To the Editor:
Activating signaling mutations are common in acute leukemia with KMT2A (previously MLL) rearrangements (KMT2A-R) [1]. When defining the genetic landscape of infant KMT2A-R acute lymphoblastic leukemia (ALL), we identified a novel FLT3N676K mutation in both infant ALL and non-infant acute myeloid leukemia (AML) [1]. FLT3N676K was the most common FLT3 mutation in our cohort and we recently showed that it cooperates with KMT2A-MLLT3 in a syngeneic mouse model [1, 2]. To study the ability of FLT3N676K to cooperate with KMT2A-MLLT3 in human leukemogenesis, we transduced human CD34+-enriched cord blood (CB) cells and followed leukemia development immunophenotypically and molecularly in NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice.
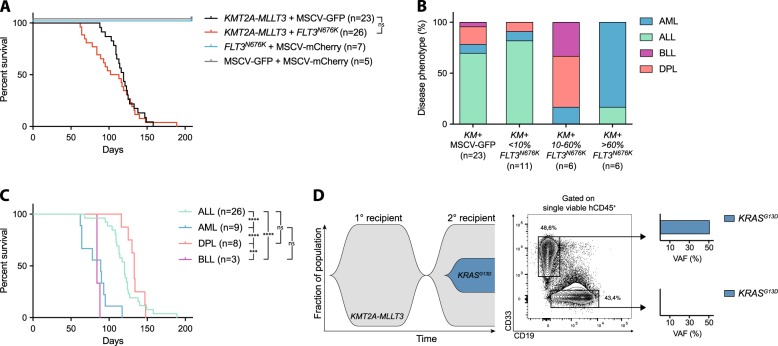
Mice that received KMT2A-MLLT3 with or without FLT3N676K developed a lethal leukemia, often with splenomegaly, thrombocytopenia, and leukocytosis, with no difference in median disease latency (107.5 and 119 days, respectively, P = 0.48) and mice receiving FLT3N676K alone showed no sign of disease (Fig. 1a, Supplementary Fig. 1A–E, and Supplementary Data 1). Leukemic mice succumbed to ALL (> 50% CD19+CD33−), AML (> 50% CD19−CD33+), double-positive leukemia (DPL, > 20% CD19+CD33+), or bilineal leukemia (BLL, <50% CD19+CD33−, < 50% CD19−CD33+, and < 20% CD19+CD33+); thus, the leukemias often coexisted with leukemia cells of another immunophenotype (Supplementary Fig. 1F, G and Supplementary Data 1) [3–5]. Previous studies have shown that retroviral overexpression of KMT2A-MLLT3 in human CB cells in NOD.CB17/Prkdcscid (NOD/SCID), NOD.Cg-PrkdcscidB2mtm1Unc (NOD/SCID-B2m), or NSG immunodeficient mice, primarily gives rise to ALL, sometimes to leukemias expressing both lymphoid and myeloid markers or bilineal leukemias, but rarely AML [3–6]. KMT2A-MLLT3-driven AML can only be generated with high penetrance, and re-transplanted in immunodeficient mice transgenically expressing human myeloid cytokines, consistent with the idea that external factors can influence the phenotype of the developing leukemia [3, 6]. In agreement, most recipients that received KMT2A-MLLT3 alone developed ALL (16/23, 69.6%) or DPL (4/23, 17.4%) and AML was rare (2/23, 8.7%) [3, 4, 6]. By contrast, five out of six recipients that received KMT2A-MLLT3+FLT3N676K and that had > 60% (n = 6, range 60.4–94.1%) of co-expressing cells, developed AML and one developed ALL (Fig. 1b, Supplementary Fig. 1H, I, and Supplementary Data 1). Thus, FLT3N676K preferentially drives myeloid expansion, similar to mutant Flt3 in a syngeneic setting [7]. FLT3 is expressed in human hematopoietic stem and progenitor cells, with the highest expression in granulocyte–macrophage progenitors (GMPs), and its signaling supports survival of those cells [8]. Combined, this suggests that FLT3N676K affects the survival of myeloid progenitors. In this context, it is interesting to note that FLT3 tyrosine kinase domain mutations are enriched in pediatric AML with KMT2A-MLLT3 [9, 10]. Most recipients with < 10% (n = 11, range 0.2–5.3%) of co-expressing KMT2A-MLLT3 + FLT3N676K cells succumbed to ALL, consistent with leukemia being driven by KMT2A-MLLT3 alone. Among those with 10–60% (n = 6, range 10.9–31.6%) of co-expressing cells, a mixture of diseases developed since leukemia could be driven both by KMT2A-MLLT3 alone and KMT2A-MLLT3 + FLT3N676K (Fig. 1b, Supplementary Fig. 1H, I, and Supplementary Data 1).
Fig. 1.
FLT3N676K alters the lineage distribution of KMT2A-MLLT3-driven leukemia. a Kaplan–Meier survival curves of NSG mice transplanted with CD34+ cord blood cells cotransduced with KMT2A-MLLT3 + FLT3N676K (n = 23), KMT2A-MLLT3 + MSCV-GFP (n = 26, of which three died and no tissue samples could be collected), FLT3N676K + MSCV-mCherry (n = 7), or MSCV-GFP + MSCV-mCherry (n = 5). b Distribution of mice that succumbed to ALL, AML, DPL, or BLL within KMT2A-MLLT3 + MSCV-GFP and KMT2A-MLLT3-mCherry + FLT3N676K-GFP recipients divided on the fraction of mCherry+GFP+ (<10%, n = 11; 10–60%, n = 6; or > 60%, n = 6) cells within hCD45+ bone marrow (BM) cells. c Kaplan–Meier survival curves of all xenograft leukemias based on their immunophenotype showed an accelerated disease for AML as compared with both ALL (P < 0.0001) and DPL (P < 0.0001). d Fish plot showing progression of one KMT2A-MLLT3 + MSCV-GFP BLL that gained a de novo KRASG13D (VAF 17% in hCD45+ BM cells) in the secondary recipient (h11.13-1) and targeted resequencing of hCD45+CD19−CD33+ and hCD45+CD19+CD33− BM cells from the secondary recipient (h11.13-1) revealed the KRASG13D mutation to reside in CD19−CD33+ leukemia cells (VAF 51%). Mantel–Cox log-rank test, ***P ≤ 0.001, ****P ≤ 0.0001, ns = not significant
Co-expression of KMT2A-MLLT3 and FLT3N676K preferentially expanded myeloid cells (P < 0.0001, Supplementary Fig. 1J) and a high proportion of CD19−CD33+ cells at sacrifice, across the cohort, correlated with accelerated disease (rs = −0.6537, P < 0.0001, Supplementary Fig. 1K, L). In agreement, AML developed with significantly shorter latency as compared with ALL and DPL (89 vs. 120 and 133 days, respectively, both P < 0.0001), but not with BLL (84 days, Fig. 1c). Further, FLT3N676K-driven AML had a tendency toward shorter survival (median latency 78 days, range 62–117 days vs. 93 days for KMT2A-MLLT3 alone, Supplementary Data 1).
To determine the evolution of phenotypically distinct leukemia cells in secondary recipients, BM cells from six primary KMT2A-MLLT3 + FLT3N676K leukemias (three each with > 60% or 20–32% of FLT3N676K-expressing cells) and from four primary KMT2A-MLLT3 leukemias, were re-transplanted. All leukemias gave rise to secondary malignancies and recipients that received BM from AML (n = 2) and ALL (n = 1) with > 60% of KMT2A-MLLT3 + FLT3N676K cells had an accelerated disease onset and maintained leukemia immunophenotype (median latency of 117 and 41 days, for the primary and secondary recipients, respectively) (Supplementary Fig. 2A-E and Supplementary Data 2). Thus, FLT3N676K circumvented the cytokine dependence normally required for myeloid cells in immunodeficient mice [3]. By contrast, all secondary recipients that received BM with 20–32% KMT2A-MLLT3 + FLT3N676K cells succumbed to ALL, irrespective of disease phenotype in the primary recipients (one AML and two BLL). Thus, the myeloid FLT3N676K-expressing cells unexpectedly decreased in size, while the KMT2A-MLLT3-expressing lymphoid cells increased to clonal dominance (Supplementary Fig. 2F, G and Supplementary Data 2). This suggests that the FLT3N676K-containing myeloid leukemia population needs to be sufficiently large to expand in secondary recipients, either because they otherwise are outcompeted by the larger population of more easily engrafted ALL cells, or because they themselves need to mediate the permissive microenvironment that allows myeloid cells to engraft. Similarly, in all but one of the secondary recipients that received BM from leukemias expressing only KMT2A-MLLT3 (two AML, one ALL, and one BLL), the disease phenotype changed and myeloid cells did not engraft (Supplementary Fig. 2H-J and Supplementary Data 2).
In the secondary recipient with maintained immunophenotype (a BLL, h11.13-1), the myeloid cells unexpectedly expanded from 38% to close to 50% (Fig. 1d and Supplementary Data 2). This suggested that the myeloid cells had acquired a de novo mutation that allowed serial transplantation, similar to what was observed for FLT3N676K. Strikingly, targeted sequencing of AML-associated genes on hCD45+ BM from this mouse identified a KRASG13D in 34% of the cells. Resequencing of hCD45+CD19−CD33+ and hCD45+CD19+CD33− BM showed that KRASG13D was present exclusively in the myeloid population and based on the variant allele frequency of 51%, that all cells carried the mutation (Fig. 1d and Supplementary Table 1, 2). Further, KRASG13D likely arose independently in h11.13-1 as no mutation was identified, at the level of our detection, in the primary (h11.13) or in a separate secondary recipient (h11.13-2) from the same primary mouse (h11.13) that developed ALL (Supplementary Table 2 and Supplementary Data 2).
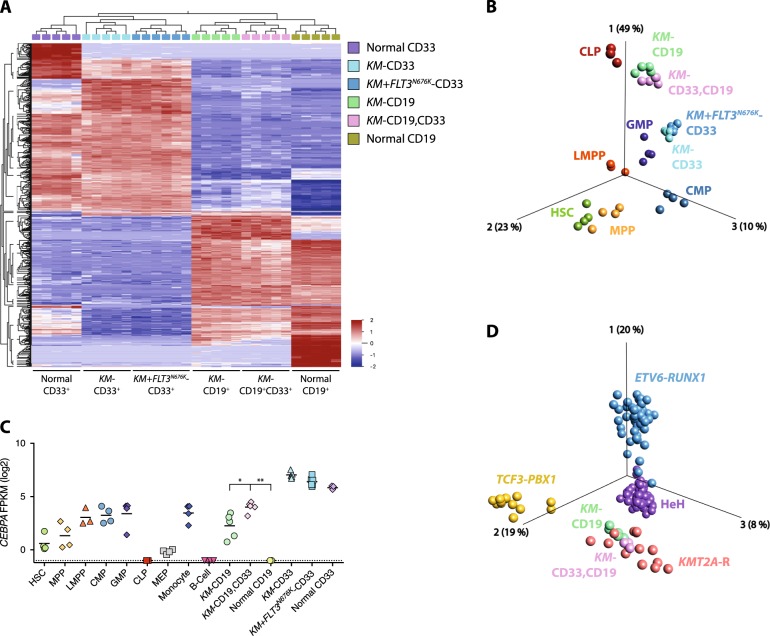
Gene expression profiling (GEP) followed by principal component analysis (PCA) showed that the leukemias segregated based on their immunophenotype, with an evident separation between leukemias and normal hematopoietic cells (Fig. 2a, Supplementary Fig. 3A, B, and Supplementary Table 3). All leukemias expressed high levels of known KMT2A-R target genes and showed enrichment of gene signatures associated with primary KMT2A-R leukemia, indicating that they maintain a GEP representative of human disease (Supplementary Fig. 3C, D and Supplementary Data 3–6). In line with the hypothesis that KMT2A-MLLT3 DPL cells are ALL cells with aberrant CD33 expression, they clustered closely with ALL cells. Both populations expressed high levels of ALL-associated cell surface markers and lymphoid transcription factors (Fig. 2a and Supplementary Fig. 3B, E-H). Further, CD33 and other AML-associated cell surface markers and key myeloid transcription factors, all showed lower expression in DPL cells as compared with normal myeloid- and AML cells (Supplementary Fig. 3G-I).
Fig. 2.
DPL cells are ALL cells with aberrant CD33 expression. a Hierarchical clustering based on multigroup comparison of myeloid CD19−CD33+ leukemia cells from KMT2A-MLLT3 + MSCV-GFP (KM-CD33) and KMT2A-MLLT3 + FLT3N676K (KM + FLT3N676K-CD33), lymphoid CD19+CD33− leukemia cells from KMT2A-MLLT3 + MSCV-GFP (KM-CD19), and double-positive CD19+CD33+ leukemia cells from KMT2A-MLLT3 + MSCV-GFP (KM-CD19,CD33), as well as normal myeloid- CD19−CD33+ (normal CD33) and lymphoid CD19+CD33− (normal CD19) cells from MSCV-GFP + MSCV-mCherry using 637 variables (P = 2.2e−15, FDR = 4.9e−14). b Supervised (1500 variables, P = 3.6e−4, FDR = 3.1e−3) PCA based on human hematopoietic stem cells (HSC), multipotent progenitors (MPP), lymphoid-primed multipotent progenitors (LMPP), common myeloid progenitors (CMP), granulocyte–macrophage progenitors (GMP), and common lymphoid progenitors (CLP) [11]. Samples with leukemia cells from KM-CD33, KM + FLT3N676K-CD33, KM-CD19, and KM-CD19,CD33 were inserted into the same PCA (still based solely on the normal populations [11]), revealing that AML cells mainly resembled GMPs and that both ALL and DPL mainly resembled CLPs. c CEBPA expression (FPKM log2) within sorted leukemia, normal populations, and within HSC, MPP, LMPP, CMP, GMP, CLP, MEP, monocytes, and B cells [11]. d Supervised (1501 variables, P = 3.3e−10, FDR = 3.1e−9) PCA based on pediatric BCP-ALL with ETV6-RUNX1, high hyperdiploid (HeH), TCF3-PBX1, or KMT2A-R [12]. Samples with leukemia cells from KM-CD19 and KM-CD19,CD33 were inserted into the same PCA (still based solely on the pediatric BCP-ALL populations [12]). Mann–Whitney U test used in (c), *P ≤ 0.05, **P ≤ 0.01, ns = not significant
By correlating the GEPs of the xenograft leukemias to those of normal hematopoietic cells [11], DPL and ALL were found to resemble normal common lymphoid progenitors (CLPs) and AML cells normal GMPs (Fig. 2b). Further, ALL and DPL cells had significantly higher expression of the transcription factor CEBPA as compared with normal lymphoid cells which lacked CEBPA expression (Fig. 2c). CEBPA drives myeloid programs and is expressed in most hematopoietic progenitors, in particular in GMPs, with CLPs lacking CEBPA expression (Fig. 2c) [11]. Finally, to link the GEPs of our xenograft leukemias to those of pediatric KMT2A-R leukemia, we utilized a dataset of pediatric B-cell precursor ALL (BCP-ALL) [12]. Multigroup comparison visualized by PCA showed that KMT2A-MLLT3 DPL and ALL mainly resembled KMT2A-R BCP-ALL (Fig. 2d), again highlighting that the xenograft leukemias resemble human leukemia.
We next studied the transcriptional changes induced by FLT3N676K in KMT2A-MLLT3 AML cells. Gene set enrichment analysis (GSEA) revealed enrichment of gene sets connected to the Myc-transcriptional network, cell cycle, and proliferation when compared with KMT2A-MLLT3 AML (Supplementary Data 7). Similar to our previous findings in a syngeneic KMT2A-MLLT3 mouse model [2], the Myc-centered program [13] was not linked to the pluripotency network (Supplementary Fig. 4A and Supplementary Data 8, 9). Since FLT3 mutations activate mitogen-activated protein kinase (MAPK) signaling [14], we investigated if FLT3N676K increased expression of MEK/ERK-pathway genes, by studying the expression of known transcriptional output genes and negative feedback regulators of the pathway [15]. Indeed, enrichment of MEK/ERK-associated genes was evident in FLT3N676K-expressing cells, suggesting that FLT3N676K allows cells to overcome normal feedback regulation, leading to sustained signaling [15] (Supplementary Fig. 4B). Finally, FLT3N676K-expressing AML showed preserved transcriptional changes to those seen in infant KMT2A-AFF1 ALL with activating mutations [1] (Supplementary Fig. 4C). Thus, FLT3N676K may circumvent the cytokine dependence seen for myeloid cells in immunodeficient mice by providing constitutive active signaling promoting cell proliferation, likely through the MAPK/ERK pathway.
Herein, we demonstrate that co-expression of KMT2A-MLLT3 and FLT3N676K in human CB cells primarily causes AML and thus alters the lineage distribution of KMT2A-MLLT3-driven leukemia. AML could only be serially transplanted with maintained immunophenotype in the presence of FLT3N676K. This is consistent with the idea that activated signaling allows myeloid cells to more efficiently engraft and maintain their self-renewal. In agreement, we identified a de novo KRASG13D in myeloid KMT2A-MLLT3-expressing cells that had expanded upon secondary transplantation. Altogether, this shows that constitutively active signaling mutations can substitute for external factors and influence the phenotype of the developing KMT2A-R leukemia, at least in xenograft models.
Accession code
Supplementary information
Acknowledgements
This work was supported by The Swedish Childhood Cancer Foundation, The Swedish Cancer Society, The Swedish Research Council, The Knut and Alice Wallenberg Foundation, BioCARE, The Crafoord Foundation, The Per-Eric and Ulla Schyberg Foundation, The Nilsson-Ehle Donations, The Wiberg Foundation, and Governmental Funding of Clinical Research within the National Health Service. Work performed at the Center for Translational Genomics, Lund University has been funded by Medical Faculty Lund University, Region Skåne and Science for Life Laboratory, Sweden.
Author contributions
AHW and AKHA designed the study and experiments; AHW, MP, AFC, ME, HS, PS, PW, CGR, JL, and HÅ performed experiments; AHW, MP, HL, and AKHA analyzed sequencing data; AHW, AKHA, AH, MJ, and RWS interpreted data; AHW and AKHA wrote the paper. MJ and RWS performed critical reading and contributed to the writing of the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41375-019-0465-1) contains supplementary material, which is available to authorized users.
References
- 1.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–7. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, et al. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat Commun. 2018;9:1770. doi: 10.1038/s41467-018-04180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–95. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sontakke P, Carretta M, Jaques J, Brouwers-Vos AZ, Lubbers-Aalders L, Yuan H, et al. Modeling BCR-ABL and MLL-AF9 leukemia in a human bone marrow-like scaffold-based xenograft model. Leukemia. 2016;30:2064–73. doi: 10.1038/leu.2016.108. [DOI] [PubMed] [Google Scholar]
- 5.Horton SJ, Jaques J, Woolthuis C, van Dijk J, Mesuraca M, Huls G, et al. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia. 2013;27:1116–26. doi: 10.1038/leu.2012.343. [DOI] [PubMed] [Google Scholar]
- 6.Barabé F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–4. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 7.Bailey E, Li L, Duffield AS, Ma HS, Huso DL, Small D. FLT3/D835Y mutation knock-in mice display less aggressive disease compared with FLT3/internal tandem duplication (ITD) mice. Proc Natl Acad Sci USA. 2013;110:21113–8. doi: 10.1073/pnas.1310559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikushige Y, Yoshimoto G, Miyamoto T, Iino T, Mori Y, Iwasaki H, et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J Immunol. 2008;180:7358–67. doi: 10.4049/jimmunol.180.11.7358. [DOI] [PubMed] [Google Scholar]
- 9.Meshinchi S, Alonzo TA, Stirewalt DL, Zwaan M, Zimmerman M, Reinhardt D, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–61. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson A, Paulsson K, Lilljebjörn H, Lassen C, Strömbeck B, Heldrup J, et al. FLT3 mutations in a 10 year consecutive series of 177 childhood acute leukemias and their impact on global gene expression patterns. Genes Chromosomes Cancer. 2008;47:64–70. doi: 10.1002/gcc.20508. [DOI] [PubMed] [Google Scholar]
- 11.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, Palffy von S, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–24. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhary C, Schwäble J, Brandts C, Tickenbrock L, Sargin B, Kindler T, et al. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106:265–73. doi: 10.1182/blood-2004-07-2942. [DOI] [PubMed] [Google Scholar]
- 15.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.