Abstract
The present study is to analyze the difference of gene methylation in early cervical adenocarcinoma and to find molecular markers for predicting the occurrence and development of cervical adenocarcinoma.
A total of 15 cases of primary cervical adenocarcinoma and 10 cases of primary cervical squamous cell carcinoma at stages IB1 or IIA1 were included in the study. Infinium MethylationEPIC BeadChip (850K) was used to screen specifically expressed genes in cervical adenocarcinoma tissues. Bisulfite sequencing polymerase chain reaction (BSP) and quantitative real-time polymerase chain reaction (qRT-PCR) were used to verify the methylation levels in cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues.
Sex determining region Y-box 1 (SOX1) and cyclin D1 (CCND1) genes participated in multiple signaling pathways, being the central nodes of gene regulatory networks. SOX1 gene, but not CCND1 gene, was a specifically methylated gene in cervical adenocarcinoma according to BSP. According to qRT-PCR, methylation level of SOX1 in cervical adenocarcinoma tissues is significantly different from that in cervical squamous cell carcinoma tissues or normal cervical tissues, and the methylation level of CCND1 in cervical adenocarcinoma tissues or cervical squamous cell carcinoma tissues is significantly different from that in normal cervical tissues.
The present study demonstrates that tumor-suppressor gene SOX1 is a methylation-specific expression gene of cervical adenocarcinoma and is expected to become a specific molecular marker for the diagnosis of cervical adenocarcinoma. However, CCND1 gene was not proven to be a specific methylation expression gene in cervical adenocarcinoma in the present study.
Keywords: CCND1, cervical adenocarcinoma, methylation, SOX1
1. Introduction
With the deepening of researches on the molecular biological mechanism of tumorigenesis, the role of epigenetics in tumorigenesis has attracted more and more concerns. DNA methylation is an important form of epigenetic modification. Genome hypomethylation and local hypermethylation are common in cancer tissues. Hypermethylation of the promoter of tumor-suppressor gene has its specific alteration pattern at all stages of the occurrence and development of many tumors, including cervical cancer. In the course of progression, hypermethylation of promoters causes inactivation of different tumor-suppressor genes. The cumulative effect of epigenetic changes plays an important role in the development of cervical cancer from normal tissues and precancerous lesions. In recent years, it has been found that epigenetic and regulatory genes are involved in the process of cervical carcinogenesis.[1] The epigenetic changes represented by DNA methylation can lead to cervical cancer.[1] The main pathogenic process is that the promoter region of tumor-suppressor gene is abnormal, leading to inactivation of target gene and regulatory gene transcription, and occurrence and progression of tumors.[2] Some molecules can be used as potential molecular markers for early detection of cervical cancer.[3]
In the present study, we used Infinium MethylationEPIC BeadChip (850K) technology from Illumina to screen abnormal methylation gene in cervical adenocarcinoma, and bisulfite sequencing PCR (BSP) and quantitative real-time polymerase chain reaction (qRT-PCR) to verify these results.
2. Materials and methods
2.1. Patients
A total of 15 cases of primary cervical adenocarcinoma and 10 cases of primary cervical squamous cell carcinoma at stages IB1 or IIA1 who underwent surgical resection at Tumor Hospital Affiliated to Xinjiang Medical University between January, 2015 and June, 2017 were included in the present study. Tumor tissues were collected from all these patients. In addition, cervical tissues from 15 contemporaneous patients with noncervical lesions who underwent total hysterectomy for other benign gynecological diseases were included as control. Tumor tissues and normal cervical tissues with a diameter of about 0.5 cm were resected in surgery and washed with aseptic saline before being dried by sterile filter paper. The tissues were frozen in liquid nitrogen and stored at −80°C. All procedures performed in the current study were approved by the Ethics Committee of Xinjiang Medical University. Written informed consent was obtained from all patients or their families.
2.2. Chip examination of methylation
Five cases of cervical adenocarcinoma tissues and 5 cases of normal cervical tissues underwent whole genome methylation examination using Infinium MethylationEPIC BeadChip (850K) (Illumina, San Diego, CA) according to the manufacturer's manual.
2.3. Bisulfite sequencing polymerase chain reaction
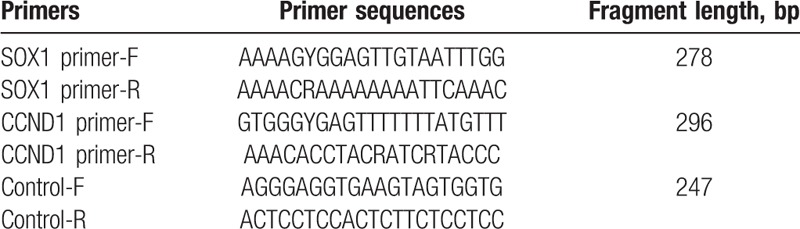
Tissue genomic DNA extraction kit (GK0122; Qiagen, Hilden, Germany) was used for the extraction of DNA from tissues samples according to the manufacturer's manual. EpiTect Fast DNA Bisulfite Kit (Qiagen, Hilden, Germany) was used to treat the extracted genomic DNA according to the manufacture's manual. Primer sequences are listed in Table 1.
Table 1.
Primer information in bisulfite sequencing polymerase chain reaction.
2.4. Quantitative real-time polymerase chain reaction
According to chip examination of methylation, sex determining region Y-box 1 (SOX1) that had significant differential expression was subjected with qRT-PCR according to the manufacture's manual (PrimeScript RT reagent Kit; RR037A; TAKARA, Dalian, China), using GAPDH as internal reference. Primer sequences are listed in Table 2. qRT-PCR was performed using TB Green Premix Ex Taq (RR420A; Takara, Dalian, China). The concentration of DNA or RNA was determined by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA). The concentration of dsDNA was calculated by OD260 × dilution times × 50/1000. For dsDNA purity evaluation, OD260/OD280 ratio was controlled within 1.6 to 1.9. The concentration of RNA was calculated by OD260 × dilution times × 40/1000. For RNA purity evaluation, OD260/OD280 ratio was controlled within 1.7 to 2.0.
Table 2.
Primer information in quantitative real-time polymerase chain reaction.

2.5. Statistical analysis
The results were analyzed using SPSS 22.0 statistical software (IBM, Armonk, NY). Counting data, grade data, and correlation were analyzed using chi-square test. Measurement data were expressed as means ± standard deviations. Normality test and homogeneity test of variance were carried out, and non-parametric analysis was used.
3. Results
3.1. Detection of methylation level in cervical adenocarcinoma tissues by whole genome methylation beadchip
To preprocess and standardize original data, body mass index method was used. The data showed no abnormal samples (Fig. 1A). Using methylation variable position (MVP) analysis, differential methylation sites between cervical adenocarcinoma and normal cervical tissues were shown (Fig. 1B). After examination of 853 307 methylation sites in cervical adenocarcinoma and normal cervical tissues, 138 653 differential methylation sites (117 843 hypermethylation sites and 20 810 hypomethylation sites) and 96 141 differential methylation genes (82 437 hypermethylation genes and 13 704 hypomethylation genes) were identified. Regulatory analysis of key gene interaction networks for 3000 genes with the most significant difference in differentially methylated region showed that SOX1 and cyclin D1 (CCND1) genes participated in multiple signaling pathways, being the central nodes of gene regulatory networks.
Figure 1.
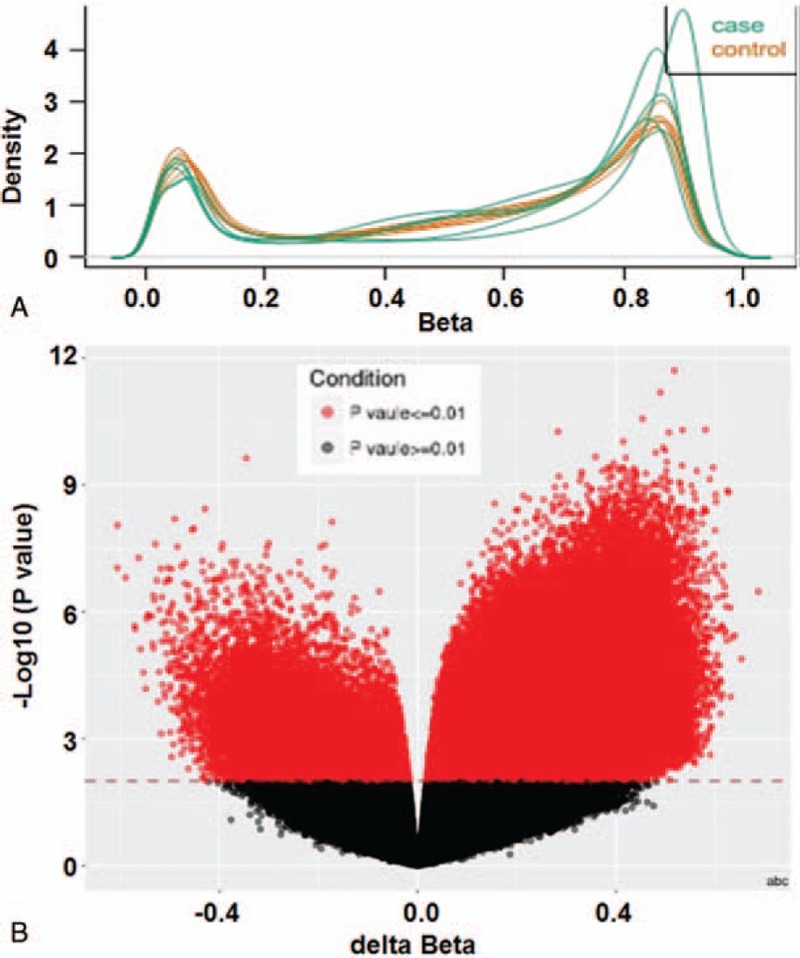
Detection of methylation level in cervical adenocarcinoma tissues. (A) Density plot of raw data (753037 probes). (B) Methylation variable position (MVP) analysis between cervical adenocarcinoma tissues and normal cervical tissues.
3.2. SOX1 gene, but not CCND1 gene, is a specifically methylated gene in cervical adenocarcinoma according to BSP
To examine the sample processing quality, absorbance at 260 and 280 nm was determined. The ratio of absorbance at 260 nm over that at 280 nm was higher than 1.7, suggesting a high purity of total DNA. To examine the integrity of total DNA (3 μL), 0.7% agarose gel electrophoresis was performed. The data showed clear DNA major band, and no dispersed band was observed (Fig. 2A), suggesting that the sample quality was suitable for downstream BSP test. According to randomly selected clone sequencing results, the percentage of methylated cytosine that was converted to thymine was low (0%), and the percentage of unmethylated cytosine that was converted to thymine was high (99.69%), suggesting that bisulfite processing results were normal. To test the methylation level of cytosine phosphate guanosine (CpG) island of SOX1 gene and CCND1 gene, BSP was performed. The data showed that the methylation level of promoter region of SOX1 gene was high (Fig. 2B), and the methylation percentages for cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues were 97.50%, 87.08%, and 41.26%, respectively. Statistical analysis showed that the methylation percentage of promoter region of SOX1 gene in cervical adenocarcinoma tissues was significantly elevated (F = 18.6400, P = .0002) (Fig. 2C). In addition, the methylation level of promoter region of CCND1 gene was not high (Fig. 2D). According to BSP, the methylation percentages of promoter region of CCND1 gene in cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues were 2.96, 1.12, and 1.52%, respectively. Statistical analysis showed that the methylation percentage of promoter region of CCND1 gene in cervical adenocarcinoma tissues was not significantly elevated (F = 0.8900, P = .4360) (Fig. 2E). The results suggest that SOX1 gene, but not CCND1 gene, is a specifically methylated gene in cervical adenocarcinoma according to BSP.
Figure 2.
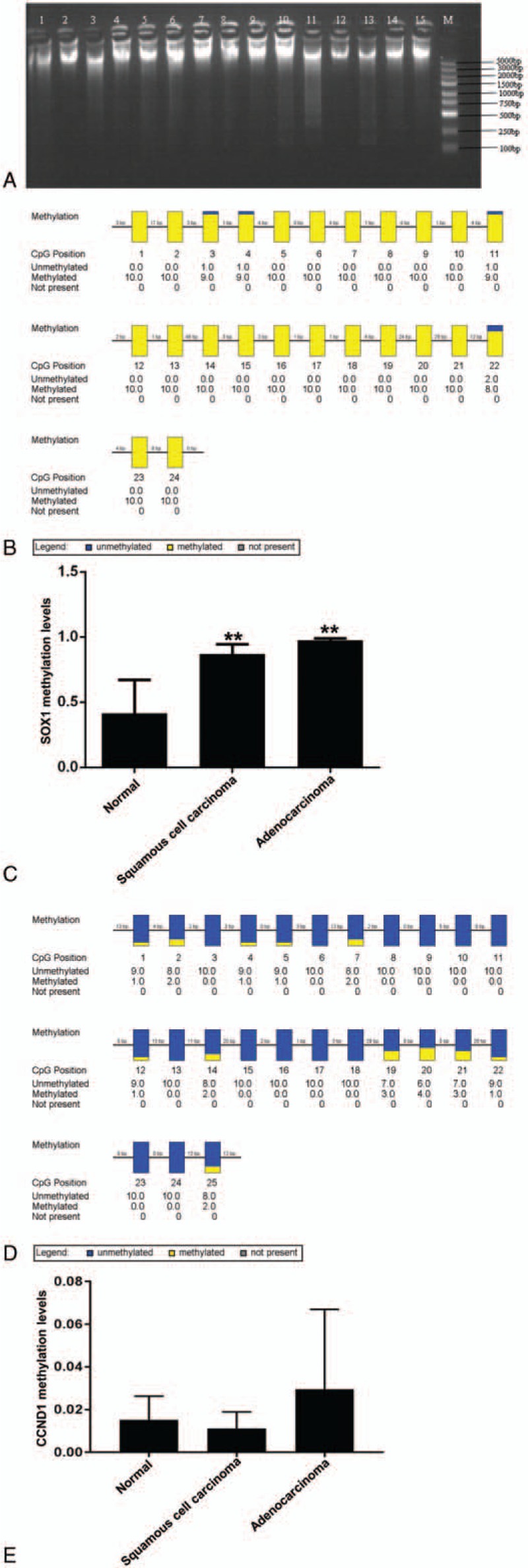
Methylation levels of promoter regions of SOX1 and CCND1 genes in cervical adenocarcinoma tissues and normal cervical tissues. (A) Total DNA agarose gel electrophoresis. M, marker (100–5000 bp); lanes 1 to 5, normal cervical tissues; lanes 6 to 10, cervical adenocarcinoma tissues; lanes 11 to 15, cervical squamous cell carcinoma tissues. (B) Aggregate plot of methylation status of SOX1 promoter region in cervical adenocarcinoma tissues. (C) Methylation levels of SOX1 promoter region in cervical adenocarcinoma, cervical squamous cell carcinoma and normal cervical tissues. ∗∗P < .05 compared with normal group. (D) Aggregate plot of methylation status of CCND1 promoter region in cervical adenocarcinoma tissues. (E) Methylation levels of CCND1 promoter region in cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues.
3.3. Expression of SOX1 and CCND1 gene methylation in cervical adenocarcinoma, squamous cell carcinoma and normal cervical tissues
To examine total RNA purity, absorbance at 260 and 280 nm was determined. The ratio of absorbance at 260 nm over that at 280 nm was higher about 1.8, suggesting a high purity of total RNA. To examine the integrity of total RNA (3 μL), 0.7% agarose gel electrophoresis was performed. The data showed that the intensity of band 28S was twice of that of band 18S, and the intensity of band 5S was the lightest (Fig. 3A), suggesting that total RNA was not degraded. qRT-PCR showed that target genes SOX1, Protein kinase C alpha type (PKC-A) (PKC-alpha), and CCND1 had typical PCR amplification curves, suggesting enough amount of product. In addition, the dissociation curves of target genes and GAPDH had single peak, suggesting that the product was pure (Fig. 3B–D). To examine the expression of methylated SOX1 and CCND1 genes, qRT-PCR was used. The ΔCt values of SOX1 gene in cervical adenocarcinoma tissues, cervical squamous cell carcinoma tissues, and normal cervical tissues were 15.0080 ± 1.8648 (F = 0.7200 ± 0.6630), 12.5183 ± 0.3462 (F = 2.3200 ± 0.4990), and 12.5717 ± 1.9868 (F = 4.2000 ± 4.1070), respectively (Fig. 3E). In addition, the ΔCt values of CCND1 gene in cervical adenocarcinoma tissues, cervical squamous cell carcinoma tissues, and normal cervical tissues were 6.1200 ± 1.6130 (F = 0.6200 ± 0.5290), 5.2200 ± 0.2020 (F = 0.7500 ± 0.1080), and 2.3600 ± 1.6960 (F = 8.7900 ± 7.6660), respectively (Fig. 3F). The expression of methylated SOX1 in cervical adenocarcinoma tissues was significantly different from that in cervical squamous cell carcinoma tissues (P < .05), but the expression of methylated CCND1 in cervical adenocarcinoma tissues was not significantly different from that in cervical squamous cell carcinoma tissues (P > .05) (Table 3). The expression of methylated SOX1 or CCND1 in cervical adenocarcinoma tissues was significantly different from that in normal cervical tissues (P < .05) (Table 4). Moreover, the expression of methylated CCND1 in cervical squamous cell carcinoma tissues was significantly different from that in normal cervical tissues (P < .05), but the expression of methylated SOX1 in cervical squamous cell carcinoma tissues was not significantly different from that in normal cervical tissues (P > .05) (Table 5). These results indicate that the methylation level of SOX1 in cervical adenocarcinoma tissues is significantly different from that in cervical squamous cell carcinoma tissues or normal cervical tissues, and the methylation level of CCND1 in cervical adenocarcinoma tissues or cervical squamous cell carcinoma tissues is significantly different from that in normal cervical tissues.
Figure 3.
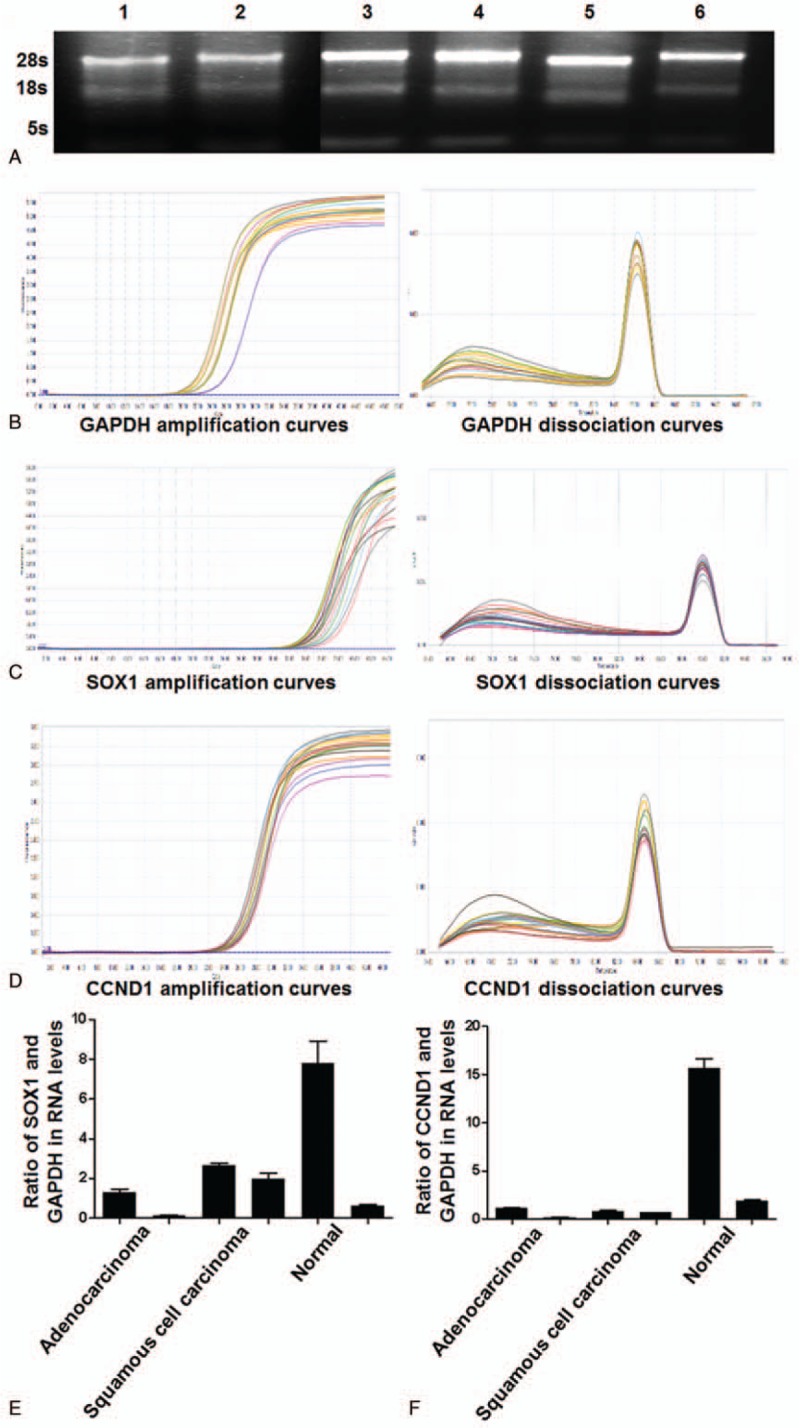
Methylation levels of SOX1 and CCND1 in cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues. (A) Agarose gel electrophoresis of total RNA. Lanes1 and 2, normal cervical tissues; lanes 3 and 4, cervical adenocarcinoma tissues; lanes 5 and 6, cervical squamous cell carcinoma tissues. (B–D) Amplification and dissociation curves of (B) GAPDH, (C) SOX1, and (D) CCND1. (E, F) Methylation levels of (E) SOX1 and (F) CCND1 in 2 cases of cervical adenocarcinoma tissues, 2 cases of cervical squamous cell carcinoma tissues, and 2 cases of normal cervical tissues.
Table 3.
Expression of SOX1 and CCND1 gene methylation in cervical adenocarcinoma and squamous cell carcinoma.

Table 4.
Expression of SOX1 and CCND1 gene methylation in cervical adenocarcinoma and normal cervical tissues.

Table 5.
Expression of SOX1 and CCND1 gene methylation in cervical squamous cell carcinoma and normal cervical tissues.

4. Discussion
The pathogenesis of cervical cancer remains unclear. Human papillomavirus (HPV) infection is subclinical and transient, and there are no cancerous or precancerous pathological lesions at the time of infection.[4] Therefore, detection of HPV infection is not enough for the screening of cervical cancer.[5] DNA methylation is the most common epigenetic alteration of key genes in cervical cancer, and it often occurs at the early stage of cervical cancer and causes abnormal gene expression in promoter region, which leads to tumorigenesis.[6] DNA methylation occurs in about 70% to 100% of cervical cancer cases, and can also be detected in 30% to 80% of precancerous cervical lesions.[7] Regardless of the status of HPV infection, hsa-mir-124, SOX1, Telomerase Reverse Transcriptase, and LIM homeobox transcription factor 1-α are molecular markers for predicting the severity of cervical lesions.[8] In precancerous cervical lesions and invasive cervical cancer, the methylation status of CDH1, p16, SOX1, and RASSFA1 genes has been found to be significantly different according to methylation-specific polymerase chain reaction.[9,10] Transcription of methylation-silenced tumor-suppressor gene at CpG site in promoter region is a key factor in the early development of tumors.[11–14] In the development of tumors, the state of methylation is not unchanged.
The SOX gene family is a family of transcription factors encoding DNA-binding domains of highly conserved high-mobility group proteins, and these transcription factors play important roles in the fine regulation of embryonic development and the normal function of stem cells.[15] SOX family members are closely related to tumorigenesis and tumor evolution. As a member of SOX family, SOX1 is regarded as a tumor-suppressor gene. Hypermethylation of gene promoter region that leads to inactivation of tumor-suppressor genes is an early event in tumorigenesis. For example, the methylation level of SOX1 gene in colorectal adenocarcinoma tissues is significantly higher than that in normal tissues.[16] It is proposed that the methylation status of paired box1 and SOX1 may be a new molecular marker for colorectal cancer screening.[17] There was a significant correlation between the down-regulation of SOX1 expression and the methylation of SOX1 promoter in primary hepatocellular carcinoma, and the methylation rate is 57.30%, being significantly higher than that in chronic hepatitis and cirrhosis. It is also discovered that DNA methylation levels of junctional adhesion moleculeA 3, SOX1, Slit guidance ligand 2, TTT, and C13ORF18 genes are significantly correlated with the prognosis of cervical intraepithelial neoplasia and can be used as predictors of cervical intraepithelial neoplasia prognosis. Therefore, SOX1 is a stable marker for tumor methylation. However, these markers are mostly in the research stage and need to be validated and evaluated by a wide range of people before they can be used in clinical practice.
In the present study, BSP showed that the promoter region of SOX1 gene in cervical adenocarcinoma tissues was hypermethylated, being consistent with the results by Illumina (850K) test and qRT-PCR.
Cyclin D1 is a regulatory protein, which plays an important role in cell cycle regulation, and its abnormal regulation can be observed in many tumors.[18] Cyclin D1 regulates cyclin-dependent kinase 4 (CDK4) to control cell cycle transition from G1 phase to S phase, which is also considered a key step in cell proliferation. It is reported that polymorphism CCND1 increases the risk of lung cancer among smokers in northern India and may be associated with overall survival in patients with small cell lung cancer.[19] A meta-analysis shows that the stronger the expression of CCND1 is, the faster the development of oral squamous cell carcinoma is, and CCND1 can be used as a biomarker for the prognosis of oral squamous cell carcinoma.[16] In addition, the epigenetic negative regulation of CCND1 by microRNA-490 is the key to glioma, and it provides a new perspective for the diagnosis, treatment, prognosis, and further transformation of glioma.[17] In the present study, BSP showed no significant difference in methylation status of CCND1 promoter among cervical adenocarcinoma, cervical squamous cell carcinoma, and normal cervical tissues, being inconsistent with the results by Illumina (850K) or qRT-PCR. However, the methylation of CCND1 gene cannot be excluded from significant indicators of cervical adenocarcinoma, considering the small number of BSP samples and detection sites.
5. Conclusions
In conclusion, the present study demonstrates that SOX1 is a methylation-specific gene in cervical adenocarcinoma. Hypermethylation of SOX1 promoter region is an important event in the occurrence and development of cervical adenocarcinoma and is expected to become a specific molecular marker for the diagnosis of cervical adenocarcinoma. However, CCND1 gene was not proven to be a specific methylation expression gene in cervical adenocarcinoma in the present study.
Acknowledgments
The authors wish to thank their department and research team for their help and dedication.
Author contributions
Data curation: Lili Yao.
Formal analysis: Min Yuan, Lili Yao.
Funding acquisition: Guzhalinuer Abulizi.
Project administration: Guzhalinuer Abulizi.
Supervision: Guzhalinuer Abulizi.
Validation: Guzhalinuer Abulizi.
Writing – original draft: Min Yuan, Lili Yao.
Writing – review & editing: Min Yuan, Lili Yao.
Footnotes
Abbreviations: BSP = bisulfite sequencing polymerase chain reaction, CCND1 = cyclin D1, CDK4 = cyclin-dependent kinase 4, DNA = deoxyribonucleic acid, HPV = human papillomavirus, MVP = methylation variable position, qRT-PCR = quantitative real-time polymerase chain reaction, SOX1 = sex determining region Y-box 1.
How to cite this article: Yuan M, Yao L, Abulizi G. Tumor-suppressor gene SOX1 is a methylation-specific expression gene in cervical adenocarcinoma. Medicine. 2019;98:38(e17225).
Funding: This work was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2017D01C396).
The authors have no conflicts of interest to disclose.
References
- [1].Esheba GE. ProExC is a novel marker for distinguishing between primary endometrial and endocervical adenocarcinomas. J Egypt Natl Cancer Inst 2013;25:87–93. [DOI] [PubMed] [Google Scholar]
- [2].Espinosa AM, Alfaro A, Roman-Basaure E, et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PloS One 2013;8:e55975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rodriguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 2008;100:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Petry KU, Schmidt D, Scherbring S, et al. Triaging Pap cytology negative, HPV positive cervical cancer screening results with p16/Ki-67 Dual-stained cytology. Gynecol Oncol 2011;121:505–9. [DOI] [PubMed] [Google Scholar]
- [5].Moelans CB, Verschuur-Maes AH, van Diest PJ. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol 2011;225:222–31. [DOI] [PubMed] [Google Scholar]
- [6].Lim EH, Ng SL, Li JL, et al. Cervical dysplasia: assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2, PAX1 and TFPI2 to improve diagnostic accuracy. Gynecol Oncol 2010;119:225–31. [DOI] [PubMed] [Google Scholar]
- [7].Huang S, Erickson B, Tang N, et al. Clinical performance of Abbott RealTime High Risk HPV test for detection of high-grade cervical intraepithelial neoplasia in women with abnormal cytology. J Clin Virol 2009;45suppl 1:S19–23. [DOI] [PubMed] [Google Scholar]
- [8].Rogeri CD, Silveira HCS, Causin RL, et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol Oncol 2018;150:545–51. [DOI] [PubMed] [Google Scholar]
- [9].Huang TH, Lai HC, Liu HW, et al. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer 2010;20:513–9. [DOI] [PubMed] [Google Scholar]
- [10].Castle PE, Stoler MH, Wright TC, Jr, et al. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol 2011;12:880–90. [DOI] [PubMed] [Google Scholar]
- [11].Lendvai A, Johannes F, Grimm C, et al. Genome-wide methylation profiling identifies hypermethylated biomarkers in high-grade cervical intraepithelial neoplasia. Epigenetics 2012;7:1268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sobti RC, Singh N, Hussain S, et al. Aberrant promoter methylation and loss of suppressor of cytokine signalling-1 gene expression in the development of uterine cervical carcinogenesis. Cell Oncol (Dordr) 2011;34:533–43. [DOI] [PubMed] [Google Scholar]
- [13].Farkas SA, Milutin-Gasperov N, Grce M, et al. Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics 2013;8:1213–25. [DOI] [PubMed] [Google Scholar]
- [14].Lin CJ, Lai HC, Wang KH, et al. Testing for methylated PCDH10 or WT1 is superior to the HPV test in detecting severe neoplasms (CIN3 or greater) in the triage of ASC-US smear results. Am J Obstet Gynecol 2011;204:21.e21-27. [DOI] [PubMed] [Google Scholar]
- [15].Shih YL, Hsieh CB, Yan MD, et al. Frequent concomitant epigenetic silencing of SOX1 and secreted frizzled-related proteins (SFRPs) in human hepatocellular carcinoma. J Gastroenterol Hepatol 2013;28:551–9. [DOI] [PubMed] [Google Scholar]
- [16].Ramos-Garcia P, Gonzalez-Moles MA, Gonzalez-Ruiz L, et al. Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: a systematic review and meta-analysis. Oral oncology 2018;83:96–106. [DOI] [PubMed] [Google Scholar]
- [17].Zhao L, Tang X, Luo R, et al. MicroRNA-490-5P targets CCND1 to suppress cellular proliferation in glioma cells and tissue through cell cycle arrest. Curr Neurovasc Res 2018;15:246–55. [DOI] [PubMed] [Google Scholar]
- [18].Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther 2002;1:226–31. [DOI] [PubMed] [Google Scholar]
- [19].Pandey A, Bahl C, Sharma S, et al. Functional role of CyclinD1 polymorphism (G870A) in modifying susceptibility and overall survival of North Indian lung cancer patients. Tumori 2018;104:179–87. [DOI] [PubMed] [Google Scholar]


