Abstract
Precise regulation of chromatin architecture is vital to physiological processes including hematopoiesis. ARID1A is a core component of the mammalian SWI/SNF complex, which is one of the ATP-dependent chromatin remodeling complexes. To uncover the role of ARID1A in hematopoietic development, we utilized hematopoietic cell-specific deletion of Arid1a in mice. We demonstrate that ARID1A is essential for maintaining the frequency and function of hematopoietic stem cells and its loss impairs the differentiation of both myeloid and lymphoid lineages. ARID1A deficiency led to a global reduction in open chromatin and ensuing transcriptional changes affected key genes involved in hematopoietic development. We also observed that silencing of ARID1A affected ATRA-induced differentiation of NB4 cells, suggesting its role in granulocytic differentiation of human leukemic cells. Overall, our study provides a comprehensive elucidation of the function of ARID1A in hematopoiesis and highlights the central role of ARID1A-containing SWI/SNF complex in maintaining chromatin dynamics in hematopoietic cells.
Subject terms: Haematopoietic stem cells, Haematopoiesis
Introduction
Hematopoiesis is a tightly regulated process that constantly generates enormous numbers of mature blood cells through a cascade of differentiation stages. Self-renewal, quiescence and differentiation of hematopoietic stem cells (HSC), as well as subsequent lineage choices into various blood cell types are precisely orchestrated by stage-specific transcriptional and epigenetic machineries. Chromatin state is transformed dynamically during differentiation [1] and epigenetic factors that remodel chromatin and facilitate establishment and maintenance of stage-specific pattern of gene expression are key to sustain normal hematopoietic development.
ARID1A (BAF250a) is a principal component of SWI/SNF (SWItch/Sucrose Non-Fermentable) family of evolutionary conserved, multi-subunit chromatin remodeling complexes. Rearrangements of chromatin structure mediated by SWI/SNF complexes are critical for modulation of gene expression, thereby, affecting a broad range of cellular processes, including proliferation and differentiation [2–5]. ARID1A resides exclusively in the BAF subclass of SWI/SNF remodelers and contains an ARID domain, which interacts with DNA in a sequence-nonspecific manner. Sequencing of cancer genomes has identified recurrent mutations of ARID1A in a wide range of tumors including gynecological, liver, gastric, and breast tumors [5–10]. ARID1A is the most frequently mutated member of the SWI/SNF family and high incidence of inactivating mutations in varied cancers along with emerging functional studies postulate ARID1A as a novel tumor suppressor [11–15]. Amongst hematological diseases, ARID1A is mutated in acute promyelocytic leukemia (APL) [16] and several lymphoid malignancies [17–26]. Nonetheless, the role of ARID1A in hematopoietic development and how its loss contributes to leukemogenesis remains elusive. Krosl et al. [27] observed increased frequency of fetal liver HSCs in ARID1A-deficient mouse embryos, possibly through regulation of the fetal liver microenvironment. However, perinatal lethality caused by constitutive deletion of ARID1A has precluded elucidation of its role in adult hematopoiesis.
In this study, we have employed hematopoietic-cell specific deletion of ARID1A in mice to address systematically its function in the hematopoietic development. We demonstrate that lack of ARID1A results in a range of hematopoietic defects including highly diminished reconstitution ability in transplantation models. Our results illustrate that ARID1A maintains frequency and quiescence of HSCs, and regulates differentiation of both myeloid and lymphoid lineages in a cell-intrinsic manner. Depletion of ARID1A leads to extensive decrease in chromatin accessibility including loss of open chromatin at promoter/enhancer regions of several key regulators of hematopoietic development.
Materials and methods
Generation of mice with hematopoietic specific deletion of Arid1a
Mice bearing floxed Arid1a allele (Arid1af/f) in which exon 9 of Arid1a gene is flanked by loxP sites have been described before [28]. Arid1af/f mice were backcrossed for five generations to C57BL/6 mice before crossing with either Vav-iCre or Mx1-Cre transgenic strains to generate hematopoietic cell-specific deletion of Arid1a. In Vav-iCre model, either Arid1af/f;Vav-iCre− or Arid1af/+;Vav-iCre− sex-matched littermates were used as controls. To induce deletion of Arid1a in adult Arid1af/f;Mx1-Cre+ mice (8–14 weeks old), five doses of 300 µg poly(I:C) (GE Healthcare) were injected intraperitoneally, every alternate day. Sex-matched Arid1af/f;Mx1-Cre− littermates were also administered poly(I:C) simultaneously and used as control in all experiments. All mice were maintained in the animal facility of Comparative Medicine Centre, National University of Singapore (NUS). All mice experiments were performed according to protocols approved by NUS Institutional Animal Care and Use Committee.
Flow cytometry and FACS sorting
Stained cells were acquired on FACS LSR II flow cytometer (BD Biosciences) and sorted on FACSAria cell sorter (BD Biosciences). Data were analyzed using FACSDIVA software (BD Biosciences). Antibodies used for flow cytometry are listed in Supplementary Table 1.
Competitive reconstitution assays
For competitive reconstitution assay with purified HSCs, we sorted LT-HSCs (CD34−Flt3− LSK) four weeks after poly(I:C) injection from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. 150 LT-HSCs (CD45.2+) from either WT or Arid1a KO mice were mixed with 300,000 BM cells (CD45.1+) from Ptprca Pepcb/BoyJ (B6.SJL) competitor mice and injected intravenously into lethally irradiated (11 Gy) B6.SJL recipient mice. Reconstitution was assessed every four weeks in peripheral blood using flow cytometry. Blood leukocytes were stained with antibodies against CD3 (T cells), CD19 (B cells), CD11b, Gr1, F4/80 (granulocytes and monocytes) along with donor (CD45.2) and competitor (CD45.1) markers.
For assays with BM cells (prior to Arid1a deletion), CD45.2-expressing BM cells from either Arid1af/f;Mx1-Cre+ or Arid1af/f;Mx1-Cre− mice were mixed in equal proportion with CD45.1-expressing (B6.SJL) competitor BM cells. Two million cells were injected into the tail vein of lethally irradiated B6.SJL recipient mice. Donor engraftment was assessed in peripheral blood 4 weeks after transplantation. Arid1a deletion was induced in recipient mice using poly(I:C) and reconstitution of different blood lineages was determined as described above.
RNA-sequencing
cDNA libraries from FACS-sorted LT-HSC were prepared using SMART-Seq v4 Ultra Low Input RNA Kit (Clontech Laboratories). poly-A selected RNA from purified CMP, GMP, and MEP populations was used for library preparation using TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer’s protocol.
Libraries were sequenced on HiSeq 4000 and 100 bp paired-end reads were aligned to murine reference transcriptome (GRCm38/mm10; Ensemble version 84) using Kallisto (version 0.43.1) [29]. Further details of RNA-seq data analysis are described in Supplementary Methods. Primers used for quantitative RT-PCR validation are listed in Supplementary Table 2.
ATAC-sequencing
ATAC-seq libraries were prepared from Lin−Kit+ BM cell as previously described [30]. Transposed DNA libraries were sequenced on HiSeq 4000. Quality of the paired end 50 bp reads was assessed using fastqc v0.11.7 and reads were then aligned to the mm10 reference genome using Bowtie2 v2.3.4.1 with the parameters --very-sensitive -X 2000 --no-mixed --no-discordant [31]. PCR duplicate reads were removed using Picard MarkDuplicates v2.17.10 (http://broadinstitute.github.io/picard). The reads were filtered using samtools v1.5 for a minimum mapping quality of 10 and only properly paired reads were retained [32]. Additionally, any reads aligning to the mitochondrial chromosome were removed. Peaks were called subsequently using macs2 v2.1.1.20160309 with the parameters --keep-dup all -f BAMPE -q 0.05, while simultaneously generating bedgraph files in Reads Per Million scale [33]. Peaks were filtered against the ENCODE blacklist regions (https://sites.google.com/site/anshulkundaje/projects/blacklists) [34]. The bedgraph files were converted to bigWig file format using the bedGraphToBigWig v4 utility. Differential binding analysis was performed using DESeq2 v1.18.1 through the DiffBind v2.6.6 bioconductor package with an FDR cut-off of 0.01 [35, 36]. Heat maps were generated using the deeptools v3.0.2 package [37]. Peaks were annotated using homer annotatePeaks.pl script [38]. Genomic profile plots for ATAC-seq signals were generated using fluff v3.0.2 [39].
Statistical analysis
Statistical analyses for all mice experiments were performed using GraphPad Prism 7 software.
Accession codes
All sequencing data generated in this study were deposited in Gene Expression Omnibus database repository. The accession numbers are: GSE125846 and GSE125848 (RNA-seq), GSE125845 (ChIP-seq), and GSE125844 (ATAC-seq).
Results
Generation of hematopoietic cell-specific deletion of Arid1a in mice
Expression analysis in FACS-sorted populations from bone marrow (BM) and spleen of C57BL/6 mice showed that Arid1a was expressed ubiquitously across different hematopoietic lineages with relatively lower expression detected in mature myeloid cells (Supplementary Figure 1a).
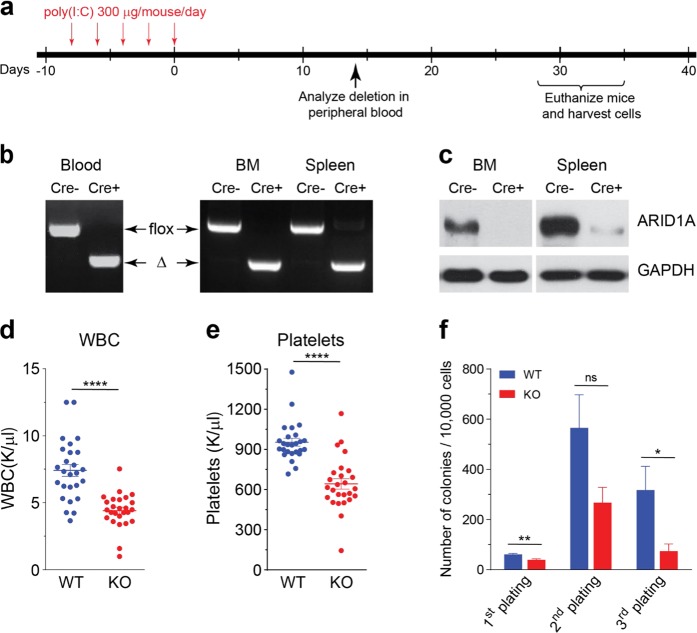
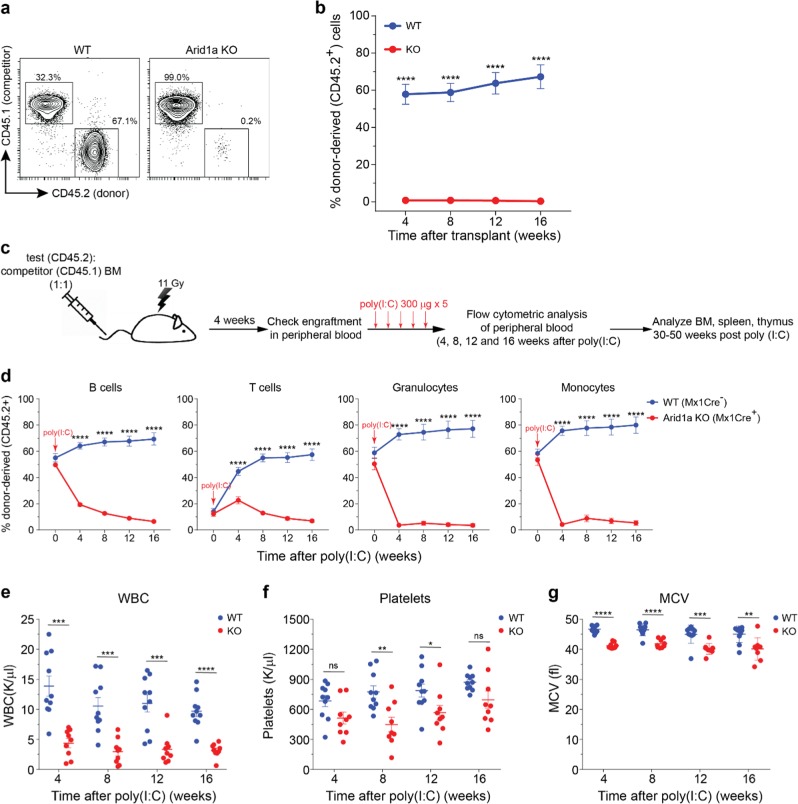
To analyze the function of ARID1A in hematopoietic development, mice carrying the Arid1a floxed allele (Arid1af/f) [28] were crossed with either Vav-iCre [40] or Mx1-Cre [41] strains. We noted that Arid1af/f;Vav-iCre+ mice were obtained at lower than expected frequency, both at birth (day 1) and weaning (day 21), suggesting perinatal mortality caused by Arid1a deletion in this model (Supplementary Table 3). Cre recombinase-mediated deletion of exon 9 and absence of ARID1A protein were verified in hematopoietic cells of the surviving Arid1af/f;Vav-iCre+ mice (Supplementary Figure 1b–c). Owing to increased mortality of Arid1af/f;Vav-iCre+ mice, more detailed analysis was performed simultaneously in mice where deletion of ARID1A was induced using the interferon inducible Mx1-Cre transgene. Administration of poly(I:C) caused rapid and complete deletion at Arid1a locus in peripheral blood and BM cells, while the deletion was ∼90% in spleen cells, which was also reflected in the residual protein levels observed in western blot analysis (Fig. 1a–c). Loss of ARID1A resulted in reduced numbers of WBCs and platelets in Arid1af/f;Mx1-Cre+ mice compared to the Arid1af/f;Mx1-Cre− mice at 4 weeks after poly(I:C) treatment (Fig. 1d, e); and a similar trend was observed in Arid1af/f;Vav-iCre+ mice (Supplementary Figure 1d-e). ARID1A-deficient BM cells exhibited decreased potential to generate myeloid colonies as both the number (Fig. 1f) and size (data not shown) of colonies was reduced in methylcellulose colony assays. Ex vivo deletion of floxed allele in Lin−Kit+ BM cells also resulted in fewer colonies (Supplementary Figure 1f-g). These initial observations suggested that depletion of ARID1A leads to perturbations in hematopoiesis.
Fig. 1.
Hematopoietic-specific deletion of Arid1a. a Schematic of workflow to induce deletion of Arid1a in adult Arid1af/f;Mx1-Cre mice using poly(I:C). b PCR analysis verifies poly(I:C)-induced deletion of Arid1a exon 9 in peripheral blood (2 weeks after poly(I:C) injection), BM and spleen (4 weeks after poly(I:C) injection) of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. Δ: deleted allele. c Western blot shows loss of ARID1A protein in BM and spleen of poly(I:C) treated Arid1af/f;Mx1-Cre mice. d, e WBC (d) and platelet (e) counts in peripheral blood of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice 4 weeks after poly(I:C) injection. f Number of colonies obtained in serial re-plating assay with total BM cells harvested from poly(I:C) treated Arid1af/f;Mx1-Cre mice (n = 9 for first plating, n = 5 for second and third platings). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001, ns = not significant
ARID1A maintains pool size and quiescence of HSCs
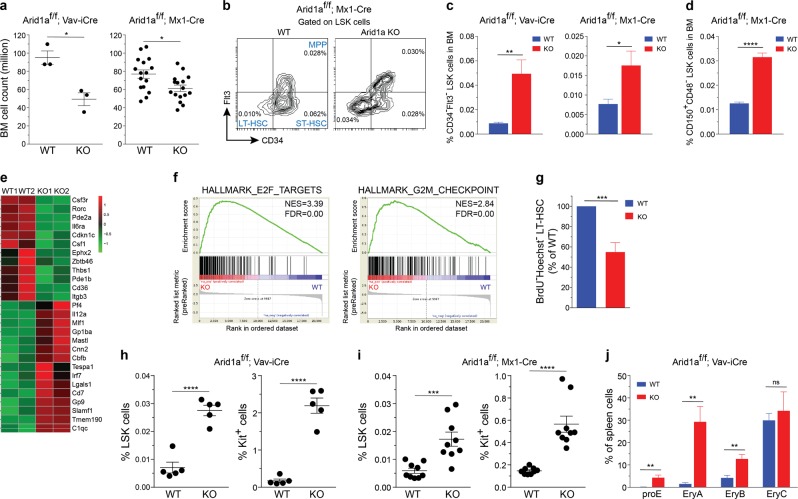
We observed that Vav-iCre mediated deletion of Arid1a early in hematopoietic development resulted in markedly reduced number of BM leukocytes; and cellularity was also decreased significantly when Arid1a was deleted in adult mice using the Mx1-Cre model (Fig. 2a). In both models of ARID1A deficiency, the frequency and numbers of Lin−Sca1+Kit+ (LSK) cells, which contains HSCs, was not significantly altered (Supplementary Figure 2a–b). Nonetheless, the proportion and absolute numbers of long-term HSCs (LTHSCs), defined either as CD34−Flt3−LSK or CD150+CD48−LSK cells were elevated in BM from both Arid1af/f;Vav-iCre+ and Arid1af/f;Mx1-Cre+ mice (4 weeks after poly(I:C) injection), compared with the control mice (Fig. 2b–d; Supplementary Figure 2c–f). We observed that loss of one Arid1a allele also resulted in increased percentage of LT-HSCs in bone marrow (Supplementary Figure 2g), suggesting a dose-dependent effect of ARID1A in maintaining HSC frequency. Global transcriptome analysis of LT-HSCs from wildtype (WT) and ARID1A-deficient BM revealed dysregulated expression of genes crucial for hematopoietic development including Csf3r, Il6ra, Csf1, and Cbfb (Fig. 2e; Supplementary Figure 2h and Supplementary Table 4). GSEA identified that genesets associated with cell cycle were significantly enriched in Arid1a KO cells compared with the WT cells (Fig. 2f). In vivo BrdU incorporation assays showed that the proportion of non-cycling HSCs (BrdU−Hoechst33342−LTHSCs) was significantly reduced in ARID1A-deficient BM, indicating loss of quiescence (Fig. 2g).
Fig. 2.
ARID1A is required to maintain HSC frequency. a BM cellularity in ARID1A-deficient mice compared with WT mice. Cell counts represent total leukocyte numbers in both femurs and tibias. For Vav-iCre model, mice were euthanized at 11–20 weeks of age; while for Mx1-Cre model, cellularity was determined 4 weeks after injection of poly(I:C). b Representative staining for LT-HSC, ST-HSC and MPP populations using CD34 and Flt3 antibodies in BM of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre– mice, four weeks after poly(I:C) injection. c Proportion of CD34–Flt3– LSK cells in BM of Arid1a deficient and control mice (n = 5 for Vav-iCre; n = 6 for Mx1-Cre). d Percentage of CD150+CD48− LSK cells in BM of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice, four weeks after poly(I:C) injection (n = 4). e Heat map depicts differential expression of known regulators of hematopoiesis affected by loss of ARID1A in LTHSCs (CD34−Flt3− LSK) compared to control (FDR < 0.1, except Cdkn1c, Csf1, Cbfb, where FDR < 0.25). LTHSC were sorted from the BM of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice, 4 weeks after poly(I:C) injection. Genes involved in hematopoiesis were curated manually from Molecular Signatures Database v6.1 and GO datasets from AmiGO 2. f GSEA plots of cell cycle-related gene signatures, HALLMARK_E2F_TARGETS and HALLMARK_G2M_CHECKPOINT, in Arid1a KO vs control LTHSCs. NES: Normalized enrichment score. g Proportion of BrdU−Hoechst33342− LTHSC in the BM of Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice (n = 6). Three weeks after administration of poly(I:C), mice were given BrdU in drinking water for one week before they were euthanized for experiment. (h-i) Proportion of LSK and Lin–Kit+ cells in the spleens of WT and Arid1a KO mice in Vav-iCre (h) and Mx1-Cre (i) models. j Frequencies of erythroid precursors in the BM of Arid1af/f;Vav-iCre+ and Arid1af/f;Vav-iCre– mice. (proE: CD71+TER119lo; EryA: CD71+TER119+FSChi; EryB: CD71+TER119+FSClo and EryC: CD71−TER119+FSClo) (n = 5). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant
Flow cytometric analysis of ARID1A-deficient spleens in both Vav-iCre and Mx1-Cre models demonstrated an expansion of progenitor/stem cell populations compared with the WT controls. This was evident from higher frequency and absolute number of Lin−Kit+ and LSK cells in the spleen of ARID1A-deficient mice (Fig. 2h, i; Supplementary Figure 3a-d). We also observed a marked elevation in erythroid precursors in spleen of Arid1af/f;Vav-iCre+ mice, although the increase was less significant in spleens of Mx1-Cre+ mice four weeks after poly(I:C) injection (Fig. 2j; Supplementary Figure 3e), indicating onset of extramedullary hematopoiesis.
Arid1a deletion leads to defects in development of myeloid and erythroid cells
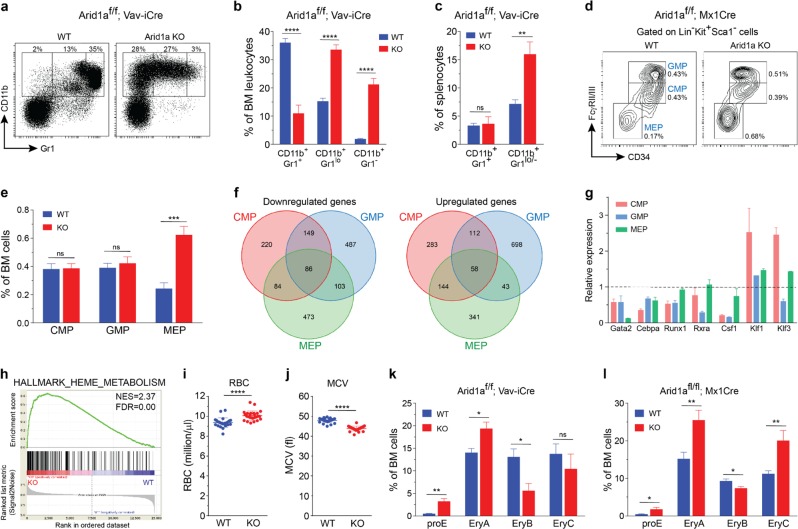
To understand how ARID1A regulates production of mature blood cells, we carefully analyzed differentiation towards myeloid and lymphoid lineages in ARID1A-deficient mice. ARID1A deficiency caused defects in granulocytic differentiation in BM of both Vav-iCre and Mx1-Cre models. This was evident by increased frequency of immature granulocytes/monocytes (CD11b+Gr1lo and CD11b+Gr1−) and a decrease in proportion of mature granulocytes (CD11b+Gr1+) in both deletion models (Fig. 3a, b; Supplementary Figure 4a–b). An increase in immature granulocytes/monocytes was also observed in the spleens of Arid1a KO mice, although the proportion of mature granulocytes was unaffected (Fig. 3c; Supplementary Figure 4c). Further analyses of myeloid progenitors in the BM of poly(I:C) treated Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice revealed that the proportion of common myeloid precursors (CMP; Lin−Kit+Sca1−CD34+FcγRII/IIIlo) and granulocyte monocyte precursors (GMP; Lin−Kit+Sca1−CD34+FcγRII/IIIhi) were unchanged in ARID1A-deficient BM; however, these populations exhibited reduced surface expression of CD34 compared with the control mice (Fig. 3d, e). Interestingly, an elevated frequency of megakaryocyte erythrocyte progenitors (MEP; Lin−Kit+Sca1−CD34−FcγRII/III−) was observed in the BM of Arid1af/f;Mx1-Cre+ mice (Fig. 3d, e). To determine the molecular basis of the observed defects in myeloid differentiation, gene expression changes were analyzed using RNA-sequencing in sorted CMP, GMP, and MEP cells isolated 4 weeks after poly(I:C) injection from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. Complete deletion of Arid1a exon 9 was evident in cells from poly(I:C) treated Arid1af/f;Mx1-Cre+ mice (Supplementary Figure 4d). Loss of ARID1A resulted in dysregulated expression of several genes either across all three populations or in a stage-specific manner (Fig. 3f; Supplementary Table 5). Importantly, altered gene expression in CMP, GMP, and MEP cells correlated well with changes in histone modifications (H3K27ac and H3K4me3) associated with transcriptional activation at respective loci (Supplementary Figure 4e-f). We used quantitative RT-PCR to validate altered expression of genes crucial for hematopoietic differentiation including Gata2, Cebpa, Runx1, Rxra, Csf1, Klf1, and Klf3 (Fig. 3g). GSEA uncovered upregulation of genes involved in metabolism of heme and erythroblast differentiation in KO CMP and MEP cells (Fig. 3h; Supplementary Figure 5a), indicating importance of ARID1A in erythropoiesis. We observed that the RBCs displayed microcytosis in peripheral blood and BM of ARID1A-deficient mice (both Mx1-Cre and Vav-iCre models) (Fig. 3i, j; Supplementary Figure 5b–d). Further examination of erythroid differentiation in BM using surface expression of CD71 and TER119 showed an accumulation of proerythroblasts (CD71+TER119lo) and altered frequencies of subsequent stages of erythrocyte maturation in the BM of mice lacking ARID1A (Fig. 3k, l; Supplementary Figure 5e–f). Collectively, these data demonstrate that ARID1A plays a pivotal role in myeloid and erythroid differentiation.
Fig. 3.
Defects in myeloid differentiation in Arid1a knockout mice. a Representative FACS profiles for BM cells from Arid1af/f;Vav-iCre+ and control mice stained with CD11b and Gr1 antibodies. b Frequencies of myeloid populations based on staining for Gr1 and CD11b markers as depicted in (a) (n = 5). c Proportion of CD11b+Gr1+ and CD11b+Gr1lo/− myeloid cells in the spleen of WT and Arid1a KO (Vav-iCre) mice (n = 5). d FACS plots depict representative staining of CMP, GMP and MEP cells in BM of poly(I:C) treated Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. e Percentages of CMP, GMP, and MEP cells in the BM of WT and Arid1a KO (Mx1-Cre) mice (n = 6). f Venn diagrams depict overlap of upregulated and downregulated genes amongst CMP, GMP and MEP cells (FDR < 0.1, mean expression > 1 & absolute log2 fold change > 0.5). RNA-sequencing was performed on cells sorted from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice 4 weeks after poly(I:C) treatment. g Quantitative RT-PCR analysis of selected genes differentially expressed (RNA-seq) in at least one of the three populations (CMP, GMP, and MEP) in the Arid1a KO mice compared with the WT mice. Cells were sorted from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice four weeks after poly(I:C) injection. Bars represent relative expression in Arid1a KO cells compared to WT cells, which were assigned a value of 1 (depicted by a dashed line), for every gene in each cell population. Results are cumulative of two independent experiments. Transcript levels of β-actin were used for normalization. h GSEA plot for geneset HALLMARK_HEME_METABOLISM in the comparison of WT and ARID1A-deficient CMP cells. NES: Normalized enrichment score. i, j Number of RBC (i) and mean corpuscular volume (MCV) (j) in peripheral blood of poly(I:C) treated Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. k, l Frequency of erythroid precursors (proE: CD71+TER119lo; EryA: CD71+TER119+FSChi; EryB: CD71+TER119+FSClo and EryC: CD71−TER119+FSClo) in BM of Arid1af/f;Vav-iCre (k) (n = 5) and Arid1af/f;Mx1-Cre (l) (n = 6) mice. Results represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant
Impaired lymphopoiesis in ARID1A-deficient mice
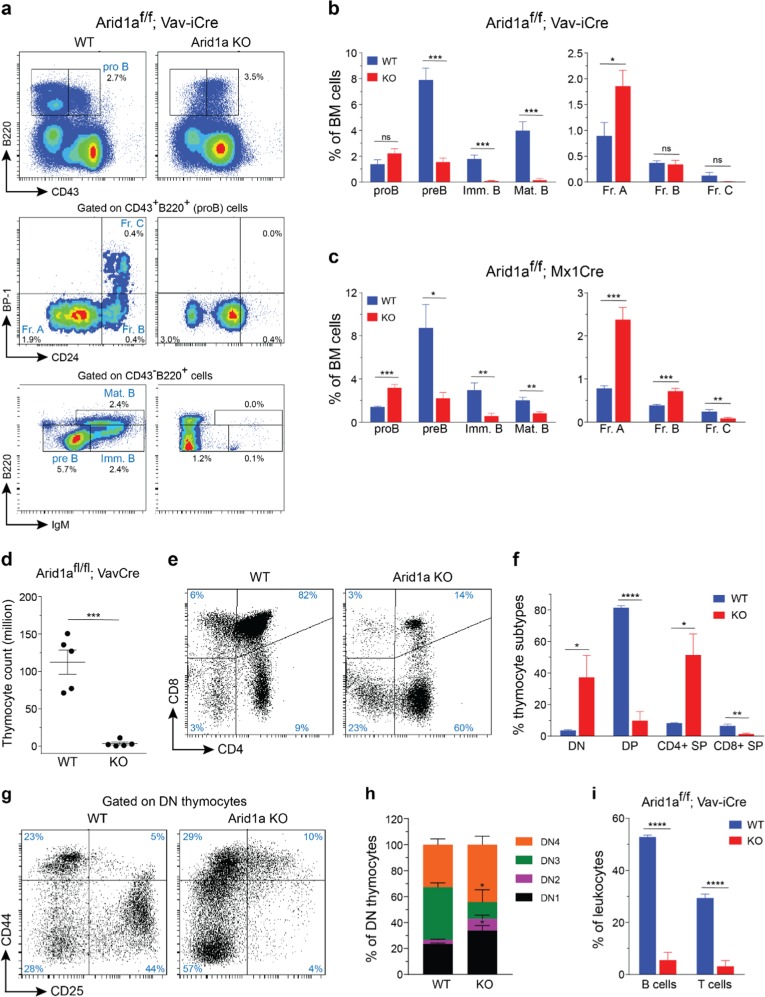
Next, we assessed the effect of ARID1A loss on the lymphoid compartment. Flow cytometric analysis revealed that the proportion of proB cells (CD43+B220+), was increased in Arid1a KO BM, in both Vav-iCre and Mx1-Cre models (Fig. 4a–c). Further fractionation of proB cells using a scheme described before [42] demonstrated a severe block in differentiation from Fraction A (CD24−BP1−) to Fraction B (CD24+BP1−) proB cells (Fig. 4a–c). This resulted in significantly lower proportions of preB (CD43−IgM−B220+), immature B (CD43−IgM+B220+), and mature B (CD43−IgM+B220hi) cells in ARID1A-deficient BM (Fig. 4a–c).
Fig. 4.
Arid1a knockout impairs lymphoid development. a Representative FACS plots show maturational stages of B cells in the BM of Arid1af/f;Vav-iCre+ and control mice. b, c Proportion of cells in different B-cell developmental stages in the BM of WT and Arid1a KO [Vav-iCre (b) (n = 5) and Mx1-Cre (c) (n = 6)] mice. proB: CD43+B220+, preB: CD43−B220+IgM−, immature B (Imm. B): CD43−B220+IgM+, mature B (Mat. B): CD43−B220hiIgM+, Fraction A (Fr. A): CD24−BP1− proB cells, Fraction B (Fr. B): CD24+BP1− proB cells, Fraction C (Fr. C): CD24+BP1+ proB cells. d Thymus cellularity in Arid1af/f;Vav-iCre+ and control (either Arid1af/f;Vav-iCre− or Arid1af/+;Vav-iCre− littermates) mice (11–20 weeks). e FACS plots depict representative staining of four major thymic populations: DN (CD4−CD8−), DP (CD4+CD8+), CD4+ and CD8+ SP cells in the thymus of Arid1af/f;Vav-iCre+ and control mice. f Frequencies of DP, DN, CD4+ SP and CD8+ SP populations in Arid1af/f;Vav-iCre+ and control mice (n = 5). g Representative flow cytometry staining for CD44 and CD25 expression within the DN population in the thymus of Arid1a KO (Vav-iCre) and control mice. h Proportions of DN1 (CD44+CD25− DN), DN2 (CD44+CD25+ DN), DN3 (CD44−CD25+ DN) and DN4 (CD44−CD25− DN) sub-populations within the DN compartment of thymus of Arid1af/f;Vav-iCre+ and control mice (n = 5). i Frequencies of B cells (CD19+) and T cells (CD3+) in the spleen of WT and Arid1a KO mice (n = 5). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant
Next, we assessed T-cell development in mice lacking ARID1A. Vav-iCre mediated deletion of Arid1a led to thymic atrophy (Fig. 4d) accompanied by severe perturbation in major T-cell compartments. CD4−CD8− double negative (DN) compartment comprises of immature thymocyte populations, and immediately precedes the CD4+CD8+ double positive (DP) stage in development. We observed a striking reduction in DP cells with a concomitant increase in the proportion of DN and CD4+ SP cells in the thymi of Arid1af/f;Vav-iCre+ mice compared with the control mice (Fig. 4e, f). Within the DN compartment, the proportions of DN1 (CD44+CD25−) and DN2 (CD44+CD25+) cells were elevated, while DN3 (CD44−CD25+) subpopulation was reduced in KO thymi, suggesting early arrest of thymocyte development (Fig. 4g, h).
We noted that Mx1-Cre mediated deletion of ARID1A was incomplete in thymoctyes 4 weeks after administration of poly(I:C) (Supplementary Figure 6a–b). Nonetheless, partial loss of ARID1A in this model caused rapid decline in thymic cellularity (Supplementary Figure 6c) and a trend towards altered frequencies of major thymic subsets (along the lines of the phenotype in the Vav-iCre deletion model) was noted (Supplementary Figure 6d-e). PCR analysis of sorted thymocyte subsets revealed a nearly complete deletion of Arid1a floxed allele in the DN fraction while the recombination was partial in DP and SP populations (Supplementary Figure 6f). These data demonstrate that ARID1A is required for normal T cell maturation in mice.
Defective maturation of the lymphoid cells led to substantially lower proportions of mature B and T cells in the spleens of ARID1A-deficient mice (Fig. 4i; Supplementary Figure 6g). Overall, our experiments establish a vital role of ARID1A in lymphoid differentiation.
ARID1A is essential for HSC function
To assess the functional activity of ARID1A-deficient HSCs, FACS-sorted LTHSCs from either poly(I:C)-treated Arid1af/f;Mx1-Cre+ or Arid1af/f;Mx1-Cre− mice were transplanted in competitive repopulation assays. We detected a striking loss of donor contribution in peripheral blood of recipient mice transplanted with Arid1a KO cells compared with WT cells (Fig. 5a, b; Supplementary Figure 7a). Proportion of donor-derived cells was markedly decreased in BM and other lymphoid organs of the recipients 16–25 weeks post-transplantation (Supplementary Figure 7b-d). This illustrated that despite an increase in frequency of the phenotypically defined HSC population, ARID1A deficiency severely impaired their reconstitution ability. To verify that ARID1A deficiency did not impede homing of the HSCs to the hematopoietic niche, ARID1A was deleted only after establishment of BM chimeras from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice (Fig. 5c). Four weeks after transplantation, we detected a similar engraftment ability of BM cells from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice (Supplementary Figure 8a). ARID1A deletion following administration of poly(I:C) resulted in rapid decline of donor-derived cells in peripheral blood of Mx1-Cre+ mice compared with the Mx1-Cre− mice (Fig. 5d). Deficiency of ARID1A affected repopulation of both lymphoid and myeloid cells in the peripheral blood (Fig. 5d). Similarly, donor-chimerism was markedly reduced in stem/progenitor population in the BM and mature blood cells in spleen and thymus of recipients transplanted with Arid1a KO cells (Supplementary Figure 8b-d). Poor reconstitution ability of Arid1a KO cells was also evident in competitive repopulation assays using BM cells from Arid1af/f;Vav-iCre+ mice compared with the control mice (Supplementary Figure 8e). To verify further cell-intrinsic effect of ARID1A deficiency on HSC function, non-competitive repopulation assays were performed. ARID1A was deleted in recipient mice four weeks after transplantation of BM from either Arid1af/f;Mx1-Cre+ or Arid1af/f;Mx1-Cre− mice. Loss of ARID1A resulted in reduction in numbers of WBCs and platelets along with emergence of smaller RBCs in the peripheral blood (Fig. 5e–g; Supplementary Figure 8f), thus recapitulating the phenotype observed in the Arid1af/f;Mx1-Cre+ mice. Collectively, these data demonstrate that ARID1A is vital for the repopulation ability of HSCs and its deficiency results in a competitive disadvantage.
Fig. 5.
ARID1A is essential for reconstitution ability of HSCs. a Representative plots show donor chimerism (CD45.2) in peripheral blood of recipients transplanted with LTHSCs (CD34−FLT3− LSK) from either control or Arid1a KO (Mx1-Cre) mice in competitive repopulation assay. b Average donor chimerism in recipient mice transplanted as in (a) and measured at 4, 8, 12, and 16 weeks post transplantation. Data are cumulative of two independent transplantation experiments (nine recipients/genotype). c Experimental design for competitive repopulation assay using total BM cells (prior to deletion of Arid1a). Lethally irradiated recipients were transplanted with BM cells (either Arid1af/f;Mx1-Cre+ or Arid1af/f;Mx1-Cre− BM mixed in equal proportion with competitor BM) and deletion of Arid1a was induced four weeks post transplantation using poly(I:C). d Frequency of donor-derived B cells, T cells, granulocytes and monocytes in peripheral blood of recipient mice at different time points post poly(I:C) administration. Two transplant experiments were performed as illustrated in (c) (11 recipients/genotype). e–g Non-competitive transplant assays. BM cells from either Arid1af/f;Mx1-Cre+ or Arid1af/f;Mx1-Cre− mice were injected into lethally irradiated mice and Arid1a deletion was induced after 4 weeks. WBC count (e), platelet count (f) and MCV (g) were determined every four weeks after poly(I:C) injection. Two independent transplantation experiments are summarised (WT = 10 recipients, Arid1a KO = 9 recipients). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant
ARID1A regulates chromatin accessibility at loci critical for hematopoiesis
To assess how ARID1A-containing SWI/SNF complex regulates chromatin accessibility in myeloid precursors, Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq) was performed on sorted Lin−Kit+ BM cells from poly(I:C) treated Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice. We observed that the biological replicates for WT and Arid1a KO samples were highly concordant (Supplementary Figure 9a-b). ATAC-seq peaks localized predominantly to intergenic and intronic regions but were also present around transcriptional start sites (TSS)/promoter regions (Supplementary Figure 9c). Integrative analysis of ATAC-seq peaks with ChIP-seq data showed that open chromatin regions were enriched within both promoter (H3K4me3) and enhancer (H3K27ac) histone marks (Supplementary Figure 9d).
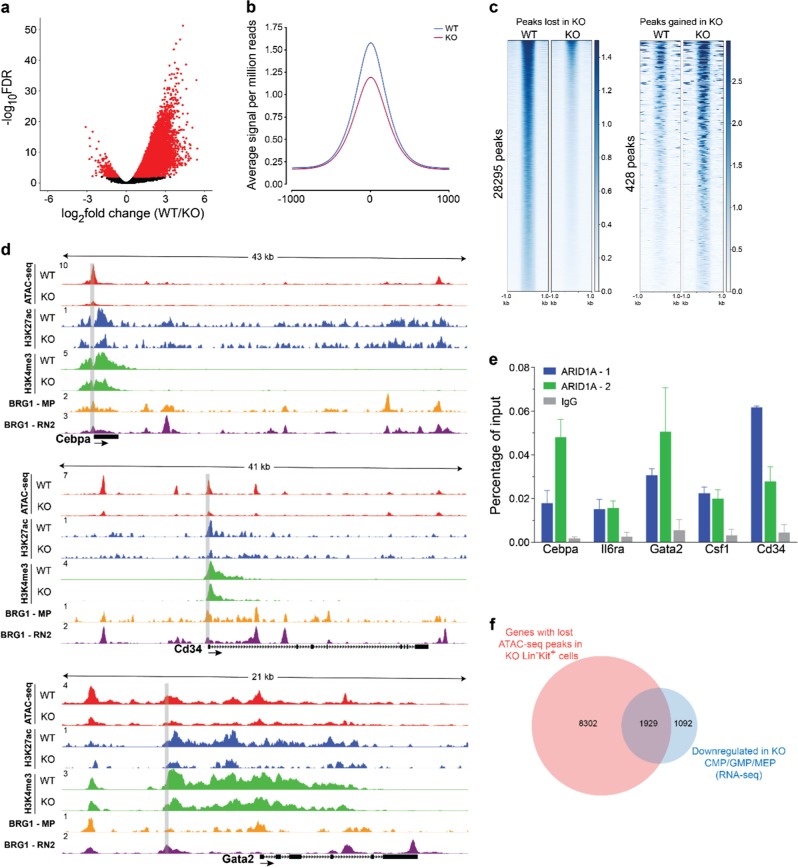
We observed reduced number of ATAC-seq peaks in Arid1a-deficient cells, suggesting global loss of open chromatin in KO cells compared with the WT cells (Fig. 6a, b; Supplementary Figure 9c). We performed differential analysis using DiffBind [36], which revealed significantly reduced signal at 28,295 loci in KO cells, while 428 peaks were gained in KO cells compared with the WT cells, thus indicating reduced chromatin accessibility upon loss of ARID1A in Lin−Kit+ BM cells (Fig. 6c; Supplementary Figure 9e and Supplementary Table 6). Peaks differentially enriched upon deletion of Arid1a were located primarily in the intergenic and intronic regions (Supplementary Figure 9f-g). Importantly, significant reduction of ATAC-seq signal occurred in Arid1a KO cells at several key loci, which harbor genes involved in hematopoiesis, including Cebpa, Cd34, Csf1, IL6ra and Gata2 (Fig. 6d and Supplementary Figure 10). Chromatin occupancy of both H3K27ac and H3K4me3, which mark transcriptionally active sites, was also reduced in Arid1a-deficient Lin− cells at these loci (Fig. 6d and Supplementary Figure 10), demonstrating a correlation between chromatin accessibility and epigenetic signatures. To assess further if ARID1A directly binds to and regulates key genes involved in hematopoiesis, we performed ChIP-qPCR analysis using two anti-ARID1A antibodies. We observed an enrichment of ARID1A binding at promoter/putative regulatory elements of all the five genes with both ARID1A antibodies compared to the IgG control (Fig. 6e). Importantly, analysis of available ChIP-seq data for BRG1 [43, 44], a catalytic subunit of SWI/SNF complex, in mouse macrophages and RN2 cells (murine MLL-AF9/NrasG12D AML cells) showed extensive overlap of its occupancy with open chromatin regions at Cebpa, Cd34, Csf1, IL6ra, and Gata2 loci (Fig. 6d and Supplementary Figure 10), thus substantiating SWI/SNF complex-mediated regulation of chromatin architecture at these loci. We noted that top four motifs identified in ATAC-seq peaks lost in Arid1a KO cells showed overlap with DNA sequences recognized by PU.1, RUNX1, GATA, and CEBPA (Supplementary Figure 9h).
Fig. 6.
ARID1A maintains chromatin accessibility in hematopoietic cells. a Volcano plot shows ATAC-seq peaks identified in WT and Arid1a KO Lin−Kit+ BM cells. Cells were sorted from Arid1af/f;Mx1-Cre+ and Arid1af/f;Mx1-Cre− mice four weeks after administration of poly(I:C). Peaks enriched significantly in either WT or Arid1a KO cells (FDR < 0.01) are depicted in red. b Average signal for all ATAC-seq peaks. c Heat maps show ATAC-seq peaks (scaled to 1 Kb) identified as either significantly closed (left) or gained (right) in KO cells using DiffBind analysis package. d Representative tracks for ATAC-seq signal (Lin−Kit+ BM cells), H3K27ac and H3K4me3 marks (both Lin− BM cells) and BRG1 ChIP-seq (macrophages (GSM2663828) and RN2 cells (GSM2092897)) at Cebpa, Cd34 and Gata2 loci. Gray rectangles encompass the region analysed for binding of ARID1A using ChIP-qPCR. e ChIP-qPCR analysis of ARID1A occupancy at the loci of Cebpa, Cd34, Il6ra, Csf1 and Gata2 genes in 32D cells using two different ARID1A antibodies (ARID1A-1: Abcam ab182560; ARID1A-2: GeneTex GTX129433). Enrichment was calculated as percentages of input. Error bars represent SEM for two independent ChIP experiments for each locus. f Venn diagram shows overlap of genes with significantly reduced ATAC-seq signal in KO Lin−Kit+ BM cells and genes downregulated in Arid1a KO CMP, GMP, and MEP cells (RNA-seq) compared with the WT cells
Next, we determined if altered chromatin accessibility observed in Lin−Kit+ BM cells corresponded with gene expression changes in CMP, GMP, and MEP populations, which collectively comprise the bulk of Lin−Kit+ cells. A substantial overlap of genes downregulated in Arid1a KO CMP, GMP, and MEP cells and those with closed chromatin in Arid1a-deficient Lin−Kit+ cells was observed (Fig. 6f), suggesting a correlation between closed chromatin and transcriptional inactivation.
Depletion of ARID1A impairs granulocytic differentiation of NB4 cells
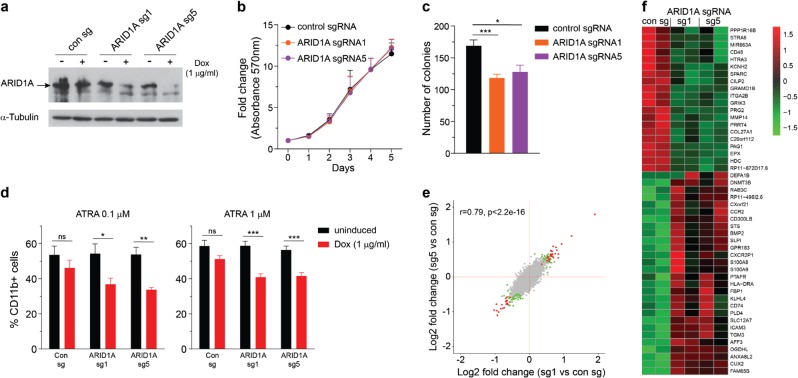
We have previously reported loss-of-function mutations of ARID1A in APL [16]. To investigate the effect of ARID1A deficiency on APL cells, we employed an inducible lentiviral vector system described before [45]. NB4 cells stably expressing Cas9 were transduced with small guide RNA (sgRNA) vector and knockout of ARID1A was induced using doxycycline (Dox) (Fig. 7a). Loss of ARID1A did not affect the viability of NB4 cells in short-term liquid culture; but ARID1A-deficient cells had decreased clonogenic potential compared with the control cells (Fig. 7b, c). To assess the effect of ARID1A deficiency on granulocytic differentiation of NB4 cells, we treated control and ARID1A-deficient cells with all-trans retinoic acid (ATRA) for 48 h and examined surface expression of CD11b using flow cytometry. Dox-treated ARID1A sgRNA-expressing cells exhibited lower CD11b expression compared to untreated cells and Dox-treated control cells (Fig. 7d). This indicates that ARID1A knockout impaired the differentiation potential of NB4 cells. RNA-sequencing revealed that 48 genes were differentially expressed (FDR < 0.1) in both ARID1A sg1- and sg5-expressing cells compared to control cells (Fig. 7e, f and Supplementary Table 7). Importantly, overall expression profiles of sg1 and sg5 targeted NB4 cells showed a high correlation (Fig. 7e). GSEA of sg1 and sg5-expressing ARID1A deficient NB4 cells showed a high overlap of enriched genesets including those involved in regulation of cell cycle, oxidative phosphorylation and MYC transcriptional network (Supplementary Figure 11a–b). Notably, genesets related to cell cycle (HALLMARK_E2F_TARGETS and HALLMARK_G2M_CHECKPOINT) were enriched in ARID1A-deficient NB4 cells, as well as Arid1a KO murine HSCs/precursors, suggesting that ARID1A mediates similar chromatin dynamics and transcriptional regulation in both murine and human cells.
Fig. 7.
Loss of ARID1A impairs granulocytic differentiation of NB4 cells. a Immunoblot shows doxycycline (Dox) induced knockout of ARID1A in NB4 cells expressing Cas9 and sgRNA targeting ARID1A. Protein lysate from NB4 cells treated with doxycycline for 7 days were analyzed. b Viability of ARID1A-deficient and control NB4 cells was determined using MTT reagent (n = 5). c Colony-forming ability of ARID1A knockout NB4 cells compared with control cells. Cells were plated in media containing methylcellulose, and colonies were enumerated after 9–11 days (n = 6). d Percentage of CD11b-expressing cells at 48 h after treatment with 0.1 (n = 5) and 1 µm ATRA (n = 7). e Correlation plot compares expression profiles of Dox-treated ARID1A sg1 and sg5 NB4 cells compared to Dox-treated control cells. Genes significantly upregulated or downregulated (FDR < 0.1; absolute log2 fold change > 0.1) in both sg1- and sg5-expressing NB4 cells are shown in red and those which are significantly altered with only one sgRNA are depicted in green. f Heat map shows differential expression of genes significantly altered in both Dox-treated ARID1A sg1 and sg5 expressing NB4 cells compared to the Dox-treated control cells (FDR < 0.1). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant
Discussion
Genetic studies using mouse models indicate that ATP-dependent SWI/SNF chromatin remodeling complex is a crucial regulator of hematopoiesis. While BAF53a, BAF47/Snf5, and BAF45a/PHF10 regulate HSC activity [46–48], other members of SWI/SNF complex play a more lineage-specific role including SMARCD2 in granulopoiesis [49, 50], Brg1 in erythroid development [51] and BAF155 in B cell development [52]. In this study, we have described in depth the role of ARID1A, a principal component of SWI/SNF, in hematopoiesis and examined how its deficiency alters chromatin structure in mouse hematopoietic cells.
In the current study, the function of ARID1A in normal hematopoiesis was elucidated by utilizing two different Cre transgenes, driven by either Vav or Mx1. Vav-iCre mediated deletion of floxed allele commences in fetal HSCs and persists through adult hematopoiesis. Mx1-Cre exerts its effect in mice following stimulation with poly(I:C). Owing to a possible developmental role of ARID1A in fetal/neonatal hematopoiesis, a large majority of ARID1A-deficient mice did not survive at weaning in our Vav-iCre model. Analysis of the hematopoietic compartment revealed largely overlapping phenotypes in both models of ARID1A deficiency, although the consequences of its loss were clearly enhanced in the Vav-iCre model. We believe that this is possibly caused by earlier deletion of Arid1a (during embryogenic hematopoiesis) in Vav-iCre model, while in Mx1-Cre model, mice were administered poly(I:C) at 8–14 weeks of age and hematopoietic parameters were analyzed four weeks later. Indeed, an efficient Mx1-Cre mediated deletion of Arid1a in BM following poly(I:C) treatment phenocopied largely the Vav-iCre model. Conversely, poly(I:C) administration caused an inefficient deletion of Arid1a in the thymus of Mx1-Cre mice which resulted in less dramatic effect on T cell maturation compared with the Vav-iCre mice.
Loss of ARID1A resulted in accumulation of LTHSCs in the BM, indicating requirement of ARID1A in maintenance of HSC pool size. This is reminiscent of increased HSC frequency observed in fetal liver of constitutive Arid1a KO embryos [27]. Results of our in vivo BrdU labeling showed elevated cycling of HSCs, implying decreased quiescence upon loss of ARID1A. Despite their increased number, Arid1a KO HSCs possessed poor multilineage reconstitution ability compared with the WT HSCs and were outcompeted in the transplantation settings. Apart from its vital role in maintaining HSC frequency and function, we uncover that ARID1A is also required for normal maturation of both myeloid and lymphoid lineages.
ATAC-seq data demonstrated that chromatin remodeling mediated by ARID1A-containing SWI/SNF complex is critical for hematopoiesis in mice. Deficiency of ARID1A led to a global decrease in open chromatin in myeloid precursors, including at several gene loci, which encode for key regulators of hematopoiesis. Notably, loss of ATAC-seq signal at these loci also correlated with reduced gene expression in myeloid precursors. One of the important downstream targets of ARID1A deficiency is GATA2, which plays an essential role in proliferation and maintenance of hematopoietic stem/progenitor cells, as well as differentiation towards multiple hematopoietic lineages [53]. Another affected gene is Cd34, protein encoded by which is expressed on the surface of HSCs and its knockout in mice leads to hematopoietic defects [54]. Similarly, Cebpa and Csf1, which are crucial regulators of myeloid differentiation [55, 56] also exhibited decreased chromatin accessibility and expression in the ARID1A-deficient cells. Therefore, we postulate that the loss of ARID1A alters chromatin dynamics and ensuing transcriptional changes manifest into impaired myeloid differentiation. Further studies are necessary to explore how loss of ARID1A effects the assembly/composition of SWI/SNF complexes and whether its closely related homolog, ARID1B, plays either an overlapping or distinctive role in chromatin remodeling in hematopoietic cells.
Evidence is emerging that ARID1A is vital for differentiation of various cell types [57–59]. Our study demonstrates that loss of ARID1A does not confer growth advantage to hematopoietic progenitors, but leads to defective differentiation of multiple hematopoietic lineages. For instance, myeloid cells in the BM and spleen of ARID1A-deficient mice displayed reduced surface expression of Gr1, indicating defects in terminal granulopoiesis. The block in myeloid differentiation caused by ARID1A deficiency may have implications in pathogenesis of APL, a subtype of AML in which truncating mutations of ARID1A were previously described [16]. In fact, silencing of ARID1A in NB4 cells did not affect significantly their growth but impaired the ATRA-induced granulocytic differentiation. Further studies are warranted to investigate whether the impaired differentiation in many hematological disorders including B-cell malignancies and APL is probably in part caused by loss-of-function mutations of ARID1A.
Following the discovery of frequent inactivating alterations of ARID1A, several approaches to therapeutically target ARID1A-deficient cancer cells have been proposed. These include increased sensitivity of ARID1A-mutated cells to inhibition of either polycomb activity, the PI3K/AKT pathway, the DNA damage checkpoint or the immune checkpoint [60]. Although these studies have focused on synthetic lethal interactions involving inactivation of SWI/SNF activity in solid tumors, particularly in ovarian cancers, which exhibit high mutational frequencies of ARID1A, similar strategies can be extended potentially to hematological malignancies harboring mutations of ARID1A. For instance, in APL cases with mutation of ARID1A, standard therapy (either ATRA or arsenic trioxide) could be combined with an approach directed at ARID1A deficiency, thus leading to potentially improved and personalized therapeutic approaches. Experimental models developed in this study including hematopoietic cell-specific Arid1a KO mice and ARID1A-deficient NB4 cells are valuable to explore these therapeutic strategies. For example, mice lacking ARID1A can be crossed with murine models of hematological malignancies for preclinical studies to test precision therapies.
Overall, this study enhances our understanding of the function of SWI/SNF complex in hematopoiesis. It demonstrates that nucleosome remodeling mediated by ARID1A is fundamental for normal hematopoietic development.
Supplementary information
Acknowledgements
We thank the staff of Comparative Medicine, NUS for their support in mice maintenance and experiments. We also acknowledge expert help and support from the FACS facility at CSI, Singapore. We also appreciate useful advice from Jiang Yanyi, Ling-Wen Ding, and Qi-Ling Zhou, CSI, Singapore. We thank the Melamed Family and Reuben Yeroushalmi for their generous support. This work was also funded by the Leukemia and Lymphoma Society, the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its Singapore Translational Research (STaR) Investigator Award to H. Phillip Koeffler (NMRC/STaR/0021/2014), the NMRC Centre Grant awarded to National University Cancer Institute of Singapore (NMRC/CG/012/2013) and the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiatives.
Author contributions
LH designed and performed research, analysed data and wrote the manuscript; VM conceived the study, designed and performed research, analysed data and wrote the manuscript; AM and PD performed bioinformatics and statistical analyses and wrote the manuscript; TWW performed research, analysed data and performed genotyping; PS, ZC, and JS performed experiments; HBMN maintained mouse colonies and performed genotyping; IL and ZW provided the Arid1a flox mouse colony; HPK conceived and supervised the study, interpreted the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lin Han, Vikas Madan
Supplementary information
The online version of this article (10.1038/s41375-019-0438-4) contains supplementary material, which is available to authorized users.
References
- 1.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–9. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–42. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 3.Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem. 2012;287:30897–905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 5.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–71. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 6.Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu RC, Wang TL. Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther. 2014;15:655–64. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda T, Banno K, Okawa R, Yanokura M, Iijima M, Irie-Kunitomi H, et al. ARID1A gene mutation in ovarian and endometrial cancers (Review) Oncol Rep. 2016;35:607–13. doi: 10.3892/or.2015.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan B, Wang TL. Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 13.Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45:1474–8. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 15.Mamo A, Cavallone L, Tuzmen S, Chabot C, Ferrario C, Hassan S, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2012;31:2090–100. doi: 10.1038/onc.2011.386. [DOI] [PubMed] [Google Scholar]
- 16.Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia. 2016;30:1672–81. doi: 10.1038/leu.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 18.Ding LW, Sun QY, Tan KT, Chien W, Mayakonda A, Yeoh AEJ, et al. Mutational Landscape of Pediatric Acute Lymphoblastic Leukemia. Cancer Res. 2017;77:390–400. doi: 10.1158/0008-5472.CAN-16-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–4. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1398–403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–98. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dietrich S, Hullein J, Lee SC, Hutter B, Gonzalez D, Jayne S, et al. Recurrent CDKN1B (p27) mutations in hairy cell leukemia. Blood. 2015;126:1005–8. doi: 10.1182/blood-2015-04-643361. [DOI] [PubMed] [Google Scholar]
- 24.Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47:1426–34. doi: 10.1038/ng.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiel MJ, Sahasrabuddhe AA, Rolland DC, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krosl J, Mamo A, Chagraoui J, Wilhelm BT, Girard S, Louis I, et al. A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood. 2010;116:1678–84. doi: 10.1182/blood-2010-03-273862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–61. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 30.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21 29 21–29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgiou G, van Heeringen SJ. fluff: exploratory analysis and visualization of high-throughput sequencing data. PeerJ. 2016;4:e2209. doi: 10.7717/peerj.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–25. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 42.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–25. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohmann AF, Martin LJ, Minder JL, Roe JS, Shi J, Steurer S, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol. 2016;12:672–9. doi: 10.1038/nchembio.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47(D1):D729–D735. doi: 10.1093/nar/gky1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–25. doi: 10.1016/S1535-6108(02)00185-X. [DOI] [PubMed] [Google Scholar]
- 47.Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120:4720–32. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- 48.Krasteva V, Crabtree GR, Lessard JA. The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling complexes is essential for hematopoietic stem cell maintenance. Exp Hematol. 2017;48:58–71 e15. doi: 10.1016/j.exphem.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Priam P, Krasteva V, Rousseau P, D’Angelo G, Gaboury L, Sauvageau G, et al. SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPvarepsilon dependent mechanism. Nat Genet. 2017;49:753–64. doi: 10.1038/ng.3812. [DOI] [PubMed] [Google Scholar]
- 50.Witzel M, Petersheim D, Fan Y, Bahrami E, Racek T, Rohlfs M, et al. Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat Genet. 2017;49:742–52. doi: 10.1038/ng.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–61. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J, Ko M, Jeon S, Jeon Y, Park K, Lee C, et al. The SWI/SNF-like BAF complex is essential for early B cell development. J Immunol. 2012;188:3791–803. doi: 10.4049/jimmunol.1103390. [DOI] [PubMed] [Google Scholar]
- 53.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2012;82:1–17. doi: 10.1016/j.critrevonc.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Cheng J, Baumhueter S, Cacalano G, Carver-Moore K, Thibodeaux H, Thomas R, et al. Hematopoietic defects in mice lacking the sialomucin CD34. Blood. 1996;87:479–90. [PubMed] [Google Scholar]
- 55.Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497:239–43. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–74. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei I, Gao X, Sham MH, Wang Z. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem. 2012;287:24255–62. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura Yoshito, Fukuda Akihisa, Ogawa Satoshi, Maruno Takahisa, Takada Yutaka, Tsuda Motoyuki, Hiramatsu Yukiko, Araki Osamu, Nagao Munemasa, Yoshikawa Takaaki, Ikuta Kozo, Yoshioka Takuto, Wang Zong, Akiyama Haruhiko, Wright Christopher V., Takaori Kyoichi, Uemoto Shinji, Chiba Tsutomu, Seno Hiroshi. ARID1A Maintains Differentiation of Pancreatic Ductal Cells and Inhibits Development of Pancreatic Ductal Adenocarcinoma in Mice. Gastroenterology. 2018;155(1):194-209.e2. doi: 10.1053/j.gastro.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 59.Lei I, West J, Yan Z, Gao X, Fang P, Dennis JH, et al. BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. J Biol Chem. 2015;290:19343–52. doi: 10.1074/jbc.M115.637389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fukumoto T, Magno E, Zhang R. SWI/SNF complexes in ovarian cancer: mechanistic insights and therapeutic implications. Mol Cancer Res. 2018;16:1819–25. doi: 10.1158/1541-7786.MCR-18-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.