SUMMARY:
The response of a cell to integrated stresses was investigated using environmental and/or genetic perturbations that disrupted labile iron homeostasis and increased oxidative stress. The effects of the perturbations were monitored as nutritional requirements, and were traced to specific enzymatic targets. A yggX gshA cyaY mutant strain required exogenous thiamine and methionine for growth. The thiamine requirement, which had previously been linked to the Fe-S cluster proteins ThiH and ThiC, was responsive to oxidative stress and was not directly affected by manipulation of the iron pool. The methionine requirement was associated with the activity of sulfite reductase, an enzyme that appeared responsive to disruption of labile iron homeostasis. Results are incorporated in a model to suggest how the activity of iron-containing enzymes not directly sensitive to oxygen, can be decreased by oxidation of the labile iron pool.
Keywords: YggX, GshA, CyaY, sulfite reductase, cobalt
INTRODUCTION:
Microorganisms such as Salmonella enterica maintain a small pool of labile iron for use in various cellular processes (Petrat et.al., 2002). The size of this pool is tightly controlled to ensure sufficient levels of cellular iron for growth while preventing its accumulation, which can be toxic. During aerobic growth, iron can participate in a series of reactions that include the Fenton reaction, resulting in the formation of the highly reactive hydroxl radical that can damage DNA (Equ. 1–3) (Keyer & Imlay, 1996; Liochev & Fridovich, 1994; Srinivasan et.al., 2000). Growing evidence has linked a number of cellular factors including YggX, glutathione, and CyaY to a role in maintaining labile iron homeostasis and thus preventing the potential toxic effects of labile iron (Ding et.al., 2007; Gralnick & Downs, 2001; Gralnick & Downs, 2003; Thorgersen & Downs, 2008; Vivas et.al., 2006).
| [Equ. 1] |
| [Equ. 2] |
| [Equ. 3] |
Cells lacking YggX exhibit defects consistent with increased oxidative stress and defects in iron homeostasis. These defects have been attributed to a role for YggX in preventing superoxide stress (Thorgersen & Downs, 2008). The connection between YggX and superoxide occurs at several levels; yggX is a member of the Sox regulon (Pomposiello & Demple, 2000), and yggX mutant strains are sensitive to the superoxide generating compound paraquat (Gralnick & Downs, 2001). Strains lacking YggX have an increased GC-TA transversion mutation frequency indicative of Fenton chemistry (Gralnick & Downs, 2003), presumably generated by superoxide (Keyer & Imlay, 1996). Mutations in the yggX and gshA loci combine to result in decreased activity of Fe-S cluster enzymes and increased sensitivity to reactive oxygen species (Thorgersen & Downs, 2008).
The main cellular reductant, glutathione, is a free thiol that accumulates to millimolar levels in the cell (Helbig et al., 2008; Jocelyn, 1972). Apart from its role as a reductant, glutathione has been shown to chelate metals (Helbig et al., 2008; Li & Manning, 1955; Perrin & Watt, 1971; Sugiura & Tanaka, 1972). Studies have linked glutathione to a role in cobalt resistance (Freeman et al., 2005; Thorgersen & Downs, 2007), and we have proposed glutathione acts as a chelator for the labile iron pool (Thorgersen & Downs, 2008). Mutants defective at the gshA locus are unable to synthesize glutathione and display several phenotypes consistent with a defect in labile iron homeostasis and oxidative stress including increased expression of Fur regulated genes, and decreased activity of Fe-S cluster proteins (Gralnick et al., 2000; Thorgersen & Downs, 2008).
CyaY is the prokaryotic homologue of frataxin, a protein that trafficks iron in the mitochondria of eukaryotes (Lutz et al., 2001; Puccio & Koenig, 2000; Yoon & Cowan, 2003). Several properties of CyaY support a role for this protein in labile iron trafficking. CyaY has an anionic surface to which both Fe2+ and Fe3+ bind (Nair et al., 2004). The Kd of Fe2+ binding to CyaY is approximately 4 μM (Bou-Abdallah et al., 2004). The iron binding properties of CyaY can be diminished by reducing agents and increased by oxidative stress. This may indicate a role for CyaY in iron homeostasis under conditions of oxidative stress (Ding et al., 2007). Double mutant strains lacking CyaY as well as either YggX or ApbC (a protein involved in Fe-S cluster synthesis) (Skovran & Downs, 2003), have decreased activity of the Fe-S cluster-containing protein NADH dehydrogenase complex I (NADH-1), suggesting Fe-S cluster metabolism is compromised in these strains (Vivas et al., 2006).
Cobalt can be toxic by competing with iron at various metabolic loci (Thorgersen & Downs, 2007). Exposure of a wild-type Salmonella strain to 160 μM cobalt generated a reduced sulfur requirement that was eliminated by iron supplementation but not by anoxic growth (Thorgersen & Downs, 2007). This requirement was traced to a defect in the enzyme sulfite reductase that contains two iron cofactors, a 4Fe-4S cluster and siroheme (Christner et al., 1983). The cobalt dependent effect on sulfite reductase was attributed to competition between iron and cobalt at the enzyme uroporphyrinogen III methylase (CysG), that inserts either iron or cobalt into Factor II to produce either siroheme or an intermediate in cobalamin synthesis, respectively (Fazzio & Roth, 1996; Goldman & Roth, 1993; Spencer et al., 1993; Stroupe et al., 2003; Thorgersen & Downs, 2007). Thus, CysG was identified as an enzyme that was sensitive to the manipulation of cellular iron pools. Low siroheme production by CysG compromises sulfite reductase (CysJI) and can generate a nutritional requirement for reduced sulfur (Thorgersen & Downs, 2007).
This study was initiated by identifying nutritional requirements of mutant strains and tracing them to the respective target enzymes. Specifically, sulfite reductase was identified as a target sensitive to oxidative stress via defects in labile iron homeostasis. Supplementing the growth medium with the defined nutritional requirements resulted in restoration of growth to the mutant strains. This supplementation resulted in cells with wild-type growth but decreased overall fitness, which could be uncovered by exposure to an external stress. In depth analysis of the effects caused by multiple combinations of perturbations to labile iron homeostasis has provided insights into this complex metabolic system.
METHODS:
Bacterial strains, media, and chemicals.
All strains used in this study are derived from S. enterica LT2 and are listed with their respective genotypes in Table 1. Standard genetic techniques were used to construct and verify multiply mutant strains. The NCE medium of Berkowitz et al. (Berkowitz et al., 1968) supplemented with 1 mM MgSO4 was used as minimal medium. Glucose (11 mM) or gluconate (11 mM) was provided as the sole carbon source. Where specified, additional sulfur sources were added to a concentration of 0.3 mM in addition to the 1 mM MgSO4 already present. Thiamine was used at a concentration of 100 nM. In an effort to control metal concentrations, all minimal media was made using Milli-Q filtered water (MQH2O)and culture tubes were used a single time. Difco nutrient broth (NB) (8 g L−1) with NaCl (5 g L−1) was used as rich media, with Difco BiTek agar added to a final concentration of 1.5% for solid media. Bismuth sulfite agar was purchased from Becton, Dickinson and Co., Sparks, MD (BD). The growth medium for nitrite reductase assays contained per liter; 2 g peptone (BD), 7 g KH2PO4, 0.05 g NH4Cl, 0.1 g NaNO2, and 11 mM glucose. The FeCl3 and CoCl2 were added to growth medium after sterilization at indicated amounts from a 0.1 M stock in 0.1 N HCl and a 0.2 M stock in MQH2O respectively. The final concentrations of antibiotics were as follows: tetracycline, 20 μg mL−1; kanamycin, 50 μg mL−1; gentamycin 6 μg mL−1; and chloramphenicol, 20 μg mL-1. All other chemicals were purchased from Sigma Chemical Co., St. Louis, Mo.
TABLE 1.
Bacterial strains
| Strain | Genotype |
|---|---|
| DM10000........... | Wild type |
| DM9673............. | yggX::Gm* |
| DM9680............. | ghsA101::Tn10d(Tc)† |
| DM9684............. | cyaY::Cm* |
| DM9715............. | ghsA101::Tn10d(Tc) cyaY::Cm |
| DM9716............ | yggX::Gm cyaY::Cm |
| DM9720............ | yggX::Gm ghsA101::Tn10d(Tc) |
| DM9742............ | yggX::Gm ghsA101::Tn10d(Tc) cyaY::Cm |
| DM10052.......... | ΔryhB1 acnA::Kn |
| DM10056.......... | yggX::Gm ΔryhB1 acnAE::Kn |
| DM10057.......... | ghsA101::Tn10d(Tc) ΔryhB1 acnA::Kn |
| DM10062.......... | cyaY::Cm ΔryhB1 acnA::Kn |
| DM10063.......... | yggX::Gm ghsA101::Tn10d(Tc) ΔryhB1 acnA::Kn |
| DM10064.......... | yggX::Gm cyaY::Cm ΔryhB1 acnA::Kn |
| DM10080.......... | ghsA101::Tn10d(Tc) cyaY::Cm ΔryhB1 acnA::Kn |
| DM10081 | yggX::Gm ghsA101::Tn10d(Tc) cyaY::Cm ΔryhB1 acnA::Kn |
| DM10287.......... | cysG1510::Tn10(Tc) |
| DM11054‡.......... | sodA101::Cm sodB111::Kn |
All strains were part of the lab stock or were generated for this study.
::Gm, ::Cm, ::Kn designations reflect insertion/deletions made by the method of Datsenko (Datsenko & Wanner, 2000).
Tn10d refers to the transposition-defective mini-Tn10 (Tn10Δ16Δ17) (Way et.al., 1984).
sodA101 and sodB111 alleles were obtained from J. Slauch (University of Illinois at Urbana-Champaign).
Growth analysis:
Growth was quantified in liquid medium, using three independent cultures. Strains were grown overnight at 37 °C in NB medium, harvested and resuspended in an equal volume of saline, and 100 μL inoculated into 5 mL of the appropriate medium in 18 X 150 mm culture tubes. Cultures were placed in an air shaker at 37 °C and growth was monitored by following the optical density at 650 nm (OD650) on a Bausch & Lomb Spectronic 20. The starting OD650 was routinely between 0.02 and 0.07 for growth curves.
Cobalt Minimal Inhibitory Concentration Determination:
Using the growth methods described above, each strain was grown in minimal glucose medium with the indicated additions at several different concentrations of CoCl2 ranging from 0 to 60 μM. A graph of CoCl2 concentration versus OD650 after 10 hrs growth was generated for each strain, and the linear portion of the toxicity curve was used to determine a minimal inhibitory concentration (MIC) for cobalt. The concentration of CoCl2 at which the cultures grew to an OD650 of 0.3 was defined as the MIC. Reported values represent the average and standard deviation for three independent cultures.
Sulfide production.
Sulfide production was used as an indirect measure of sulfite reductase activity when cobalt was involved. This was necessary since the detection of sulfide in the sulfite reductase assay is inhibited by cobalt. Overnight 2 mL NB cultures were grown at 37 °C. A two microliter aliquot of the overnight culture was poked into a bismuth sulfite plate containing 0.3 mM cysteine (Becton, Dickinson and Co., Sparks, MD) prepared with increasing amounts of CoCl2 (50 – 250 μM) (Goldman & Roth, 1993). After a 12 hr incubation at 37 °C the diameter of the black Bi2S3 zone surrounding the inoculation site and indicative of sulfide production was measured.
Enzyme assays:
Enzyme assays were performed with three independent cultures for each strain. Protein concentrations were determined using the Bradford assay (Bradford, 1976).
(i). Sulfite reductase assays.
Overnight 2 mL NB cultures grown at 37 °C were inoculated 200 μL into 5 mL minimal NCE glucose medium containing 0.15 mM djenkolic acid dihydrocloride as the sole source of sulfur. Cultures were harvested and washed with 1X NCE minimal salts at an OD650 of 0.4–0.6. Cell pellets were resuspended in 300 μL of 0.05 M KPO4 buffer pH 7.7 and were lysed by sonication. Assays were performed using the method of Dreyfuss (Dreyfuss & Monty, 1963) detecting sulfide production over time. Sulfide was assayed colorimetrically by the method of Siegel (Siegel, 1965). Values are reported as specific activity (μmol sulfide produced min−1 mg−1 protein).
(ii). Nitrite reductase assays.
Whole cells were prepared from anaerobically grown cultures for nitrite reductase assays as described previously (Thorgersen, 2007). Assays used the method of Cole (Cole & Ward, 1973) detecting nitrite consumption over time. Nitrite was assayed colorimetrically by the method of Snell as modified by Cole (Cole & Ward, 1973; Snell & Snell, 1949).
(iii). Aconitase assays.
Overnight 2 mL NB cultures grown at 37 °C were inoculated 150 μL into 5 mL minimal NCE gluconate medium supplemented with 0.2% casamino acids, 100 nM thiamine, and 50 μM FeCl3 with or without 20 μM CoCl2. Cell free extracts were generated, and assays were performed as previously described (Skovran & Downs, 2000). Values are reported as specific activity (ΔA240 min−1 mg−1 protein [average ± standard deviation]).
(iv). NADH dehydrogenase complex I assays.
NADH dehydrogenase complex I (NDH-1) activity was assayed as an adaptation from Skovran and Zambrano described by Vivas (Skovran et al., 2004; Vivas et al., 2006; Zambrano & Kolter, 1993). Values are reported as specific activity (ΔA340 min−1 mg−1 protein).
(v). Succinate dehydrogenase assays.
Succinate dehydrogenase activity was assayed in the extracts generated for the aconitase assays above, as described previously (Skovran & Downs, 2000). Values are reported as specific activity (ΔA600 min−1 mg−1 protein).
RESULTS:
Loss of YggX, GshA, and/or CyaY generates a growth requirement for thiamine and/or reduced sulfur.
Growth analysis showed that lesions in the yggX, gshA and cyaY loci interact to generate and/or enhance nutritional requirements that indicate defects in iron homeostasis. Data in Table 2 showed that strains with a lesion in one locus grew with wild-type proficiency in minimal medium. Growth of double mutant strains lacking gshA was compromised; with the yggX gshA mutant showing no detectable growth on minimal medium. The addition of thiamine restored growth to this strain, as has been previously reported (Gralnick et al., 2000). Growth of the gshA cyaY double mutant was enhanced by thiamine, suggesting a similar but less severe cellular defect (data not shown). Introduction of a mutation in the third locus exacerbated the nutritional defect, resulting in a requirement for both thiamine and methionine to achieve full growth.
TABLE 2.
Strains lacking YggX, GshA, and CyaY exhibit growth defects in minimal glucose medium with cobalt*.
| Strain | Minimal | Thi | Met | Thi, Met | Co | Co, Thi | Co, Met | Co, Thi, Met |
|---|---|---|---|---|---|---|---|---|
| Wild type | 0.25 ± 0.03† | n.d.§ | n.d. | n.d. | 0.24 ± 0.03 | n.d. | n.d. | n.d. |
| yggX | 0.30 ± 0.03 | n.d. | n.d. | n.d. | 0.24 ± 0.05 | n.d. | n.d. | n.d. |
| gshA | 0.23 ± 0.02 | n.d. | n.d. | n.d. | 0.12 ± 0.02 | 0.24 ± 0.05 | 0.21 ± 0.02 | 0.25 ± 0.03 |
| cyaY | 0.26 ± 0.03 | n.d. | n.d. | n.d. | 0.25 ± 0.03 | n.d. | n.d. | n.d. |
| yggX gshA | N.G.‡ | 0.16 ± 0.02 | N.G. | 0.28 ± 0.02 | N.G. | 0.14 ± 0.04 | N.G. | 0.20 ± 0.02 |
| yggX cyaY | 0.27 ± 0.03 | n.d. | n.d. | n.d. | N.G. | 0.20 ± 0.02 | 0.29 ± 0.02 | 0.27 ± 0.03 |
| gshA cyaY | 0.17 ± 0.03 | n.d. | n.d. | n.d. | N.G. | 0.11 ± 0.03 | 0.16 ± 0.03 | 0.22 ± 0.02 |
| yggX gshA cyaY | N.G. | N.G. | N.G. | 0.27 ± 0.02 | N.G. | N.G. | N.G. | 0.25 ± 0.03 |
Cultures were grown at 37oC with shaking in minimal NCE glucose (11mM) medium. The following concentrations were used; thiamine (100 nM), CoCl2 (10 μM), and methionine (0.3 mM).
Values are averages of the specific growth rate (± standard deviations) of three independent cultures. Specific growth rate (μ) was calculated by the equation ln(X/X0)/T, where X is A650, X0 is A650 at time zero, and T is time (in hours).
N.G. No Growth
n.d. Not Determined
Growth of the triple mutant strain was tested under numerous conditions to evaluate the effects of oxygen and iron on the growth requirements for thiamine and methionine. Growth in standing cultures, which limits aeration of the culture, eliminated both the thiamine and methionine requirements of the triple mutant strain (Fig. 1(a)). This contrasted with the sulfur requirement induced by cobalt which was not corrected by anoxic growth (Thorgersen & Downs, 2007). When 1 mM FeCl3 was added, growth of the triple mutant strain in minimal glucose medium required thiamine but not methionine (Fig. 1(b)).
Figure 1:
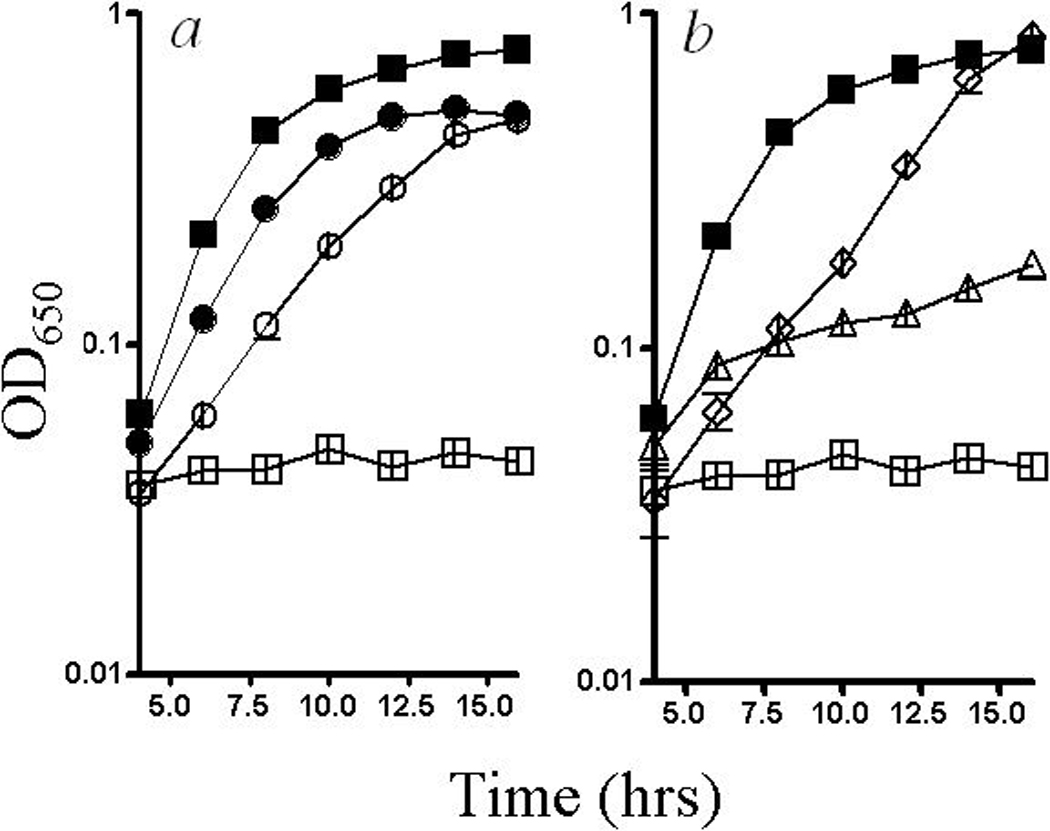
Growth defects in a yggX gshA cyaY strain. Wild type (filled) and yggX gshA cyaY (open) strains were grown in minimal glucose medium. a) Growth conditions were with (squares) or without (circles) aeration. b) Growth media included no additions (squares), thiamine and iron (diamonds), and methionine and iron (triangles).
At levels not affecting the wild type, cobalt potentiated growth defects in strains lacking YggX, GshA, and/or CyaY. Data in Table 2 showed that, while wild type was unaffected, 10 μM cobalt exacerbated the nutritional requirement(s) in the gshA mutant and eliminated growth in each of the multiply mutant strains. In the majority of strains, the addition of either thiamine or methionine restored significant growth. In both the yggX gshA mutant and the triple mutant strain, the addition of cobalt had little to no effect on the growth requirements, suggesting that in these strains the major target(s) of cobalt was damaged by consequences of the mutant background.
Growth medium impacts the minimal inhibitory concentration (MIC) of cobalt in the mutant strains.
The sensitivity of the mutant strains was characterized by defining the MIC of cobalt for the relevant strains in several media (Table 3). The MIC of cobalt was defined as the concentration of cobalt at which a strain was unable to grow to an OD650 of 0.3 within 10 hrs (see Materials and Methods). The data in Table 3 is complementary to the data in Table 2, since it allows conclusions about the fitness, or ability to survive additional perturbation by cobalt, of strains with similar growth rates. Of the single mutant strains, the gshA mutant was most sensitive to cobalt. This effect has been reported and it was hypothesized that some of the toxic effects of cobalt are mediated through depletion of the free thiol pool (Thorgersen & Downs, 2007). The ability of the double and triple mutant strains to withstand a cobalt challenge was significantly decreased. Although growth rate of the triple mutant strain in the presence of 10 μM cobalt was restored to wild-type levels by thiamine and methionine (Table 2), this strain had a lower MIC of cobalt in the presence of both nutrients, than do other strains with the same growth rate. One interpretation of this result is that cobalt provides an additional stress on the metabolic network that cannot be adapted to in the absence of one or more of the mutant loci.
TABLE 3.
Cobalt minimal inhibitory concentrations of mutant strains*.
| Strain | Relevant Genotype | Minimal | Thi | Met | Thi, Met |
|---|---|---|---|---|---|
| DM1000 | Wild type | 18.1 ± 0.2† | 20.4 ± 1.5 | 38.5 ± 0.8 | 43.6 ± 1.9 |
| DM9673 | yggX | 14.9 ± 1.2 | 19.0 ± 2.8 | 30.6 ± 3.3 | 44.8 ± 0.3 |
| DM9680 | gshA | 4.0 ± 1.7 | 9.3 ± 1.0 | 25.1 ± 0.7 | 36.5 ± 0.2 |
| DM9684 | cyaY | 17.2 ± 1.3 | 15.4 ± 1.0 | 43.1 ± 1.5 | 41.8 ± 2.5 |
| DM9720 | yggX gshA | N.G.‡ | 3.1 ± 3.1 | N.G. | 29.3 ± 1.9 |
| DM9716 | yggX cyaY | 4.0 ± 2.0 | 8.5 ± 0.7 | 20.0 ± 5.4 | 36.0 ± 0.7 |
| DM9715 | gshA cyaY | N.G. | 6.1 ± 0.3 | 13.8 ± 1.1 | 37.7 ± 1.4 |
| DM9742 | yggX gshA cyaY | N.G. | N.G. | N.G. | 13.1 ± 1.1 |
Cultures were grown at 37oC with shaking in minimal NCE glucose (11mM) medium. The following concentrations were used; thiamine (100 nM), and methionine (0.3 mM).
Values are the concentration of CoCl2 (μM) at which strains were unable to grow above an OD650 of 0.3 after 10 hrs.
NG: no growth, cultures failed to reach an OD650 of 0.3 in minimal glucose medium with no CoCl2.
The methionine requirement linked to sulfite reductase activity.
Exogenous cobalt impacts sulfite reductase activity (and thus methionine synthesis) by decreasing siroheme synthesis and impacting Fe-S cluster occupancy (Thorgersen & Downs, 2007). Sulfite reductase was assayed to extend our understanding of the interplay between the mutant loci and cobalt (Table 4). Among the single mutants, only the lack of gshA affected sulfite reductase activity, decreasing it by ~50%. Among the double mutants the yggX gshA strain showed a defect significantly different than the relevant parental strains. However, the triple mutant strain again had a severe defect, with 15 % activity found in the wild type. When standing conditions were used to decrease aeration of the growing cultures, the sulfite reductase activity of wild type and the triple mutant strain were indistinguishable (5.5 ± 0.1 and 5.6 ± 0.1 μM sulfide produced min−1 mg−1 protein respectively). Consistently, the activity of another siroheme containing enzyme, nitrite reductase, in cells grown under anoxic conditions was no different between the triple mutant strain and wild type. Thus the mutant effects were negated by lowering the oxygen content of the cultures, which distinguished the mutant effects from those caused solely by cobalt, which were not corrected by anoxic growth conditions (Thorgersen & Downs, 2007).
TABLE 4.
Sulfite reductase activity of mutant strains.
| Strain | Relevant genotype | Sulfite reductase activity |
|---|---|---|
| DM10000 | Wild type | 7.1 ± 0.9* (1.00)† |
| DM9673 | yggX | 6.0 ± 1.2 (0.85) |
| DM9680 | gshA | 3.2 ± 0.6 (0.46) |
| DM9684 | cyaY | 5.7 ± 1.0 (0.81) |
| DM9720 | yggX gshA | 1.9 ± 0.3 (0.27) |
| DM9716 | yggX cyaY | 5.2 ± 0.9 (0.74) |
| DM9715 | gshA cyaY | 2.8 ± 0.5 (0.40) |
| DM9742 | yggX gshA cyaY | 1.1 ± 0.4 (0.15) |
Values are specific activity (μmol sulfide produced min−1 mg−1 protein [average ± standard deviation]) of three independent cultures.
Values are relative activity obtained by dividing the activity of the relevant strain by the activity of the wild-type parent.
Bismuth sulfite agar plates were used to assess the sensitivity of sulfite reductase to cobalt in the mutant strains (Fig. 2). The black zone surrounding the inoculation site (indicative of sulfide production) of all the strains was measured in bismuth sulfite plates containing cysteine and increasing concentrations of cobalt after 24 hours. Sulfide production in the triple mutant strain was low with no cobalt present, and undetectable with the lowest concentration of cobalt used (50 μM). All strains showed a decrease in sulfide production caused by increasing cobalt, but in general the strains lacking cyaY showed the largest effect by cobalt. In other conditions tested in this study, the loss of CyaY alone did not have a visible effect, however, CyaY appeared to be required for a normal response to a cobalt challenge as measured by sulfide production. This result could indicate that the role of CyaY allows some level of metal specificity that impacts the iron containing cofactors of sulfite reductase, and supports the general model that CyaY is involved in iron trafficking in vivo. A cysG mutant is unable to make siroheme. As expected this strain had no sulfite reductase activity and provided a control for any other cellular activities that might produce sulfide.
Figure 2:
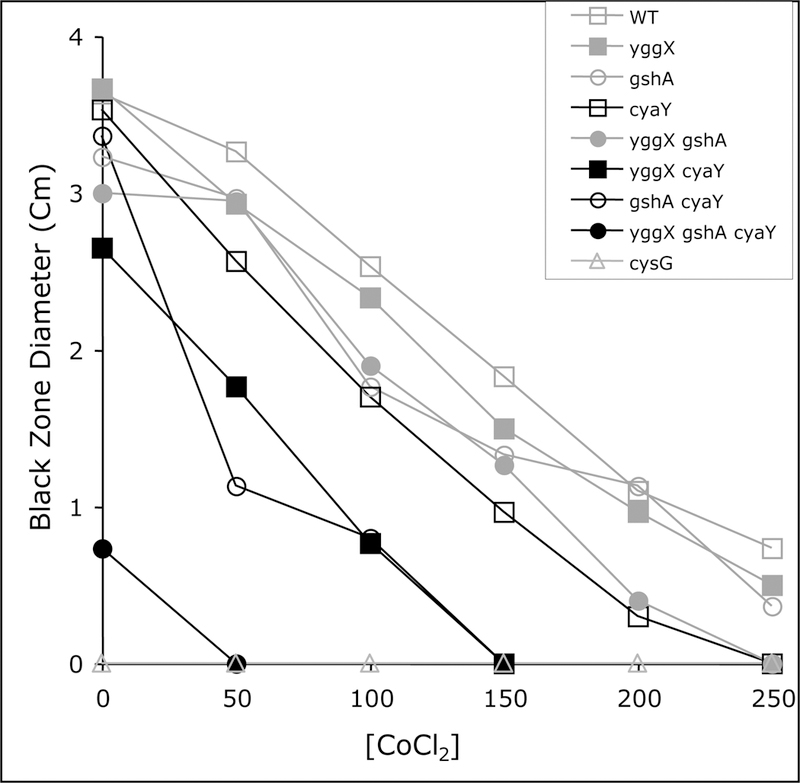
Cobalt decreases sulfide production. Sulfide production was measured indirectly in strains lacking YggX, GshA, and/or in the presence of cobalt. A 2 μl sample of an overnight NB culture of the indicated strain was pierced into bismuth sulfite plates supplemented with 0.3 mM cysteine containing increasing amounts of CoCl2. The diameter of the black zone radiating from the spot was measured after 24 h of incubation at 37°C, as an indication of sulfide production.
Superoxide dismutase mutants in Salmonella enterica.
Labile iron homeostasis and oxidative stress are intertwined, such that disruption of the labile iron pool can result in oxidative stress, and oxidative stress can disrupt the labile iron pool. Therefore, if the triple mutant strain was experiencing oxidative stress, the nutritional requirements of the strain should be replicated by exposing a wild-type strain to oxidative stress. A S. enterica strain lacking both cytoplasmic superoxide dismutases (sodA sodB) was constructed and the nutritional requirements were assessed. The sodA sodB strain failed to grow in minimal glucose medium aerobically, but grew anaerobically. Supplementation with multiple combinations of amino acids including those that satisfy the growth requirement of the equivalent E. coli strain (Carlioz & Touati, 1986) (branch chain, sulfur containing, and aromatic) failed to restore significant growth. NB and minimal glucose supplemented with casamino acids (0.2%) supported growth of the sodA sodB mutant strain with growth rates of 0.23 and 0.32 respectively. However, the mutant had an extended growth lag (3–6 hr) compared to wild type. The sodA sodB mutant in our S. enterica background appeared to experience an amount of superoxide stress that would prevent the analysis of subtle nutritional requirements, and this mutant was not further characterized herein.
Superoxide can generate similar effects as the loss of YggX, GshA, and CyaY.
A wild-type Salmonella strain was exposed to increasing concentrations of the superoxide generating compound paraquat to determine if the hierarchy of nutritional requirements seen with the mutant strains appeared. (Fig. 3(a)). Exposure to 4 μM paraquat effectively prevented growth of wild-type S. enterica in minimal glucose medium. If methionine and thiamine were added to the growth medium, the cells attained significant growth in the presence of 4 μM paraquat. The effect of thiamine on growth was significant and had previously been noted (Dougherty & Downs, 2006), but methionine alone had only a slight effect.
Figure 3:
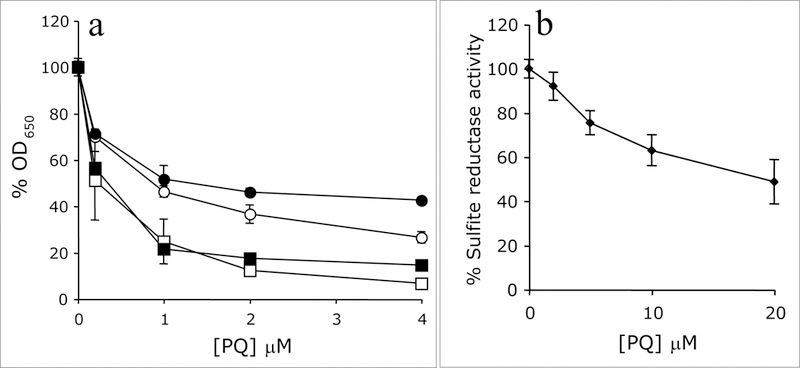
Superoxide generates nutritional requirements and decreases sulfite reductase activity. a) Wild-type Salmonella enterica was grown in the presence of increasing concentrations of paraquat in defined media. Media was with (filled) or without (open) methionine and with (circles) or without (squares) thiamine. Thus, strains represented with open squares contained no supplements and those with filled circles contained both. Growth is reported as the percent growth compared to growth in the absence of paraquat after a 24 hr incubation period. b) The specific activity of sulfite reductase in cell-free extracts of wild-type cells grown in defined medium with increasing concentrations of paraquat is represented.
Sulfite reductase activity was assayed from wild-type cells grown in the presence of increasing concentrations of paraquat (Fig. 3(b)). As the concentration of paraquat increased, the specific activity of sulfite reductase decreased. At 20 μM paraquat, the activity of sulfite reductase in a wild-type strain was 48.8% of the activity seen for cells grown without paraquat suggesting that either directly or indirectly, superoxide stress could damage sulfite reductase. The effect of paraquat on the methionine growth requirement was subtle, indicating that effects of superoxide on sulfite reductase activity were not large enough to generate a growth requirement. This scenario was supported by the relatively weak effect of PQ on the activity of sulfite reductase in vitro. The idea that several enzymes could have significantly lowered activity, but not result in a nutritional requirement led us to test several different Fe-S cluster proteins that may have distinguishable defects in the triple mutant strain.
Fe-S cluster proteins are compromised in stains lacking YggX, GshA, and CyaY.
The enzyme sulfite reductase contains a buried 4Fe-4S cluster and siroheme cofactor both involved in electron transfer (Christner et al., 1983). The enzyme ThiH, which has previously been linked to the thiamine auxotrophy of a yggX gshA mutant strain (Gralnick et al., 2000; Martinez-Gomez et al., 2004), is a member of the SAM radical superfamily of proteins characterized by a solvent-exposed oxygen-labile 4Fe-4S cluster (Berkovitch et al., 2004; Kriek et al., 2007; Layer et al., 2003). The activities of three other Fe-S cluster-containing proteins with different properties were assayed in the mutant strain backgrounds to understand the depth and impact these mutations have on Fe-S cluster/iron metabolism. Significantly each enzyme seemed to be affected differently.
Aconitase B is a member of the dehydratase family of proteins characterized by a solvent exposed 4Fe-4S cluster involved in substrate binding (Gardner & Fridovich, 1991). These clusters are known to be sensitive to multiple forms of oxidative stress (Flint et al., 1993; Jang & Imlay, 2007). Aconitase B was assayed in a ryhB acnA background to negate regulatory effects of Fur in the mutant strains, and eliminate activity of the oxygen stable aconitase A protein (Masse & Gottesman, 2002; Thorgersen & Downs, 2007). Western hybridization experiments determined that similar levels of AcnB protein were present in all strains. The activity of AcnB in the mutant backgrounds displayed different properties than the activity of sulfite reductase (Table 5). The gshA single mutant was not overly decreased in activity compared to the other single mutant strains, and the relative aconitase activity in the triple mutant strain compared to wild type was significantly higher (41% as opposed to 15%) than with other enzymes.
TABLE 5.
Strains lacking YggX, GshA, and CyaY have decreased activity of Fe-S cluster enzymes.
| Strain | Relevant genotype* | AcnB activity | SDH activity | NDH-1 activity |
|---|---|---|---|---|
| DM10052 | wild type | 3950 ± 331† (1.00)‡ | 2850 ± 254 (1.00) | 4470 ± 327 (1.00) |
| DM10056 | yggX | 2780 ± 241 (0.70) | 2150 ± 148 (0.75) | 3460 ± 289 (0.78) |
| DM10057 | gshA | 3020 ± 158 (0.76) | 1620 ± 421 (0.57) | 2810 ± 167 (0.63) |
| DM10062 | cyaY | 2660 ± 212 (0.67) | 2100 ± 245 (0.74) | 4090 ± 379 (0.92) |
| DM10063 | yggX gshA | 1510 ± 111 (0.38) | 550 ± 154 (0.19) | 970 ± 223 (0.22) |
| DM10064 | yggX cyaY | 2100 ± 113 (0.53) | 1400 ± 76 (0.49) | 2060 ± 204 (0.46) |
| DM10080 | gshA cyaY | 1790 ± 193 (0.45) | 1160 ± 131 (0.41) | 1990± 300 (0.45) |
| DM10081 | yggX gshA cyaY | 1640 ± 192 (0.41) | 690 ± 287 (0.24) | 320 ± 163 (0.07) |
all strains are lacking ryhB acnA
Values are specific activity (ΔA min−1 mg−1 protein [average ± standard deviation]) of three independent cultures.
Values are relative activity obtained by dividing the activity of the relevant strain by the activity of the ryhB acnA parent.
The NADH dehydrogenase complex 1(NDH-1), encoded by the nuo genes, contains nine Fe-S clusters and is part of the electron transport chain (Friedrich & Bottcher, 2004; Hinchliffe & Sazanov, 2005). Succinate dehydrogenase (SDH) contains three Fe-S clusters (Yankovskaya et al., 2003). Both succinate dehydrogenase (SDH) and NDH-1 contain buried Fe-S clusters involved in electron transfer, like sulfite reductase. Unlike sulfite reductase, SDH and NDH-1 do not contain siroheme. The activities of SDH, NDH-1, like sulfite reductase, were lowest in the gshA mutant strain as compared to in the other single mutant strains. Significantly, only NDH-1 had lower activity in the triple mutant strain as compared to the yggX gshA strain, indicating CyaY may play a significant role in the maturation of this complex when other components of the system are compromised.
DISCUSSION:
Labile iron homeostasis and oxidative stress can be viewed as a cycle. Perturbations that increase labile iron result in oxidative stress through Fenton chemistry. Likewise, oxidative stress damages oxygen labile Fe-S cluster proteins increasing intracellular labile iron levels, resulting in Fenton chemistry (Keyer & Imlay, 1996). A yggX gshA cyaY triple mutant strain displays phenotypes including nutritional requirements and decreased activity of iron containing enzymes, which indicate that this strain experiences increased oxidative stress and has defects in iron homeostasis.
Requirements for thiamine and methionine are generated by distinct mechanisms.
The nutritional requirements of the triple mutant strain (thiamine and methionine) result from distinct targets that display different sensitivities to oxidative stress and disruption of the iron pool. The thiamine requirement appears to be directly caused by oxidative stress, and the methionine requirement appears to be caused by disruption of iron homeostasis, which can be caused indirectly by oxidative stress. In support of this model, both requirements in the triple mutant strain can be suppressed by anoxic growth (Fig. 1(a)), but iron supplementation specifically suppressed the methionine requirement and not the thiamine requirement (Fig. 1(b)). Also, cobalt can disrupt iron homeostasis in the absence of oxygen, and was shown to cause a requirement for reduced sulfur but not thiamine in the absence of oxygen (Thorgersen & Downs, 2007). Further, cobalt generated a thiamine requirement only in strains lacking either YggX or GshA, which have increased oxidative stress (Gralnick & Downs, 2001; Thorgersen & Downs, 2008). Consistently, the superoxide generating compound paraquat caused a strong thiamine requirement but only a slight demand for increased methionine (Fig. 3(a)).
The enzyme targets that are the cause of the nutritional requirements have properties that are consistent with the above hypothesis. The thiamine auxotrophy in a yggX gshA strain was previously observed and was linked to the 4Fe-4S cluster proteins ThiH and ThiC (Dougherty & Downs, 2006; Gralnick et al., 2000; Martinez-Gomez et al., 2004). Both ThiH and ThiC belong to the family of SAM radical proteins that contain a solvent-exposed oxygen-labile Fe-S cluster and have no activity when assayed in the presence of oxygen (Kriek et al., 2007; Leonardi & Roach, 2004; Martinez-Gomez & Downs, 2008). Proteins in this family are known to be sensitive to superoxide (Berkovitch et al., 2004; Layer et al., 2003). The methionine requirement was traced to sulfite reductase, which contains an Fe-S cluster buried within the structure of the protein (Christner et al., 1983). This enzyme retains activity when assayed in the presence of oxygen (Dreyfuss & Monty, 1963).
Insights into the roles of YggX, glutathione, and CyaY in iron homeostasis
Each of the single and multiple mutant strains disrupted in yggX, gshA, and/or cyaY displayed differences in nutritional requirements, sensitivity to cobalt, and enzyme activities that provide insights into the role of these components in cellular metabolism. Of the single mutant strains, the strain that cannot synthesize glutathione (gshA), displayed the most severe defects including sensitivity to cobalt and decreased activity in all the Fe-S cluster enzymes tested with the exception of AcnB. Loss of GshA sensitized the strain so that cobalt targeted both the methionine and thiamine nutritional requirements (Table 3). Our current model suggests glutathione participates as a labile iron chelator in the cell. In this scenario, the loss of glutathione would increase the pool of iron available for Fenton chemistry (Thorgersen & Downs, 2008). Such a role for glutathione would position it at the intersection of the labile iron pool and oxidative stress. Such a central role could explain the strong phenotypic consequences of a gshA mutation seen in this and other studies.
YggX has been proposed to impact labile iron homeostasis through an interaction with superoxide (Thorgersen & Downs, 2008). In this study, the yggX mutant strain was sensitive to cobalt in a thiamine specific manner. Our hypothesis that the thiamine requirement is directly linked with oxidative stress is consistent with the proposed model for YggX and this observation.
The defects associated with the cyaY mutant strain were subtle and consisted of a slight sensitivity to cobalt and decreased sulfite reductase activity in the presence of cobalt. Despite the apparently small impact, disruption of cyaY had significant effects in combination with gshA and/or yggX mutations emphasizing the complexity of the integrated system. In the absence of CyaY sulfide production was highly sensitive to cobalt. This result suggests that the presence of CyaY aided in metal selectivity, an observation consistent with the current model of CyaY as a labile iron trafficking protein.
Other Fe-S cluster protein targets of direct/indirect oxidative stress
The observation that several of the mutant strains remained sensitive to cobalt even when their nutritional requirements were satisfied indicated that these strains were less fit and suggested that additional targets were compromised in the mutant backgrounds. Consistently, the activities of the Fe-S cluster enzymes tested, AcnB, SDH, and NDH-1, were decreased in several of the mutant strains. Of these enzymes, AcnB retained the most activity in the mutant backgrounds. This result was unexpected since AcnB is a member of the dehydratase family of proteins and is known to contain a solvent-exposed 4Fe-4S cluster that is oxygen labile (Gardner & Fridovich, 1992).
The enzymes sulfite reductase, SDH, and NDH-1 have Fe-S clusters that are involved in electron transfer and that are buried within the protein (Christner et al., 1983; Friedrich & Bottcher, 2004; Hinchliffe & Sazanov, 2005; Yankovskaya et al., 2003). These enzymes retain activity in the presence of oxygen yet their activities in the mutant strains were significantly decreased. We propose that these enzymes are targets sensitive specifically to defects in labile iron homeostasis and that the observed sensitivity of these enzymes to oxidative stress is indirect, occurring via the labile iron pool. This conclusion is supported by the above discussion linking the methionine requirement to a disruption in iron homeostasis rather than oxidative stress since the methionine requirement was traced to the enzyme sulfite reductase. Sulfite reductase, SDH, and NDH-1 also share the characteristic that their activities are lower in the gshA strain as compared to the other single mutant strains.
The study of integrated metabolic systems can be complicated by the existence of indirect effects. Iron homeostasis in particular is difficult to study due to its connection to oxidative stress through Fenton chemistry. Studying multiple targets impacted by oxidative stress and perturbations to iron homeostasis in a yggX gshA cyaY strain has provided insights that will facilitate the dissection of connections linking iron homeostasis to the rest of the metabolic network.
ACKNOWLEDGEMENTS:
We would like to thank James Slauch for providing phage lysates containing the sodA and sodB insertion mutations. This work was supported by the competitive grants program of the National Science Foundation (MCB-0445654). Funds were also provided from a 21st Century Scientists Scholars Award from the J.M. McDonnell fund to DMD. M.P.T. was supported by a Biotechnology Traineeship from the NIH (T32 GM08349), and the Jerome J. Stefaniak Predoctoral Fellowship from the Department of Bacteriology.
REFERENCES:
- Berkovitch F, Behshad E, Tang KH, Enns EA, Frey PA & Drennan CL (2004). A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase. Proc Natl Acad Sci USA 101, 15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ, Roth J & Ames BN (1968). Procedure for identifying nonsense mutations. J Bacteriol 96, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM & Dennis Chasteen N (2004). Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J Mol Bio 341, 605–615. [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carlioz A & Touati D (1986). Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? Embo J 5, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner JA, Janick PA, Siegel LM & Munck E (1983). Mossbauer studies of Escherichia coli sulfite reductase complexes with carbon monoxide and cyanide. Exchange coupling and intrinsic properties of the [4Fe-4S] cluster. J Biol Chem 258, 11157–11164. [PubMed] [Google Scholar]
- Cole JA & Ward FB (1973). Nitrite reductase-deficient mutants of Escherichia coli K12. J Gen Microbiol 76, 21–29. [DOI] [PubMed] [Google Scholar]
- Datsenko KA & Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Yang J, Coleman LC & Yeung S (2007). Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J Biol Chem 282, 7997–8004. [DOI] [PubMed] [Google Scholar]
- Dougherty MJ & Downs DM (2006). A connection between iron-sulfur cluster metabolism and the biosynthesis of 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate in Salmonella enterica. Microbiol 152, 2345–2353. [DOI] [PubMed] [Google Scholar]
- Dreyfuss J & Monty KJ (1963). Coincident Repression of the Reduction of 3’-Phosphoadenosine 5’-Phosphosulfate, Sulfite, and Thiosulfate in the Cysteine Pathway of Salmonella Typhimurium. J Biol Chem 238, 3781–3783. [PubMed] [Google Scholar]
- Fazzio TG & Roth JR (1996). Evidence that the CysG protein catalyzes the first reaction specific to B12 synthesis in Salmonella typhimurium, insertion of cobalt. J Bacteriol 178, 6952–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH, Tuminello JF & Emptage MH (1993). The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem 268, 22369–22376. [PubMed] [Google Scholar]
- Freeman JL, Persans MW, Nieman K & Salt DE (2005). Nickel and cobalt resistance engineered in Escherichia coli by overexpression of serine acetyltransferase from the nickel hyperaccumulator plant Thlaspi goesingense. Appl Environ Microbiol 71, 8627–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T & Bottcher B (2004). The gross structure of the respiratory complex I: a Lego System. Biochim Biophys Acta 1608, 1–9. [DOI] [PubMed] [Google Scholar]
- Gardner PR & Fridovich I (1991). Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem 266, 19328–19333. [PubMed] [Google Scholar]
- Gardner PR & Fridovich I (1992). Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J Biol Chem 267, 8757–8763. [PubMed] [Google Scholar]
- Goldman BS & Roth JR (1993). Genetic structure and regulation of the cysG gene in Salmonella typhimurium. J Bacteriol 175, 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick J, Webb E, Beck B & Downs D (2000). Lesions in gshA (encoding gamma-L-glutamyl-L-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar typhimurium LT2. J Bacteriol 182, 5180–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick J & Downs D (2001). Protection from superoxide damage associated with an increased level of the YggX protein in Salmonella enterica. Proc Natl Acad Sci USA 98, 8030–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick JA & Downs DM (2003). The YggX protein of Salmonella enterica is involved in Fe(II) trafficking and minimizes the DNA damage caused by hydroxyl radicals: residue CYS-7 is essential for YggX function. J Biol Chem 278, 20708–20715. [DOI] [PubMed] [Google Scholar]
- Helbig K, Bleuel C, Krauss GJ & Nies DH (2008). Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol 190, 5431–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P & Sazanov LA (2005). Organization of iron-sulfur clusters in respiratory complex I. Science 309, 771–774. [DOI] [PubMed] [Google Scholar]
- Jang S & Imlay JA (2007). Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocelyn PC (1972). Biochemistry of the sulfhydryl group. New York: Academic Press. [Google Scholar]
- Keyer K & Imlay JA (1996). Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA 93, 13635–13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek M, Martins F, Leonardi R, Fairhurst SA, Lowe DJ & Roach PL (2007). Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J Biol Chem 282, 17413–17423. [DOI] [PubMed] [Google Scholar]
- Layer G, Moser J, Heinz DW, Jahn D & Schubert WD (2003). Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. Embo J 22, 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi R & Roach PL (2004). Thiamine biosynthesis in Escherichia coli: In vitro reconstitution of the thiazole synthase activity. J Biol Chem 279, 17054–17062. [DOI] [PubMed] [Google Scholar]
- Li NC & Manning RA (1955). Some metal complexes of sulfur-containing amino acids. J Am Chem Soc 77, 5225–5227. [Google Scholar]
- Liochev SI & Fridovich I (1994). The role of O2.- in the production of HO.: in vitro and in vivo. Free Radic Biol Med 16, 29–33. [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W & Herrmann JM (2001). The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol 307, 815–825. [DOI] [PubMed] [Google Scholar]
- Martinez-Gomez NC, Robers M & Downs DM (2004). Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. J Biol Chem 279, 40505–40510. [DOI] [PubMed] [Google Scholar]
- Martinez-Gomez NC & Downs DM (2008). ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry, IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E & Gottesman S (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99, 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair M, Adinolfi S, Pastore C, Kelly G, Temussi P & Pastore A (2004). Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure 12, 2037–2248. [DOI] [PubMed] [Google Scholar]
- Perrin DD & Watt AE (1971). Complex formation of zinc and cadmium with glutathione. Biochim Biophys Acta 230, 96–104. [DOI] [PubMed] [Google Scholar]
- Petrat F, de Groot H, Sustmann R & Rauen U (2002). The chelatable iron pool in living cells: a methodically defined quantity. Biol Chem 383, 489–502. [DOI] [PubMed] [Google Scholar]
- Pomposiello PJ & Demple B (2000). Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol 182, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccio H & Koenig M (2000). Recent advances in the molecular pathogenesis of Friedreich ataxia. Hum Mol Genet 9, 887–892. [DOI] [PubMed] [Google Scholar]
- Siegel LM (1965). A Direct Microdetermination for Sulfide. Anal Biochem 11, 126–132. [DOI] [PubMed] [Google Scholar]
- Skovran E & Downs DM (2000). Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar typhimurium: implications for thiamine synthesis. J Bacteriol 182, 3896–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovran E & Downs DM (2003). Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J Bacteriol 185, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovran E, Lauhon CT & Downs DM (2004). Lack of YggX results in chronic oxidative stress and uncovers subtle defects in Fe-S cluster metabolism in Salmonella enterica. J Bacteriol 186, 7626–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell FD & Snell CT (1949). Colorimetric Methods of Analysis, 3rd edn. New York: Van Nostrand. [Google Scholar]
- Spencer JB, Stolowich NJ, Roessner CA & Scott AI (1993). The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett 335, 57–60. [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Liba A, Imlay JA, Valentine JS & Gralla EB (2000). Yeast lacking superoxide dismutase(s) show elevated levels of “free iron” as measured by whole cell electron paramagnetic resonance. J Biol Chem 275, 29187–29192. [DOI] [PubMed] [Google Scholar]
- Stroupe ME, Leech HK, Daniels DS, Warren MJ & Getzoff ED (2003). CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat Struct Biol 10, 1064–1073. [DOI] [PubMed] [Google Scholar]
- Sugiura Y & Tanaka H (1972). Iron-sulfide chelates of some sulfur-containing peptides as model complex of non-heme iron proteins. Biophys Res Comm 46, 335–340. [DOI] [PubMed] [Google Scholar]
- Thorgersen MP & Downs DM (2007). Cobalt targets multiple metabolic processes in Salmonella enterica. J Bacteriol 189, 7774–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgersen MP & Downs D (2008). Analysis of yggX and gshA mutants provides insights on the labile iron pool in Salmonella enterica. J Bacteriol, accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas E, Skovran E & Downs DM (2006). Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J Bacteriol 188, 1175–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Davis MA, Morisato D, Roberts DE & Kleckner N (1984). New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32, 369–379. [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G & Iwata S (2003). Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704. [DOI] [PubMed] [Google Scholar]
- Yoon T & Cowan JA (2003). Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc 125, 6078–6084. [DOI] [PubMed] [Google Scholar]
- Zambrano MM & Kolter R (1993). Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol 175, 5642–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]


