEZ257, which we previously used to study PixELLs containing VioE/VioC (Fig. 4). Indeed, we found that flux through both enzymatic branches could be switched with light: PV/V levels were highest in the light when VioE/VioC PixELLs were dissociated, and DV levels were highest in the dark (Fig. 5b). However, unlike the results obtained from our linear pathway (Figs. 3 and 4), we did not observe a change in PDV levels in the branched-enzyme scenario (Fig. 5b). PDV is produced non-enzymatically from PTDV, so the observation of constant PDV levels suggests that the PTDV intermediate levels are no longer changed by light-triggered clustering. This observation may reflect the balance of two competing effects. VioE-VioC clustering is expected to simultaneously increase the consumption of PTDV by VioC but decrease its encounter frequency with VioD; these two effects may balance such that combined flux through both enzymatic pathways is unchanged.
Taken together, our data demonstrate that the light-induced assembly/disassembly of enzyme-containing membraneless organelles can be used to shunt metabolic flux towards a product of interest and away from competing branches. We observe similar deoxyviolacein results with both light-induced optoClusters and darkness-induced PixELLs, demonstrating that our results are robust to off-target, light-dependent processes such as photo-degradation of metabolites or unintended manipulation of endogenous light-sensitive biochemical reactions. In future studies, the bidirectional control afforded by these two systems could also be useful to enhance different sets of reactions under light and dark conditions, thereby reversibly switching cells between ‘growth’ and ‘production’ phases33.
Discussion
Metabolic engineers have previously used physical subcellular compartmentalization for metabolic regulation22. Here, we demonstrate that recent advances in light-controlled protein clustering enable us to assemble and disassemble synthetic compartments in yeast. We developed strategies to tune expression levels while mitigating cell-to-cell variability. Using these strategies, we show that two different optogenetic clustering systems achieve light-dependent changes in the formation of violacein pathway products and limit the concentration of intermediate metabolites. Our work thus demonstrates that light-controlled synthetic organelles can indeed be harnessed to dynamically regulate flux through a metabolic pathway.
Nevertheless, developing optogenetic control over enzyme clustering for metabolic engineering is not without its challenges. Reversible clustering activity occurs only at an intermediate range of fusion protein expression levels, and the propensity to cluster can be strongly influenced by fusion to particular metabolic enzymes. For instance, we found that VioE-fused optoClusters and PixELLs exhibit constitutive clustering, even at expression levels that support light-induced assembly/disassembly of VioC clusters. In general, constitutive clustering would be expected for metabolic enzymes like VioE that exhibit an intrinsic tendency to homo-oligomerize40.
To address these challenges, we developed a method to sample a wide range of expression levels for our fusion proteins, while also ensuring tight control of expression levels within each transformed colony (Fig. 2). We also established five light-dependent clustering systems for use in budding yeast (Cry2, Cry2olig, optoDroplets, optoClusters and PixELLs) that each have different biophysical properties and which may be optimal for different applications. Additional strategies may prove useful to fine-tune cluster behavior for specific applications, including tuning the density of enzymes within the synthetic organelles (e.g. by mixing enzyme-fused and non-enzyme-fused optoClusters in a single cell)15, making targeted mutations to eliminate a metabolic enzyme’s homo-oligomerization interfaces, or varying the length and sequence of the disordered domain.
Despite these challenges, the use of light-dependent synthetic organelle formation for metabolic control shows considerable promise. The 6-fold increase in DV titers that we observe by co-localizing VioC and VioE into PixELLs matches the theoretical maximum enhancement for co-localizing two consecutive enzymes in a metabolic pathway15, and corresponds to an 18-fold increase in DV-to-PDV production ratio. Moreover, the benefit to pathway efficiency is predicted to increase steeply as the number of co-localized enzymes increases. A theoretical analysis suggests that three-enzyme clusters could exhibit metabolic flux enhancements up to 110-fold relative to the diffuse-enzyme case15. Future work that extends controllable enzyme clustering to longer pathways could thus enhance production even more dramatically than what was reported here.
A second important consequence of enzyme co-localization is that the accelerated conversion of pathway intermediates is expected to lower their intracellular steady-state concentration. Complex metabolic pathways often contain intermediates that can be siphoned off to alternative undesirable products, lowering yields41. Intermediates may also be toxic; for instance, in the production of the antimalarial drug artemisinin it is vital to prevent accumulation of the toxic intermediate isopentenyl pyrophosphate16,42. Our results suggest that optogenetic clustering can lower the overall cellular level of intermediate metabolites produced by the clustered enzymes. Although the PTDV intermediate in our pathway is not amenable to direct quantification, measurements of its auto-oxidized product, PDV, suggest that clustering can drive a two- to three-fold decrease in intracellular PTDV concentration at steady state (Figs. 3c and 4c). Moreover, comparing PDV levels in two variants of the light-switchable violacein pathway, one with an enzymatic branch point and one without (Figs. 4 and 5), suggests that clustering at non-branched steps is most effective for depleting intermediates.
In conclusion, we offer a new method for metabolic flux enhancement using light-inducible enzyme compartmentalization into synthetic organelles. Using deoxyviolacein biosynthesis as a model system, we demonstrate that light-switchable organelles can shift flux in both linear and branched metabolic pathways. Our results reveal not only that optogenetics can be harnessed for post-translational metabolic control, but also that phase separation and aggregation are sufficient to shift metabolic pathway flux, opening the door to a deeper understanding of natural metabolisms as well as new opportunities for metabolic engineering.
Online Methods
Assembly of DNA constructs
Ligations and one-step isothermal assembly reactions were performed using previously described methods33. Cry2 and PixELL constructs were obtained from previously published sources (Addgene constructs 111507, 111506, and 111503) and amplified using PCR with homology arms for Gibson assembly24,25. Backbones were either amplified by PCR or cut using available enzymes from the pJLA vector system20. All constructs were sequenced by Genewiz (Supplementary Table 1). Sequences of optogenetic systems are available in Supplementary Sequences.
Strain construction and descriptions
Further description of the constructs and strains described below can also be found in Supplementary Tables 1 and 2 and Supplementary Fig. 11.
Construction of optoDroplet, optoCluster and PixELL expressing strains.
To create optoDroplet and optoCluster strains, we integrated multiple copies of constructs containing different combinations of FUSN, Cry2 and Cry2olig (pNS1, pNS2, pNS3, pNS4) fused to fluorescent proteins and selected on increasing levels of zeocin (400 mg/L, 800 mg/L, 1,200 mg/L, and 1,600 mg/L, which corresponded to an increasing number of integration events). The resulting strains are yNS47, yNS48, yNS49, and yNS50, respectively. We observed that only constructs with the FUSN tag (pNS1, pNS3) formed visible phase-separated bodies when induced with light. To construct PixELL-expressing strains, we integrated a single copy of FUSN-Citrine-PixE (EZ-L498 to make YEZ231) into the HIS3 locus and multiple copies of FUSN-FusionRed-PixD (EZ-L499 to make YEZ232, selected on 1,200 mg/L zeocin) into the δ-sites in the yeast genome. No phase separation was observed when only one component, PixD or PixE alone, was used (YEZ234).
Screening of other clustering constructs.
To test other light-inducible clustering tags, we constructed plasmids pNP1-Drop, pNP3-Drop, pNP7, and EZ-L477. These plasmids represent all combinations of fusions of FUSN-FusionRed-Cry2 and FusionRed-Cry2olig with either VioE or VioC. We screened 24 colonies of yNS21 + pNP1-Drop + pNP3-Drop (yNS34drop), 24 colonies of yNS21 + pNP2-Drop + pNP3-Drop (yNS36drop), and 24 colonies of yNS21 + pNP7+ EZ-L477 (YEZ250). None of these combinations yielded a higher production of DV in the light than in the dark.
Why might optoClusters and PixELLs exhibit strong metabolic shifts while optoDroplets do not? One possibility is that optoDroplets exhibit weaker light-switchable clustering and thus total enzyme redistribution. The optoCluster system includes an additional point mutation that favors Cry2 oligomerization and clustering. Also, the PixELL system is made up of two Pix proteins, so whichever of the two is limiting in expression tends to exhibit near-complete redistribution in/out of clusters. These differences could lead optoDroplets to have a lower total shift from a diffuse to clustered enzyme distribution.
A second possibility is that although variability is quite low within a colony, the expression levels (and thus, number of clusters) of the optogenetic tools can still vary substantially between colonies. By screening more than 24 colonies or testing additional [Zeo]max values, it may be still be possible to optimize optoDroplet-enzyme expression to shift metabolic flux.
Deoxyviolacein pathway control using optoClusters.
We constitutively expressed VioA and VioB by integrating pNS5 into BY4741 to generate yNS21. Using δ-integration, we then expressed a VioC (pNP1 or pNP2) and a VioE (pNP3 or pNP4) plasmid, both fused to FUSN-Cry2olig tags at either the N or C termini, in all possible combinations for a total of 4 yeast constructs. (See Methods, See Supplementary Tables 1 and 2). This made strains yNS34 (pNP1, pNP3), yNS34cterm (pNP1, pNP4), yNS36 (pNP2, pNP3), and yNS36cterm (pNP2, pNP4). After selecting in 800 mg/L of zeocin and screening multiple colonies of each combination, we observed an increased production of DV relative to PDV in response to light activation in strains yNS34 and yNS36.
Deoxyviolacein pathway control using PixELLs.
To redirect flux towards DV with the PixELL system, we integrated EZ-L528 (for expression of VioA and VioB, required to produce the IPA imine dimer precursor metabolite from tryptophan; see Supplementary Figure 3 and Tables 1 and 2) into BY4741 to make YEZ282. We then integrated one copy of a FUSN-Citrine-PixE-VioE fusion (EZ-L526) under a strong, constitutive promoter, PPGK1, to make YEZ255. We expressed various levels (at 400 mg/L, 800 mg/L, 1,200 mg/L, and 1,600 mg/L of zeocin) of a FUSN-FusionRed-PixD-VioC fusion (EZ-L527) by using δ-integration (YEZ257, Fig. 4b, see Supplementary Tables 1 and 2). For colonies where [Zeo]max = 1,200 mg/L, we observed a higher level of DV production when the culture was grown in the dark than when the DV production when the same culture was grown in the light (Fig. 4c). The best colony showed a 6.1-fold change from light to dark conditions with consistent decreases in PDV titer (Fig. 4d). This effect was not observed for colonies where [Zeo]max was 400 mg/L, 800 mg/L or 1,600 mg/L of zeocin. We controlled for the effects of clustering by integrating EZ-L499 into YEZ255, resulting in YEZ281, a strain that clusters PixD and PixE upon blue light stimulation but lacks VioC and thus produces no DV. We also added pNS7 to YEZ281 to control for constitutive non-localizing VioC control, making YEZ512.
Diverting flux away from VioD using PixELLs.
To test the effect of the PixELL system on a metabolic branchpoint containing a competing enzyme, we added VioD to the existing system. We added EZ-L859 to YEZ257 to make YEZ511. We saw that in YEZ511, the entire system produced more products of the violacein pathway (PDV, DV, proviolacein, and violacein). However, we also saw the intended effect, which was a shift from more DV production in the light to more proviolacein and violacein production in the dark. This dependence on light condition of proviolacein and violacein production was not seen in our control strains.
Shortening the FUSN tag for diverting flux using PixELLs.
As the size of the tags are large and could complicate protein activity and/or expression, we tested a tag of reduced size for chemical production. We did this by first limiting the size of the FUSN domain to the first 93 amino acids. We named this iteration sFUS. We first tested to see how sFUS functions with fluorescent proteins. We added EZ-L767 (sFUS-FR-PixD) to YEZ231 and selected on 1,200 mg/L of zeocin to make YEZ555. We then wanted to test minimizing the tag on DV production. We did this by both shortening FUS to sFUS and removing the fusion red protein from the VioC expression construct. We integrated EZ-L786 into YEZ255 to make YEZ553 and selected on 1,200 mg/L of zeocin to make YEZ553. We controlled for the effects of clustering by integrating EZ-L767 into YEZ255, resulting in YEZ554, a strain that clusters PixD and PixE upon blue light stimulation but lacks VioC and thus produces no DV.
Yeast strains and transformations
Strain transformations were performed using standard lithium acetate protocols. For zeocin selection assays, the DNA added was varied between 100 μg and 3 mg per transformation depending on the target zeocin concentration32,33. For experiments with a target [Zeo]max = 400 mg/L, we transformed 70 μL of competent cells using 100 μg of DNA; for [Zeo]max = 800 mg/L, we transformed them with 500 μg of DNA; for [Zeo]max = 1,200 mg/L, we used 1.5 mg of DNA; and for [Zeo]max = 1,600 mg/L, we transformed 3 mg of DNA. Colonies were grown on both the [Zeo]max concentration and one level higher (800 mg/L for [Zeo]max = 400 mg/L). Colonies that appear on the [Zeo]max concentration plate but not on the higher concentration plate were selected as colonies with that [Zeo]max.
Fluorescence Microscopy
To prepare culture for microscopy, yeast strains were cultured overnight at 30 °C from a single colony, in the appropriate synthetic complete media with 2% glucose (e.g. SC-ura + 2% glucose for a strain that contained a URA3 plasmid), in a 24-well plate covered with aluminum foil. Synthetic media was used to avoid the high auto-fluorescence of YPD. The following day, cultures were diluted 1:20 and allowed to grow for 2 hours at 30°C. Wells of a 96-well glass-bottomed plate (Sigma CLS4580) were coated with 50 μL of 1 mg/mL Concanavalin A (Sigma) dissolved in 20 mM sodium acetate. After washing wells with ddH2O, yeast cultures were transferred and spun down at 1000 RPM for 3 min. All imaging was carried out using a 60X oil immersion objective (NA 1.4) on a Nikon TI Eclipse microscope with a CSU-X1 confocal spinning disk, an EM-CCD camera, and appropriate laser lines, dichroics and filters. Blue light photoactivation was carried out by exciting with a 488 nm laser line or illumination from a 450 nm LED. To quantify the kinetics of cluster assembly/disassembly (Supplementary Fig. 3) we measured the background-subtracted mean μ and standard deviation σ of pixel intensities within each cell and at each timepoint. We then calculated the signal-to-noise ratio (SNR) of pixel intensities (defined as SNR = μ / σ), a sensitive and reproducible metric reflecting the extent of clustering25.
Analysis of cluster number and assembly/disassembly kinetics
We observed light-dependent organelle formation in yeast made to express three of our optogenetic systems: optoClusters, optoDroplets and PixELLs. Yet in each of these cases, the extent and timescale of clustering differed, an observation that we sought to describe more quantitatively using live-cell imaging in each case. We imaged yeast strains yNS47 (OptoDroplets), yNS49 (OptoClusters) and YEZ232 (PixELLs) in the FusionRed channel during cycles of 450 nm blue light illumination or darkness. We then quantified the extent of clustering by analyzing the number of clusters per cell and the kinetics of cluster assembly/disassembly using changes in the pixel-to-pixel signal-to-noise ratio (SNR, which measures the homogeneity of cluster intensities (i.e., lower SNR = more clustering). We found that on average we observed between 1–4 clusters per cell across these three systems, with fewer PixELLs and OptoDroplets per cell, and more OptoClusters per cell under clustering conditions (Supplementary Fig. 3a). However, the number of clusters certainly depends on a large number of parameters, including the length of time of clustering (due to processes such as ripening and fusion events) and the expression level of the constructs, so these results should be taken as indicative of results in our conditions, not universal properties of these optogenetic tools.
We also measured the kinetics of cluster assembly/disassembly, observing fast light-induced changes in all three systems. These changes worked in opposing directions depending on the optogenetic system used. For instance, we observed light-induced assembly over ~5 min in OptoCluster/OptoDroplet cells (Supplementary Fig. 3b–c), and light-induced disassembly within 30 sec in PixELL-expressing cells (Supplementary Fig. 3d). In contrast, dark-induced reversion occurs on different timescales for each optogenetic system: PixD switches back to its dark-state conformation with a half-life of ~5 sec2, whereas Cry2 switches back in ~2 min and Cry2olig in ~20 min3. Matching these optogenetic dark-state kinetics, we found that Cry2-based OptoDroplets dissociated in minutes, Cry2olig-based OptoClusters were not fully dissociated even after 30 min in the dark, and PixD-based PixELLs reassembled in minutes after dark incubation (Supplementary Fig. 3b–d).
Assessing and correcting for violacein product photobleaching
An optogenetic system requires continuous illumination with blue light which raises the possibility of light-induced photobleaching or degradation. To measure the photobleaching and/or degradation of PDV, DV, proviolacein, and violacein under blue light stimulation, we measured the production of PDV, DV, proviolacein, and violacein in strains constitutively expressing violacein enzymes without optogenetic control (yNS51, MZW342, MZW375, MZW377, and MZW378) under lit and dark conditions. In four of these strains (MZW342, MZW375, MZW377 and MZW378), expression of the violacein pathway enzymes was under the control of a β-estradiol inducibler promoter. We thus added β-estradiol to a final concentration of 1 μM throughout the fermentation1. For all light stimulation experiments we used the same blue light source under identical conditions.
We found that DV is degraded slowly and at a constant rate by blue light, so that illuminated samples always exhibited a proportionally smaller DV peak by HPLC (Supplementary Fig. 5). Individual points represent yNS51, MZW342, MZW375, MZW377, and MZW378, five strains with different DV production levels. We thus normalized all DV measurements performed after blue light illumination using the standard curve produced by these control strains (Supplementary Fig. 5). We observed no photobleaching by blue light for PDV, proviolacein, or violacein in these assays. No differences in growth rate or maximum optical density were observed from these strains when cultured in the light or dark
Light panel set ups
All light illumination for fermentations were performed with blue LED panels (HQRP New Square 12” Grow Light Blue 517 LED 14W), placed 40 cm from cell cultures. At these heights, the light panel outputs ranged from 73 μmoles/m2/s to 82 μmoles/m2/s, based on measurements taken using a quantum meter (Model MQ-510 from Apogee Instruments).
Yeast fermentations
8 colonies were chosen from each zeocin selection plate for optoCluster and PixELL fermentations to screen for DV and PDV production. Each colony’s [Zeo]max was noted and the colony was saved for future use by plating onto an agar plate. Colonies were used to inoculate 1 mL of SC-his + 2% glucose media in 24-well plates and grown overnight at 30 °C, 200 RPM, and under ambient light conditions. Each culture was then diluted into 2 different plates and grown for 20 hours, with one plate grown under continuous blue light and the other wrapped in tinfoil and grown in the dark. Each culture was then centrifuged at 1,000 RPM for 5 minutes under ambient light and cell pellets were resuspended in 1 mL of fresh SC-his + 2% glucose media. The plates were then incubated under their respective light conditions for 96 hours at 30 °C. Each 1 mL sample was then transferred to a 1.5 mL microcentrifuge tube and centrifuged at 1,000 RPM for 10 minutes under ambient light. The supernatants were discarded and pellets were then resuspended in 1 mL of methanol (see Extraction and Quantification of Violacein Pathway Products). After identification of the colonies exhibiting the highest light-dependent fold-change in metabolic yield, 4 replicates of the best colonies were then performed and quantified using the same protocol.
Extraction and quantification of deoxyviolacein pathway products
To quantify the DV, PDV, proviolacein, and violacein produced by the fermentations, the cell pellets obtained from centrifuging 1 mL fermentation samples (see Yeast Fermentation, above) were resuspended in 1 mL of 100% methanol and incubated at 95 °C for 15 minutes, vortexing for 2–5 seconds halfway through. Cells were then centrifuged at 13,000 RPM for 5 min and approximately 800 μL of supernatant was transferred to a new microcentrifuge tube. The new microcentrifuge tube was again centrifuged at the same conditions and transferred to HPLC vials for analysis. Filtration of extracts was avoided because products are trapped by the filter membrane. Extracts were run on an Alltech Alltima C18 column (250 × 4.6 mm, 5 μm particle size) on an Agilent 1200 Series LC system using 0.1% trifluoroacetic acid in acetonitrile (Solvent A), 0.1% trifluoroacetic acid in water (Solvent B), and the following method: start at 5% Solvent A; from 0–10 min, linear increase of Solvent A from 5% to 95%; hold at 95% Solvent A from 10–13 min; Linear decrease of Solvent A from 95% to 5% A from 13–13.5 min. The flow rate was 0.9 mL/min and products were monitored with an Agilent diode array detector (DAD) at 565 nm. Product identities were confirmed using an Agilent 6120 Quadrupole mass spectrometer, using electrospray ionization in positive mode. Retention times were 10.04 min for proviolacein (m/z [M+H]+ of 328), 10.84 min for prodeoxyviolacein (m/z [M+H]+ of 312), 10.95 min for violacein (m/z [M+H]+ of 344), and 12.25 min for deoxyviolacein (m/z [M+H]+ of 328).
Normalization for photobleaching of deoxyviolacein
To normalize for the degradation of any violacein pathway product under blue light, we performed fermentation and product analysis on 5 strains that produced constant amounts of deoxyviolacein-pathway chemicals: MZW342, MZW375, MZW377, MZW378, and yNS51 (Supplementary Table 2). Concentrations of DV, PDV, proviolacein, and violacein were measured for cultures that were fermented under identical conditions except in either blue light or in the dark. These fermentations mimicked the ones we performed to control assembly or disassembly of synthetic organelles – 96 hours of fermentations after resuspension in new media. Although PDV, proviolacein, and violacein levels were consistent for dark and light conditions, DV exhibited significant but consistent degradation under blue light (Supplementary Fig. 5). Least squares regression of the data reveals that DV measurements of samples fermented under blue light scaled linearly DV measurements of samples fermented in the dark. To compensate for this degradation, we corrected all DV measurements for cultures fermented under blue light by a factor of 3.453.
Measurement of fluorescence protein levels
To measure the protein concentrations of the relevant clustered enzymes, we grew cells from each of YEZ281, YEZ257, yNS34, and yNS36 in liquid SC-his overnight in the dark (tin foiled). Each culture was then diluted into 2 different plates in quadruplicates to 0.1 OD600 in fresh SC-his and grown for 20 hours, with one plate grown under continuous blue light and the other wrapped in tinfoil and grown in the dark. Each culture was then centrifuged at 1,000 RPM for 5 minutes under ambient light and cell pellets were resuspended in 1 mL of fresh SC-his + 2% glucose media.
The plates were then incubated under their respective light conditions for 96 hours at 30°C. Cells were then diluted 1:10 into fresh SC-his + 2% glucose media. Fluorescence and optical density (OD600) measurements were taken using a TECAN plate reader (infinite M200PRO). The excitation and emission wavelengths used for citrine fluorescence measurements were 485 nm and 535 nm, respectively, using an optimal gain for all measurements. The excitation and emission wavelengths used for fusion red fluorescence measurements were 570 nm and 615 nm, respectively, using an optimal gain for all measure. To process fluorescence data, the background fluorescence from the media was first subtracted from values. Then, the fluorescence/OD600 values of cells lacking a fluorescence protein construct were subtracted from the fluorescence values (fluorescence/OD600) of each sample to normalize for light bleaching of the media and cell contents. Thus, reported values were calculated according to the following formula.
YEZ140 was used as the no-fluorescence control. All fluorescence measurements were done at the end of experiments or on samples taken from experimental cultures.
Yeast cell fixing and staining with 4′,6-diamidino-2-phenylindole (DAPI)
We previously found that PixeLLs and OptoDroplets/OptoClusters show an increased propensity to cluster in the nucleus of mammalian cells24,25, raising the possibility that light-controlled clusters are also predominantly localized to the nucleus in yeast. To determine if this was the case, we set out to fix and stained PixELL-expressing cells and co-stained them with DAPI to mark the cell nucleus.
Yeast strains YEZ232 and YEZ555 were inoculated into SC-his + 2% glucose medium and grown to an OD600 of 2. From these cultures, 900 μL was transferred to a 1.5 mL microcentrifuge tube containing 100 μL of 37% (w/v) formaldehyde. This mixture was incubated at room temperature for 90 minutes before spinning down at 7,500 g for 1 minute and washing with 0.3 mL of Dulbecco’s phosphate-buffered saline (DPBS) solution two times. After resuspension in 0.3 mL of DPBS, 0.7 mL of 200 Proof ethanol was added to permeabilize and incubated at room temperature for 30 minutes. Cells were then spun down again and resuspended in 300 μL of DPBS, diluted 10-fold into new DPBS and mixed 1:1 with 100 ng/mL DAPI in DPBS solution. Cells were then plated and imaged based on our standard microscopy procedure (see Fluorescence Microscopy section).
Statistics
Statistical significance was determined using a standard t test for p values. T scores were calculated by the formula: . P values were calculated using a degree of freedom of 3 and a one-sided t-test calculator.
Supplementary Material
Acknowledgements
We thank all members of the Toettcher and Avalos laboratories for helpful comments. We also thank J. Dueber for kindly providing violacein enzyme plasmids. E.Z. thanks Siyang Han for her support and would also like to ask: Will you marry me? This work was supported by the Maeder Graduate Fellowship in Energy and the Environment (to EMZ), NIH grant DP2EB024247 (to JET) and The Pew Charitable Trusts, the U.S. DOE Office of Biological and Environmental Research, Genomic Science Program Award DESC0019363, and NSF CAREER Award CBET-1751840 (to JLA), and a Schmidt Transformative Technology grant (to JET and JLA).
Footnotes
Competing financial interests statement
Some of the authors are co-inventors on patent applications harnessing optogenetics for metabolic engineering (J.L.A., J.E.T., E.M.Z.; patent application WO2017177147A1), and establishing optogenetic control of protein clustering (J.E.T.; patent application US20170355977A1).
Data availability statement
All plasmids, strains, and raw data will be made available upon request to the corresponding authors.
References
- 1.Nielsen J & Keasling JD Engineering Cellular Metabolism. Cell 164, 1185–1197 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Keasling JD Manufacturing Molecules Through Metabolic Engineering. Science 50, 1355 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ajikumar PK et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalwani MA, Zhao EM & Avalos JL Current and future modalities of dynamic control in metabolic engineering. Curr. Opin. Biotechnol 52, 56–65 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Tomala K & Korona R Evaluating the fitness cost of protein expression in saccharomyces cerevisiae. Genome Biol. Evol 5, 2051–2060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockman IM & Prather KLJ Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab. Eng 28, 104–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan SZ & Prather KL Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol 41, 28–35 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Thomik T, Wittig I, Choe JY, Boles E & Oreb M An artificial transport metabolon facilitates improved substrate utilization in yeast. Nat. Chem. Biol 13, 1158–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Lin JL, Zhu J & Wheeldon I Synthetic Protein Scaffolds for Biosynthetic Pathway Colocalization on Lipid Droplet Membranes. ACS Synth. Biol 6, 1534–1544 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Pedley AM & Benkovic SJ A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci 42, 141–154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French JB et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y et al. Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat. Commun 8, 15212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanaswamy R et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci 106, 10147–10152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kistler HC & Broz K Cellular compartmentalization of secondary metabolism. Front. Microbiol 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellana M et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol 32, 1011–1018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George KW et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab. Eng 47, 60–72 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Dueber JE et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol 27, 753–759 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Li T, Chen X, Cai Y & Dai J Artificial Protein Scaffold System (AProSS): An efficient method to optimize exogenous metabolic pathways in Saccharomyces cerevisiae. Metab. Eng 49, 13–20 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Lau YH, Giessen TW, Altenburg WJ & Silver PA Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun 9, 1311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avalos JL, Fink GR & Stephanopoulos G Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol 31, 335–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLoache WC, Russ ZN & Dueber JE Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun 7, 11152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammer SK & Avalos JL Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol 13, 823–832 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Toettcher JE, Voigt C a, Weiner, O. D. & Lim, W. a. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods 8, 35–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin Y et al. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dine E, Gil AA, Uribe G, Brangwynne CP & Toettcher JE Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Syst 6, 655–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taslimi A et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun 5, 4925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura H et al. Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions. Nat. Mater 17, 79–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS & Schaffer DV Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–52 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Gil AA et al. Photoactivation of the BLUF Protein PixD Probed by the Site-Specific Incorporation of Fluorotyrosine Residues. J. Am. Chem. Soc 139, 14638–14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shemiakina II et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun 3, 1204 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Yuan J & Ching CB Combinatorial Assembly of Large Biochemical Pathways into Yeast Chromosomes for Improved Production of Value-added Compounds. ACS Synth. Biol 4, 23–31 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Zhao EM et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan H & Bauer CE PixE promotes dark oligomerization of the BLUF photoreceptor PixD. Proc. Natl. Acad. Sci 105, 11715–11719 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brachmann CB et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Giaever G & Nislow C The yeast deletion collection: A decade of functional genomics. Genetics 197, 451–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Entian KD & Kötter P 25 Yeast Genetic Strain and Plasmid Collections. Methods in Microbiology 36, 629–666 (2007). [Google Scholar]
- 38.Lee ME, Aswani A, Han AS, Tomlin CJ & Dueber JE Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res 41, 10668–10678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bracha D et al. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 175, 1467–1480.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan KS, Balibar CJ, Turo KE, Walsh CT & Drennan CL The violacein biosynthetic enzyme VioE shares a fold with lipoprotein transporter proteins. J. Biol. Chem 283, 6467–6475 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin VJJ, Pitera DJ, Withers ST, Newman JD & Keasling JD Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nature 21, 796–802 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Ro D et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 3–6 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.