Abstract
Integrating single-cell trajectory analysis with pooled genetic screening could reveal the genetic architecture that guides cellular decisions in development and disease. We applied this paradigm to probe the genetic circuitry that controls epithelial-to-mesenchymal transition (EMT). We profiled epithelial cells undergoing a spontaneous, spatially determined EMT in the presence or absence of TGF-β via single-cell RNA-seq. Pseudospatial trajectory analysis identified continuous waves of gene regulation, as opposed to discrete “partial” stages of EMT. KRAS was connected to exit from the epithelial state and acquisition of a fully mesenchymal phenotype. A pooled single-cell CRISPR-Cas9 screen identified EMT-associated receptors and transcription factors, including regulators of KRAS, whose loss impeded progress along EMT. Inhibiting the KRAS effector MEK, and its upstream activators EGFR and MET, demonstrates that interruption of key signaling events reveals regulatory “checkpoints” in the EMT continuum that mimic discrete stages and reconciles opposing views of the program that controls EMT.
Introduction
During EMT, cells dissolve strong contacts and leave organized sheets, shifting from apical-basal to front-rear polarity. As they become mesenchymal, their motility and ability to break down extracellular matrix enables them to invade surrounding tissue1,2. EMT is fundamental to development3, wound healing4,5, and the metastatic dissemination of tumor cells2,5,6.
Several studies identified discrete intermediate “stages” of EMT based on expression of a handful of marker genes7-9. However, recent single-cell mass cytometry and RNA-seq analyses of breast cancer cells suggest that they fall along a continuum10,11. As such, it remains unclear whether or not cells exist in functionally discrete states during EMT, and the genetic circuitry that controls the transition remains incompletely defined. Partial EMT is implicated in renal fibrosis12,13 and pancreatic ductal adenocarcinoma14 and is positively correlated with tumor grade and metastatic potential in head and neck squamous cell carcinoma15. Characterizing the nature of intermediate EMT would have an immediate impact on our understanding of disease.
Here, we apply single-cell RNA sequencing (scRNA-seq) coupled with unsupervised machine learning techniques16,17 to analyze a “pseudospatial”18 model of EMT and identify regulators of its progression. We analyze a two-dimensional (2D) model system of spontaneous, confluence-dependent EMT in human mammary epithelial cells19. Cells fell continuously along a gradient of EMT progression, revealing distinct waves of gene regulation. We incorporate a pooled single-cell CRISPR-Cas9 screen into our pseudospatial trajectory analysis to determine the dependency of EMT-associated signaling events on progression along the EMT continuum. These experiments uncover a hierarchy of transcription factors and cell surface receptors that drive cells through EMT. Loss-of-function of one of several surface receptors slow progress through EMT, explaining how cells transiting through a continuous process appear to be in one of several discrete stages in some experimental systems.
Results
Pseudospatial trajectory analysis of spontaneous EMT
To define the transcriptional program executed by normal human cells undergoing EMT, we devised an in vitro system in which cells from an epithelial colony migrate into unoccupied margins of the plate (Fig. 1a). We seeded MCF10A mammary epithelial cells19 within cloning rings as a high-confluence patch in the center of a tissue culture dish. We then removed the rings after which cells at the border sense adjacent, unoccupied space and spontaneously undergo an EMT. The spontaneous EMT in this system is analogous to that observed for MCF10A cells upon wounding in scratch-wound healing assays20,21. Cells at the periphery of the patch acquired a spindle-like morphology and formed leading and protruding edges consistent with acquisition of a mesenchymal phenotype (Supplementary Fig. 1). Cells collected from a single well of our assay expressed levels of e-cadherin and vimentin protein spanning a dynamic range that included those cultured at low or high confluence (Supplementary Fig. 1c-d). We dissected the patch to isolate “inner” cells (2,440 cells) and “outer” cells (2,564 cells). Inner and outer fractions were dissociated into single-cell suspensions and subjected to scRNA-seq on the 10X Chromium platform (Fig. 1a and Supplementary Table 1).
Figure 1: Pseudospatial trajectory reconstruction of spontaneous EMT reveals the transition as a continuum of epithelial-mesenchymal states.
a) Schematic of spontaneous confluence-dependent EMT assay, cell isolation and pseudospatial trajectory reconstruction using Monocle2. Red circle denotes the area that defines inner and outer cells for macro-dissection. b-c) t-SNE embedding of cells from our spontaneous EMT assay. Cells are colored by the fraction from which they were isolated (b) or expression of the mesenchymal marker VIM (c). d) Trajectory of inner and outer MCF10A cells upon spontaneous EMT progression. Arrow denotes progression of pseudospace. Insert: density of cells across pseudospace. e) Left: Stitched brightfield images of an MCF10A colony at the end of our spontaneous EMT assay (2,000 μm scale bar). Right, top to bottom: E-cadherin and DAPI staining of cells from the center, middle and edge of the MCF10A colony (50 μm scale bar, representative fields from 6 images across 3 independent samples). f) Expression of epithelial and mesenchymal markers across pseudospace. Cells are colored as in b. g) Hierarchical clustering of kinetic curves for dynamically regulated genes across pseudospace for all 5,004 cells in our experiment (likelihood ratio test, FDR q < 1 × 10−10 and AUC > 10). Rows represent row centered dynamics of gene expression. h) Gene-set analysis using the Gene Ontology Biological Processes and MSigDB Hallmarks gene-set collections of gene clusters from g (hypergeometric test FDR, q < 0.05). i) Expression of epithelial and mesenchymal markers across pseudospace in primary human mammary epithelial cells (HuMEC). Cells are colored as in b.
Unsupervised clustering with t-distributed stochastic neighbor embedding (t-SNE) separated cells from inner and outer fractions (Fig. 1b), and expression of the mesenchymal marker VIM varied in a reciprocal gradient to the epithelial markers CDH1 and DSP across embedded cells (Fig. 1c, Supplementary Fig. 2). However, we did not observe separated clusters of cells along this axis of epithelial and mesenchymal marker expression, suggesting continual progression along an EMT rather than a sequence of discrete stages.
Individual cells at similar radii from the center of a patch could be in different stages of EMT, analogous to how cells proceed asynchronously through temporal processes such as differentiation. To resolve cellular heterogeneity and recover the program that characterizes a cell’s progress through EMT, we ordered cells using Monocle16,17. Monocle organized cells along a linear pseudospatial18,22 trajectory, with cells from inner and outer fractions concentrated at the beginning and end of its axis, respectively (Fig. 1d; Supplementary Fig. 3). Simulated sampling from the ends of the continuum and repeating our analysis excluded the possibility that this continuity was an artifact of trajectory analysis (Supplementary Fig. 4).
Classic markers of EMT varied in expression over the trajectory. Protein and mRNA levels of the epithelial marker E-cadherin (CDH1) decreased as cells radiated from the center of the colony and over the pseudospatial trajectory, consistent with a spontaneous spatially determined EMT (Fig. 1e-f). Conversely, mRNA levels of VIM increased sharply in cells in the second half of the trajectory (Fig. 1f). Partial or intermediate EMT has classically been defined as co-expression of epithelial and mesenchymal traits23,24. Accordingly, cells positive for both CDH1 and VIM mRNA were most frequent in the second half of the trajectory (Supplementary Fig. 5). Population level average expression of two epithelial markers, CDH1 and CRB3, did not vary drastically between inner and outer fractions (Supplementary Fig. 6), highlighting the value of single-cell techniques to capture the dynamics of gene regulatory changes associated with EMT.
We next identified genes regulated during EMT by performing differential expression analysis parameterized by each cell’s position along the trajectory (Supplementary Table 2). Clustering the 1,105 differentially expressed genes (DEGs) (likelihood ratio test; FDR q < 1 × 10−10; AUC > 10 in at least one quantile, see Methods and Supplementary Table 3) revealed six groups of genes with similar kinetics. We performed gene-set analysis using the Gene Ontology biological processes25,26 and MSigDB hallmarks molecular signature27 gene-set collections. Genes in cluster 6 were upregulated and enriched for roles in translational regulation or EMT, while those in downregulated cluster 1 were linked to epidermis development. Cluster 5, highly expressed in the outermost regions of the pseudospatial trajectory, was associated with regulation of the cell cycle, consistent with relief of contact-mediated inhibition of proliferation (Fig. 1g-h and Supplementary Table 4).
Gene-set analysis identified pathways upstream of pseudospace-dependent gene expression. Cluster 1 was enriched for genes repressed by active KRAS signaling28,29, including some with roles in EMT. For example, keratin 1 (KRT1) was expressed in cells at the epithelial end of the trajectory but silenced as cells approached the border of the patch (Supplementary Fig. 7). Keratins traffic E-cadherin to the cell membrane, while vimentin does not30, and the shift in cytoskeletal filament composition from keratin- to vimentin-containing is integral to EMT31. The EMT-associated cluster 6 included the unfolded protein response (UPR) transcriptional regulator ATF4 whose increased expression preceded upregulation of genes in cluster 5, which was enriched for genes associated with the UPR (cluster 5 and 6, Fig. 1g-h and Supplementary Fig. 8), consistent with a recent study demonstrating that induction of EMT elicits protective activation of the UPR32.
Repeating our spatial EMT assay and single-cell transcriptional profiling using primary human mammary epithelial cells (HuMEC) identified a similar linear pseudospatial trajectory and distribution of inner and outer cells (Supplementary Fig. 9a-b, Supplementary Table 5). Dynamics of epithelial and mesenchymal marker expression was comparable albeit with decreased CDH1 downregulation and more drastic upregulation of FN1 (Fig. 1i and Supplementary Fig. 9c). Having identified a spatial EMT in another epithelial cell type we sought to understand how this phenotype changes in response to a strong inducer of EMT.
Pseudospatial trajectory alignment elucidates TGF-β-driven full EMT
Activation of the transforming-growth factor β (TGF-β) pathway leads to a powerful induction of EMT33,34. We repeated our pseudospace experiment, this time treating cells with TGF-β to promote mesenchymal conversion in MCF10A cells7. We sequenced transcriptomes of 2,121 inner and 2,116 outer colony cells which, although segregated in t-SNE space, did not form coherent clusters and for which expression of FN1 and VIM continuously vary (Supplementary Fig. 10). Thus, adding a strong extracellular signal promoting EMT did not drive cells into discrete stages. We therefore constructed a pseudospatial trajectory for TGF-β as well (Supplementary Fig. 11).
To compare cells from spontaneous and TGF-β-driven EMT trajectories, we used trajectory alignment35-37, a technique that employs Dynamic Time Warping38,39 to map cells onto a common pseudospatial axis (Fig. 2a). Along the aligned axis, cells treated with TGF-β expressed CDH1 and CRB3 with similar kinetics to those undergoing confluence-mediated EMT (Fig. 2b), consistent with reports that maintenance of cell-cell contacts prevents TGF-β stimulation from fully repressing an epithelial phenotype40. However, TGF-β exposure is sufficient to drive expression of mesenchymal genes even in cells within the epithelial core. Additionally, only cells treated with TGF-β and positioned at the trajectory’s outer extreme expressed robust levels of FN1 and CDH2, suggesting a full E- to N-cadherin switch. Exposure of HuMEC cells to TGF-β similarly led to a robust increase in VIM and FN1 at the beginning of the trajectory (Supplementary Fig. 12c); however, expression of CDH2 was not apparent. A broader gene-set analysis comparing normalized average expression scores41 showed that TGF-β drove MSigDB Hallmark EMT genes higher and GO-BP epidermis development genes lower in both MCF10A and HuMEC cells (Supplementary Fig. 13).
Figure 2: Alignment of spontaneous and TGF-ß-driven EMT pseudospatial trajectories identifies discrete waves along the EMT continuum.
a) Dynamic time warping of pseudospatial trajectories allows for comparison of the dynamics of EMT progression along a common axis. b) Epithelial and mesenchymal markers expression across warped pseudospace (cells are colored by treatment). c) Hierarchical clustering of kinetic curves for dynamically regulated genes that vary significantly between spontaneous (5,004 cells) and TGF-ß-driven (4,237 cells) EMT trajectories (likelihood ratio test, FDR, q < 1 × 10−10 and ∣ΔAUC∣ > 0.02). Rows represent row centered dynamics of gene expression. At left: Gene-set analysis on gene clusters using the Oncogenic Signatures gene-set collection (hypergeometric test FDR, q < 0.05). Red and blue arrows denote association with increased or decreased activity, respectively. At right: Gene-set analysis on gene clusters using the GO-BP and Hallmarks gene-set collections (hypergeometric test FDR, q < 0.05). d-f) Pseudospatial expression dynamics of EMT-associated genes that increase in expression at the end of spontaneous and TGF-ß-driven trajectories (d), towards the end of the TGF-ß-driven trajectory (e) and towards the middle of the TGF-ß-driven trajectory (f). g-h) Boxplots of early and late EMT scores of MCF10A cells at early and late positions in pseudospatial trajectories (Mock = 1,020 cells, TGF-ß = 772 cells) and HNSCC tumors (6 = 80 cells, 20 = 321 cells, 5 = 41 cells, 18 = 140 cells, 22 = 119 cells, 25 = 54 cells, 17 = 330 cells, 16 = 56 cells). Boxplots depict the median score (bold line within box) with lower and upper hinges depicting the 25th and 75th percentiles, respectively. i) Density of cells across EMT trajectories after k-nearest neighbor projection of HNSCC tumor cells to MCF10A cells under spontaneous and TGF-ß-driven conditions.
To identify genes responsive to TGF-β, we tested for differential expression as a function of TGF-β treatment, subtracting changes attributable to pseudospatial position. This analysis identified 1,328 genes in 10 clusters with distinct TGF-β-dependent dynamics (Fig. 2c, likelihood ratio test; FDR q < 1 × 10−10 and ∣ΔAUC∣ > 0.02, see Methods, Supplementary Fig. 14 and Supplementary Table 6). For example, cluster 5 contained cell-cycle-related genes upregulated along both trajectories (Fig. 2c and Supplementary Table 7). Cluster 4 contained genes upregulated towards the end of the spontaneous trajectory but maintained at high levels throughout the TGF-β-mediated trajectory (Fig. 2c). This cluster included 2 EMT-associated genes, one of which, NNMT, is a marker of the metabolic changes that accompany EMT42 (Fig. 2d). In contrast, clusters 6 and 8 contained EMT genes that peaked at the middle or end of the TGF-β-driven trajectory, respectively (Fig. 2c,e and f), but were unaltered or induced to a lesser degree in the spontaneous trajectory. Therefore, cells at comparable positions in spontaneous vs. TGF-β-mediated EMT continua as defined by epithelial markers display pronounced transcriptional differences.
To explore which molecular regulators are responsible for shared and distinct patterns of spontaneous and TGF-β-mediated gene regulation during EMT we performed gene-set analysis using the MSigDB Oncogenic Signature gene-set collection. This gene-set collection is composed of genes whose expression increases or decreases as a function of perturbing signaling pathways43. Cluster 8 included genes upregulated as cells treated with TGF-β undergo EMT but are weakly altered during spontaneous EMT. These were enriched for genes expressed in response to KRAS signaling28, including genes with roles in EMT, such as CXCL1 and CXCL2, which induce cellular migration44,45. (Supplementary Fig. 15). Conversely, cluster 10 included epithelial marker genes downregulated early in spontaneous EMT and expressed at low levels in cells treated with TGF-β (e.g. KRT4 and KRT16) (Supplementary Fig. 16). These and several others are known to be repressed by active KRAS signaling28. This observation, together with pathway analysis of spontaneous EMT, implies that KRAS signaling is sustained throughout both spontaneous and TGF-β-driven transitions, suggesting it governs multiple points of the EMT continuum in normal cells.
Single-cell flow cytometric profiling of TGF-β-induced EMT described the transition as a three-state process7,46. In contrast to this “discrete” view, we observed a continuous trajectory over which cells were distributed and along which many genes, including classic markers of epithelial and mesenchymal states, exhibit smooth changes in expression. Few cells undergoing spontaneous EMT expressed high levels of some mesenchymal markers, raising the possibility that we failed to capture some discrete, physiologically important “stages” of EMT. However, exposing cells to TGF-β also drives them over a continuum, albeit one with different spatial patterns of transcriptional regulation.
To investigate whether, in vivo, tumor cells transit through an EMT continuum similar to the one observed in vitro, we re-analyzed single-cell RNA-seq data from patients with head and neck squamous cell carcinoma (HNSCC)15. The most mesenchymal tumor, as ranked by Puram et al., expressed EMT genes at similar levels to cells at the outer end of our TGF-β-driven pseudospatial trajectory (Supplementary Fig. 17). Genes that make up early and late waves of KRAS-associated EMT in vitro (cluster 10 and 8, respectively, Fig. 2C) were expressed in a manner consistent with their partial EMT phenotypes assigned by Puram et al. (Fig. 2g-h). To confirm that the similarity between our in vitro model and the tumor cells was not limited to known EMT genes, we projected tumor cells onto our spontaneous and TGF-β-driven trajectories based on full transcriptome signatures using a nearest-neighbor matching algorithm47 (Methods). Most tumor cells mapped to the end of our spontaneous EMT trajectory. In contrast, tumor cells projected more uniformly over the TGF-β-driven trajectory (Fig. 2i). Individual tumors covered a substantial range of the trajectory, suggesting our TGF-β-driven model captures much of the transcriptional diversity present in a single patient sample. Finally, we tested whether Monocle 2 could reconstruct pseudospatial trajectories directly from tumor cells. For three of the four tumors with sufficient cells for Monocle analysis, the algorithm recovered a linear trajectory (Supplementary Fig. 18) with similar expression kinetics to in vitro trajectories (Supplementary Fig. 18). Taken together, these analyses suggest that the waves of gene regulation that occur during EMT in vitro occur to varying extents in vivo.
A pooled loss-of-function screen identifies genes regulating EMT progression
We reasoned that certain regulators control passage through parts of the EMT continuum and a lack of one or more of these signals leads to accumulation of cells at “discrete” EMT “stages”. To identify regulators of progression along the continuum, we devised a high-throughput loss-of-function screen to probe the architecture of pathways with known involvement in EMT. Several groups recently devised methods for coupling CRISPR-based screens and a scRNA-seq readout, i.e. by capturing the identity of the single-guide RNA(s) (sgRNAs) that a cell received in conjunction with its gene expression profile48-52. Here we used a modified version of CROP-Seq52, which does not rely on the pairing of sgRNAs with distally located barcodes. We recently showed that this design is preferable to alternatives, avoiding template switching between sgRNAs and associated barcodes during lentiviral co-packaging53.
We selected 16 cell surface receptors and 24 transcription factors for screening via CROP-seq in our 2D EMT system (Fig. 3a). These targets include receptors reported to activate KRAS (EGFR, MET, FGFR1, FGFR2, ITGAV, ITGB1 and ITGB3)54-57 along with others that drive Wnt, Notch, and other pathways (Fig. 3b). Transcription factors that activate or repress EMT genes included both well-characterized (SNAI1/2, TWIST1/2, ZEB1/2) and recently reported (FOXD3, GATA6, SOX9) regulators1. We repeated our in vitro EMT assay with a mixture of cells edited with sgRNAs to one of the 40 genes (or non-targeting controls, NTC) and subjected them to scRNA-seq after culture with TGF-β (12,337 cells) or without (17,093 cells). Unsupervised clustering analysis of cells treated with TGF-β identified prominent, clearly demarcated clusters of cells that retained expression of the epithelial markers CDH1 and CRB3 and failed to upregulate FN1 and VIM (Fig. 3c and Supplementary Fig. 19a-b). Cells expressing sgRNAs targeting TGFBR1 and TGFBR2 were enriched in these clusters (Fisher’s exact test; FDR p < 1 × 10−7) (Fig. 3d-e and Supplementary Fig. 19c), while NTC sgRNAs were largely absent from them. Importantly, this distribution was not a result of the number of TGFBR1 and TGFBR2 sgRNA cells in our screen (Fig. 3f). Cells with sgRNAs against TGFBR1 and TGFBR2 expressed lower levels of FN1 and VIM than those with NTC sgRNAs, indicating a failure to activate a TGF-β-driven EMT (Fig. 3g) confirming that CROP-seq can be used to identify molecular phenotypes along the EMT continuum.
Figure 3: Multiplexed loss-of-function screening of EMT-associated genes recovers deficiencies in TGF-ß-induced EMT.
a) Schematic of pooled approach to determine regulators of distinct EMT states. Red circle in right panel denotes the area that defines the boundary between inner and outer cells for macro-dissection. b) Collection of EMT associated cell surface receptors and transcription factors included in our CROP-seq screen. c-d) t-SNE embedding of sgRNA containing cells from our TGF-ß-exposed CROP-Seq experiment colored by EMT marker expression (c) or expression of NTC or TGFBR2 sgRNAs (d). e) Fraction of cells within VIM low clusters expressing NTC, TGFBR1 or TGFBR2 sgRNAs from our TGF-ß-exposed CROP-Seq screen. f) Total number of cells expressing NTC, TGFBR1 or TGFBR2 sgRNAs from our TGF-ß-exposed CROP-Seq screen. g) Expression of FN1 and VIM across cells expressing a sgRNA to NTC (943 cells), TGFBR1 (219 cells) or TGFBR2 (2299 cells) from our TGF-ß-exposed CROP-Seq experiment. Point within the violin depicts the mean expression level for each group with violin spanning the minimum and maximum expression value across cells.
We next sought to organize edited cells into a pseudospatial trajectory. We compared NTC cells from inner and outer fractions, which revealed 1,197 and 761 DEGs in the spontaneous and TGF-β-driven EMT, respectively, more than 80% of which were also found in unedited EMT experiments (Supplementary Fig. 20a-d). Pseudospatial trajectories reconstructed from NTC cells aligned to unedited trajectories with only minimal warping (Supplementary Fig. 20e-g). We then provided Monocle 2 with all edited cells, which constructed trajectories along which EMT marker genes were expressed with kinetics similar to unedited cells (Supplementary Fig. 21). Differential expression analysis identified 978 and 4,079 genes that varied across genotypes along spontaneous and TGF-β-driven trajectories, respectively (Supplementary Fig. 22 and Supplementary Tables 8 and 9).
We hypothesized that loss of surface receptors that transduce signals important for EMT, or the transcription factors they drive, would alter a cell’s progress along the trajectories. To determine whether loss-of-function of EMT-associated targets altered their progression along pseudospace, we divided the trajectory into bins according to the density of cells along spontaneous and TGF-β-driven EMT trajectories resulting in 7 and 8 bins, respectively. We then tested whether cells carrying sgRNAs against each target altered their distribution over these ‘regions’ of the aligned trajectories, relative to NTCs. We determined empirical false-discovery rates of these tests by comparing enrichments of knockout cells to a random sampling of NTC cells (Supplementary Fig. 23, see Methods for details).
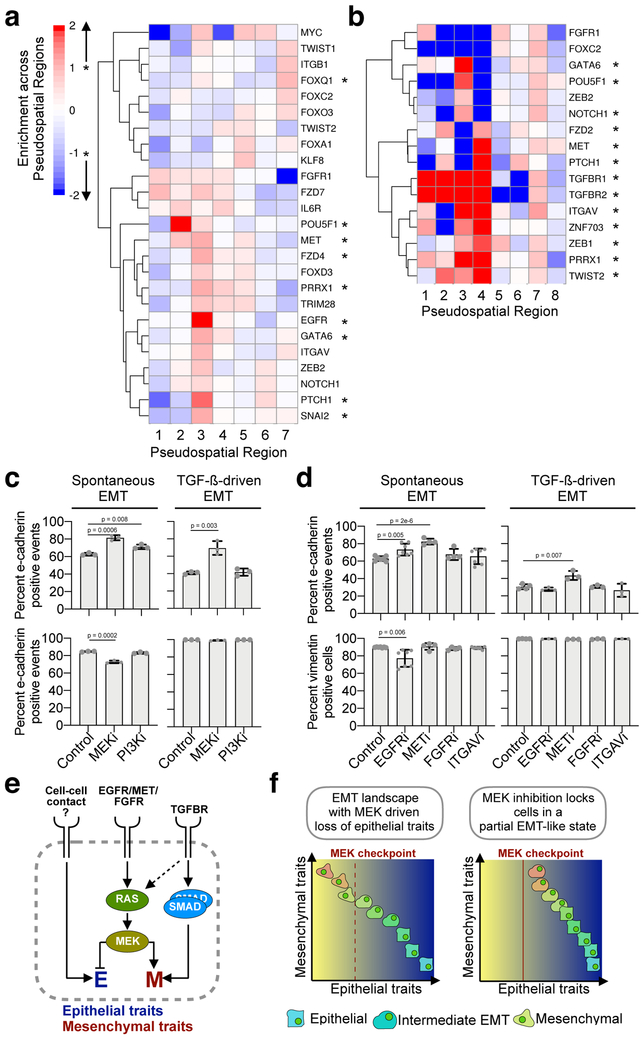
Of the 40 genes tested, 30 significantly shifted the cells’ pseudospatial positions when targeted via CROP-seq, with 11 overlapping between conditions (Fig. 4a-b and Supplementary Fig. 23). Some targets were modestly enriched (less than two-fold) at a given pseudospatial position. For example, in the spontaneous EMT trajectory, cells with sgRNAs targeting FZD7 were enriched at region 1, near the epithelial core of the trajectory, and region 3 (Fig. 4a). Other gene knockouts induced strong, focal accumulation of cells at one or two positions along the EMT continuum (Fig. 4a-b). Loss of EGFR induced focal accumulation at region 3 (Supplementary Fig. 24a). Similarly, cells with sgRNAs against MET were enriched in regions 2 and more strongly in region 3. The majority of significantly enriched targets accumulated in region 3 directly preceding a decrease in the total number of CDH1 single-positive cells and an increase in CDH1/VIM double-positive cells (Supplementary Fig. 25).
Figure 4: Accumulation of knockout cells across spontaneous and TGF-ß-driven EMT trajectories identifies regulators of discrete checkpoints across the EMT continuum.
a-b) Enrichment of knockouts whose distribution is significantly altered across pseudospace, and therefore EMT progression, in our spontaneous (11,908 cells) (a) and TGF-ß-driven (9,951 cells) (b) conditions. The distribution of cells expressing sgRNAs against EMT genes was compared to the distribution of NTC controls using Chi square (empirically determined FDR < 10%). For targets whose distribution is altered enrichment across each region was determined by calculating the odds ratio. c) Percent E-cadherin (top panels) or vimentin (bottom panels) positive cells in MCF10A colonies exposed to MEK (U0126) and PI3K (LY294002) inhibition after spontaneous (left panels) or TGF-ß-driven (right panels) EMT. Error bars denote standard deviation from the mean (n = 3, two-tailed Student’s t test). d) Percent E-cadherin (top panels) or vimentin (bottom panels) positive cells in MCF10A colonies exposed to EGFR (Erlotinib), MET (Crizotinib), FGFR (Infigratinib) and ITGAV (Cilengitide) inhibition after spontaneous (left panels) or TGF-ß-driven (right panels) EMT. Error bars denote standard deviation from the mean (at left: spontaneous EMT control/EGFRi/ITGAVi n = 7, METi/FGFRi n = 4 independent samples; at right: TGF-ß-driven EMT control n = 4, EGFRi/METi/FGFRi/ITGAVi n = 3 independent samples, two-tailed Student’s t test). e) Inferred EMT regulatory network and putative regulators identified in this study. f) Model depicting the MEK dependent EMT regulatory checkpoint created and its effects on the development of intermediate EMT phenotypes.
Edited cells across the TGF-β-treated trajectory had a distinct set of genes from those that control progression through spontaneous EMT, reflecting the direct activation of EMT-associated transcription factors by SMAD signaling58. The pseudospatial regions encompassing the first half of the trajectory were strongly enriched for TGFBR1 and TGFBR2 knockouts (region 1-4, Fig. 4b and Supplementary Fig. 24b). As in spontaneous EMT, loss of numerous genes in TGF-β-treated cells concentrated them at defined pseudospatial positions. ZEB1, proposed to effect an irreversible switch to a mature mesenchymal state7, GATA6, NOTCH1 and POU5F1 were concentrated beginning in region 3, suggesting that this position in the trajectory coincides with a decision point cells pass through during EMT.
Of the seven receptors in our screen known to activate Ras/MAPK signaling, five (EGFR, MET, ITGAV, ITGB1, and FGFR1) altered the distribution of cells over the trajectory, and all but MET concentrated them at just one or two regions. Interestingly, only MET and ITGAV did so during spontaneous and TGF-β-driven EMT. In the spontaneous EMT trajectory, early accumulation of cells expressing sgRNAs against the receptor tyrosine kinases EGFR and MET55,56, suggested that one or both are responsible for the early wave of KRAS activity associated with exit from the epithelial state. In the TGF-β-mediated EMT trajectory regions 3 and 4 displayed robust accumulation of cells expressing sgRNAs against the ITGAV integrin and regions 1, 5 and 7 were enriched for cells expressing sgRNAs against the tyrosine kinase FGFR159. Integrins function as heterodimers between ɑ and β subunits and ɑvβ1 heterodimers have been shown to mediate TGF-β signaling during fibrosis60, a process where EMT has an important role12,13,61. These precede the terminal EMT state in our TGF-β trajectory and may contribute to the KRAS-associated late EMT signature identified by our dynamic time warping analysis (Fig. 2c).
To understand how KRAS signaling drives cells through EMT, we performed our in vitro assay in the presence of small molecules that block RAS signaling. RAS exerts its regulatory program via activation of the RAF/MEK/ERK and PI3K/AKT pathways62,63. We therefore tested whether loss of MEK (via U0126 treatment) or PI3K signaling (via LY294002 treatment) is sufficient to block exit from the epithelial state and/or acquisition of mesenchymal phenotypes. Doses of both drugs were chosen to minimize effects on cell viability (Supplementary Fig. 26). We used flow cytometry to determine the proportion of cells expressing the early EMT markers E-cadherin and vimentin and the mature mesenchymal markers N-cadherin and cytoplasmic fibronectin. Upon spontaneous EMT, inhibition of PI3K activity led to a modest increase in cells expressing E-cadherin (Fig. 4c). In contrast, MEK inhibition led to a pronounced increase in E-cadherin and an accompanying decrease in vimentin (Fig. 4c-d). Inhibiting MEK prevented downregulation of E-cadherin even in the presence of TGF-β yet had no effect on the proportion of cells expressing vimentin, N-cadherin or fibronectin. Treatment of HuMEC cells with U0126 also decreased the induction of vimentin under spontaneous and TGF-β-driven EMT and decreased fibronectin accumulation after TGF-β exposure (Supplementary Fig. 27).
To map the upstream regulators of this MEK-induced EMT program, we treated MCF10A undergoing spontaneous and TGF-β-driven EMT with small molecules targeting RTKs and integrins from the genetic screen (EGFR-erlotinib; MET-crizotinib; FGFR-infigratinib; ITGAV-cilengitide). Inhibiting EGFR led to an increase in E-cadherin-positive cells and a decrease in vimentin-positive cells only in spontaneous EMT, consistent with EGFR knockout inducing accumulation in pseudospace only in the absence of TGF-β (Fig. 4d). Conversely, MET inhibition led to increases in E-cadherin-positive cells in both spontaneous and TGF-β-driven conditions, reflecting pausing of knockout cells along both EMT trajectories (Fig. 4d). FGFR and ITGAV inhibition did not significantly alter CDH1 and VIM levels suggesting that they lead to accumulation by alteration of other signaling pathways. We further examined the role of EGFR in regulating the transition into spontaneous EMT by treating cells with a higher dose of erlotinib and expanding our panel of marker proteins. In addition to confirming EGFR’s regulation of E-cadherin and vimentin we observed that blocking EGFR signaling decreased the level of crumbs3 and desmoplakin during spontaneous EMT (Supplementary Fig. 28a-b). Brightfield images of spontaneous EMT colonies showed a decrease in cells undergoing individual migration, a key phenotypic characteristic of cells transitioning into a mesenchymal state (Supplementary Fig. 28c).
Although inhibiting MEK was not sufficient to prevent activation of the mesenchymal program in MCF10A in the presence of TGF-β, cells co-expressed E-cadherin and high levels of vimentin, N-cadherin and fibronectin protein (Fig. 4c and Supplementary Fig. 29). This suggests that activation of the RAF/MEK/ERK pathway is required for downregulation of the epithelial program in normal mammary epithelial cells, but that alternate pathways can activate the mesenchymal program when RAF/MEK/ERK signaling is blocked.
Lastly, we explored how expression of factors that alter the accumulation along EMT in MCF10A relate to the diverse EMT phenotypes observed in HNSCC tumors. Hierarchical clustering of the mean expression level of cell surface receptors identified a strong relationship between receptor expression and the extent of EMT across tumor samples (Supplementary Fig. 30). Expression of FZD2, FZD7, FGFR1 and PTCH1 was inversely correlated with levels of EMT genes. With the exception of PTCH1, edited cells lacking these genes were enriched at the beginning of our EMT trajectories (Supplementary Fig. 30). Conversely, tumors expressing high levels of EMT genes (Supplementary Fig. 30) also expressed MET, ITGAV, ITGB1, TGFBR1 and TGFBR2.
Discussion
The integration of single-cell trajectory analysis and pooled genetic screening has the potential to map the genetic circuits that control progression across biological transitions. Understanding the regulation of EMT is a fundamental goal in developmental and cancer biology and has the potential to yield new therapeutic opportunities for intervention in cancer. In contrast to numerous reports of “partial”, “hybrid”, or “intermediate” EMT stages, both our analysis and recent scRNA-seq and mass cytometry studies of a cancer line10,11 indicate that cells are organized along a continuum during EMT.
Our CRISPR/scRNA-seq loss-of-function screen reconciles these two conflicting views of gene regulation in EMT. Previously, we showed that a loss-of-function mutation can create a branch from the wild-type trajectory on which cells execute an alternative gene expression program64. Here, we show that transcription factor and signaling receptor gene knockouts can cause cells to accumulate at defined points along the EMT continuum, implying the existence of a sequence of “checkpoints” to progress through it. Therefore, although cells fall along a transcriptional continuum during EMT, genetic insults that disable key signaling pathways could enrich a particular gene expression profile within a cell population, creating the impression of a stable intermediate phenotype. Consistent with this finding, recent single-cell profiling of head and neck squamous cell carcinoma found evidence for diverse partial EMT states at the leading edge of tumors15, which could arise from genetic heterogeneity amongst cancer cells. Our analysis suggests that local variation in signaling in key pathways could also contribute substantially to a tumor’s EMT phenotype.
Several large modules of genes with distinct, wave-like patterns of regulation during spontaneous- or TGF-β-mediated EMT were enriched for targets of KRAS, which may therefore be involved throughout the EMT continuum. KRAS signaling can be initiated via various upstream signals, making it difficult to pinpoint which drive signaling at each point on the continuum. Focal accumulation of cells lacking particular effectors of KRAS signaling early in spontaneous (EGFR and MET) and late in TGF-β-mediated (FGFR2 and ITGAV) EMT suggests that the cell responds to a sequence of cues to execute steps in the program. TGF-β and RAF/MEK/ERK are known to be involved in EMT, but how the two pathways interact during the process is not clear. Here, we show that in the absence of exogenous TGF-β, inhibiting RAF/MEK/ERK is sufficient to block exit from the epithelial state and prevent activation of the mesenchymal gene expression program (Fig. 4e). However, when cells are exposed to exogenous TGF-β, this pathway can “shortcut” MEK to activate the mesenchymal program directly. Further, we find that loss of MEK activity can lock cells in a partial EMT-like state where cells co-express E-cadherin and high levels of early and late mesenchymal markers. Taken together, these observations point to the existence of “checkpoints” in the EMT continuum at which cells can arrest and accumulate, creating the impression of discrete stages in bulk cell assays (Fig. 4f).
Our study combines single-cell trajectory analysis with high-throughput pooled loss-of-function screening, which constitutes a powerful approach for identifying upstream signals of pathways that regulate cellular phenotypes. We expect that this methodology will shed light on the genetic architecture that governs not just EMT but diverse biological processes in development and disease. More generally, the observation that interrupting a signaling pathway can enrich a particular transcriptional state within a cell population will inform ongoing debates surrounding the definitions of cell type and state and the delineation of human cellular ontology.
Materials and Methods
Cell culture
MCF10A breast epithelial cells were purchased from ATCC and used within 10 passages. Human mammary epithelial cells (HuMEC) were purchased from ThermoFisher Scientific and passage 4 cells used for all experiments. Cas9-expressing MCF10A (MCF10A-Cas9) were generated by transduction with lentiCas9-blast lentivirus (Addgene) and selected with 10 μg/ml blasticidin (ThermoFisher Scientific) 72 hours post-transduction. Cells were cultured at 37 ˚C and 5% CO2 in MCF10A media composed of DMEM/F12 (ThermoFisher Scientific) containing 10% fetal bovine serum (ThermoFisher Scientific), 1% penn-strep (ThermoFisher Scientific), 10 ng/ml EGF (LC Labs), 5 μg/ml insulin (ThermoFisher Scientific), 10 ng/ml cholera toxin (List Labs) and 1 μg/ml hydrocortisone (Sigma).
2D in vitro EMT assay
Prior to cell seeding the cloning area of a 4.7 mm diameter cloning ring (Sigma) was marked on the bottom of plates and 2.5 × 105 MCF10A, HuMEC or MCF10A-Cas9 cells were seeded within cloning rings placed in the center of the marked well of a 6-well tissue culture dish and cells allowed to adhere overnight. Cloning rings were then removed, and wells were washed twice with 3 ml of Dulbecco’s PBS (ThermoFisher Scientific) to remove non-adhered cells and MCF10A media added. For TGF-β-treated cells, 4 ng/ml TGF-β (Pepro Tech) was added to media and TGF-β was replenished every 48 hours. 7 days post removal of cloning rings, inner and outer cell fractions were collected by scraping away outer and inner cells, respectively, using a cell lifter (Costar) and remaining cells were dissociated using TrypLE (ThermoFisher Scientific). There area from which cells were scraped was determined by the outer diameter of the previously marked cloning ring and wells were inspected under a dissecting microscope to assess purity of the fraction.
Crystal violet and E-cadherin immunofluorescence staining
MCF10A colonies were rinsed with DPBS, fixed by incubating with 4% paraformaldehyde (EM grade, Electron Microscopy Sciences) for 20 minutes followed by incubation with pure ethanol for 10 minutes at room temperature. For crystal violet staining, fixed colonies were incubated in 0.05% w/v crystal violet (Sigma) in water for 20 minutes and excess crystal violet removed by washing 5× for 5 minutes with DPBS. For E-cadherin staining, fixed colonies were blocked by washing 3× for 5 minutes in IF buffer (0.1% triton x-100 (Sigma) and 2% BSA (Fisher Scientific) in PBS). Colonies were then incubated in IF buffer containing mouse anti-E-cadherin antibody (Cell Signaling) for 2 hours at room temperature and washed with IF buffer. For imaging, colonies were incubated for 1 hour in IF buffer containing Alexa-488 conjugated goat anti-mouse IgG in IF buffer (Invitrogen), washed with IF buffer and 5 μg/ml Hoechst 33342 (Invitrogen) added to colonies. Brightfield imaging of crystal violet stained whole colonies was performed on a Zeiss Axio Observer by stitching whole well 10× images according to manufacturer’s instructions (Carl Zeiss Microimaging). Immunofluorescence imaging of E-cadherin stained colonies was performed by taking representative fields from the center middle and edge of the colony.
Flow cytometry for EMT marker protein levels
MCF10A cells were plated at the center of wells in 6-well plates as previously described. 2 hours after plating, colonies were washed with PBS and medium was replaced. After 7 days, cells were harvested using TrypLE, washed twice with PBS, resuspended in 500 μl of PBS, fixed by addition of 5 ml of ice-cold ethanol added dropwise while vortexing and samples stored at −80 °C. Fixed samples were washed and blocked in PBS containing 1% bovine serum albumin (BSA) (Sigma). Samples were split in 2 and one aliquot incubated overnight with mouse anti-cytoplasmic fibronectin antibody (Abcam, ab6328) and rabbit anti-e-cadherin antibody (Cell Signaling, 3195) and the other incubated with rabbit anti-vimentin antibody (Cell Signaling, 5741) and mouse anti-n-cadherin antibody (Cell Signaling, 14215). For spontaneous EMT, cells treated with 500 nM erlotinib (Supplementary Fig. 28), fixed cells were incubated with a mix of rabbit anti-e-cadherin antibody (Cell Signaling, 3195), rat anti-CRB3 antibody (Abcam, ab180835) and mouse anti-desmoplakin I+II antibody (Abcam, ab16434) or a mix of rabbit anti-vimentin antibody (Cell Signaling, 5741) and mouse anti-pan-keratin antibody (Cell Signaling, 4545). Antibody incubations were performed in PBS containing 1% BSA and 0.1% triton X-100 (Sigma). Samples were washed 3 times with PBS containing 0.1% triton X-100, incubated for 1 hour with goat anti-rabbit Alexa 647 and goat anti-mouse Alexa 488 secondary antibodies in PBS containing 1% BSA and 0.1% triton X-100, washed 3 times with PBS containing 0.1% triton X-100 and resuspended in PBS for analysis on an LSRII flow cytometer (BD Biosciences) as depicted in Supplementary Figure 32.
Small-molecule inhibition of KRAS-MEK-ERK pathway activators and flow cytometry for EMT markers
The MEK inhibitor U0126, the PI3K inhibitor LY294002, the EGFR inhibitor erlotinib, the MET inhibitor crizotinib and the FGFR inhibitor infigratinib were purchased from LC Laboratories and resuspended in DMSO. The ITGAV inhibitor cilengitide was purchased from Selleck Chemicals as a 10 mM solution in DMSO. To determine the highest inhibitor concentration that does not have a negative effect on cell viability we seeded MCF10A cells at 2.5 × 104 cells per well in 96-well plates. After allowing cells to attach overnight wells exposed for 96 hours with increasing doses of each inhibitor or DMSO vehicle alone as shown in Supplementary Figure 23. The highest concentration of each inhibitor that were cells exhibit 90% or higher percent control growth was used to determine the effect of target inhibition on the induction of a spontaneous and TGF-β-driven EMT. MCF10A and HuMEC cells were plated at the center of wells in 6-well plates as previously described. 24 hours after plating, colonies were washed with PBS and cells were pretreated for 1 hour in media with or without 1 μM U0126, 1 μM LY294002, 100 nM erlotinib, 1 μM crizotinib, 1 μM infigratininb or 10 μM cilengitide. After pre-incubation, medium was replaced with medium with or without 4 ng/ml TGF-β1 as well as any inhibitor with which cells were pretreated. TGF-β1 was replenished every 48 hours. After 7 days, samples were harvested and processed for flow cytometry of EMT marker protein levels as described above.
Construction of single-cell RNA libraries and sequencing
Single-cell suspensions of inner and outer cells from Mock and TGF-β-treated MCF10A and HuMEC cells were washed and resuspended in PBS containing 0.04% ultrapure BSA (ThermoFisher Scientific) at 1 × 106 cells/ml. For pseudospatial experiments in the absence or presence of TGF-β presented in Figures 1 and 2, 2,000-3,000 cells were captured on the Chromium platform (10X Genomics) using one lane per fraction. Single-cell mRNA libraries were built using the single-cell 3’ solution V1 kit, libraries sequenced on an Illumina NextSeq 500/550 using 75 cycle high output kits (Read 1 = 64, Read 2 = 5, Index 1 = 14 and Index 2 = 8) and data preprocessed using the Cell Ranger 1.3.1 pipeline (10X Genomics). CROP-seq pseudospatial libraries were generated in a similar fashion capturing 7,000-9,000 cells per fraction. The aggregation option in Cell Ranger was used to normalize libraries to equivalent number of mean reads per cell specifically: 47,905 and 30,636 mean reads per cell for initial MCF10A and HuMEC pseudospatial experiments, respectively, and 43,557 mean reads per cell for CROP-seq experiments. The percent of reads mapping to the transcriptome for all samples was between 77.8% and 84.1%. We observed a median of 12,380 and 8,672 unique molecular identifiers (UMI) per cell for initial MCF10A and HuMEC pseudospatial experiments, respectively, and 13,951 median UMIs per cells for CROP-seq experiments. Additional metrics for each individual scRNA-seq library can be found in Supplementary Table 1 and Supplementary Figure 31.
t-SNE embedding
We performed PCA on a matrix composed of cells and gene expression values for genes expressed in more than 50 cells, reduced dimensions to the top 25 principal components and t-distributed stochastic neighbor embedding (t-SNE) was initialized in this PCA space to reduce to 2 t-SNE dimensions using the reduceDimension function in Monocle2 specifying num_dim = 25, max_component = 2, norm_method = log and reduction_method = tSNE. To visualize the gene expression level of EMT markers in t-SNE space the gene expression levels of CDH1, DSP and VIM in every cell was normalized by the library size of each cell (the Size_Factor in Monocle2), a pseudocount of 0.1 was added and values log10 normalized.
Pseudospatial reconstruction of single-cell transcriptomes
Trajectories were constructed according to the procedure recommended in the Monocle 2 documentation (http://cole-trapnell-lab.github.io/monocle-release/docs/#constructing-single-cell-trajectories). Briefly, genes used to order cells were selected by comparing the inner and outer cell fractions in the assay. For each cell type (MCF10A/HuMEC), differential gene expression analysis was performed between differentialGeneTest() function in Monocle 217,64. Each gene was fit with a generalized linear model (GLM) via the formula “y ~ cell_fraction”, specifying a simple two-group contrast between the fractions. The response (the size-factor adjusted UMIs for the gene) was modeled as a negative binomially distributed random variable. Testing for significant genes was conducted by comparing each gene’s model against a reduced model “y ~ 1” via likelihood ratio test.
The top differentially expressed genes (likelihood ratio test, FDR, q < 1 × 10−10 and absolute of the log2 fold-change > 1) were chosen as “ordering genes” to recover pseudospatial trajectories using the setOrderingFilter(), reduceDimension() and orderCells() functions in Monocle 2 using default parameters with the exception of setting ncenter = 500 during dimensionality reduction. Expression of key EMT markers across pseudospace was visualized using the plot_genes_in_pseudotime function in Monocle2 specifying a minimum value of 0.1 (min_expr = 0.1).
Detection and visualization of spatially dependent genes
To extract and visualize genes that vary over a trajectory (beyond the variability one would expect across unordered cells), we used the procedure recommended by the Monocle 2 documentation (http://cole-trapnell-lab.github.io/monocle-release/docs/#finding-genes-that-change-as-a-function-of-pseudotime). To identify changes in gene expression across pseudospatial trajectories we fit splines with 3 degrees of freedom to capture the dynamics of gene expression over psedusopace and tested for differential gene expression analysis using a full model of “y ~ sm.ns(pseudospace, df=3)”, which encode a cell’s position on the trajectory as a continuous covariate. To further filter genes by those with the largest effect size we divided pseudospace into 5 quantiles, calculated the area under the curve (AUC) for each gene at each quantile and filtered differentially expressed genes to those having an AUC > 10 in at least 1 quantile and an FDR of < 1 × 10−10. Differentially expressed genes were variance stabilized and scaled, clustered and visualized using the pheatmap function from the R package pheatmap specifying ward.D2 as the clustering method. To identify biological processes and pathways enriched in clusters of differentially expressed genes across pseudospace we performed hypergeometric testing using the piano R package specifying genes expressed in more than 50 cells as the background set.
Calculation of aggregate gene expression scores
To determine the extent to which cells in different samples activate certain gene expression modules we calculated for each cell a normalized aggregate expression score for defined gene-sets. For a matrix of genes and cells we log10 normalized gene expression for the defined genes in each set after library size normalization and addition of a pseudocount of 1. For each cell we then calculated the mean normalized expression level of genes in the gene-set and mean centered and variance scaled mean normalized expression values across all cells. The compare_means function from the ggpubr packaged was used to determine the significance in changes in scores between endpoints in MCF10A and HuMEC EMT trajectories (Supplementary Fig. 13) specifying wilcox.test as the method and using the Holm procedure (holm) to correct for multiple hypothesis testing.
Dynamic time warping of pseudospatial trajectories
Alignment of Mock and TGF-β-treated trajectories for MCF10A, MCF10A-Cas9 and HuMEC pseudospatial trajectories was performed as in 35 setting the Mock and TGF-β-treated cell trajectories as the reference and query, respectively. Briefly, to arrive at a common pseudospatial axis, trajectories were aligned based on the intersect of genes used for ordering Mock and TGF-β-driven trajectories where pseudospace values were scaled from 0-100, smoothed splines were fit to each gene using the genSmoothCurves function in Monocle2, splines were variance stabilized and scaled prior to alignment using the dtw function from the DTW R package using the following options: step pattern = rabiner Juang step pattern (type = 3 and slope.weighting = c), open.begin and open.end = FALSE. To identify genes that describe the differences in the interaction between pseudospace and TGF-β treatment across Mock and TGF-β-driven trajectories we performed differential gene expression analysis using a full model of y~pseudospace*treatment and a reduced model of y~pseudospace. We isolated differentially expressed genes with the largest differences between treatments by dividing pseudospace into 5 equally spaced quantiles, calculating the area under the curve (AUC; calculated using spline interpolation) for each treatment within each quantile and identifying genes with a relative difference in auc (relative AUC difference = abs(AUC1-AUC2)/sum(AUC1+AUC2)) larger than 0.02 in at least 1 quantile and an FDR < 1 × 10−10.
Pre-processing of human neck and squamous cell carcinoma (HNSCC) dataset
Processed data from the single-cell RNA sequencing of HNSCC tumors described in Puram et al.15 were downloaded from the GEO Omnibus database (GSE103322) and a Monocle2 Cell Data Set (cds) object was created using gene expression and metadata available in GSE103322_HNSCC_all_data.txt.gz specifying a lower detection limit of 0.1 and choosing tobit as the expression family. Expression values were then converted to mRNA per cell using the Census64 algorithm implemented in the relative2abs function in Monocle2 after which a new cds object was created specifying negbinomial.size as the expression family. For all analyses normal cell types and cancer cells from lymph node metastases were excluded. Additionally, only cells that were not processed using the Maxima reverse transcriptase enzyme were chosen for analysis as use of Maxima enzyme was found by Puram et al. to introduce a batch effect. HNSCC tumor samples that had at least 40 cells after applying the exclusion criteria described above where chosen for further analysis.
K-nearest neighbor projection of HNSCC tumor cells onto spontaneous and TGF-β-driven MCF10A EMT trajectories
We used PCA to reduce the dimensions of a matrix composed of HNSCC tumor cells and MCF10A cells from either our spontaneous or TGF-β-driven EMT condition and gene expression values for genes expressed in more than 50 cells to the top 20 principal components. For each HNSCC tumor sample we determined the top 20 nearest neighbors from either our spontaneous or TGF-β-driven EMT condition using the k-nearest neighbor search implemented in the FNN R package using the get.knnx function specifying kd_tree as the search algorithm. Finally, the mean pseudospace values for the top 20 MCF10A nearest neighbors was used to assign a pseudospace value for each HNSCC tumor cell.
Reconstruction of HNSCC tumor cell trajectories and alignment to MCF10A EMT trajectories
We chose the four tumor samples with the highest number of cells after applying the exclusion criteria described above (HNSCC17, HNSCC18, HNSCC20 and HNSCC22). Reconstruction of HNSCC trajectories was performed as described above for MCF10A and HuMEC cells with the exception that genes expressed in at least 50 cells across all tumors were used as feature genes for the setOrderingFilter function in Monocle2. Dynamic time warping of HNSCC and either spontaneous or TGF-β-driven EMT trajectories was performed as previously described with the exception that alignment genes from either spontaneous or TGF-β-driven EMT trajectories were set as alignment genes and the open.begin and open.end parameters of the dtw function in the DTW R package were set to TRUE to allow alignment of HNSCC tumor samples anywhere along the MCF10A trajectories. The dtwPlot function from the DTW R package was used to visualize the alignment of HNSCC trajectories to either MCF10A spontaneous or TGF-β-driven EMT trajectories.
Cloning, lentiviral packaging and transduction of CROP-seq libraries
CROP-Seq lentiviral vector (Addgene) was prepped for sgRNA library insertion as described in 52. Briefly, vector was digested using BsmBI (New England Biolabs) and Fast Alkaline Phosphatase (ThermoFisher Scientific). Oligonucleotides (IDT), each containing an sgRNA and homology for Gibson ligation, were designed as follows:
[U6 homology]-[sgRNA]-[sgRNA backbone homology]
5’-tatcttGTGGAAAGGACGAAACACC[G]-[20bp sgRNA]-gttttagagctaGAAAtagcaagttaaaataagg-3’
where addition of the G immediately upstream of the sgRNA ensures transcription from pol III promoters.
Oligonucleotides (overall design and individual sgRNA sequences can be found in Supplementary Table 10) were made double stranded by PCR with primers against the invariant regions. 10 fmols of digested vector and 200 fmols of double stranded oligonucleotides to the digested CROP-Seq vector were ligated using the In-Fusion HD kit (Clontech) by incubation at 50 ˚C for 1 hour. Libraries were then transformed into stellar competent cells (Clontech), transformations were diluted in 250 μl of LB, spread onto 6 LB agar plates containing ampicillin and bacteria culture at 30 ˚C for 24 hours. Resulting colonies were scraped with LB, pooled and vector recovered using a DNA midi kit (Qiagen). Lentivirus was generated by transfecting HEK293T in MCF10A media lacking penn-strep cells with our CROP-Seq library using the ViraPower lentiviral packaging mix (ThermoFisher Scientific) according to manufacturer’s instructions. Collected lentiviral supernatant was filtered using a 45 μm steriflip vacuum filter (Fisher Scientific). MCF10A-Cas9 cells were transduced with increasing amounts of our CROP-Seq lentiviral library and selected with puromycin, retaining transduced cells that had an approximate MOI of 0.3.
Enrichment of sgRNA containing transcripts and genotype assignment
For CROP-Seq experiments, a nested PCR was performed on 5-10 ng of unsheared cDNA to enrich for sgRNAs positioned on the 3’ UTR of the puromycin resistance gene transcripts. All oligonucleotide sequences used for enrichment of sgRNA containing transcripts can be found in Supplementary Table 10. Briefly, PCR reactions were performed using a P7 reverse primer equivalent to the one introduced by the oligo containing beads in the 10X Chromium Single cell 3’ solution V1 (5’- CAAGCAGAAGACGGCATACGA -3’). For the first PCR the forward primer directed towards the beginning of the U6 promoter was:
5- TTTCCCATGATTCCTTCATATTTGC -3
For the second PCR the forward primer binds at the beginning of the sgRNA and adds the standard Nextera R1 sequence:
5- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGcTTGTGGAAAGGACGAAACAC -3
In the final PCR amplicons were indexed with standard Nextera P5 index primers:
5- AATGATACGGCGACCACCGAGATCTACAC[10bp Index]TCGTCGGCAGCGTC -3
A 1× Ampure cleanup was performed after each PCR with 1/5th of PCR1 added to PCR2 and 1/25th of PCR2 added to PCR3. Libraries were sequenced as spike-ins with transcriptome scRNA-Seq libraries. Fnal cellular barcodes and UMIs were extracted from position sorted BAM files output by Cell Ranger 1.3.1. We then attempt to find a perfect match for sequences preceding the sgRNA (GTGGAAAGGACGAAACACCG) or use a striped Smith-Watterman alignment to locate the sequence within an error tolerance of 2 bp shorter than the expected sequence. For each match or alignment, the sgRNA sequence is extracted and compared to a whitelist of all sgRNA within an edit distance of half the minimum distance between any pair of guides in our sgRNA library tracking matches for each cell. Chimeric sequences were removed by the approach detailed in 48. sgRNA sequences with over 3 reads accounting for more than 7.5% of sgRNA reads assigned to a given cell were assigned to each cell. These assignments were combined with the filtered gene expression matrix created by Cell Ranger to assign high quality cells.
t-SNE embedding and distribution of knockout cells across t-SNE space
We performed PCA on a matrix composed of cells each of which contain only one guide from our CROP-Seq screen and gene expression values for genes expressed in more than 50 cells and reduced dimensions to 25 principal components. t-distributed stochastic neighbor embedding (t-SNE) was initialized in this PCA space to reduce to 2 t-SNE dimensions. We then performed density peak clustering in this t-SNE space. A Chi-square test was performed to determine whether the distribution for a sgRNA and targets in the t-SNE was significantly different compared to NTC at an FDR cutoff of 5%. Knockouts whose distribution was significantly different from NTC were subjected to further analysis. For each sgRNA we derived a weight to estimate the functional editing rate using an expectation minimization approach by first modeling the t-SNE distribution as a mixture of cells with functional and non-functional edits where the mixing parameter is the relative functional edit rate for the sgRNA; estimating the weighted average of the empirical t-SNE distribution for each guide; and estimating relative functional edit rate as the one that maximizes the observed t-SNE distribution. Weighted contingency tables were then generated containing the t-SNE clusters and weighted cell counts across clusters. Fisher’s exact test was then used to identify knockouts enriched across t-SNE clusters at an FDR of 5%. Chi square and Fisher’s exact test were performed using R functions chisq.test and fisher.test, respectively.
Reproducibility of spatially dependent differential gene expression within our pooled CROP-seq screen
Differential gene expression analysis between isolated cell fractions was performed for cells expressing non-targeting control guide RNAs from our pooled CROP-seq screen under spontaneous and TGF-β-driven EMT conditions using a full model of “y ~ cell_fraction”. The overlap of differentially expressed genes was then compared to initial spatial experiments in MCF10A. The correlation (Pearson’s r) of beta coefficients between cells from our initial spatial experiments and cells expressing non-targeting control guide RNAs across the full list of original spatial DEGs was calculated using the cor.test function in R.
Trajectory reconstruction of CROP-Seq loss of function screen and calculating knockout enrichment across pseudospace
Spontaneous and TGF-β-driven EMT trajectories of our CROP-Seq loss-of-function screen were individually constructed and aligned as described above. To identify changes in gene expression along pseudospace upon gene editing we subsetted cells expressing sgRNAs against a target or non-targeting controls, estimated gene-level dispersions and performed a differential gene expression analysis using a full model of y ~ pseudospace*genotype and a reduced model of y ~ pseudospace. After repeating for all targets, P values were corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. Differential gene expression analysis of the effect of each individual knockout on gene expression over pseudospace is dependent on the rate of non-functional edits, the penetrance of the resulting phenotype and the number of cells expressing sgRNAs against a particular target in our screen. To overcome these challenges and identify changes in the distribution of edited cells along EMT, cells were then clustered according to where cells accumulated along pseudospace into pseudospatial regions along spontaneous and TGF-β pseudospace coordinates using the density peak algorithm17 implemented in Monocle2 using the clusterCells function. As not all guide RNAs are equally efficient and not all edited cells contain an edit that leads to loss of function we used an expectation-maximization approach to estimate the functional edit rate of every guide RNA relative the most efficient guide RNA for a target as described in 53. Briefly, we modeled the distribution of guide RNA containing cells across pseudospatial regions as a mixture of cells with a functional edit and the distribution of cells expressing non-targeting control guide RNAs with the mixing parameter being the functional edit rate for a particular guide RNA. For the expectation step of our model we estimate the distribution of cells with a functional edit as the weighted average of the relative functional edit rate. Lastly, for the maximization step, we choose the relative function edit rate for a guide RNA as that which maximizes the likelihood of the observed distribution of cells expressing a guide RNA across pseudospatial regions under the mixture model. We then used these guide RNA weights to arrive at weighted cell counts of guide RNA containing cells across pseudospatial regions.
We assessed whether the distribution of guide RNA containing cells as well as a random subset of non-targeting control cells was significantly different compared to the larger pool of non-targeting control expressing cells across pseudospatial regions using a Chi-square test. To determine an empirical FDR, we repeated this procedure for 1,000 iterations and calculated the rate at which cells expressing guide RNAs against a target were identified as more significantly distributed across regions compared to the random subset of non-targeting control cells. To obtain a score for the enrichment of differentially distributed targets (FDR < 0.1) across pseudospatial regions we calculated the odds ratio for each target-region pair using fisher.test in R using the presence or absence of non-targeting control cells in the region and background as failures in our contingency tables. Targets accumulated in at least one region at an enrichment score (log2 of the odds ratio) of 1 or higher were regarded as strongly enriched. Finally, hierarchical clustering of the enrichment score across pseudospatial regions for differentially distributed targets was used to visualize accumulation of cells across pseudospace using the pheatmap function in the pheatmap package specifying ward.D2 as the clustering method.
Mean expression of enriched targets in HNSCC tumor cells
Expressed cell surface receptors and transcription factors were identified as those expressed across a minimum of 50 cells across tested HNSCC tumors. To determine the mean expression levels of enriched targets from our CRISPR-Cas9 screen in HNSCC tumors we log10 normalized gene expression for the defined genes after library size normalization and addition of a pseudocount of 1. Then the mean expression level across all cells for a given tumor were averaged. Results were visualized as a heatmap of enriched target gene expression levels using the pheatmap function in the pheatmap R package. Expression profiles were clustered specifying ward.D2 as the clustering method. A column annotation was added depicting the relative partial EMT rank for every tumor as observed by Puram et al.
Statistical methods
Differential gene expression analyses were performed using the differential gene test implemented in Monocle2 and test results corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. Wilcoxon rank sum test was used to determine statistical significance of the differences in aggregate gene expression scores for cells across various treatments with correction for multiple hypothesis testing performed using the Holm procedure. Two-tailed Student’s t tests were used to determine statistical significance of changes in EMT marker protein expression measured via flow cytometry.
Data Availability
Data are available on GEO under accession number GSE114687. Data will also be provided via the Github repository described in “Code availability”.
Code Availability
Code can be found on Github at https://github.com/cole-trapnell-lab/pseudospace.
Reporting Summary
Additional information on experimental design can be found in the Life Sciences Reporting Summary associated with this article.
Supplementary Material
Acknowledgements
We thank all members of the Trapnell and Shendure laboratories for helpful discussions during the course of this study and feedback on our manuscript, particularly S. Srivatsan, L. Saunders, H. Pliner and J. Packer. We would like to thank N.M. Cruz for feedback on our manuscript. J.L.M.F. would like to thank S.V. McFaline-Cruz for support. J.L.M.F. was supported by T32HL007828 and T32HG000035. A.J.H. was supported by an NSF Graduate Research Fellowship. J.S. and C.T. are supported by NIH grants U54DK107979 and the Paul G. Allen Frontiers Group. C.T. is supported by NIH grant DP2HD088158, RC2DK114777, R01HL118342 and is partly supported by an Alfred P. Sloan Foundation Research Fellowship. J.S. is supported by NIH grants DP1HG007811, R01HG006283 and is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lamouille S, Xu J & Derynck R Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nieto MA Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Sauka-Spengler T & Bronner-Fraser M A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Li M et al. Epithelial-mesenchymal transition: An emerging target in tissue fibrosis. Exp. Biol. Med. 241, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieto MA, Angela Nieto M, Huang RY-J, Jackson RA & Thiery JP EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J et al. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 7, ra91 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Lu M, Jolly MK, Levine H, Onuchic JN & Ben-Jacob E MicroRNA-based regulation of epithelial–hybrid–mesenchymal fate determination. Proceedings of the National Academy of Sciences 110, 18144–18149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong T et al. An Ovol2-Zeb1 Mutual Inhibitory Circuit Governs Bidirectional and Multi-step Transition between Epithelial and Mesenchymal States. PLoS Comput. Biol. 11, e1004569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnaswamy S, Zivanovic N, Sharma R, Pe’er D & Bodenmiller B Learning Edge Rewiring in EMT from Single Cell Data. bioRxiv 155028 (2017). doi: 10.1101/155028 [DOI] [Google Scholar]
- 11.van Dijk D et al. MAGIC: A diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. (2017). doi: 10.1101/111591 [DOI] [Google Scholar]
- 12.Grande MT et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 21, 989–997 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lovisa S et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 21, 998–1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aiello NM et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell 45, 681–695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puram SV et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu X et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scialdone A et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 289–293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarrió D et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68, 989–997 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez LG, Wu X & Guan J-L Wound-Healing Assay. in Cell Migration 023–030 [Google Scholar]
- 21.Vuoriluoto K et al. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 30, 1436–1448 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Joost S et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 3, 221–237.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliekelman MJ et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 75, 1789–1800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George JT, Jolly MK, Xu J, Somarelli J & Levine H Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. (2017). doi: 10.1158/0008-5472.CAN-16-3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Gene Ontology Consortium & The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbie DA et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchernitsa OI et al. Transcriptional basis of KRAS oncogene-mediated cellular transformation in ovarian epithelial cells. Oncogene 23, 4536–4555 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Toivola DM, Tao G-Z, Habtezion A, Liao J & Omary MB Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 15, 608–617 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Huang RY-J, Guilford P & Thiery JP Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J. Cell Sci. 125, 4417–4422 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Feng Y-X et al. Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4, 702–715 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Miettinen PJ, Ebner R, Lopez AR & Derynck R TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127, 2021–2036 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CaulIn C, Scholl FG, Frontelo P, Gamallo C & Quintanilla M Chronic Exposure of Cultured Transformed Mouse Epidermal Cells to Transforming Growth Factor- \ 1 Induces an Epithelial-Mesenchymal Transdifferentiation and a Spindle Tumoral Phenotype. Cell Growth and Differentiation-Publication American Association for Cancer Research 6, 1027–1036 (1995). [PubMed] [Google Scholar]
- 35.Cacchiarelli D et al. Aligning Single-Cell Developmental and Reprogramming Trajectories Identifies Molecular Determinants of Myogenic Reprogramming Outcome. Cell Syst 7, 258–268.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Alpert A, Moore LS, Dubovik T & Shen-Orr SS Alignment of single-cell trajectories to compare cellular expression dynamics. Nat. Methods (2018). doi: 10.1038/nmeth.4628 [DOI] [PubMed] [Google Scholar]
- 37.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vintsyuk TK Speech discrimination by dynamic programming. Cybern. Syst. Anal. 4, 52–57 (1968). [Google Scholar]
- 39.Rabiner L & Juang BH Fundamentals of speech recognition. (PTR Prentice Hall, 1993). [Google Scholar]
- 40.Masszi A et al. Integrity of Cell-Cell Contacts Is a Critical Regulator of TGF-β1-Induced Epithelial-to-Myofibroblast Transition. Am. J. Pathol. 165, 1955–1967 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaul YD et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell 158, 1094–1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo P-L, Shen K-H, Hung S-H & Hsu Y-L CXCL1/GROα increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-κB/HDAC1 epigenetic regulation. Carcinogenesis 33, 2477–2487 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Al-Alwan LA et al. Differential roles of CXCL2 and CXCL3 and their receptors in regulating normal and asthmatic airway smooth muscle cell migration. J. Immunol. 191, 2731–2741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian X-J, Zhang H & Xing J Coupled Reversible and Irreversible Bistable Switches Underlying TGFβ-induced Epithelial to Mesenchymal Transition. Biophys. J. 105, 1079–1089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaitin DA et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Xie S, Duan J, Li B, Zhou P & Hon GC Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill AJ et al. On the design of CRISPR-based single-cell molecular screens. Nat. Methods (2018). doi: 10.1038/nmeth.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark EA & Hynes RO Ras Activation Is Necessary for Integrin-mediated Activation of Extracellular Signal-regulated Kinase 2 and Cytosolic Phospholipase A2but Not for Cytoskeletal Organization. J. Biol. Chem. 271, 14814–14818 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Citri A & Yarden Y EGF–ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Peschard P & Park M From Tpr-Met to Met, tumorigenesis and tubes. Oncogene 26, 1276–1285 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Ornitz DM & Itoh N The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, Lamouille S & Derynck R TGF-β-induced epithelial to mesenchymal transition. Cell Res. 19, 156–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmad I, Iwata T & Leung HY Mechanisms of FGFR-mediated carcinogenesis. Biochim. Biophys. Acta 1823, 850–860 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Reed NI et al. The v 1 integrin plays a critical in vivo role in tissue fibrosis. Sci. Transl. Med. 7, 288ra79–288ra79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalluri R & Neilson EG Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112, 1776–1784 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simanshu DK, Nissley DV & McCormick F RAS Proteins and Their Regulators in Human Disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karnoub AE & Weinberg RA Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu X et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on GEO under accession number GSE114687. Data will also be provided via the Github repository described in “Code availability”.