Abstract
Background
Multiple sclerosis (MS) is a common autoimmune disease of the central nervous system (CNS), and is associated with genetic factors. FOXP3 gene polymorphism has been reported as the risk factor for MS, however, previous studies have showed conflicting results. The purpose of this study is to investigate the association between FOXP3 gene polymorphism and the susceptibility to MS.
Methods
Pubmed, Embase, library of Cochrane, and Web of Science were used to search the eligible articles from January 1980 up to October 2018. The odds ratio (ORs) and its 95% confidence intervals (CI) were used to evaluate the strength of association. Allele model, homozygote model, heterozygote model, dominant model, and recessive model were used to evaluate the association between FOXP3 gene polymorphism and MS.
Results
A total of 5 studies contained 1276 MS patients and 1447 controls (for rs3761548) and 600 MS patients and 640 controls (for rs2232365) were enrolled in this meta-analysis. The association showed significant differences in allele and dominant model for rs3761548 polymorphism. In addition, a clear tendency to significance was detected in homozygote and recessive model for rs3761548 (P = .052). Subgroup analysis indicated a significant risk of MS in all genotype models but heterozygotes in Asians.
Conclusion
FOXP3 gene polymorphism rs3761548 was associated with a higher MS risk, especially in Asians. This conclusion needs to be validated in more large samples and multiracial studies.
Level of evidence
Level III diagnostic study.
Keywords: FOXP3 gene, meta-analysis, multiple sclerosis, single nucleotide polymorphism
1. Introduction
Multiple sclerosis (MS) is an immune-related central nervous disease, which manifests as variable symptoms including optic neuritis, fatigue, spasticity, neuro-urological dysfunction, paresthesia and hypesthesia, and headache.[1] In pathophysiology, this disease was characterized by inflammation, demyelination, proliferation of astrocytes, and varying degrees of axonal degeneration.[2] Meanwhile, variant cells and their markers such as T cells, clonal expansion of B cells, and their antibody products were found in MS patients, indicating the involvement of abnormalities of T-cells especially CD4 and CD8 T cells and B-cells response.[3,4] It was considered that the occurrence and progression of MS may attribute to the virus infection, vitamin deficiency, mitochondrial dysfunction, and oxidative stress.[5] However, up to now, the exact reason for MS has not been determined.
Recently, more and more studies focus on the effect of single nuclei polymorphisms (SNPs) on some autoimmune disease including MS.[6] Previous literatures have reported several genetic variants (IL-6, HLA-DRB1, and STAT4) were correlated with development of MS.[2,7,8] The transcription factor forkhead box P3 (FOXP3) which belongs to the fork-winged helix family and encoded by the FOXP3 gene was regarded as an important molecular marker that regulate T-reg cell development and function.[9] The prior study demonstrated that CD4+CD25+Foxp3+ T-regs played a crucial role in preventing autoimmunity and undesirable T cell responses.[10] Moreover, mutation of the FOXP3 gene has been shown to be a risk in unexplained recurrent spontaneous abortions, Grave's disease, and allergic rhinitis.[11–13] Currently, some studies reported two key SNPs (rs3761548 and rs2232365) on FOXP3 gene may be involved in the development of MS.[14,15] However, recent studies still obtained conflicting results that may lead to inconsistent outcomes.
In this meta-analysis, we collected data from several databases and combined them for analyzing to investigate the overall influence of FOXP3 gene polymorphism on the risk of MS.
2. Methods
The authors performed the meta-analysis according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). Meanwhile, since the present study is based on data from published studies, the informed consent of the patients and the ethical approval were not required.
2.1. Study strategy
Pubmed, Embase, Cochrane library, and Web of Science were used for the potential studies searching. (Polymorphism OR “single-nucleotide polymorphism” OR “SNP” OR “mutant” OR “mutation” OR “variant” OR “variation”) AND (“FOXP3” OR “transcription factor forkhead box P3” OR “rs3761548” OR “rs2232365”) AND (“multiple sclerosis” OR “multiple sclerosis”) were used as the key words or Mesh terms for searching. The final search was conducted up to October 1, 2018 and the language of searched literatures was not restricted.
2.2. Inclusion and exclusion criteria
The included studies met the following criteria:
-
1.
case-control;
-
2.
the study evaluated the association between FOXP3 gene polymorphism and MS;
-
3.
the study contained sufficient data of genotype frequency.
The exclusion criteria were as follows:
-
1.
studies without controls groups;
-
2.
studies with only abstracts and reviews;
-
3.
studies without specific distribution of genotype;
-
4.
studies reported animal models.
The 9-point Newcastle-Ottawa Scale (NOS) was used for quality assessment of the included studies.
2.3. Data extraction
Two independent reviewers (YJ Z and B P) assessed and reviewed all identified studies in terms of inclusion and exclusion criteria. Conflicts were reached to agreement via the discussion with the third authors (HL Y). Authors, date of publication, race, sample size, genotype and allele distribution of cases and controls were all extracted and recorded from the enrolled studies.
2.4. Statistical analysis
This meta-analysis preformed the data analyzing by using Stata version 11.0 (Stata Corp, College Station, TX, USA). The odds ratio (OR) and 95% confidence interval (CI) were used to assess the relationship between rs3761548 and rs2232356 polymorphism and the risk of MS. The allelic contrast, dominant, recessive, and homozygotes models were all conducted in this study. The evaluation of heterogeneity of studies was using the Q test and was quantified using I2. The fixed-effects model was used when I2 < 50%. In contrast, the random-effects model was applied when I2 > 50%. The subgroup analysis was performed stratified by ethnicity. The Begg and Egger's test were used for the assessment of publication bias and the sensitivity analysis was conducted by assessing the change of combined ORs values after elimination of each individual study. A P value equal to or <.05 was considered the threshold for statistical significance.
3. Results
3.1. Literatures search
Based on the database of Pubmed, Embase, Cochrane library, Web of Science, and other resources, a total of 95 articles were relevant to the search terms. After elimination of duplicated studies and exclusion of improper studies via titles and abstracts, 18 articles remained for full text reviewing. Then, 13 articles were excluded for following reasons (lack of sufficient data of distribution of genotype, animal models, and comment/editorial articles). Ultimately, 5 articles were eligible in quantitative synthesis (current meta-analysis). The specific flow chart was shown in Figure 1.
Figure 1.
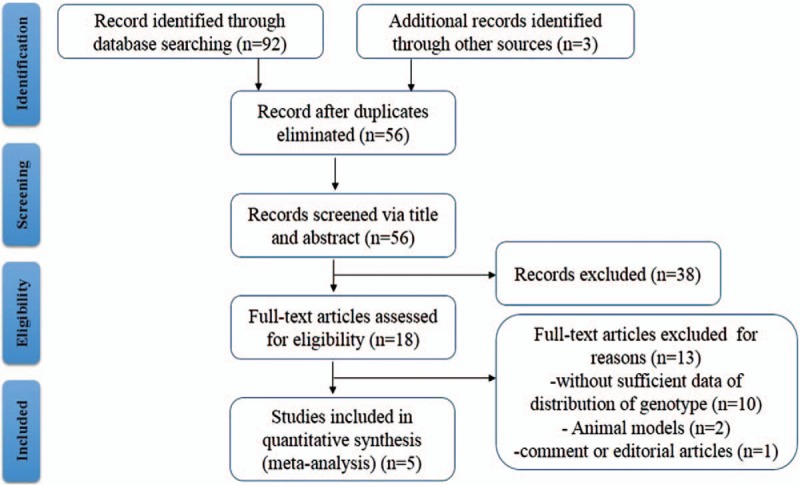
Flow diagram of the selection of literature.
3.2. Characteristic of eligible studies
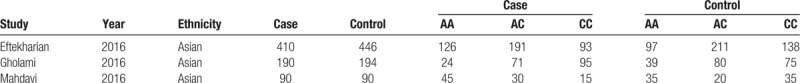
For the 5 included studies,[16–20] 4 studies reported the comparison of rs3761548 polymorphism, and 3 studies stated comparison of rs2232365 polymorphism between MS and controls. The analysis of rs3761548 polymorphism contained 1276 MS patients and 1447 controls. The scores of NOS scale of all included studies were larger than 6 points, indicating a good quality (Table 1). There was a significant difference in the sex of patients and controls (OR: 0.76, 95% CI: 0.65–0.90, P = .001), and no significant difference in age (OR: 0.06, 95% CI: −0.01–0.14, P = .088) (Table 2). The analysis of re2232365 polymorphism contained 600 MS patients and 640 controls. Here, there was no significant difference in both sex (OR: 0.77, 95% CI: 0.48–1.25, P = .295) and age between patients and controls (OR: 0.08, 95% CI: −0.03 to 0.19, P = .156) (Table 3).
Table 1.
The scores of each study based on Newcastle-Ottawa scale.

Table 2.
Characteristics of eligible studies for SNPs of rs3761548.
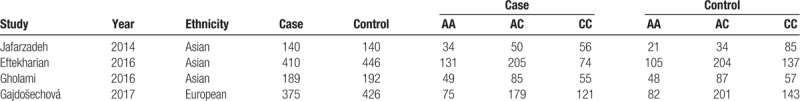
Table 3.
Characteristics of eligible studies for SNPs of rs2232365.
3.3. Quantitative synthesis
For rs3761548 polymorphism, there was a significantly decreased risk of MS under allele model (OR: 0.76, 0.58–1.00, P = .049) and dominant model (OR: 0.77, 0.64–0.94, P = .008) (Table 4). No significant association was detected between rs37651548 and MS under the homozygote model (OR: 0.63, 0.40–1.00, P = .052), heterozygote model (OR: 0.89, 0.72–1.10, P = .276), and the recessive model (OR: 0.67, 0.45–1.01, P = .056) (Figs. 2 and 3). Further, subgroup analysis was performed by ethnicity. There were significant differences in all genetic models but heterozygote model in Asians (Table 5). In contrast, no significant difference was detected in European population under any genetic model (Figs. 4 and 5).
Table 4.
Meta-analysis of the association between FOXP3 polymorphism and multiple sclerosis.

Figure 2.
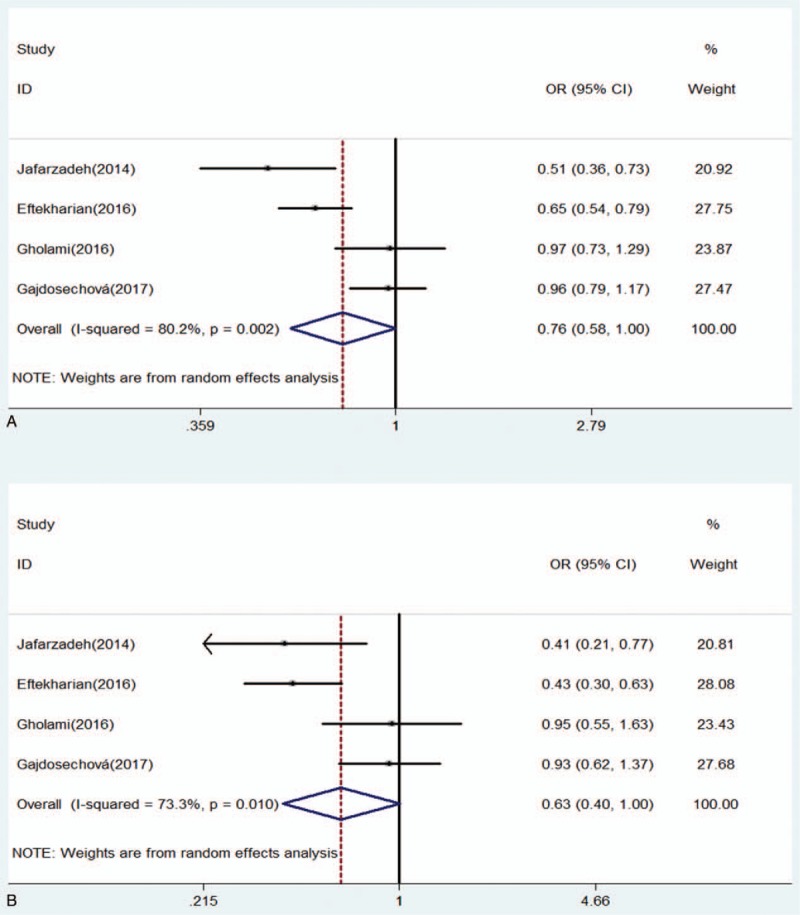
Forest of association between rs3761548 polymorphism of FOXP3 gene and risk of MS under (A) allele model (C/A); (B) homozygote model (CC/AA). CI = confidence interval, OR = odd ratio.
Figure 3.
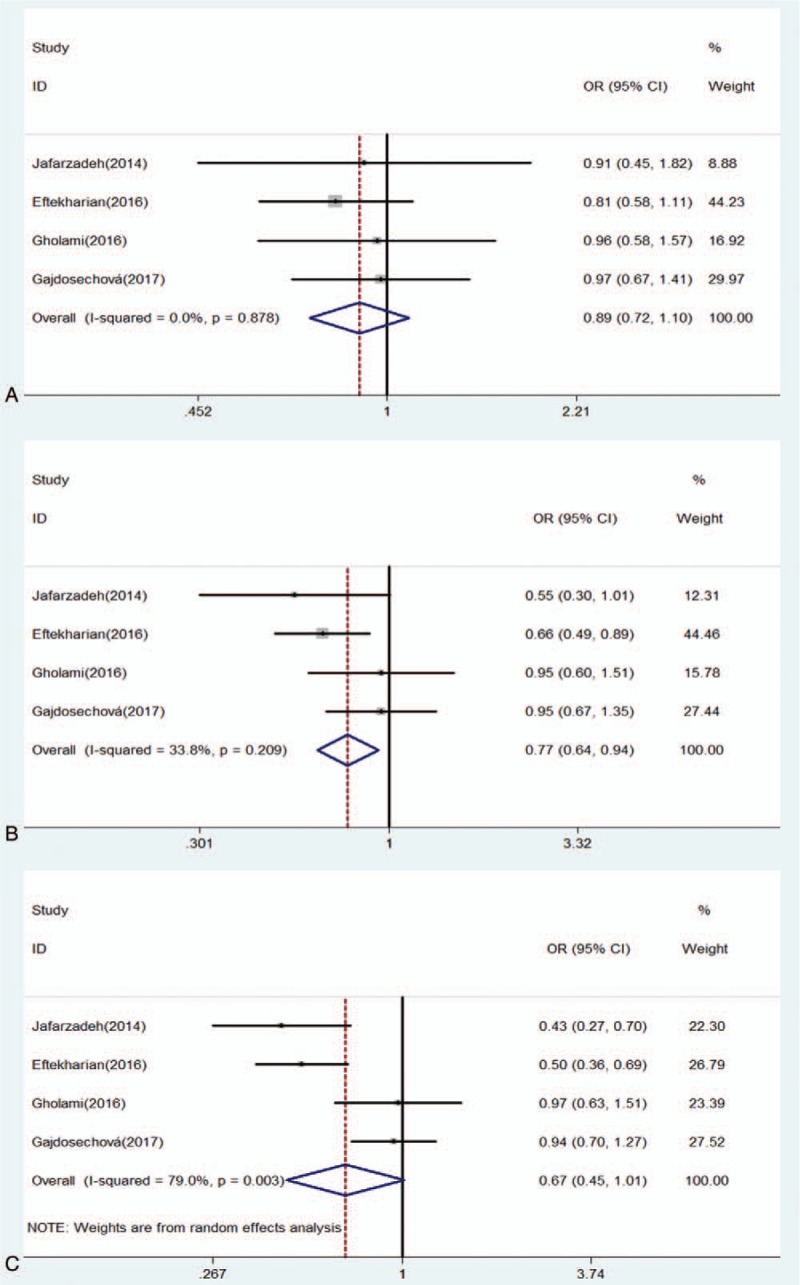
Forest of association between rs3761548 polymorphism of FOXP3 gene and risk of MS under (A) heterozygote model (CA/AA); (B) dominant model (CC + CA/AA); (C) recessive model (CC/CA + AA).
Table 5.
Subgroup-analysis of the association between rs3761548 polymorphism and multiple sclerosis.

Figure 4.
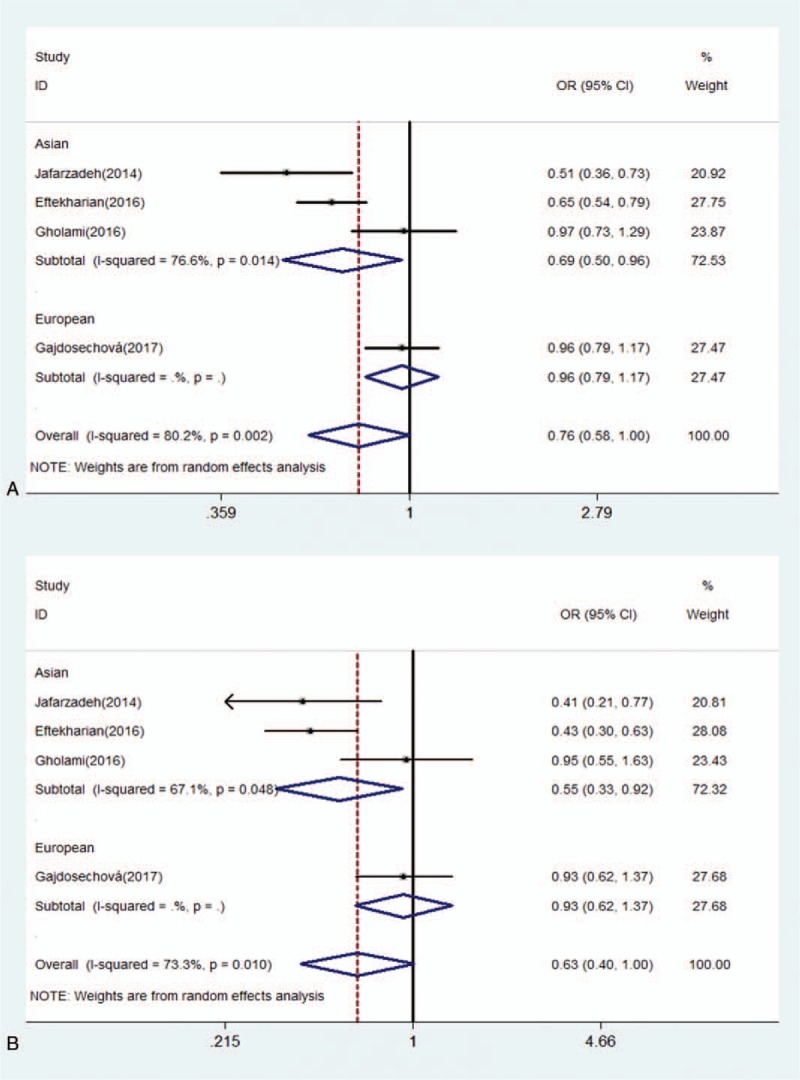
Forest of association between rs3761548 polymorphism of FOXP3 gene and risk of MS for subgroup analysis by ethnicity under (A) allele model (C/A); (B) homozygote model (CC/AA).
Figure 5.
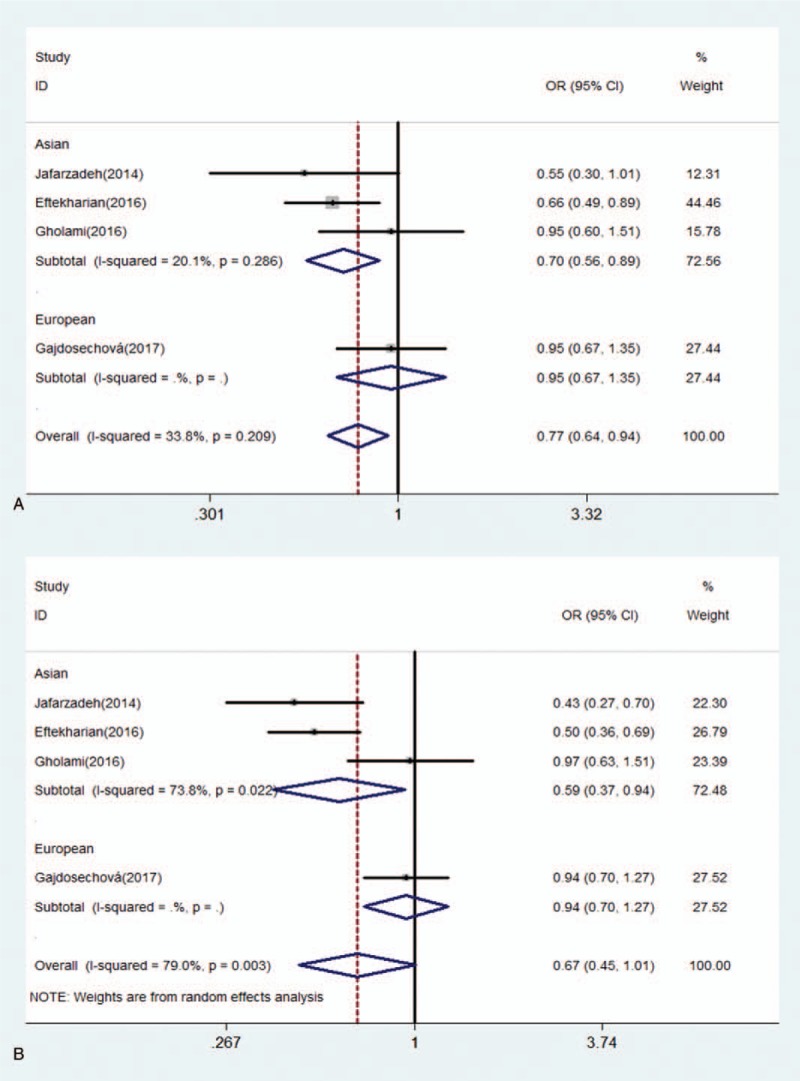
Forest of association between rs3761548 polymorphism of FOXP3 gene and risk of MS for subgroup analysis by ethnicity under (A) dominant model (CC + CA/AA); (B) recessive model (CC/CA + AA).
For rs2232365 polymorphism, we did not find a significant difference in the distribution of genotypes between MS and controls under allele model (OR: 1.22, 0.68–2.18, P = .503), homozygote model (OR: 1.40, 0.52–3.77, P = .507), heterozygote model (OR: 1.37, 0.67–2.79, P = .383), dominant model (OR: 1.40, 0.63–3.09, P = .411), and recessive model (OR: 1.15, 0.61–2.16, P = .663) (Figs. 6 and 7).
Figure 6.
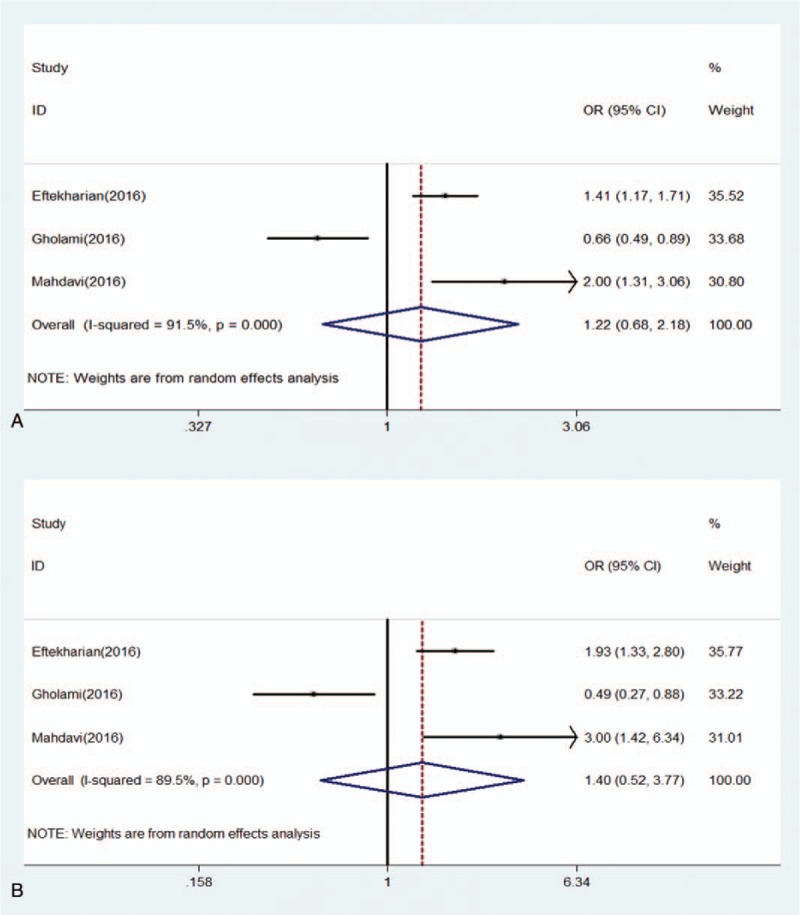
Forest of association between rs2232365 polymorphism of FOXP3 gene and risk of MS under (A) allele model (A/C); (B) homozygote model (AA/CC).
Figure 7.
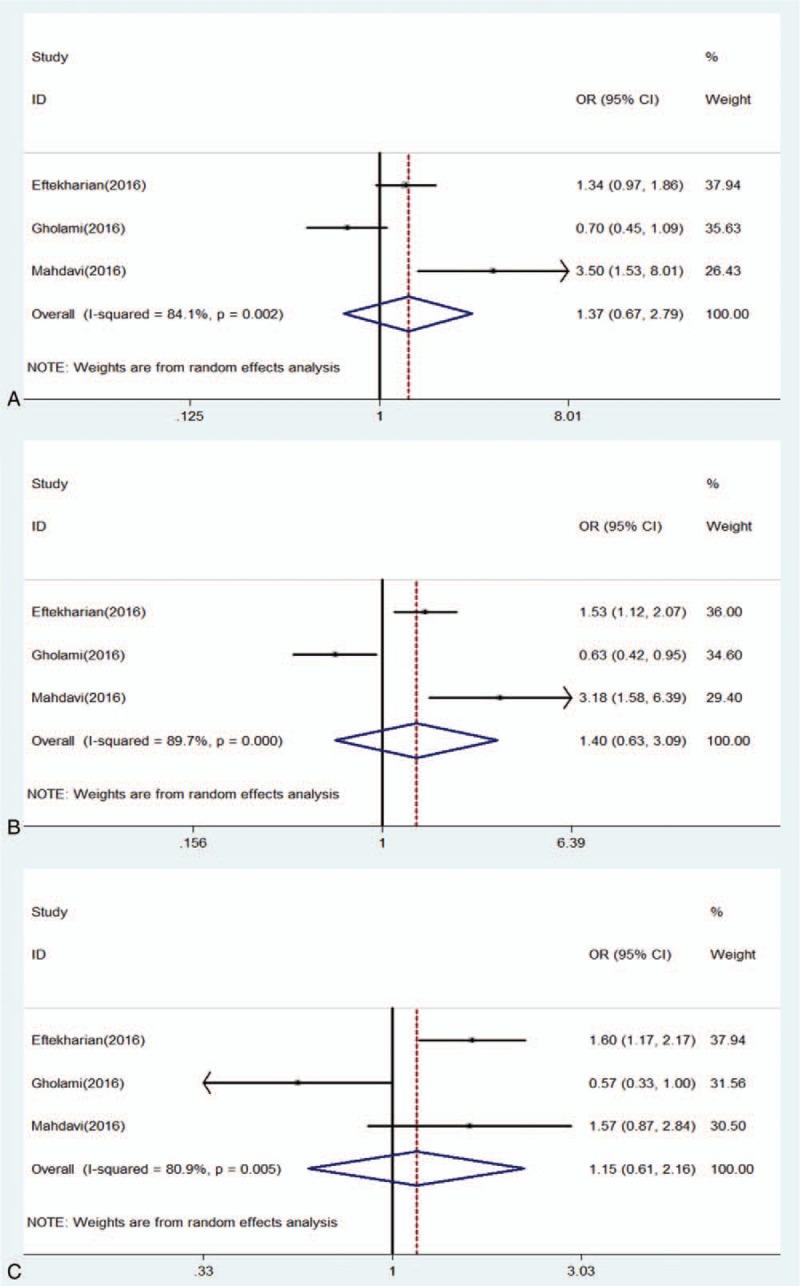
Forest of association between rs2232365 polymorphism of FOXP3 gene and risk of MS under (A) heterozygote model (AC/CC); (B) dominant model (AA + AC/CC); (C) recessive model (AA/AC + CC).
3.4. Publication bias
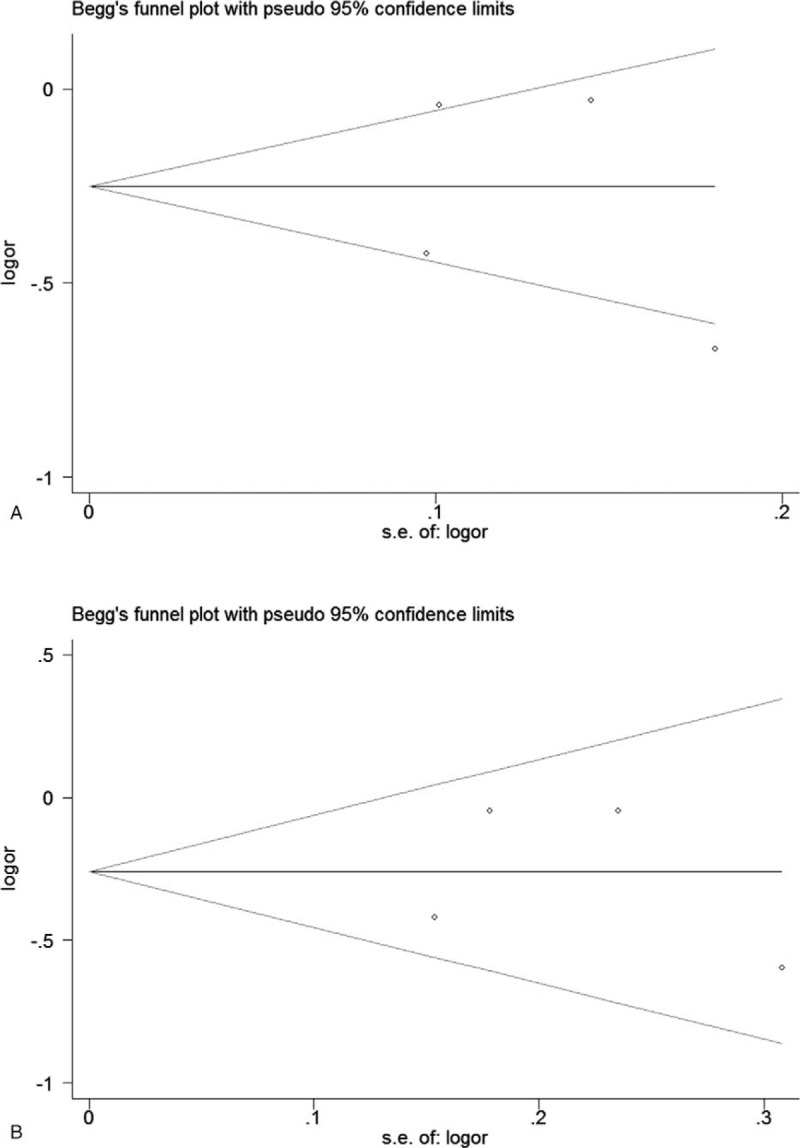
We performed the Begg funnel plot and Egger's test to assess the potential publication bias in the identified studies. The results showed no evidence of publication bias in this meta-analysis (Egger test: P = .733 under allele model for rs3761548, P = .925 under dominant model for rs3761548) (Fig. 8).
Figure 8.
Sensitive analysis of association between rs3761548 polymorphism of FOXP3 gene and risk of MS under (A) allele model (C/A); (B) homozygote model (CC/AA).
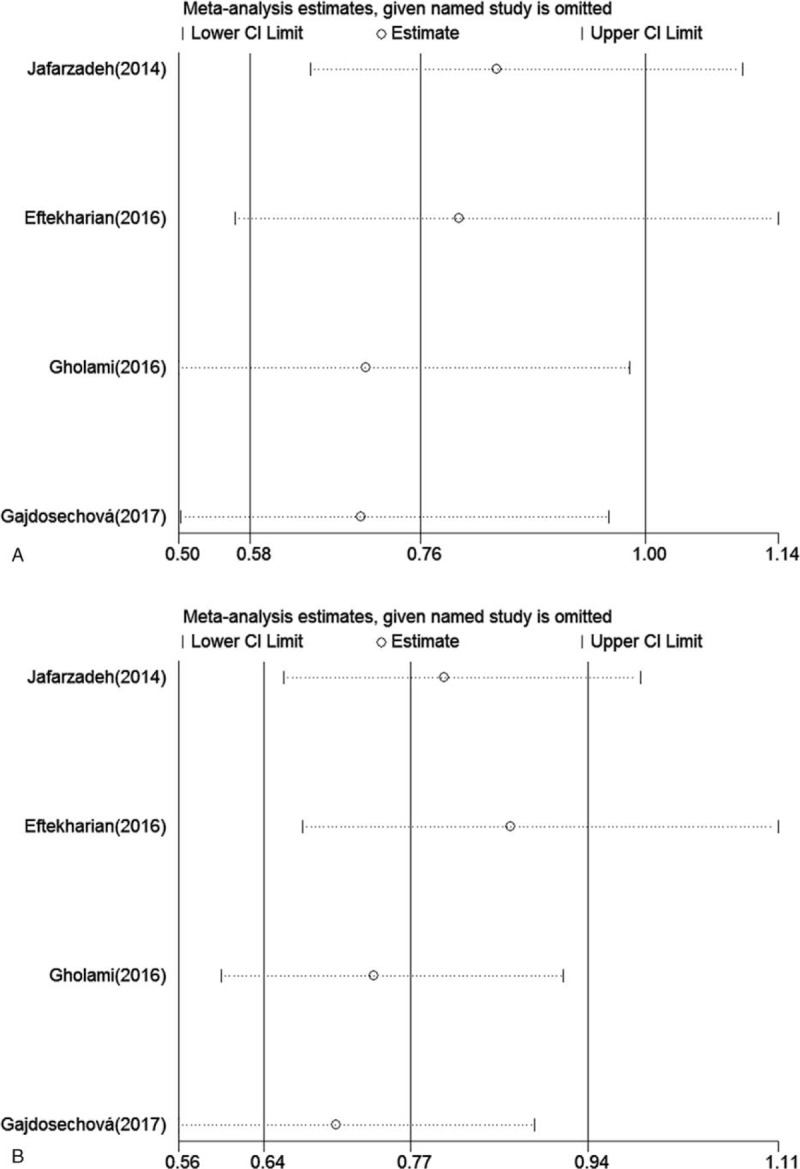
3.5. Sensitive analysis
Sensitive analysis was conducted to assess the influence of one study on the pooled ORs value (allele model and dominant model for rs3761548), and whether the results can be reverted by eliminating the individual study. The result did not alter after deleting each study, indicating the stability of the results of this meta-analysis (Fig. 9).
Figure 9.
Begg funnel plot for the potential missing studies under (A) dominant model (CC + CA/AA); (B) recessive model (CC/CA + AA).
4. Discussion
As a kind of chronic inflammatory demyelinating disorder of the central nervous system, MS usually occurs in young adults and causes torturous symptoms and complications.[21] Unfortunately, until now, no exact etiology of this disease was found. It was widely believed that MS may be the result of the interaction between genetic and environmental factors together.[22] Lots of potential genetic polymorphisms have also been identified and proved to be associated with the development of MS. A prior case-control study investigated the distribution of the Vitamin D receptor (VDR) gene in MS and revealed five VDR genetic polymorphisms as the risk factors for MS susceptibility.[23] Similar results were also reported between HLA alleles variant and MS.[24]
FOXP3 gene was located on the p arm of the X chromosome and contained 11 coding exons and 3 non-coding exons in the 5’ upstream region in humans.[25] Meanwhile, this gene was mainly expressed in CD4+ CD25+ thymocytes and CD4+ CD25+ peripheral T cells, which play a crucial role in immune regulation.[26] Several studies had reported that FOXP3 acted as a key target in the generation and maturation of T-reg cells, indicating the mutation of this gene may cause the dysfunction of T-reg cells even reversed the process of naïve T cells transfer to T-reg cells.[27] Further, recessive X-linked mutations of FOXP3 gene have been identified as a potential reason for immune-dysregulation, which means that even mild alterations of FOXP3 expression may result in common autoimmune diseases.[28] Due to its X-linked characteristic, the genetic polymorphism of FOXP3 has been regarded as a marked risk factor in breast cancer among different ethnic populations (Chinese, Israeli, and Indians).[29] Similarly, the balance between Th1 and Th2 cells in the maternal uterus may be broken because of mutation of FOXP3 gene, which can harm the fetus during pregnancy and subsequently result in a re-iterant abortion.[30] Additionally, such disturbance of immune system function and aberrant immune response can affect other organs in body and cause several immune-related disorders including autism, Wilms’ tumor, thyroid cancer, recurrent infertility and so on.[31–34] In recent studies, FOXP3 gene polymorphism was correlated with MS. However, these studies were small sample sizes and had low statistical power which may lead to contradictory results. Hence, we performed this meta-analysis by combining the independent studies and estimating the overall effect to overcome the individual limitation and to draw more convinced conclusions.
In present meta-analysis, after searching and reviewing 95 potential articles, we finally consolidated a total of 5 eligible studies to seek the variant polymorphism of two representative genes (rs3761548 and rs2232365) in MS susceptibility. To our limited knowledge, this study is the first meta-analysis that investigated the correlation between FOXP3 polymorphism and MS. Our results revealed that the variant allele and dominant model for rs3761548 can significantly decrease the risk of MS compared with the wild genotype. Meanwhile, under homozygote and recessive model, the difference between MS patients and controls also showed a marginal trend toward significance (P = .052 and P = .056). Considering the discrepancy of ethnicity between articles, we also performed stratified analysis categorized by race (3 studies from Asia and 1 study from Europe). The outcomes of subgroup analysis showed a significant association between rs3761548 gene polymorphism and MS under all genetic models except for heterozygotes in Asians. Above data supported the hypothesis that FOXP3 gene polymorphism (rs3761548) was associated with MS. On the other hand, no significant trend was identified for the occurrence of MS in each genetic model of rs2232365 gene. Moreover, either the sensitive analysis or the publication bias analysis showed no statistical significance in both the allele and dominant model for rs3761548. Hence, we speculated that rs3761538 polymorphism in FOXP3 gene may have an association with the susceptibility to MS, especially for Asians.
Although the genetic polymorphism in the promoter region of FOXP3 gene including rs3761548 has been studied for several years, the specific impacts of these SNPs were still unclear. It was widely considered that T-regs acted as an important component in immune system and inflammatory response, which implied that defective T-regs could lead to various autoimmune diseases.[10] At the same time, previous studies found that FOXP3 was an indispensable molecular in the development and function of T-reg cells.[35] Therefore, some opinions hypothesized that the mutation of FOXP3 can affect T-reg function and thereby lead to immune-related diseases. With modifying the transcription and altering the binding specificity of transcription factors, rs3761548 polymorphism was able to affect the expression of FOXP3 genes.[25] In addition, the allelic alteration of rs3761548C > A can disturb the binding of E47 and c-Myb on the FOXP3 promoter site, resulting in the defective transcription of FOXP3.[36] Furthermore, another study suggested that rs3761548 polymorphism was located in the core sequence of the putative binding site for transcription factor, specificity protein 1 (Sp1), indicating the mutation of C to A may obstruct the interaction between Sp1 and FOXP3 promoter subsequent disturb the expression of FOXP3 gene.[37] Our preliminary findings revealed that rs3761538 polymorphism in FOXP3 may be a vital factors in development of MS, which provide a new insight to understanding the genetic factors of MS susceptibility. Moreover, with the development of gene therapy, modification of rs3761538 polymorphism may be regarded as a potential therapeutic target in future.
4.1. Limitations and future directions
Considering several limitations of this meta-analysis, the results should be interpreted carefully. First, due to the insufficient data, this study did not investigate the influence of the other three SNPs of FOXP3 (rs3761547, rs3761549, and rs2280883) on MS susceptibility. Second, we could not perform the subgroup analysis by gender and stages of MS due to the limited data of included studies. Third, the number of the enrolled studies in this meta-analysis was small due to the limited number of reports about the FOXP3 gene on MS, which may introduce some bias. Fourth, the heterogeneity of rs2232365 was at a high level because of only three identified studies. Last, four of the five eligible studies were from Asians, which may limit our conclusion. For future studies, more attention should be paid to several research directions:
-
1.
more SNPs polymorphism of FOXP3 should be analyzed and summarized;
-
2.
gender, age, and other parameters can be considered as subgroup variables for data processing;
-
3.
other ethnic populations including European and American subjects, should be performed to confirm the effect of FOXP3 gene polymorphism on MS risk.
5. Conclusion
Based on the limited data, our meta-analysis preliminary indicated that rs3761548 polymorphism was associated with the elevated susceptibility of MS especially among Asian populations. However, more well-designed studies with large sample sizes and multiple ethnicities studies are needed to corroborate our conclusions.
Acknowledgments
We thank all the participants for their contribution to this work.
Author contributions
YJ Z and JX Z contributed equally to this work. YJ Z and B P designed the study. JX Z and H L did the literature search, study quality assessment and data extraction. YJ Z and JX Z performed the statistical analysis and drafted the tables and figures. YJ Z wrote the first draft of this analysis, and HL Y helped to finish the final version. All authors approved the conclusions of our study.
Data curation: Junxin Zhang, Hao Liu.
Methodology: Fan He.
Writing – original draft: Yijian Zhang.
Writing – review & editing: Angela Chen, Huilin Yang, Bin Pi.
Footnotes
Abbreviations: CI = confidence intervals, FOXP3 = transcription factor forkhead box P3, MS = multiple sclerosis, OR = odd ratio.
How to cite this article: Zhang Y, Zhang J, Liu H, He F, Chen A, Yang H, Pi B. Meta-analysis of FOXP3 gene rs3761548 and rs2232365 polymorphism and multiple sclerosis susceptibility. Medicine. 2019;98:38(e17224).
YZ and JZ contributed equally to this study.
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Availability of data and material: All data generated or analyzed in this work are included in the published version.
Competing interests: All authors confirmed no competing interest in this work.
This work is supported by the Jiangsu Provincial Clinical Orthopedics Center.
References
- [1].Gebhardt M, Kropp P, Hoffmann F, et al. Headache at the time of first symptom manifestation of multiple sclerosis: a prospective, longitudinal study. Eur Neurol 2018;80:115–20. [DOI] [PubMed] [Google Scholar]
- [2].Benesova Y, Vasku A, Bienertova-Vasku J. Association of interleukin 6, interleukin 7 receptor alpha, and interleukin 12B gene polymorphisms with multiple sclerosis. Acta Neurol Belgica 2018;118:493–501. [DOI] [PubMed] [Google Scholar]
- [3].Avila M, Bansal A, Culberson J, et al. The role of sex hormones in multiple sclerosis. Eur Neurol 2018;80:93–9. [DOI] [PubMed] [Google Scholar]
- [4].Xia ZL, Qin QM, Zhao QY. A genetic link between CXCR5 and IL2RA gene polymorphisms and susceptibility to multiple sclerosis. Neurol Res 2018;40:1040–7. [DOI] [PubMed] [Google Scholar]
- [5].Safizadeh B, Hoshyar R, Mehrpour M, et al. The role of expression and activity of 15-Lipoxygenase isoforms and related cytokines in patients with Multiple Sclerosis and healthy controls. J Neuroimmunol 2018;325:32–42. [DOI] [PubMed] [Google Scholar]
- [6].Wang H, Pardeshi LA, Rong X, et al. Novel variants identified in multiple sclerosis patients from Southern China. Front Neurol 2018;9:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mohajer B, Abbasi N, Pishgar F, et al. HLA-DRB1 polymorphism and susceptibility to multiple sclerosis in the Middle East North Africa region: A systematic review and meta-analysis. J Neuroimmunol 2018;321:117–24. [DOI] [PubMed] [Google Scholar]
- [8].Nageeb RS, Omran AA, Nageeb GS, et al. STAT4 gene polymorphism in two major autoimmune diseases (multiple sclerosis and juvenile onset systemic lupus erythematosus) and its relation to disease severity. Egyptian J Neurol Psychiatry Neurosurg 2018;54:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cekin N, Pinarbasi E, Bildirici AE, et al. FOXP3 rs3761548 polymorphism is associated with knee osteoarthritis in a Turkish population. Int J Rheum Dis 2018. [DOI] [PubMed] [Google Scholar]
- [10].Hashemi V, Farrokhi AS, Tanomand A, et al. Polymorphism of Foxp3 gene affects the frequency of regulatory T cells and disease activity in patients with rheumatoid arthritis in Iranian population. Immunol Lett 2018;204:16–22. [DOI] [PubMed] [Google Scholar]
- [11].Zhang G, Zhang D, Shi W, et al. The impact of FOXP3 polymorphism on the risk of allergic rhinitis: a meta-analysis. Ann Hum Genet 2017;81:284–91. [DOI] [PubMed] [Google Scholar]
- [12].Mishra S, Srivastava A, Mandal K, et al. Study of the association of forkhead box P3 (FOXP3) gene polymorphisms with unexplained recurrent spontaneous abortions in Indian population. J Genet 2018;97:405–10. [PubMed] [Google Scholar]
- [13].Shehjar F, Afroze D, Misgar RA, et al. Association of FoxP3 promoter polymorphisms with the risk of Graves’ disease in ethnic Kashmiri population. Gene 2018;672:88–92. [DOI] [PubMed] [Google Scholar]
- [14].Wawrusiewicz-Kurylonek N, Chorazy M, Posmyk R, et al. The FOXP3 rs3761547 gene polymorphism in multiple sclerosis as a male-specific risk factor. Neuromol Med 2018;20:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Isik N, Yildiz Manukyan N, Aydin Canturk I, et al. Genetic susceptibility to multiple sclerosis: the role of FOXP3 gene polymorphism. Noro psikiyatri arsivi 2014;51:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jafarzadeh A, Jamali M, Mahdavi R, et al. Circulating levels of interleukin-35 in patients with multiple sclerosis: evaluation of the influences of FOXP3 gene polymorphism and treatment program. J Mol Neurosci 2015;55:891–7. [DOI] [PubMed] [Google Scholar]
- [17].Eftekharian MM, Sayad A, Omrani MD, et al. Single nucleotide polymorphisms in the FOXP3 gene are associated with increased risk of relapsing-remitting multiple sclerosis. Hum Antibodies 2016;24:85–90. [DOI] [PubMed] [Google Scholar]
- [18].Gholami M, Darvish H, Ahmadi H, et al. Functional Genetic Variants of FOXP3 and Risk of Multiple Sclerosis. Iranian Red Crescent Medical Journal 2017;19:e34597.doi: 10.5812/ircmj.34597. [Google Scholar]
- [19].Rad MR, Safa A. FOXP3 polymorphism rs2232365 and its association with multiple sclerosis susceptibility. Tehran Univ Med J 2016;74:425–32. [Google Scholar]
- [20].B DG, Javor J, Čierny D, et al. Association of FOXP3 polymorphisms rs3761547 and rs3761548 with multiple sclerosis in the Slovak population. Activitas Nervosa Superior Rediviva 2017;59:9–15. [Google Scholar]
- [21].Javor J, Shawkatova I, Durmanova V, et al. TNFRSF1A polymorphisms and their role in multiple sclerosis susceptibility and severity in the Slovak population. Int J Immunogenet 2018;doi: 10.1111/iji.12388. [DOI] [PubMed] [Google Scholar]
- [22].Ghavimi R, Alsahebfosoul F, Salehi R, et al. High-resolution melting curve analysis of polymorphisms within CD58, CD226, HLA-G genes and association with multiple sclerosis susceptibility in a subset of Iranian population: a case-control study. Acta Neurol Belgica 2018;doi: 10.1007/s13760-018-0992-y. [DOI] [PubMed] [Google Scholar]
- [23].Krenek P, Benesova Y, Bienertova-Vasku J, et al. The impact of five VDR polymorphisms on multiple sclerosis risk and progression: a case-control and genotype-phenotype study. J Mol Neurosci 2018;64:559–66. [DOI] [PubMed] [Google Scholar]
- [24].Al Jumah M, Kojan S, Al Shehri AM, et al. HLA class II polymorphism in Saudi patients with multiple sclerosis. HLA 2018;91:17–22. [DOI] [PubMed] [Google Scholar]
- [25].Hanel SA, Velavan TP, Kremsner PG, et al. Novel and functional regulatory SNPs in the promoter region of FOXP3 gene in a Gabonese population. Immunogenetics 2011;63:409–15. [DOI] [PubMed] [Google Scholar]
- [26].Yang G, Zhou H, Hickford JG. Polymorphism of the ovine FOXP3 gene (FOXP3). Vet Immunol Immunopathol 2011;140:303–6. [DOI] [PubMed] [Google Scholar]
- [27].Jahan P, Sreenivasagari R, Goudi D, et al. Role of Foxp3 gene in maternal susceptibility to pre-eclampsia—a study from South India. Scand J Immunol 2013;77:104–8. [DOI] [PubMed] [Google Scholar]
- [28].Ban Y, Tozaki T, Tobe T, et al. The regulatory T cell gene FOXP3 and genetic susceptibility to thyroid autoimmunity: an association analysis in Caucasian and Japanese cohorts. J Autoimmun 2007;28:201–7. [DOI] [PubMed] [Google Scholar]
- [29].Jahan P, Ramachander VR, Maruthi G, et al. Foxp3 promoter polymorphism (rs3761548) in breast cancer progression: a study from India. Tumour Biol 2014;35:3785–91. [DOI] [PubMed] [Google Scholar]
- [30].Naderi-Mahabadi F, Zarei S, Fatemi R, et al. Association study of forkhead box P3 gene polymorphisms with unexplained recurrent spontaneous abortion. J Reprod Immunol 2015;110:48–53. [DOI] [PubMed] [Google Scholar]
- [31].Arishima T, Sasaki S, Isobe T, et al. Maternal variant in the upstream of FOXP3 gene on the X chromosome is associated with recurrent infertility in Japanese Black cattle. BMC Genet 2017;18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jiang W, Zheng L, Xu L, et al. Association between FOXP3 gene polymorphisms and risk of differentiated thyroid cancer in Chinese Han population. J Clin Lab Anal 2017;31: doi: 10.1002/jcla.22104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ozawa PM, Ariza CB, Losi-Guembarovski R, et al. Wilms’ tumor susceptibility: possible involvement of FOXP3 and CXCL12 genes. Mol Cell Pediatr 2016;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Safari MR, Ghafouri-Fard S, Noroozi R, et al. FOXP3 gene variations and susceptibility to autism: a case-control study. Gene 2017;596:119–22. [DOI] [PubMed] [Google Scholar]
- [35].Kagoya Y, Saijo H, Matsunaga Y, et al. Arginine methylation of FOXP3 is crucial for the suppressive function of regulatory T cells. J Autoimmun 2018;doi: 10.1016/j.jaut.2018.09.011. [DOI] [PubMed] [Google Scholar]
- [36].Oda JM, Hirata BK, Guembarovski RL, et al. Genetic polymorphism in FOXP3 gene: imbalance in regulatory T-cell role and development of human diseases. J Genet 2013;92:163–71. [DOI] [PubMed] [Google Scholar]
- [37].Song P, Wang XW, Li HX, et al. Association between FOXP3 polymorphisms and vitiligo in a Han Chinese population. Brit J Dermatol 2013;169:571–8. [DOI] [PubMed] [Google Scholar]


