To the Editor:
Chimeric antigen receptors (CARs) have undoubtedly revolutionized immunotherapy, especially in the B-cell acute lymphoblastic leukemia (ALL) arena where over 80% of complete remissions are observed in refractory/relapsed (R/R) B-cell ALL patients treated with CD19-directed CAR T-cells (CARTs) [1]. However, despite holding an unprecedented promise, several issues still have to be resolved before CARTs can be expanded to novel targets and/or malignancies or even provided as first-line treatment in B-cell ALL [2]. For instance, toxicities such as cytokine release syndrome and immune escape mechanisms including loss of the antigen under CART-mediated pressure remain major concerns, urging further research on the mechanisms underlying CARTs cytotoxicity.
In this sense, loss of CD19 antigen is frequently observed after CD19-directed CARTs therapy in B-cell ALL [3, 4], but is particularly common in MLL-rearranged (MLLr) B-cell ALL, an aggressive subtype of B-cell ALL (dismal in MLL-AF4+ infants) associated with lymphoid-to-myeloid lineage switch [3, 5, 6]. We read with interest the work recently published in Leukemia by Li et al. reporting a novel CAR targeting both CD19 and CD133 [7]. This study proposes to use a bi-specific CAR targeting both CD19 and CD133 antigens in a Boolean OR-gate approach for MLLr B-cell ALL as a strategy to avoid and treat CD19- relapses. The authors reasoned that CD133, encoded by PROM1 gene, is a specific marker for MLLr leukemia because PROM1 is an MLL target, especially in MLL-AF4 B-cell ALL [8–10]. They went on and performed in vitro assays showing than CD19/CD133 bi-specific CAR triggers robust cytotoxicity against CD19 + CD133 + and CD19-CD133+ B-cell lines [7], thus suggesting it may help in reducing subsequent lineage switch in MLLr B-cell ALL.
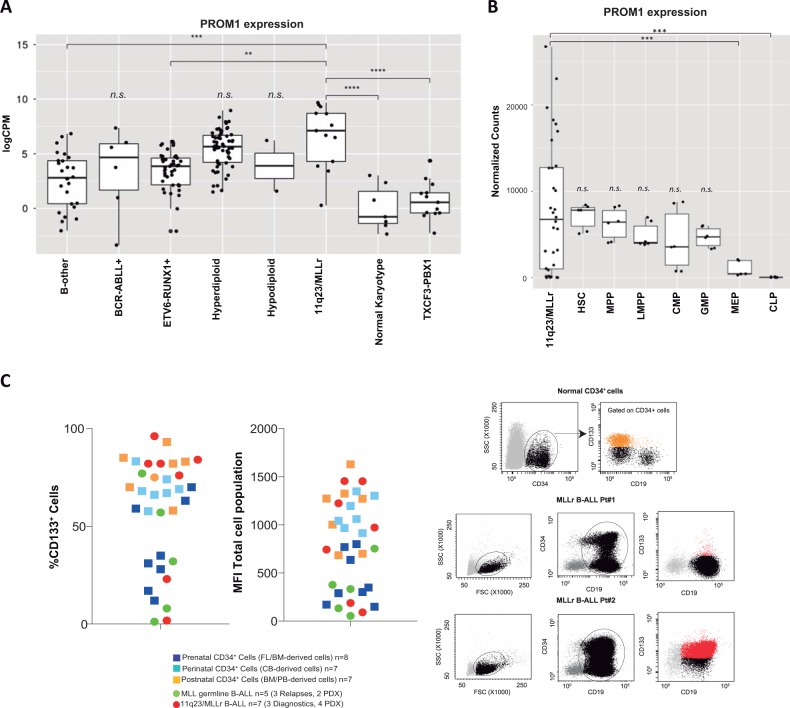
A major drawback for CD133 as target in immunotherapy is its expression in hematopoietic stem and progenitor cells (HSPCs), which would likely exert “on-target off-tumor” myeloablative, life-threatening toxicity [11, 12]. Because B-cell ALL is molecularly heterogeneous and can be diagnosed during infancy, childhood and adulthood, we have characterized PROM1/CD133 expression in a large cohort of cytogenetically distinct B-cell ALL subgroups (n = 212 patients) as well as in different subpopulations of normal CD34+ HSPCs obtained across hematopoietic ontogeny from 22-weeks old human fetal liver (FL, prenatal), cord blood (CB, perinatal), and adult G-CSF-mobilized peripheral blood/bone marrow (PB/BM, postnatal). An initial analysis of publicly available RNA-seq data [13] from 170 diagnostic B-cell ALL patients confirmed that PROM1 is overexpressed in patients with MLLr B-cell ALL, although its expression is not significantly higher than in other cytogenetic subgroups (Fig. 1a). We then analyzed PROM1 during HSPC development and observed that PROM1 is highly expressed in early normal hematopoietic stem cells (HSC) and multipotent progenitors (MPP) with its expression decreasing from the lymphoid-primed multipotent progenitors (LMPP) onwards with its expression being marginal at later stages of myeloid differentiation (megakaryocyte-erythroid progenitors, MEP) and common lymphoid progenitors (CLP) [14] (Fig. 1b). Importantly, 70% (22/32) of 11q23/MLLr B-cell patients (both MLL-AF4 and MLL-AF9) express equal (9/32) or lower (13/32) PROM1 levels that HSCs and MPPs, which raises doubts about the suitability of PROM1 as a target for B-cell ALL immunotherapy [15].
Fig. 1.
Characterization of CD133/PROM1 expression in B-cell ALL and normal HSPCs. a Expression level of PROM1 in the indicated cytogenetic subgroups of B-cell ALL (n = 170 patients at diagnosis) determined by RNA-seq represented in log2(CPM) scale, with CPM = counts per million [13]. b RNA-seq analysis comparing the expression of PROM1 in 11q23/MLLr B-cell ALL (n = 29 patients) with that in distinct fractions of Lin-CD34 + CD38-CD19- non-lymphoid normal HSPCs (HSC hematopoietic stem cells, MPP multipotent progenitors, LMPP lymphoid-primed multipotent progenitors, CMP common myeloid progenitors, GMP granulocyte-monocyte progenitor, MEP megakaryocyte-erythroid progenitors) and in common lymphoid progenitors (CLP) [14]. Data shown as normalized counts. The boxes define the first and third quartiles. The horizontal line within the box represents the median. c Frequency (left) and mean fluorescence intensity (MFI, middle) of CD133+ BM blasts/cells in MLLr (n = 7) and non-MLL B-cell ALLs (n = 5) primary diagnostic/relapse samples or primografts (PDXs), and normal CD34+ HSPCs derived from FL (n = 8), CB (n = 7) and adult PB/BM (n = 7). Representative FACS dot plots for CD133 in normal CD34+ HSPCs (upper right) and BM samples from two independent MLLr B-cell ALL patients (bottom right)
FACS clinical immunophenotyping provides a priori a more rapid and feasible clinically relevant diagnostic information than RNA-seq during the decision-making process. Thus, we next FACS-analyzed the expression of CD133 (PROM1 gene product) in the cell surface of BM-derived primary blasts and primografts (PDXs) obtained from 11q23/MLLr (n = 7) and non-MLL (n = 5) B-cell ALL patients, and in comparison with healthy prenatal (22 weeks old FL), perinatal (CB) and adult (PB/BM) CD34+ HSPCs (Fig. 1c). Consistent with the RNA-seq data, the expression of CD133 in 11q23/MLLr blasts is intermingled with that observed in CD34+ HSPCs across hematopoietic ontogeny (Fig. 1c).
Our data demonstrates that PROM1/CD133 is similarly expressed between MLLr B-cell ALL primary blasts and normal non-lymphoid HSPCs across ontogeny, thus indicating that “on-target, off-tumor” toxic/myeloablative effects are likely to occur if used in a bi-specific CAR approach where CD133 antigen will be constantly targeted regardless of the co-expression of CD19 in the same cell. Our data therefore raises concerns about using CD133 as a target for MLLr B-cell ALL immunotherapy. An alternative to circumvent HSPC toxicity would be to engineer dual CAR T-cells with one CAR engaging an antigen (i.e., CD19) mediating T-cell activation and another CAR engaging a second antigen (i.e., CD133) mediating T-cell co-stimulation [16]. Unfortunately, although such a CD19/CD133 dual CAR might be likely safe due to its cytotoxicity being restrained only to cells co-expressing CD19 and CD133, its specific cytotoxic performance will be poor since not the entire MLLr B-cell ALL blast population is CD19 + CD133+ (Fig. 1c). Another alternative approach to prevent HSPC toxicity would be to have in place a potent molecular switch (i.e., iCas9) to eliminate CAR133-expressing T-cells as necessary [17]. Further long-term in vivo studies using both primary B-cell ALL cells and normal HSCPs remain to be conducted to elucidate the efficacy versus the myeloablative toxicity of a CAR CD133 [18, 19].
Acknowledgements
We thank the Interfant treatment protocol and local physicians for contributing patient samples: Dr. Ronald W Stam (Princess Maxima Centre, Utrech), Dr. Mireia Camos and Dr. Jose Luis Fuster (Spanish Society of Pediatric Hematoncology), Dr. Paola Ballerini (A. Trousseau Hospital, Paris). We also thank Prof. Paresh Vyas (Oxford Univeristy, UK) and Prof. Kajsa Paulsson (Lund University, Sweden) for facilitating access to their RNA-seq database. This work has been supported by the European Research Council (CoG-2014-646903, PoC-2018-811220) to PM, the Spanish Ministry of Economy and Competitiveness (MINECO, SAF-SAF2016-80481-R, BIO2017-85364-R) to PM and EE, the Generalitat de Catalunya (SGR330, SGR102 and PERIS) to PM and EE, the Spanish Association against cancer (AECC-CI-2015) to CB, and the Health Institute Carlos III (ISCIII/FEDER, PI14-01191) to CB. PM also acknowledges financial support from the Obra Social La Caixa-Fundaciò Josep Carreras. SRZ and TV are supported by a Marie Curie fellowships. OM is supported by the Catalan Government through a Beatriu de Pinos fellowship. MB is supported by MINECO through a PhD scholarship. PM is an investigator of the Spanish Cell Therapy cooperative network (TERCEL).
Data availability
All genomic data is already publicly available. A full data availability will be provided.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Clara Bueno, Email: cbueno@carrerasresearch.org.
Pablo Menéndez, Email: pmenendez@carrerasresearch.org.
References
- 1.Maude SL, et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med. 2018;378:439–48. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghorashian S, Amrolia P, Veys P. Open access? Widening access to chimeric antigen receptor (CAR) therapy for ALL. Exp Hematol. 2018;66:5–16. doi: 10.1016/j.exphem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Gardner R, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood. 2016;127:2406–10. doi: 10.1182/blood-2015-08-665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sotillo E, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5:1282–95. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanjuan-Pla A, et al. Revisiting the biology of infant t(4;11)/MLL-AF4 + B-cell acute lymphoblastic leukemia. Blood. 2015;126:2676–85. doi: 10.1182/blood-2015-09-667378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiland J, et al. BCP-ALL blasts are not dependent on CD19 expression for leukaemic maintenance. Leukemia. 2016;30:1920–3. doi: 10.1038/leu.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, et al. TanCAR T cells targeting CD19 and CD133 efficiently eliminate MLL leukemic cells. Leukemia. 2018;32:2012–6. doi: 10.1038/s41375-018-0212-z. [DOI] [PubMed] [Google Scholar]
- 8.Guenther MG, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–8. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak AB, Nixon AM, Moffat J. The mixed lineage leukemia (MLL) fusion-associated gene AF4 promotes CD133 transcription. Cancer Res. 2012;72:1929–34. doi: 10.1158/0008-5472.CAN-11-3589. [DOI] [PubMed] [Google Scholar]
- 10.Wuchter C, et al. Impact of CD133 (AC133) and CD90 expression analysis for acute leukemia immunophenotyping. Haematologica. 2001;86:154–61. [PubMed] [Google Scholar]
- 11.Menendez P, et al. The composition of leukapheresis products impacts on the hematopoietic recovery after autologous transplantation independently of the mobilization regimen. Transfusion. 2002;42:1159–72. doi: 10.1046/j.1537-2995.2002.00190.x. [DOI] [PubMed] [Google Scholar]
- 12.Yin AH, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–12. [PubMed] [Google Scholar]
- 13.Paulsson K, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47:672–6. doi: 10.1038/ng.3301. [DOI] [PubMed] [Google Scholar]
- 14.Quek L, et al. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213:1513–35. doi: 10.1084/jem.20151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agraz-Doblas A, Bueno C, Bashford-Rogers R, Anindita R, Schneider P, Bardini M, et al. Unravelling the cellular origin and clinical prognosis markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica. 2018; in press. [DOI] [PMC free article] [PubMed]
- 16.Aldoss I, et al. Redirecting T cells to eradicate B-cell acute lymphoblastic leukemia: bispecific T-cell engagers and chimeric antigen receptors. Leukemia. 2017;31:777–87. doi: 10.1038/leu.2016.391. [DOI] [PubMed] [Google Scholar]
- 17.Diaconu I, et al. Inducible Caspase-9 selectively modulates the toxicities of CD19-Specific chimeric antigen receptor-modified T cells. Mol Ther. 2017;25:580–92. doi: 10.1016/j.ymthe.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, Lassailly F, Tettamanti S, Spinelli O, et al. Chimeric antigen recptors against CD33/CD123 antigens efficiently target primary acute myeloid cells in vivo. Leukemia. 2014;28:1596–605. doi: 10.1038/leu.2014.62. [DOI] [PubMed] [Google Scholar]
- 19.Kenderian SS, Ruella M, Shestova O, Klichincky M, Aikawa V, Morrissette JJD, et al. CD33-directed chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia. 2015;29:1637–47. doi: 10.1038/leu.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genomic data is already publicly available. A full data availability will be provided.