To the Editor
Multiple myeloma (MM) is the third most common hematological malignancy, after Non-Hodgkin Lymphoma and Leukemia. MM is generally preceded by Monoclonal Gammopathy of Undetermined Significance (MGUS) [1], and epidemiological studies have identified older age, male gender, family history, and MGUS as risk factors for developing MM [2].
The somatic mutational landscape of sporadic MM has been increasingly investigated, aiming to identify recurrent genetic events involved in myelomagenesis. Whole exome and whole genome sequencing studies have shown that MM is a genetically heterogeneous disease that evolves through accumulation of both clonal and subclonal driver mutations [3] and identified recurrently somatically mutated genes, including KRAS, NRAS, FAM46C, TP53, DIS3, BRAF, TRAF3, CYLD, RB1 and PRDM1 [3–5].
Despite the fact that family-based studies have provided data consistent with an inherited genetic susceptibility to MM compatible with Mendelian transmission [6], the molecular basis of inherited MM predisposition is only partly understood. Genome-Wide Association (GWAS) studies have identified and validated 23 loci significantly associated with an increased risk of developing MM that explain ~16% of heritability [7] and only a subset of familial cases are thought to have a polygenic background [8]. Recent studies have identified rare germline variants predisposing to MM in KDM1A [9], ARID1A and USP45 [10], and the implementation of next-generation sequencing technology will allow the characterization of more such rare variants.
In this study, we sought to explore the involvement of rare germline genetic variants in susceptibility to MM.
Within our discovery cohort of peripheral blood samples (see Supplementary Methods) from 66 individuals from 23 unrelated families analyzed by WES, DIS3 (NM_014953) was the only gene in which putative loss-of-function variants were observed in at least two families. An additional cohort of 937 individuals (148 MM, 139 MGUS, 642 unaffected relatives and eight individuals with another hematological condition) from 154 unrelated families (including the individuals in the discovery cohort) were screened for germline variants in DIS3 using targeted sequencing (Supplementary Table S1). In total, we detected DIS3 germline putative loss-of-function variants in four unrelated families. The DIS3 genotypes for the identified variants were concordant between WES and targeted sequencing (where available) and independently confirmed by Sanger sequencing on DNA extracted from uncultured whole blood. The variant allele frequencies (VAF) were close to 50%, as expected of a germline variant (Supplementary figure S1).
The DIS3 gene, located in 13q22.1, encodes for the catalytic subunit of the human exosome complex, and is recurrently somatically mutated in MM patients [4, 5, 11, 12]. The somatic variants are predominantly missense variants localized in the RNB domain mainly abolishing the exoribonucleolytic activity [4, 13], and are often accompanied by LOH or biallelic inactivation due to 13q14 deletion, implying a tumor suppressor role for DIS3 in MM [5, 12, 13].
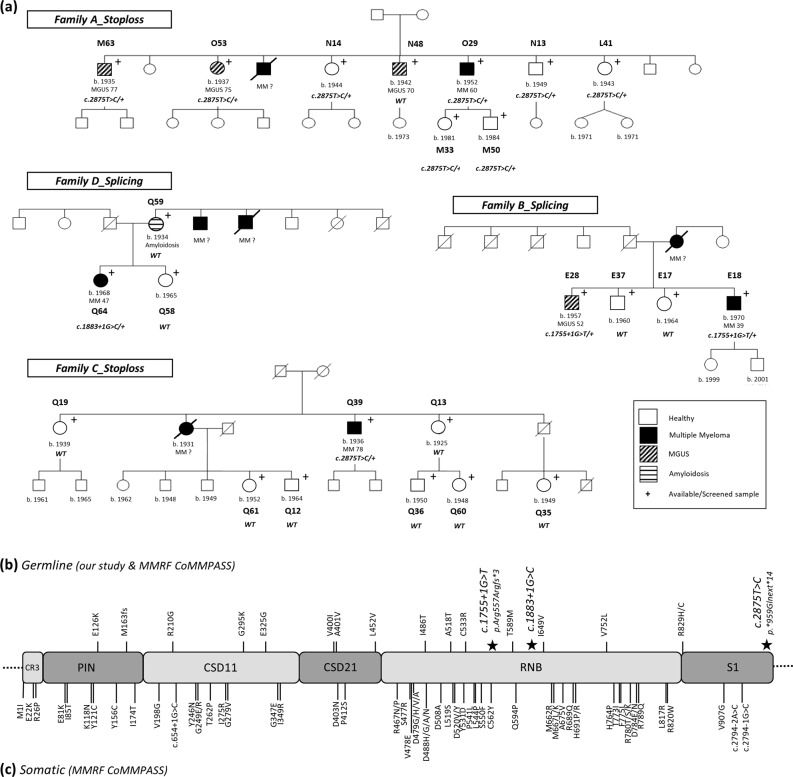
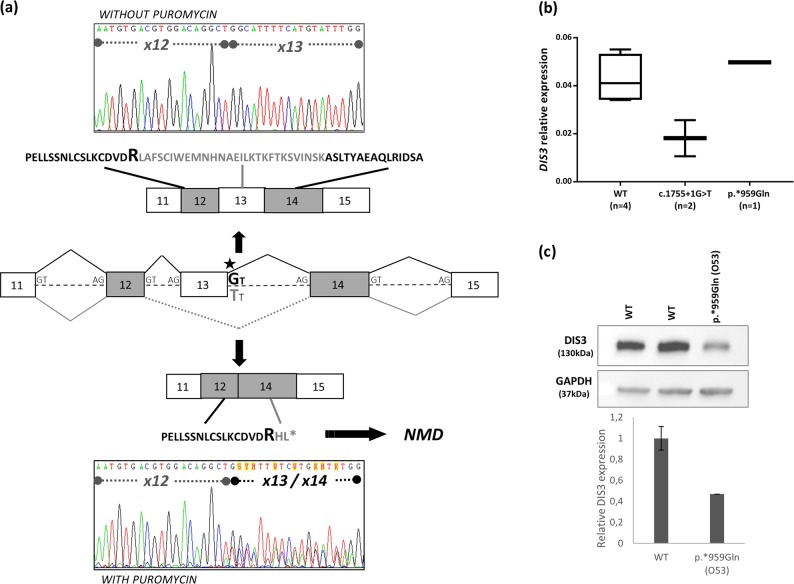
The first DIS3 variant, observed in 2 affected siblings (1 MGUS and 1 MM case) from family B (Fig. 1a), was located in the splice donor site of exon 13 (c.1755+1G>T; chr13: 73,345,041; GRCh37/hg19, rs769194741) (Supplementary Figure S1a). It is predicted to abolish the splice donor site and cause skipping of exon 13, introducing a premature termination codon (p.Arg557Argfs*3) and result in a truncated DIS3 protein that lacks part of the exonucleolytic active RNB and S1 domains (Fig. 1b, c). The presence of this variant in two siblings, implying Mendelian segregation, is consistent with a germline, rather than somatic, origin. We investigated whether a DIS3 transcript from the variant allele is generated but is subsequently eliminated by Nonsense Mediated Decay (NMD) by incubating Lymphoblastoid Cell Lines (LCLs) derived from the two c.1755+1G>T allele carriers with and without puromycin, which suppresses NMD. The mRNA transcript corresponding to the variant allele was clearly present in LCLs treated with puromycin in both carriers, whereas not detectable in untreated LCLs (Fig. 2a), consistent with the variant allele being transcribed but subsequently degraded via the NMD pathway. In line with this observation, analysis of DIS3 mRNA expression by qRT-PCR showed an average 50% reduced expression in the c.1755+1G>T carriers (range 40.7−61.4%) as compared to non-carriers (Fig. 2b). A second splicing variant (c.1883+1G>C; chr13: 73,342,922; GRCh37/hg19) located in the splice donor site of exon 14 within the RNB domain was identified in a MM case from family D (Fig. 1a, b, Supplementary figure S1c). However, the individual’s mother (Q59), affected with amyloidosis, did not carry the variant, implying that MM in the allele carriers’ maternal uncles is unlikely to be explained by this DIS3 variant. Whether the mRNA transcript encoded by this germline variant undergoes NMD could not be explored due to lack of appropriate material (LCLs, RNA).
Fig. 1.
DIS3 variants in MM cases. a Pedigrees from families carrying a germline DIS3 variant. Available samples for screening are marked with a “+” symbol. Families A and C carry the p.*959Glnext*14 (c.2875T>C) stop-loss variant. Family B carries the c.1755+1G>T splicing variant and family D carries the c.1883+1G>C splicing variant. The genotype of all screened individuals is shown on each pedigree. WT: wild type. b, c Schematic representation of identified germline and somatic variants in the distinct DIS3 protein domains. b Germline variants were identified through WES and targeted resequencing in families with reoccurrence of MM/MGUS as well as in a collection of sporadic MM cases (MMRF CoMMpass Study). The DIS3 variants discussed in the present study are depicted with a star on the upper part of the figure. c Somatic DIS3 variants were identified in sporadic MM cases from the MMRF CoMMpass Study. We observe that in contrast to the clustering of somatic DIS3 missense variants in the RNB and PIN domains, germline variants are scattered throughout the gene and consist of splicing, stop-loss and missense variants
Fig. 2.
DIS3 c.1755+1G>T splicing variant results in nonsense-mediated mRNA decay (NMD) and affects mRNA expression, while the c.2875C>T (p.*959Glnext*14) stop-loss variant affects protein levels. a LCLs from patients E18 and E28 (not shown) carrying the c.1755+1G>T splicing variant were cultured with and without puromycin. The chromatogram from treated cells (with puromycin) showed a mixture of the wild-type and mutant transcript lacking exon 13, which was not detected in the non-treated cells (without puromycin). Thus, the mutant transcript is degraded by NMD. b Box plot representing the relative DIS3 mRNA expression in c.1775+1G>A (n=2) and p.*959Glnext*14 (n = 1) carriers compared to non-carriers (n = 4). All reactions were performed in triplicates. c Western blot with an anti-DIS3 antibody was performed in LCLs from one p.*959Glnext*14 carrier and two wild-type individuals (anti-GAPDH antibody as internal control). The relative DIS3 expression in the p.*959Glnext*14 carrier was reduced by 50% compared to non-carriers, suggesting that the mutant allele is translated but degraded shortly after
A third DIS3 variant disrupting the wild-type termination codon (stop-loss) (c.2875T>C; p.*959Gln; chr13:73,333,935; GRCh37/hg19, rs141067458) (Fig. 1b, Supplementary Figure S1b) was identified in two unrelated families (A and C, Fig. 1a). This variant is expected to result in a putative read-through variant and a DIS3 protein with an additional 13 amino acids in the C-terminus (p.*959Glnext*14). It was detected in 3 out of 4 affected siblings (2 MGUS (M63, O53) and 1 MM case (O29)), as well as 5 unaffected relatives (N14, N13, L41, M33 M50) from family A. The Mendelian segregation of this variant in this pedigree is also consistent with germline origin. An additional MM case from family C carried the variant, while we were unable to assess the other MM-afflicted family member (Fig. 1a). As expected of a stop-loss variant, NMD was not observed (data not shown), and gene expression analysis showed no effect on DIS3 mRNA levels (Fig. 2b). However, western blot analysis demonstrated that DIS3 protein levels were markedly lower (~50%) in the p.*959Glnext*14 carrier (O53, family A) compared to non-carriers (Fig. 2b, c).
Next, we sought to determine if rare, putative deleterious variants in DIS3 were more frequent in an independent series of MM cases compared to unaffected individuals. We performed mutation burden tests between 781 MM cases and 3534 controls from the MMRF CoMMpass Study with WES data available. After testing for systemic bias in this dataset (see Supplementary Methods, Supplementary Figure S2), we undertook a burden test for association between functional DIS3 variants and MM. DIS3 putative functional variants (truncating and likely deleterious missense variants, see Supplementary Methods) were more frequent among MM patients (30/781) than controls (72/3534) (OR = 1.92 95%CI:1.25–2.96, p = 0.001). Although the p.*959Glnext*14 stop-loss variant was recurrently found in 10/781 MM cases and 15/3534 controls (OR = 3.07 95%CI:1.38 to 6.87, p = 0.0007), it did not entirely explain the excess of DIS3 variants among cases as there is evidence for association with other putative functional variants (Supplementary Figure S3a). We additionally genotyped the p.*959Glnext*14 stop-loss variant in an independent series of sporadic MM cases and controls from the IMMEnSE Consortium. While this variant was very rare in this series (8/3020 MM cases relative to 3/1786 controls), there was a consistent but non-significant association between this variant and MM (OR = 3.15 95% CI: 0.74–13.43 p = 0.122).
To explore the functional consequence of germline DIS3 variants, we compared MM tumor transcriptomes from patients harboring germline (n = 21) and somatic (n = 96) DIS3 putative functional variants to non-carriers (n = 655). Differential expression analyses showed an enrichment of pathways associated with global ncRNA processing and translational termination in germline DIS3 carriers including ncRNA processing, ncRNA metabolic process, translational termination, and RNA metabolism. Among somatic DIS3 carriers, significantly enriched pathways include interferon alpha/beta signalling, mRNA splicing, mRNA processing and transcription (Supplementary Figure S3b, Supplementary tables S3 and S4a–d). These findings are consistent with the proposed DIS3 role in regulating mRNA processing [14] and more specifically mRNA decay, gene expression and small RNA processing [15]. We also observed that, several long-intergenic non-protein coding RNAs, non-coding and antisense RNAs were significantly enriched among DIS3 carriers (Supplementary table S5a, b) supporting previous studies that demonstrate an accumulation of transcripts from non-protein coding regions, snoRNA precursors and certain lncRNAs in DIS3 mutant cells, along a general deregulation of mRNA levels probably due to the sequestration of transcriptional factors from the accumulated nuclear RNAs [16].
To our knowledge, this is the first observation of germline DIS3 likely deleterious variants in familial MM and our results suggest that the involvement of DIS3 in MM etiology may extend beyond somatic alterations to germline susceptibility. We reported rare germline DIS3 variants in ~2.6% of our cohort of families with multiple cases of MM and MGUS (4/154). The germline variants described here are predicted to have loss-of-function impact on DIS3. Consistent with this, the 1755+1G>T (rs769194741) splicing variant induces NMD and results in reduced DIS3 mRNA expression, supporting the proposal that DIS3 is acting as a tumor suppressor gene in MM [13]. Moreover, the c.2875T>C (rs141067458) stop-loss variant (p.*959Glnext*14) results in reduced DIS3 protein expression suggesting that the mutant allele is translated but degraded shortly after. Notably, in contrast to the clustering of somatic DIS3 mutations in the PIN and RNB domains, germline variants identified both in familial and sporadic MM cases are scattered throughout the gene (Fig. 1b, c). Despite the fact that these variants do not segregate perfectly with MM in the identified families and the rarity of DIS3 germline likely deleterious variants limits our statistical power, the subsequent mutation burden and transcriptome analyses provided supportive data towards DIS3 acting as an “intermediate-risk” MM susceptibility gene.
Supplementary information
18-LEU-1250_RevisedManuscript_SupplementaryData
Acknowledgements
This work was supported by the French National Cancer Institute (INCA) and the Fondation Française pour la Recherche contre le Myélome et les Gammapathies (FFMRG), the Intergroupe Francophone du Myélome (IFM), NCI R01 NCI CA167824 and a generous donation from Matthew Bell. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD018522. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank the Association des Malades du Myélome Multiple (AF3M) for their continued support and participation. Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: James D. McKay, Charles Dumontet
Contributor Information
James D. McKay, Phone: +33 4 72 73 80 93, Email: mckayj@iarc.fr
Charles Dumontet, Phone: +33 4 78 77 72 36, Email: charles.dumontet@chu-lyon.fr.
Supplementary information
The online version of this article (10.1038/s41375-019-0452-6) contains supplementary material, which is available to authorized users.
References
- 1.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113:5418–22. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan GJ, Davies FE, Linet M. Myeloma aetiology and epidemiology. Biomed Pharmacother. 2002;56:223–34. doi: 10.1016/S0753-3322(02)00194-4. [DOI] [PubMed] [Google Scholar]
- 3.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2013;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan GJ, Johnson DC, Weinhold N, Goldschmidt H, Landgren O, Lynch HT, et al. Inherited genetic susceptibility to multiple myeloma. Leukemia. 2014;28:518–24. doi: 10.1038/leu.2013.344. [DOI] [PubMed] [Google Scholar]
- 7.Went M, Sud A, Försti A, Halvarsson BM, Weinhold N, Kimber S, et al. Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma. Nat Commun. 2018;9:3707. doi: 10.1038/s41467-018-04989-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halvarsson BM, Wihlborg AK, Ali M, Lemonakis K, Johnsson E, Niroula A, et al. Direct evidence for a polygenic etiology in familial multiple myeloma. Blood Adv. 2017;1:619–23. doi: 10.1182/bloodadvances.2016003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Xiaomu, Calvo-Vidal M. Nieves, Chen Siwei, Wu Gang, Revuelta Maria V., Sun Jian, Zhang Jinghui, Walsh Michael F., Nichols Kim E., Joseph Vijai, Snyder Carrie, Vachon Celine M., McKay James D., Wang Shu-Ping, Jayabalan David S., Jacobs Lauren M., Becirovic Dina, Waller Rosalie G., Artomov Mykyta, Viale Agnes, Patel Jayeshkumar, Phillip Jude, Chen-Kiang Selina, Curtin Karen, Salama Mohamed, Atanackovic Djordje, Niesvizky Ruben, Landgren Ola, Slager Susan L., Godley Lucy A., Churpek Jane, Garber Judy E., Anderson Kenneth C., Daly Mark J., Roeder Robert G., Dumontet Charles, Lynch Henry T., Mullighan Charles G., Camp Nicola J., Offit Kenneth, Klein Robert J., Yu Haiyuan, Cerchietti Leandro, Lipkin Steven M. Germline Lysine-Specific Demethylase 1 (LSD1/KDM1A) Mutations Confer Susceptibility to Multiple Myeloma. Cancer Research. 2018;78(10):2747–2759. doi: 10.1158/0008-5472.CAN-17-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waller RG, Darlington TM, Wei X, Madsen MJ, Thomas A, Curtin K, et al. Novel pedigree analysis implicates DNA repair and chromatin remodeling in multiple myeloma risk. PLoS Genet. 2018;14:e1007111. doi: 10.1371/journal.pgen.1007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weißbach S, Langer C, Puppe B, Nedeva T, Bach E, Kull M, et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br J Haematol. 2015;169:57–70. doi: 10.1111/bjh.13256. [DOI] [PubMed] [Google Scholar]
- 12.Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018;132:587–97. [DOI] [PMC free article] [PubMed]
- 13.Lionetti M, Barbieri M, Todoerti K, Agnelli L, Fabris S, Tonon G, et al. A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget. 2015; 6. 10.18632/oncotarget.4674. [DOI] [PMC free article] [PubMed]
- 14.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 15.Robinson S, Oliver A, Chevassut T, Newbury S. The 3’ to 5’ exoribonuclease DIS3: from structure and mechanisms to biological functions and role in human disease. Biomolecules. 2015;5:1515–39. doi: 10.3390/biom5031515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczepińska T, Kalisiak K, Tomecki R, Labno A, Borowski LS, Kulinski TM, et al. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015;25:1622–33. doi: 10.1101/gr.189597.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
18-LEU-1250_RevisedManuscript_SupplementaryData