Abstract
Background and objective
Direct-acting antivirals (DAA) facing resistance continue to be used in some areas worldwide. Thus, identifying hepatitis C virus (HCV) genotypes/subtypes and loci with certain prevalent resistance-associated substitutions (RASs) deserves attention. We investigated the global and regional frequencies of naturally occurring RASs among all confirmed HCV subtypes (n=86) and explored co-occurring and mutually exclusive RAS pairs within and between genes NS3, NS5A, and NS5B.
Methods
A total of 213,908 HCV sequences available as of July 10, 2019 were retrieved from the NCBI nucleotide database. After curation, 17,312 NS3, 8,478 NS5A, and 25,991 NS5B sequence fragments from DAA-naïve patients were screened for RASs. MEGA 6.0 was used to translate aligned nucleotide sequences into amino acid sequences, and RAS pairs were identified by hypergeometric analysis.
Results
RAS prevalence varied significantly among HCV subtypes. For example, D168E, highly resistanct to all protease inhibitors except voxilaprevir, was nearly absent in all subtypes except in 43.48% of GT5a sequences. RASs in NS3 exhibiting significantly different global distribution included Q80K in GT1a with the highest frequency in North America (54.49%), followed by in Europe (22.66%), Asia (6.98%), Oceania (6.62%), and South America (1.03%). The prevalence of NS3 S122G in GT1b was highest in Asia (26.6%) and lowest in Europe (2.64%). NS5A L28M, R30Q, and Y93H in GT1b, L31M in GT2b, and NS5B C316N in GT1b was most prevalent in Asia. A150V in GT3a, associated with sofosbuvir treatment failure, was most prevalent in Asia (44.09%), followed by Europe (31.19%), Oceania (24.29%), and North America (19.05%). Multiple mutually exclusive or co-occurring RAS pairs were identified, including Q80K+R155K and R155K+D168G in GT1a and L159F+C316N and R30Q (NS5A)+C316N (NS5B) in GT1b.
Conclusion
Our data may be of special relevance for those countries where highly effective antivirals might not be available. Considering the specific RASs prevalence will help the clinicians to make optimal treatment choices. The RASs pairs would benefit anti-HCV drug development.
Keywords: hepatitis C virus, direct-acting antiviral, resistance-associated substitution, subtype
Introduction
Infection with hepatitis C virus (HCV) is a global public health problem. Between 130 and 170 million people are HCV chronically infected1 and up to 4 million individuals are newly infected with HCV annually.2 Persistent HCV infection induces high risk for developing severe liver diseases, such as liver cirrhosis and hepatocellular carcinoma.2,3 HCV is an enveloped positive-sense single-stranded RNA virus whose replication can be robust; model-based calculations indicate production of 1012 virions/day.4,5 This high-level replication and a lack of viral RNA polymerase proofreading contribute to HCV’s genetic divergence. The virus is currently classified into seven major genotypes and 86 subtypes according to the International Committee on Taxonomy of Viruses (June 2017) (https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification) with ~30% divergence at the genotype level and ~15% divergence at the subtype level.6 HCV clearance, which is associated with reduced rates of de novo hepatocellular carcinoma,7 is strongly dependent on HCV genotype/subtype when induced by interferon-based antiviral therapy with sustained virologic response (SVR) rates of approximately 50–80%.8–10
HCV therapy has been revolutionized with the advent of direct-acting antivirals (DAA) that directly target HCV gene products, including NS3 protease inhibitors, NS5A inhibitors, nucleos(t)ide inhibitors (NI), and non-nucleoside inhibitors (NNI) of the NS5B RNA-dependent RNA polymerase.11 Generally, DAA based regimens yield highly promising SVR rates (>90%). However, virologic failure still occurs and has been associated with the emergence of HCV variants with resistance-associated substitutions (RASs), which impair drug susceptibility. Notably, RASs can occur naturally in a genotype/subtype-dependent manner before DAA-induced selective pressure occurs.12–17 For example, the SVR rate of daclatasvir/asunaprevir was severely attenuated due to baseline RASs (65.4% with RASs vs 94.3% without RASs).18 Moreover, due to the negative effects of RAS Q80K on the efficacy of simeprevir, clinical guidelines recommend pre-treatment screening in patients infected with HCV subtype GT1a.19,20 Thus, assessing the prevalence of naturally occurring RASs in different HCV genotypes/subtypes and determining their global geographic distribution will help optimize the selection of therapeutic regimens.
Several studies have assessed the prevalence of naturally occurring RASs in HCV genes NS3, NS5A, and/or NS5B from DAA-naïve patients but have focused on particular subtypes21–25 or have used HCV sequence databases26–28 containing a relatively small number of sequences covering very few subtypes. Welzel et al29 performed the largest study to date including 46 subtypes across 5 geographic regions, but the RAS distributions determined in that study were based on clinical trials from regional medical centers in primarily developed countries and thus may not reflect the global HCV RAS landscape.
To address this knowledge gap, the aims of our current study were to (1) investigate the global and regional prevalence of HCV RASs among all confirmed HCV subtypes (n=86) by mining HCV sequences in NCBI nucleotide database and Los Alamos HCV database, (2) explore the RASs pairs showing significant more or less co-occurrence.
Materials and methods
HCV datasets
HCV genomic sequences available as of July 10, 2019 were retrieved from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide) in GenBank (full) format using the following searching criteria: the title contained the words “hepatitis C virus” and the organism was “hepatitis C virus”. The following information was extracted for each sequence: accession number, times of sampling and publication, HCV genotype/subtype, geographic region, and treatment. If the above-mentioned parameters were not available, we extracted the information from publications linked with the sequences. A total of 213,908 HCV genomic sequences were ultimately retrieved. Only one sequence from any duplicate sets and the sequence obtained from the last visit for patients with multiple visits was retained for further analysis. Sequence exclusion criteria were as follows: (1) no NS3, NS5A, or NS5B fragments present; (2) low quality; (3) from non-human hosts; (4) different clone sequences from the same patient; (5) sequences that encoded non-functional proteins; (6) sequences without any available subtype information or with mixed-genotypes; (7) groups of sequences with ambiguous DAA treatment information (eg, “some DAA-treated patients”) that did not specify which patients/sequences were DAA-treated; and (8) sequences from DAA-treated patients. Sequences were confirmed to be from DAA-treated patients based on the NCBI database description and/or the linked publications. Finally, 17,312 NS3, 8,478 NS5A, and 25,991 NS5B sequences were retained for further analysis.
All nucleotide sequences were aligned using the Los Alamos HCV Database (LANL; http://hcv.lanl.gov/components/sequence/HCV/search/searchi.html), which also provided curated HCV subtype and geographic region information for some sequences. All sequences were aligned against the H77 reference sequence (GenBank accession no. NC_004102). The aligned nucleotide sequences in FASTA format were downloaded and then translated into their corresponding amino acid sequences with MEGA 6.0 software (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA) and manually checked and edited as necessary. The MEGA 6.0 output table was further analyzed with R (version 2.10.0) to calculate allele frequencies for each RAS. We focused only on the defined genomic regions relevant to drug resistance, including the first 630 amino acids in NS3, the first 100 amino acids in NS5A, and all 591 amino acids in NS5B.
RASs
RASs were defined by a combination of substitutions summarized in three review papers,30–32 and others recently reported associated with DAA treatment failure and/or conferred a ≥2-fold change in susceptibility compared with a reference strain via in vitro replicon assays.33–39
NS3 RASs included V36A/G/L/M, Q41K/R, F43C/I/L/S/V, T54A/S, V55A/I, Y56F/H/N, Q80G/H/K/L/R, S122D/G/N/R/T, S138T, R155C/G/I/K/L/N/Q/S/T/W, A156G/H/K/L/M/S/T/V, V158A, A166T, D168/A/C/E/F/G/H/I/K/L/N/Q/R/S/T/V/Y, I170T/V, and L175M.
NS5A RASs included K24/A/E/G/N/Q/R, S24F/H/T, Q24K/T, T24A/S, K26E, M28A/G/I/K/S/T/V, L28A/F/I/M/S/T/V, F28C/M/S/V,Q30D/E/G/H/I/K/L/N/R/S/T/Y, R30C/E/G/H/K/N/Q/S/T, A30G/H/K/V, L30A/F/G/H/Q/R/S, L31F/I/M/P/V/W, M31F/I/L/V, P32A/L/Q/R/S, S38F, Q54H, H58D/L/N, P58A/D/G/L/R/S/T, T58A/D/G/H/L/N/S, E62D/L, A92K/P/T, C92A/K/N/R/S/T, E92K, Y93C/F/H/I/L/N/R/S/T/W, and T93A/H/I/N/S.
NS5B RASs included A150V, L159F, G188D, K206E, E237G, N244I, S282G/R/T, M289I/L, L314H, C316F/H/N/Y, L320F, V321A/I, S368T, A395G, N411S, M414I/T/V, N444K, C445F, E446K/Q, Y448C/H, C451S, A553T/V, G554S, S556G/N/R, G558R, D559G/N, Y561H, and S565F.
Statistical analysis
Differences in RAS prevalence among geographic regions were determined using Fisher’s exact test. Probabilities (P-values) of observing a pair of RASs together in no fewer or no greater than n sequences by random chance were calculated using the hypergeometric test. Statistical analyses were performed using R (version 2.10.0).
A P value <0.05 was considered to be statistically significant.
Results
Prevalence of naturally occurring NS3 RASs in 86 HCV subtypes
The prevalence of naturally occurring NS3 RASs in different HCV subtypes is shown in Table 1. Majority of NS3 RASs were absent or have very low frequencies (<0.5%), and only several RASs including Q80K, S122G/T/N and D168E were observed in a high rate of sequences in a subtype dependent manner (Figure S1A). The RAS Q80K confers low-level resistance to simeprevir in vitro and is associated with a reduced treatment response in vivo. We found Q80K-positive sequences in 31.74% of HCV subtype GT1a sequences (2277/7178) but only 1.14% of sequences in subtype GT1b (81/7176). This RAS was found frequently in subtypes GT1d (86.67%, 13/15), 5a (100%, 46/46), and 6a (98.28%, 402/409) but was very rare in subtype 3a (0.24%, 2/820). The RAS was also observed in 16.67% (1/6) of subtype 1i sequences. All GT4 and other GT1, 3, 5, 6, 7 subtypes harbored no Q80K-positive sequences. Q80R, which confers resistance to simeprevir/asunaprevir/faldaprevir, was rarely present in G1a (0.49%, 35/7178), G1b (0.3%, 21/7176), 3i (16.67%, 1/6), 4d (1.45%, 1/69), and 6a (0.24%, 1/409). Similarly, R155K, which carries variants associated with resistance to protease inhibitors such as simeprevir, asunaprevir, paritepravir, vaniprevir, and faldaprevir, was rarely present in 0.96% (69/7164) of GT1a, 16.67% (1/6) 1h, and 0.25% (2/805) 3a sequences. A156L/T/V, the only RASs conferred high resistance (>100-fold) to voxilaprevir (a potent pan-genotypic second generation of protease inhibitor), were not detected except in 1a, and 1b with frequencies <0.05%. The RASs at position D168, highly resistant to all protease inhibitors except voxilaprevir, were rare (approximately 1%) in nearly all HCV subtypes, whereas D168E occurred in 43.48% (20/46) of GT5a sequences. RASs at position 122, which confer resistance to simeprevir, asunaprevir and/or voxilaprevir in GT1a and/or GT1b, was highly prevalent in GT5a (122T, 73.9%), GT6a (122N, 76.3%) and GT1b (122G, 9.34%). S122R (confers resistance to simeprevir and asunaprevir) was exclusively detected in GT2 and with GT2 subtype-specific frequencies (i.e., 100% in 2b, 2c, 2d, 2e, 2f, 2i, 2j, 2l, 2m, 2q and 2t but only 1.89% (1/53) in 2a and 0% in 2r and 2u). These present of these RASs may limit the use of some inhibitors for treating the corresponding subtypes. V36L, associated with resistance to asunaprevir, paritepravir, and faldaprevir, was uncommon in GT1a (1.50%) and 1b (0.96%) but more frequent (13.33% to 100%) in five GT1 subtypes including 1d, 1e, 1g, 1i, and 1l. This RAS was also found in almost all GT2, 3, 4, 5, and 7 sequences, as well in several GT6 subtypes. Another asunaprevir/paritepravir/faldaprevir RAS, V36M, was only observed in GT1a (0.48%) and 1b (0.03%). T54S was infrequent in GT1a (3.02%) and 1b (2.01%). The frequency of V170A was extremely low (<0.1%) in GT1a or GT1b but significantly varied among other subtypes with respect to frequency. Lastly, three RASs (Q41R, F43L/S, Y56H) were only found in GT1a or GT1b and at an extremely low prevalence. (<0.1%)
Table 1.
(Continued).
| Sub-type | S122 DGNRT |
R155 CGIKLNQSTW |
A156 GHKLMSTV |
V158A | A166T | D168 Anyone |
I170 TV |
L175M |
|---|---|---|---|---|---|---|---|---|
| 1a | 6.43G,0.85N,0.18T,0.01R | 0.96K, 0.01M, 0.04T, 0.01S | 0.04T | 0.01 | . | 0.23E, 0.01G,0.01N | 5.49V,0.11T | 2.32 |
| 1b | 9.34G,4.46T,3.2N,0.04R | . | 0.01T | . | 0.03 | 0.67E, 0.03Q,0.01V | 66.1V,0.15T | 96.4 |
| 1c | 80N | . | . | . | . | . | 80V | . |
| 1d | 6.67T | . | . | . | . | . | 100V | 100 |
| 1e | 73.3N,20.0T | . | . | . | . | . | 93.3V | . |
| 1g | 20N | . | . | . | . | . | 100V | . |
| 1h | 16.7T | 16.67K | . | . | . | . | 100V | 100 |
| 1i | . | . | . | . | . | . | 100V | 100 |
| 1j | 100N | . | . | . | . | . | 100V | . |
| 1k | 100T | . | . | . | . | . | . | . |
| 1l | 22.2T, 11.1N |
. | . | . | . | . | 88.9V | . |
| 1m | 50T | . | . | . | . | . | . | . |
| 1n | . | . | . | . | . | . | . | 50 |
| 2a | 1.89R | . | . | . | . | . | 1.89V | 1.89 |
| 2b | 100R | . | . | . | . | . | 1.80V | . |
| 2c | 100R | . | . | . | . | . | . | . |
| 2d | 100R | . | . | . | . | . | . | . |
| 2e | 100R | . | . | . | . | . | . | . |
| 2f | 100R | . | . | . | . | . | . | . |
| 2i | 100R | . | . | 25 | . | . | . | . |
| 2j | 100R | . | . | . | . | . | . | . |
| 2k | 75R | . | . | . | . | . | 25V | 25 |
| 2l | 100R | . | . | . | . | . | 100V | . |
| 2m | 100R | . | . | . | . | . | . | . |
| 2q | 100R | . | . | . | . | . | . | . |
| 2r | 100T | . | . | . | . | . | . | . |
| 2t | 100R | . | . | . | . | . | . | . |
| 2u | . | . | . | . | . | . | . | . |
| 3a | 0.37T | 0.25K | 0.12G | . | 5.59T | 98.9Q,0.50R,0.13H/K | 4.41V | 0.13 |
| 3b | 0.73T | 5.07I, 4.35M | . | . | . | 100Q | . | . |
| 3d | . | . | . | . | . | 100Q | . | . |
| 3e | . | . | . | . | . | 100Q | . | . |
| 3g | . | . | . | . | . | 100Q | . | . |
| 3h | . | . | . | . | . | 100Q | . | . |
| 3i | . | . | . | . | . | 100Q | 100V | . |
| 3k | . | . | . | . | . | 100Q | . | . |
| 4a | 96.7T | . | . | . | . | . | 90.6V | . |
| 4b | 50T | . | . | . | . | . | 100V | . |
| 4c | 100T | . | . | . | . | . | 100V | . |
| 4d | 95.7T | . | . | . | 1.75 | . | 98V | . |
| 4f | 100T | . | . | . | . | . | 100V | . |
| 4g | 100T | . | . | . | . | . | 100V | . |
| 4k | 100T | . | . | . | . | . | 100V | . |
| 4l | 100T | . | . | . | . | . | 100V | . |
| 4m | . | . | . | . | . | . | 75V | . |
| 4n | 100T | . | . | . | . | . | 100V | . |
| 4o | 100T | . | . | . | . | . | 100V | . |
| 4p | 100T | . | . | . | . | . | 100V | . |
| 4q | 100N | . | . | . | . | . | 100V | . |
| 4r | 100T | . | . | . | . | . | 100V | . |
| 4s | 100N | . | . | . | . | 100T | 100V | . |
| 4t | 100T | . | . | . | . | . | 100V | . |
| 4v | 75N | . | . | . | . | . | 100V | . |
| 4w | 100T | . | . | . | . | . | 100V | . |
| 5a | 73.9T | . | . | . | . | 43.5E | 8.7V | . |
| 6a | 76.3N,0.49D/G | . | . | . | 0.49 | 1.71E | 0.24V | 100 |
| 6b | 100T | . | . | . | . | . | . | 100 |
| 6c | . | . | . | . | . | . | 100V | 100 |
| 6d | 100T | . | . | . | . | . | 100V | 100 |
| 6e | 80T | . | . | . | . | . | 100V | 100 |
| 6f | 100T | . | . | . | . | . | 100V | 100 |
| 6g | 100N | . | . | . | . | 50E | 100V | 100 |
| 6h | 100N | . | . | . | . | . | 80V,20A | 100 |
| 6i | . | . | . | . | . | . | 100V | 100 |
| 6j | 100N | . | . | . | . | . | 100V | 100 |
| 6k | 100T | . | . | . | . | . | 100V | 100 |
| 6l | 25N,25T | . | 12.5V | . | . | . | 100V | 100 |
| 6m | 100T | . | . | . | . | . | . | 100 |
| 6n | 100N | . | . | . | . | . | 100V | 100 |
| 6o | 100T | . | . | . | . | . | 100V | 100 |
| 6p | 100T | . | . | . | . | . | 100V | 100 |
| 6q | 100T | . | . | . | . | . | 100V | 100 |
| 6r | 100T | . | . | . | . | . | 100V | 100 |
| 6s | 66.7T | . | . | . | . | . | 100V | 100 |
| 6t | 100T | . | . | . | . | . | 100V | 100 |
| 6u | 100T | . | . | . | . | . | 100V | 100 |
| 6v | 100T | . | . | . | . | . | 100V | 100 |
| 6w | . | . | . | . | . | . | 100V | 100 |
| 6xa | 100T | . | . | . | . | . | 100V | 100 |
| 6xb | 100T | . | . | . | . | . | 50V | 100 |
| 6xc | 100T | . | . | . | . | . | . | 100 |
| 6xd | 66.7N | . | . | . | . | . | 100V | 100 |
| 6xe | 100T | . | . | . | . | . | 100V | 100 |
| 6xf | 100T | . | . | . | . | . | 100V | 100 |
| 7a | 100G | . | . | . | . | 100Q | 100V | 100 |
| 7b | . | . | . | . | . | 100Q | . | . |
Notes: Data are presented as %. The dot denotes 0%. aNot all the positions were analyzed using the same number of sequences. The first figure is the lowest number, and the second figure is the largest number. n.a.: not applicable because of different natural amino acid sequences in the respective HCV genotype/subtype (NS3 V170 and M175 are the dominant amino acids in GT1b).
Table 1.
Prevalence of naturally occurring NS3 RASs in 86 HCV subtypes
| Sub-type | No. of seqa | V36 AGLM |
Q41 KR |
F43 CILSV |
T54 AS |
V55AI | Y56 FHN |
Q80 GHKLR |
|---|---|---|---|---|---|---|---|---|
| 1a | 7071, 7180 | 1.50L, 0.48M, 0.01A/G | 0.01R | 0.01L, 0.01S | 3.02S, 0.01A | 2.25A,1.82I | 0.67F,0.06H | 31.74K,1.14L,0.49R,0.01G,0.01H |
| 1b | 6849, 7133 | 0.96L, 0.03M, 0.01A | 0.04R,0.03K | 0.04L | 2.01S | 0.44A,021I | 26F | 3.68L,1.14K,0.30R,0.03H,0.01G |
| 1c | 5 | . | . | . | . | . | . | . |
| 1d | 15 | 13.33L | . | . | 66.7S | . | 6.67F | 86.67K |
| 1e | 15 | 100L | . | . | 100S | 6.67I | . | . |
| 1g | 5 | 20L | . | . | 80S | . | . | . |
| 1h | 6 | . | . | . | . | . | . | . |
| 1i | 6 | 16.67L | . | . | . | . | 16.7F | 16.67K |
| 1j | 1 | . | . | . | . | . | . | . |
| 1k | 1 | . | . | . | . | . | . | . |
| 1l | 9 | 22.22L | . | . | . | . | 11.1F | 77.8L |
| 1m | 2 | . | . | . | . | . | . | . |
| 1n | 2 | . | . | . | . | . | 50F | 50L |
| 2a | 53 | 98.11L | . | . | . | . | 3.77F | 98.1G,1.89Q |
| 2b | 111 | 100L | . | . | . | . | 3.6F | 100G |
| 2c | 9, 46 | 100L | . | . | . | . | 100F | 100G |
| 2d | 1 | 100L | . | . | . | . | 100F | 100G |
| 2e | 1 | 100L | . | . | 100A | . | 100F | 100G |
| 2f | 2 | 100L | . | . | . | . | 100F | 100G |
| 2i | 4 | 100L | . | . | . | . | 100F | 100G |
| 2j | 6 | 83.3L,16.7M | . | . | . | . | 100F | 100G |
| 2k | 4 | 75L | . | . | . | . | 75F | 75G |
| 2l | 4 | 100L | . | . | . | . | . | 100G |
| 2m | 4 | 100L | . | . | . | . | 100F | 100G |
| 2q | 2 | 100L | . | . | . | . | 100F | 100G |
| 2r | 1 | 100L | . | . | . | . | 100F | 100G |
| 2t | 1 | 100L | . | . | . | . | 100F | 100G |
| 2u | 1 | 100L | . | . | . | . | . | 100G |
| 3a | 780, 820 | 99.6L | . | . | 0.24S | 0.24I | 0.24F | 0.24K |
| 3b | 138 | 100L | . | . | 1.45S | . | . | . |
| 3d | 1 | 100L | . | . | . | . | . | . |
| 3e | 1 | 100L | . | . | . | . | . | . |
| 3g | 2 | 100L | . | . | . | . | . | . |
| 3h | 2 | 100L | . | . | . | . | . | . |
| 3i | 6 | 100L | . | . | . | . | . | 16.67R |
| 3k | 2 | 100L | . | . | . | . | . | . |
| 4a | 83, 92 | 97.8L | . | . | 6.52S | . | . | . |
| 4b | 2 | 100L | . | . | . | . | . | . |
| 4c | 1, 2 | 100L | . | . | . | . | . | . |
| 4d | 49, 69 | 100L | . | . | . | . | . | 1.45R |
| 4f | 11 | 100L | . | . | . | . | . | . |
| 4g | 3 | 100L | . | . | . | . | . | . |
| 4k | 4 | 100L | . | . | . | . | . | . |
| 4l | 3 | 100L | . | . | . | . | . | . |
| 4m | 4, 5 | 100L | . | . | . | . | . | . |
| 4n | 3, 4 | 100L | . | . | . | . | . | . |
| 4o | 4 | 100L | . | . | . | . | . | . |
| 4p | 1 | 100L | . | . | . | . | . | . |
| 4q | 1 | 100L | . | . | . | . | . | . |
| 4r | 6 | 100L | . | . | . | . | . | . |
| 4s | 1 | 100L | . | . | . | . | . | . |
| 4t | 1 | 100L | . | . | . | . | . | . |
| 4v | 4 | 100L | . | . | . | . | . | . |
| 4w | 2 | 100L | . | . | . | . | . | . |
| 5a | 46 | 100L | . | . | . | . | 100F | 100K |
| 6a | 409 | . | . | . | . | . | 0.73F | 98.28K,0.24L,0.24R |
| 6b | 2 | . | . | . | . | . | . | . |
| 6c | 2 | . | . | . | . | . | . | . |
| 6d | 1,2 | 50L | . | . | . | . | . | . |
| 6e | 10 | . | . | . | . | . | . | . |
| 6f | 22 | 100L | . | . | . | . | . | . |
| 6g | 2 | 100L | . | . | . | . | . | . |
| 6h | 5 | . | . | . | . | . | . | . |
| 6i | 3 | . | . | . | . | . | . | . |
| 6j | 3 | . | . | . | . | . | . | . |
| 6k | 8 | 12.5L | . | . | . | . | . | . |
| 6l | 8, 9 | 11.11L | . | . | . | . | . | . |
| 6m | 4 | . | . | . | . | . | . | . |
| 6n | 25 | . | . | . | . | . | . | . |
| 6o | 3 | . | . | . | . | . | . | . |
| 6p | 2 | . | . | . | . | . | . | . |
| 6q | 3 | . | . | . | . | . | . | . |
| 6r | 3 | . | . | . | . | . | 66.7F | . |
| 6s | 3 | 100L | . | . | . | . | 100F | . |
| 6t | 4 | 100L | . | . | . | . | 50F | . |
| 6u | 3 | . | . | . | . | . | . | . |
| 6v | 6 | . | . | . | . | . | . | . |
| 6w | 3 | 100L | . | . | . | . | . | . |
| 6xa | 3 | . | . | . | . | . | 100F | . |
| 6xb | 2 | . | . | . | . | . | . | . |
| 6xc | 1 | . | . | . | . | . | . | . |
| 6xd | 3 | 100L | . | . | . | . | . | 33.3L |
| 6xe | 2 | . | . | . | . | . | . | . |
| 6xf | 2 | 50L | . | . | . | . | . | . |
| 7a | 2 | 100L | . | . | . | . | . | . |
| 7b | 1 | 100L | . | . | 100S | . | . | . |
Prevalence of naturally occurring NS5A RASs in 86 HCV subtypes
The prevalence of naturally occurring NS5A RASs in different HCV subtypes is shown in Table 2. Similar to NS3, most RASs were absent or have very low frequencies (<0.5%) (Figure S1B). RAS Y93H was associated with reduced NS5A-targeted DAA efficacy, with or without L31M/V/I, in GT1b-infected patients.40 Y93H appeared in sequences of subtypes GT1a (0.41%, 12/2928), 1b (4.25%, 80/1882), 1c (25%,1/4), 1m (50%, 1/2), 3a (1.35%, 14/1114), 4a (3.33%,1/30), 4b (50%,1/2), 4g (33.33%,1/3), 7a (100%,2/2) and 7b (100%,1/1). Other substitutions at this position, such as Y93C/F/N/S, were uncommon in GT1a, 1b, 2a, 3a, and 6a (0.03%-1.92%), but were prevalent in other subtypes, including GT1c, 1g, 1m, 4w, 6e, 6m, 6n, 6o, 6u, 6v, and 6xe (15.38%-100%). L31M, which confers resistance to daclatasvir/omibitasvir/ledipasvir, has been associated with reduced elbasvir/grazoprevir efficacy in patients with HCV-GT1a infection.17 L31M was rare in GT1a (0.65%,19/2928) and 1b (2.63%,49/1865) sequences and absent in GT3a, 5a, and all GT6 subtypes except in one GT6a sequence. In contrast, this RAS was frequently detected (≥50%) in subtypes G1d, 1e, 1l, 1m, 3b, and a majority of GT2 and 4 subtypes. A30K, which is associated with daclatasvir resistance, was only detected in 2.25% of GT3a sequences but was found in nearly 100% of sequences from other GT3 subtypes. The most commonly observed RAS in GT1b was Q54H (26.76%, daclatasvir resistance). RASs L28M (daclatasvir/ombitasvir resistance) and R30Q (daclatasvir resistance) were identified in 2.37% and 4.66% of GT1b sequences, respectively.
Table 2.
(Continued).
| Subtype | L31 FIMPVW |
P32 ALQRS |
Q54 H |
H58 DLNc |
E62 DL |
A92KPTd | Y93 CFILNRSTW |
Y93 H |
|---|---|---|---|---|---|---|---|---|
| 1a | 0.65M | 0.03L,0.07S | 96.52 | 0.10D,0.10N | 1.4D | 0.51P | 0.24C,0.17N, 0.07F, 0.03S | 0.41 |
| 1b | 2.63M,0.27I,0.05F | 0.05S | 26.76 | 3.43S,0.8T,0.48L,0.48A,0.05R | 0.05D,0.21L | 1.28T | 0.05C, 0.05F, 0.05S | 4.25 |
| 1c | . | . | 75.00 | . | 50D | . | 25N | 25.00 |
| 1d | 100M | . | . | . | . | . | . | . |
| 1e | 50M | . | 50.00 | . | . | 100T | . | . |
| 1g | . | . | . | . | . | . | 100F | . |
| 1h | . | . | . | . | . | . | . | . |
| 1i | . | . | 100 | . | . | . | . | . |
| 1j | . | . | . | . | . | . | . | . |
| 1k | . | . | . | . | . | . | . | . |
| 1l | 100M | . | 33.33 | . | . | . | . | . |
| 1m | 50M | . | . | . | 50D | . | 50.00C | 50.00 |
| 1n | . | . | . | . | 100D | . | . | . |
| 2a | 82.7M | . | . | . | . | . | 1.92F | . |
| 2b | 64.8M | . | . | . | . | 0.69A,0.69S | . | . |
| 2c | 11.1M | . | . | . | . | . | . | . |
| 2d | . | . | . | . | . | . | . | . |
| 2e | 100M | . | . | . | . | 100S | . | . |
| 2f | 100M | . | . | . | . | 50S | . | . |
| 2i | 100M | . | . | . | . | . | . | . |
| 2j | 100M | . | . | . | . | . | . | . |
| 2k | 75M | . | 25.00 | . | . | 25A | . | . |
| 2l | . | . | . | . | 50L | 100S | . | . |
| 2m | 100M | . | . | . | . | 100S | . | . |
| 2q | 50M | . | . | . | . | . | . | . |
| 2r | 100M | . | . | . | . | . | . | . |
| 2t | . | . | . | . | . | . | . | . |
| 2u | 100M | . | . | . | . | . | . | . |
| 3a | 0.18P | . | . | 0.09L | 3.32L | . | 0.36C, 0.09F | 1.35 |
| 3b | 84.2M,5.26V | . | . | . | 36.8D | . | . | . |
| 3d | 100M | . | . | . | . | . | . | . |
| 3e | . | . | . | . | . | . | . | . |
| 3g | 50M,50V | . | . | . | . | . | . | . |
| 3h | . | . | . | . | . | . | . | . |
| 3i | . | . | . | . | . | . | . | . |
| 3k | 100M | . | . | . | 100L | . | . | . |
| . | . | . | . | . | . | . | . | |
| 4a | 3.33V | . | 100 | . | 10D | . | . | 3.33 |
| 4b | . | . | 100 | . | 50D | 50P | 50T | 50.00 |
| 4c | . | . | 100 | . | . | . | . | . |
| 4d | . | . | 100 | . | . | . | . | . |
| 4f | . | . | 100 | . | . | . | . | . |
| 4g | . | . | 100 | . | . | . | 33.3R | 33.33 |
| 4k | 66.7L | . | 100 | . | . | . | . | . |
| 4l | . | . | 100 | . | . | . | . | . |
| 4m | . | . | 100 | . | . | . | . | . |
| 4n | 33.3L | . | 100 | . | . | . | . | . |
| 4o | . | . | 100 | . | 25D | . | . | . |
| 4p | . | . | 100 | . | . | . | . | . |
| 4q | . | . | 100 | . | . | . | . | . |
| 4r | 85.7L | . | 100 | . | . | . | . | . |
| 4s | 100L | . | 100 | . | . | . | . | . |
| 4t | . | . | 100 | . | . | . | . | . |
| 4v | . | . | 100 | . | . | . | . | . |
| 4w | . | . | 100 | . | . | . | 100S | . |
| L31FIMPVW | . | . | . | . | . | . | . | |
| 5a | . | . | . | . | . | . | . | . |
| 6a | 0.86M | . | 98.28 | . | 1.72L | . | 0.86I, 0.86S | . |
| 6b | . | . | 100 | 100S | . | . | . | . |
| 6c | . | . | 100 | 100G | . | 100P | . | . |
| 6d | . | . | . | 100S | . | . | 100I | . |
| 6e | . | . | . | 7.69H | . | . | 15.38S | . |
| 6f | . | . | 94.44 | 16.7S,5.56L | . | . | . | . |
| 6g | . | . | . | . | . | . | . | . |
| 6h | . | . | 100 | 20S | . | . | . | . |
| 6i | . | . | 100 | . | 66.7D | . | . | . |
| 6j | . | . | 75.00 | 50A | 50D | . | . | . |
| 6k | . | . | 25.00 | . | 87.5D | . | . | . |
| 6l | . | . | . | . | 100D | . | . | . |
| 6m | . | . | 100 | . | . | . | 100S | . |
| 6n | . | . | 100 | . | . | . | 100S | . |
| 6o | . | . | . | 100A | . | . | 50S | . |
| 6p | . | . | 100 | . | . | . | . | . |
| 6q | . | . | 100 | . | . | . | . | . |
| 6r | . | . | . | 33.3S | . | . | . | . |
| 6s | . | . | 100 | . | . | . | . | . |
| 6t | . | . | . | 100G | . | . | . | . |
| 6u | . | . | . | . | . | . | 100S | . |
| 6v | . | . | . | . | . | . | 100S | . |
| 6w | . | . | 100 | . | 66.7D | . | . | . |
| 6xa | . | . | 100 | . | . | . | . | . |
| 6xb | . | . | . | . | 100D | . | . | . |
| 6xc | . | . | . | . | . | . | . | . |
| 6xd | . | . | 100 | 33.3A | . | . | . | . |
| 6xe | . | . | 100 | . | . | . | 100S | . |
| 6xf | . | . | . | . | . | . | . | . |
| 7a | . | . | . | . | 100L | . | . | 100 |
| 7b | . | . | . | . | . | . | . | 100 |
Notes: Data are presented as %. The dot denotes 0%. aNot all the positions were analyzed using the same number of sequences. The first figure is the lowest number, and the second figure is the largest number. n.a. not applicable because of different natural amino acid sequences in the respective HCV genotype/subtype (NS5A K24, M28, Q30, and H58 are the dominant amino acids in GT1a; F28 is the dominant amino acid in subtype 2a and L28 in subtype 2b; A30 is the dominant amino acid in GT3; T93 is the dominant amino acid in GT6). bFor GT1b, Q24K was screened; for GT3 and GT2 except GT2a, S24FHT were screened. cFor GT1b, P58A/D/G/L/R/S/T was screened; for GT6, T58A/D/G/H/L/N/S were screened. dFor GT2, C92A/K/N/R/S/T was screened; for GT3, E92K were screened.No sequences harbored S38F. K26E was found in 0.11% of 1a, and 0.27% of 3a sequences.
Table 2.
Prevalence of naturally occurring NS5A RASs in 86 HCV subtypes
| Subtype | No.of seqa | K24 AEGNQRb |
M28 AGIKSTV |
F28CS | L28 AFIMSTV |
Q30 DEGHIKLNRSTY |
R30 CEGHKNQST |
A30 GHKV |
|---|---|---|---|---|---|---|---|---|
| 1a | 2855, 2928 | 0.46R,0.21E,0.32Q | 3.79V, 0.38T,0.10I | n.a. | n.a. | 1.30L, 0.79H, 0.44R | n.a. | n.a. |
| 1b | 1817, 1882 | 1.93K | n.a. | n.a. | 0.11F,2.37M,0.11V | n.a. | 4.66Q,0.59K,0.05H | n.a. |
| 1c | 4 | . | 50V | . | 50M,50V | . | 100Q | . |
| 1d | 1 | . | . | . | . | 100R | . | . |
| 1e | 2 | 50R | . | . | 100M | . | 100Q | . |
| 1g | 2 | 50R | . | . | . | 50.00R | 50.00Q | . |
| 1h | 3 | 100Q | . | . | . | 100R | . | . |
| 1i | 1 | 100Q | . | . | . | 100R | . | . |
| 1j | 1 | . | . | . | 100M | . | 100Q | . |
| 1k | 1 | . | 100A | . | 100A | . | 100Q | . |
| 1l | 3 | 66.67G | . | . | 100M | 33.33R | 66.67Q | . |
| 1m | 2 | . | . | . | 100M | 50S | 50S,50Q | . |
| 1n | 2 | . | . | . | 100M | . | 100Q | . |
| 2a | 52 | . | . | . | 100F | 100K | 100K | 100K |
| 2b | 145 | . | . | . | 4.83F | 99.3K | 99.3K | 99.3K |
| 2c | 9 | . | . | 33.3C | 66.67F | 22.2R,77.8K | 77.8K | 77.8K |
| 2d | 1 | . | . | . | . | 100K | 100K | 100K |
| 2e | 1 | . | . | . | 100F | 100K | 100K | 100K |
| 2f | 2 | 50F | 50S | 50S | 50F,50S | 100K | 100K | 100K |
| 2i | 4 | . | . | . | 100F | 100K | 100K | 100K |
| 2j | 6 | . | . | . | 66.67F | 100K | 100K | 100K |
| 2k | 4 | 25Q | . | . | 25.00F | 25R,75K | 75K | 75K |
| 2l | 2 | . | . | . | . | 100K | 100K | 100K |
| 2m | 4 | . | . | . | . | 50R,50K | 50K | 50K |
| 2q | 2 | . | . | . | . | 100K | 100K | 100K |
| 2r | 1 | . | . | . | 100F | 100K | 100K | 100K |
| 2t | 1 | . | . | . | . | 100K | 100K | 100K |
| 2u | 1 | . | . | . | 100F | 100K | 100K | 100K |
| 3a | 1112, 1114 | 1.08A,0.09T | 0.27V,0.09T | . | 0.09T,99.2M | 1.26L, 0.90T,0.45S | 0.90T,0.45S | 2.25K,0.81V |
| 3b | 19 | . | . | . | 100M | 5.26R,84.2K | 84.2K | 84.2K |
| 3d | 1 | . | . | . | 100M | 100K | 100K | 100K |
| 3e | 1 | . | . | . | 100M | 100K | 100K | 100K |
| 3g | 2 | 50T | . | . | 100M | 100K | 100K | 100K |
| 3h | 2 | . | . | . | 100M | . | . | . |
| 3i | 6 | . | . | . | 100M | 16.67R,83.3K | 83.3K | 83.3K |
| 3k | 2 | 100G | . | . | 50M | 100K | 100K | 100K |
| . | . | . | . | . | . | . | . | . |
| 4a | 30 | . | 6.67V | . | 10M,6.67V | 86.67L,3.33R,3.33H3.33S | 3.33H,3.33S | 3.33H |
| 4b | 2 | . | . | . | . | 50.00S | 50.00S | . |
| 4c | 1 | . | . | . | . | 100R | . | . |
| 4d | 11 | . | . | . | . | 90.91R | . | . |
| 4f | 6 | . | . | . | . | 66.67R | 33.33Q | . |
| 4g | 3 | . | . | . | . | 66.67L,33.33R | . | . |
| 4k | 3 | . | . | . | . | 100R | . | . |
| 4l | 3 | . | . | . | . | 100R | . | . |
| 4m | 4 | . | . | . | . | 100S | 100S | . |
| 4n | 3 | . | . | . | . | 100R | . | . |
| 4o | 4 | . | . | . | 100M | 100T | . | . |
| 4p | 1 | . | . | . | . | 100R | . | . |
| 4q | 1 | . | . | . | . | 100R | . | . |
| 4r | 7 | . | 42.86V,28.6I | . | 28.6I,28.6M,42.7V | 100R | . | . |
| 4s | 1 | . | . | . | . | 100R | . | . |
| 4t | 1 | . | . | . | . | 100R | . | . |
| 4v | 4 | . | . | . | . | 100R | . | . |
| 4w | 2 | . | . | . | 100M | 100S | 100S | . |
| . | . | . | . | . | . | . | . | . |
| 5a | 23 | 100Q | . | . | . | . | 100Q | . |
| 6a | 116 | 89.7Q,6.03R | 2.59V | . | 23.3F,2.59V | 100R | . | . |
| 6b | 2 | . | 100T | . | 100T | 100R | . | . |
| 6c | 2 | . | 100V | . | 100V | 50T | 50T | . |
| 6d | 1 | 100R | 100V | . | 100V | 100T | 100T | . |
| 6e | 13 | 15.38R | 46.15V | . | 53.9M,46.2V | 100S | 100S | . |
| 6f | 18 | . | 61.11T,38.89A | . | 61.1T,38.9A | 100S | 100S | . |
| 6g | 2 | . | . | . | 100M | 50.00S | 50.00S | . |
| 6h | 5 | . | 100V | . | 100V | . | . | . |
| 6i | 3 | . | 66.67V | . | 66.67V | . | . | . |
| 6j | 4 | . | 100V | . | 100V | . | . | . |
| 6k | 8 | 25R | 100V | . | 100V | . | . | . |
| 6l | 7 | . | 100V | . | 100V | . | . | . |
| 6m | 4 | . | 100V | . | 100V | 100S | 100S | . |
| 6n | 9 | . | 100V | . | 100V | 100S | 100S | . |
| 6o | 4 | . | . | . | . | . | . | . |
| 6p | 3 | . | 66.67V | . | 33.3M,66.7V | 100S | 100S | . |
| 6q | 3 | . | 100V | . | 100V | 100S | 100S | . |
| 6r | 3 | . | 33.33G,33.33T | . | 33.33T | . | . | . |
| 6s | 3 | 33.33R | . | . | 33.3M | 100S | 100S | . |
| 6t | 4 | . | 100V | . | 100V | 100S | 100S | . |
| 6u | 2 | . | 100V | . | 100V | 100S | 100S | . |
| 6v | 4 | . | 100V | . | 100V | 100S | 100S | . |
| 6w | 3 | . | 100T | . | 100T | . | . | . |
| 6xa | 3 | . | 100V | . | 100V | 100S | 100S | . |
| 6xb | 2 | . | 100V | . | 100V | . | . | . |
| 6xc | 1 | . | 100V | . | 100V | . | . | . |
| 6xd | 3 | . | . | . | 66.67F | 100R | . | . |
| 6xe | 2 | . | 100V | . | 100V | 100S | 100S | . |
| 6xf | 2 | 50R | 100V | . | 100V | . | . | 50V |
| 7a | 2 | . | . | . | . | 100S | 100S | . |
| 7b | 1 | . | . | . | . | 100S | 100S | . |
Prevalence of naturally occurring NS5B NI-specific and NNI-specific RASs in 86 HCV subtypes
The prevalence of naturally occurring NI-specific NS5B RASs and NNI-specific RASs in different HCV subtypes is shown in Table 3. Except for a few RASs with high rates, others have very low rates (Figure S1C). A150V has recently been found to be associated with a reduced response to treatment with sofosbuvir and ribavirin, with or without pegylated interferon in GT3a infected patients.34 A150V is highly prevalent in sequences of GT3a (31.5%, 103/327). L159F was found in 11.19% (297/2655) of GT1b sequences but in only 0.09% (2/2346) of GT1a sequences. S282T, the only known variant conferring sofosbuvir resistance in vitro, rarely appeared in GT1a (0.19%, 10/5182), 1b (0.15%, 11/7440), 2b (0.22%, 1/455), 3a (0.03%, 1/3003) and 4a (0.35%, 3/857).
Table 3.
(Continued).
| Sub type | NS5B NNI RAS position | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C316 NYHF |
S368T | N411S | M414 TIV |
C445 F |
E446 KQ |
Y448 CH |
A553 ITV |
G554 SD |
S556 GNR |
G558R | D559GN | |
| 1a | 0.02N, 0.09Y | . | . | 0.11T | . | 0.11Q, 0.04K | 0.07H, 0.04C | 0.06V | . | 0.79G,0.34R, 0.11N | 0.42 | . |
| 1b | 43.09N, 0.09Y,1.19H | 0.03 | . | 0.14T, 0.32I,0.03V | 0.67 | 98.46Q | 0.26H, 0.03C | . | 0.09D | 11.77G,0.83N | 0.09 | 0.05N |
| 1c | . | . | . | . | . | . | . | . | . | . | . | . |
| 1d | . | . | . | . | . | 100Q | . | . | . | . | . | . |
| 1e | 10.17N | . | . | . | . | . | . | . | . | . | . | . |
| 1g | 5.26Y, 2.63H | . | . | . | . | 20Q | . | . | . | . | . | . |
| 1h | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 1i | . | . | . | . | . | 100Q | . | . | . | . | . | . |
| 1j | . | . | . | . | . | . | . | . | . | . | . | . |
| 1k | . | . | . | . | . | . | 100H | . | . | . | . | . |
| 1l | 2.50H | . | . | . | . | . | . | . | . | . | . | . |
| 1m | . | . | . | . | . | . | . | . | . | . | . | . |
| 1n | . | . | . | . | . | . | . | . | . | . | . | . |
| 2a | 0.14Y | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 2b | . | . | . | . | 100 | . | . | 95.37V | . | 100G | . | . |
| 2c | . | . | . | . | 100 | . | . | 87.5V, 6.25I | 100S | 100G | . | . |
| 2d | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 2e | . | . | . | . | 100 | . | . | 87.5V | . | 100G | . | . |
| 2f | . | . | . | . | 100 | . | . | 75V 25I |
. | 100G | . | . |
| 2i | . | . | . | . | 100 | . | . | 100 | 100S | 100G | . | . |
| 2j | . | . | . | . | 100 | . | . | 50V, 33.3I, 16.7T | . | 100G | . | . |
| 2k | 3.12H | . | . | . | 90.9 | 9.09Q | . | 88.89V | . | 88.9G | . | . |
| 2l | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 2m | . | . | . | . | 100 | . | . | 100V | 100S | 100G | . | . |
| 2q | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 2r | 7.69N, 7.69Y | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 2t | . | . | . | . | 100 | . | . | 100V | . | . | . | . |
| 2u | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 3a | 0.03Y | . | 0.34 | . | 99.7 | 0.12Q | . | 100V | 1.04S | 97.8G | . | 1.11N |
| 3b | . | . | . | . | 100 | 0.99Q | 0.33H | 100V | . | 100G | . | . |
| 3d | . | . | . | . | 100 | 100Q | . | 100V | . | 100G | . | . |
| 3e | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 3g | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 3h | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 3i | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 3k | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4a | 0.48N | . | . | 91.4V,7.76I | 100 | . | . | 100V | . | 100G | . | . |
| 4b | 14.29N, 7.14H | . | . | 50V | 100 | . | . | 100V | . | 100G | . | . |
| 4c | 5.66Y, 3.77H | . | . | 100I | 100 | . | . | 100V | . | 100G | . | . |
| 4d | 0.16Y | . | . | 2.80T, 96.8I | 100 | . | . | 100V | . | 100G | . | . |
| 4f | 81.61N | . | . | 83.3V,16.7I | 100 | . | . | 100V | . | 100G | . | . |
| 4g | 6.25H | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4k | 0.79Y, 4.76H | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4l | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4m | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4n | 4.35Y | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4o | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4p | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4q | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4r | 60.3H,0.79N | . | . | 9.09I | 100 | . | . | 100V | . | 75N | . | . |
| 4s | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4t | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4v | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 4w | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 5a | . | . | . | . | 98.1 | . | . | 100V | 5.41S | 97.3G | . | . |
| 6a | . | . | . | . | 100 | . | 0.41H | . | . | 0.84R | . | . |
| 6b | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6c | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6d | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6e | 0.72Y | 2.33 | . | . | 98.2 | . | . | . | . | 5.26G,94.7R | . | . |
| 6f | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6g | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6h | . | . | . | . | 100 | . | . | . | . | 85.71G | . | . |
| 6i | . | . | 11.1 | . | 100 | . | . | . | . | . | . | . |
| 6j | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6k | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6l | . | . | . | . | 100 | . | . | . | . | . | . | 10.0G |
| 6m | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6n | . | . | . | 1.16V | 100 | . | 1.16H | . | . | 12.5R | . | . |
| 6o | . | . | . | . | 100 | . | . | 25V | . | 75R | . | . |
| 6p | . | . | . | . | 100 | . | . | 50V | . | 100R | . | . |
| 6q | . | . | . | . | 100 | . | . | . | . | 100R | . | . |
| 6r | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6s | . | . | . | . | 100 | . | . | 100V | . | 100R | . | . |
| 6t | . | . | . | . | 100 | . | . | . | . | 25R | . | . |
| 6u | . | . | . | . | 100 | . | . | . | . | 100R | . | . |
| 6v | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6w | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6xa | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6xb | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6xc | . | . | . | . | 100 | . | . | . | . | 100R | . | . |
| 6xd | . | . | . | . | 100 | . | . | . | . | . | . | 33.3G |
| 6xe | . | . | . | . | 100 | . | . | . | . | . | . | . |
| 6xf | . | . | . | . | 100 | . | . | . | . | 100R | . | . |
| 7a | . | . | . | . | 100 | . | . | 100V | . | 100G | . | . |
| 7b | . | . | . | . | 100 | . | . | . | 100S | 100G | . | . |
Notes: Data are presented as %. The dot denotes 0%. C451S were detected in 1.05% of 1b sequences, G188D in 0.82% of 3a, N224I in 3.06% of 3a and in 3.45% of 6k, A395G in 0.75% of 4a, and Y561H in 0.10% of 1b. L314H, N444K, S565F were not found. aNot all the positions were analyzed using the same number of sequences. The first figure is the lowest number, and the second figure is the largest number.
Table 3.
Prevalence of naturally occurring NS5B RASs in 86 HCV subtypes
| Sub type | No.of seqa | NS5B NI RAS position | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A150 V |
L159 F |
K206 E |
E237G | S282 TGR |
M289 IL |
L320 F |
V321AI | ||
| 1a | 1433, 5412 | . | 0.09 | 0.12 | 1.54 | 0.19T,0.02G,0.02R | 0.10L | 0.02 | 0.23I,0.02A |
| 1b | 2090, 7462 | 0.08V | 11.2 | 0.03 | 0.06 | 0.15T,0.26R,0.03G | 0.01I | 0.03 | 1.83I,0.01A |
| 1c | 3, 27 | . | . | . | . | . | . | . | . |
| 1d | 1, 27 | . | . | . | . | . | . | . | . |
| 1e | 2, 59 | . | . | . | . | . | . | . | . |
| 1g | 2, 41 | . | . | . | . | . | . | . | 2.7I |
| 1h | 3, 30 | . | . | . | . | . | . | . | . |
| 1i | 1, 6 | . | . | . | . | . | . | . | . |
| 1j | 1, 4 | . | . | . | . | . | . | . | . |
| 1k | 1, 8 | . | . | . | . | . | . | . | 100I |
| 1l | 3, 40 | . | . | . | . | . | . | . | 2.5I |
| 1m | 2, 5 | . | . | . | . | . | . | . | . |
| 1n | 2, 2 | . | . | . | . | . | . | . | 100I |
| 2a | 28, 742 | 13.8 | . | . | . | . | 1.49I, 0.54L | 0.14 | . |
| 2b | 97, 460 | 1.03 | . | . | 3.02 | 0.22T | 7.73I | . | 0.44I |
| 2c | 13, 286 | . | . | . | . | . | 0.35I | . | 0.78I |
| 2d | 1, 9 | . | . | . | . | . | . | . | . |
| 2e | 1, 25 | . | . | . | . | . | 100L | . | . |
| 2f | 2, 14 | . | . | . | . | . | . | . | . |
| 2i | 4, 84 | . | . | . | . | . | 1.19I, 2.38L | . | . |
| 2j | 6, 89 | . | . | 16.7 | 4.76 | . | 1.12I | . | . |
| 2k | 9, 72 | . | . | . | . | . | 1.39I | . | 3.17I |
| 2l | 2, 20 | . | . | . | . | . | 5.00I, 90.0L | . | . |
| 2m | 3, 21 | . | . | . | . | . | . | . | . |
| 2q | 2, 5 | . | . | . | . | . | 20.00I | . | . |
| 2r | 1, 13 | 100 | . | . | . | . | . | . | 46.2I |
| 2t | 1, 1 | . | . | . | . | . | . | . | . |
| 2u | 1, 1 | . | . | . | . | . | . | . | . |
| 3a | 90, 3155 | 31.5 | . | 7.38 | 0.45 | 0.2R,0.03T | 0.07L | 0.03 | 0.13I |
| 3b | 20, 672 | 2.63 | . | 7.92 | 0.67 | 0.15G | . | . | . |
| 3d | 1, 3 | . | . | 100 | . | . | . | . | . |
| 3e | 1, 4 | . | . | . | . | . | . | . | . |
| 3g | 2, 8 | . | . | . | . | . | . | . | . |
| 3h | 2, 12 | . | . | . | . | . | 100L | . | . |
| 3i | 5, 20 | . | . | 33.3 | . | . | . | . | . |
| 3k | 2, 57 | . | . | 55.6 | . | . | . | . | . |
| 4a | 42, 857 | . | . | . | 10.05 | 0.35T,0.12R | 0.95L | 0.12 | 0.12I |
| 4b | 2, 14 | . | . | . | . | . | 7.14L | . | 7.14I |
| 4c | 1, 59 | . | . | . | . | 1.69G | . | . | 3.77I |
| 4d | 25, 672 | . | . | . | 0.95 | 0.18R | 0.18I, 0.18L | . | . |
| 4f | 5, 87 | . | . | . | 2.38 | . | . | . | . |
| 4g | 3, 16 | . | . | . | . | . | . | . | 6.25I |
| 4k | 3, 127 | . | . | . | . | 1.57G | 0.79L | . | 7.14I |
| 4l | 5, 31 | . | . | . | 9.68 | . | 7.69L | . | . |
| 4m | 4, 29 | . | . | 25 | . | . | . | . | . |
| 4n | 2, 31 | . | . | . | 3.23 | . | . | . | . |
| 4o | 4, 44 | . | . | . | . | . | 2.38I | . | . |
| 4p | 1, 16 | . | . | . | . | . | . | . | . |
| 4q | 1, 15 | . | . | . | . | . | 26.7L | . | . |
| 4r | 3, 126 | . | . | . | 1.15 | . | . | . | 60.3I |
| 4s | 1, 3 | . | . | . | . | . | . | . | . |
| 4t | 1, 22 | . | . | . | . | . | . | . | . |
| 4v | 2, 6 | . | . | . | 66.67 | . | 16.7L | . | . |
| 4w | 2, 2 | . | . | . | . | . | . | . | . |
| 5a | 37, 419 | . | . | . | . | . | 1.19I | 0.25 | . |
| 6a | 118, 1290 | . | . | . | . | 0.08R | 1.16L | . | . |
| 6b | 2, 9 | . | . | . | . | . | 77.8L | . | . |
| 6c | 2, 12 | . | . | . | . | . | 100L | . | . |
| 6d | 1, 13 | . | . | . | . | . | 100L | . | . |
| 6e | 16, 139 | . | . | . | . | . | 98.6L | . | . |
| 6f | 5, 121 | . | . | . | . | . | 98.4L | . | 0.87I |
| 6g | 2, 5 | . | . | . | . | . | . | . | . |
| 6h | 5, 42 | . | . | . | . | . | . | . | . |
| 6i | 4, 38 | . | . | . | . | . | . | . | . |
| 6j | 4, 16 | . | . | . | . | . | 50.0L | . | . |
| 6k | 8, 31 | . | . | . | . | . | 100L | . | . |
| 6l | 7, 42 | . | . | 14.3 | . | . | 100L | . | . |
| 6m | 4, 21 | . | . | 87.5 | . | . | . | . | . |
| 6n | 8, 196 | . | . | 6.45 | . | . | 94.9L | . | 0.54I |
| 6o | 4, 13 | . | . | . | . | . | 100L | . | . |
| 6p | 2, 15 | . | . | . | . | . | 6.67I, 93.3L | . | . |
| 6q | 3, 18 | . | . | . | . | . | 100L | . | 16.7I |
| 6r | 3, 11 | . | . | . | . | . | 100L | . | . |
| 6s | 3, 5 | . | . | . | 100 | . | 100L | . | . |
| 6t | 4, 4 | . | . | . | . | . | 100L | . | . |
| 6u | 2, 43 | . | . | . | . | . | 4.65L | . | . |
| 6v | 4, 6 | . | . | . | . | . | . | . | . |
| 6w | 3, 4 | . | . | . | . | . | 100 L | . | . |
| 6xa | 3, 4 | . | . | . | . | . | . | . | . |
| 6xb | 2, 2 | . | . | . | . | . | 100L | . | . |
| 6xc | 1, 1 | . | . | . | . | . | 100L | . | . |
| 6xd | 3, 3 | . | . | . | . | . | 33.3L | . | . |
| 6xe | 2, 2 | . | . | 50 | . | . | 100L | . | . |
| 6xf | 2, 4 | . | . | . | . | . | 100L | . | . |
| 7a | 2, 2 | . | . | . | . | . | . | . | . |
| 7b | 1, 2 | . | . | . | . | . | . | . | . |
All observed NNI-specific RASs are associated with dasabuvir. C316N was common in sequences of GT1b (43.09%, 3179/7377), GT4f (81.61%, 71/87), 4b (14.29%, 2/14), and 1e (10.17%,6/59). C316H was observed in GT1b (1.19%, 88/7377) and 5–10% of several GT4 subtypes but was more prevalent in GT4r (60.32%, 76/126). The frequency of S556G was higher in GT1b than in GT1a (11.77% vs.0.79%) and was found in 6h (85.71%, 6/7), 6e (5.26%, 1/19), and GT2, 3, 4, 5 and 7 subtypes (nearly 100%). However, this RAS was absent in other GT1 subtypes and GT6 subtypes, although S556N, a closely related variant, was harbored by GT4r (75%, 3/4). S556R was found in GT1a (0.34%) and in several GT6 subtypes (6a, 6e, 6n, 6o, 6p, 6q, 6s, 6t, 6u, 6xc, and 6xf).
Geographical distribution of RASs
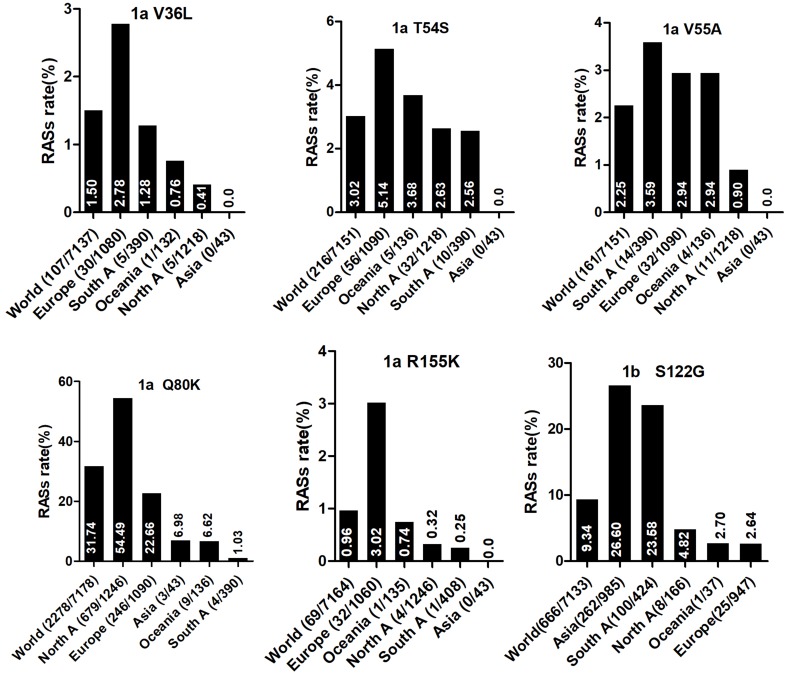
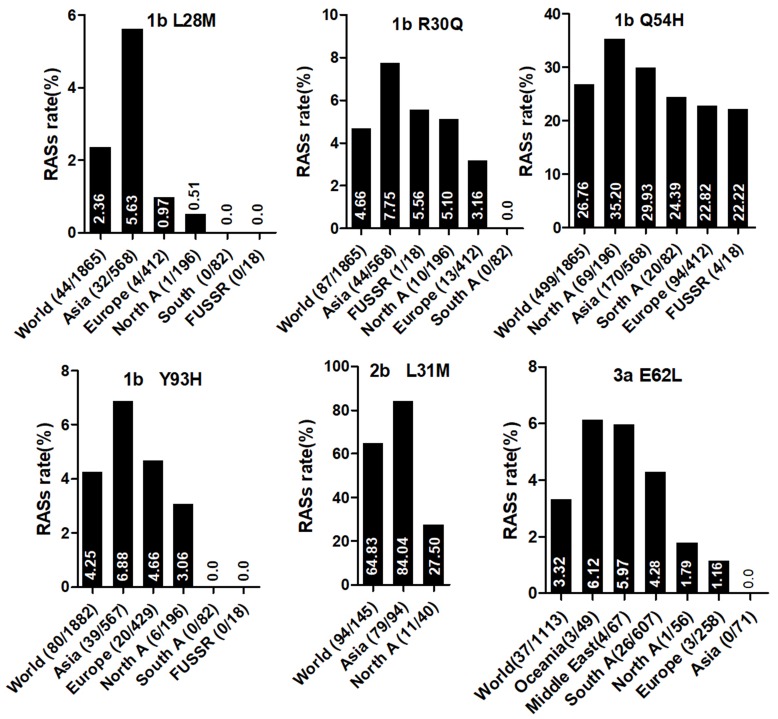
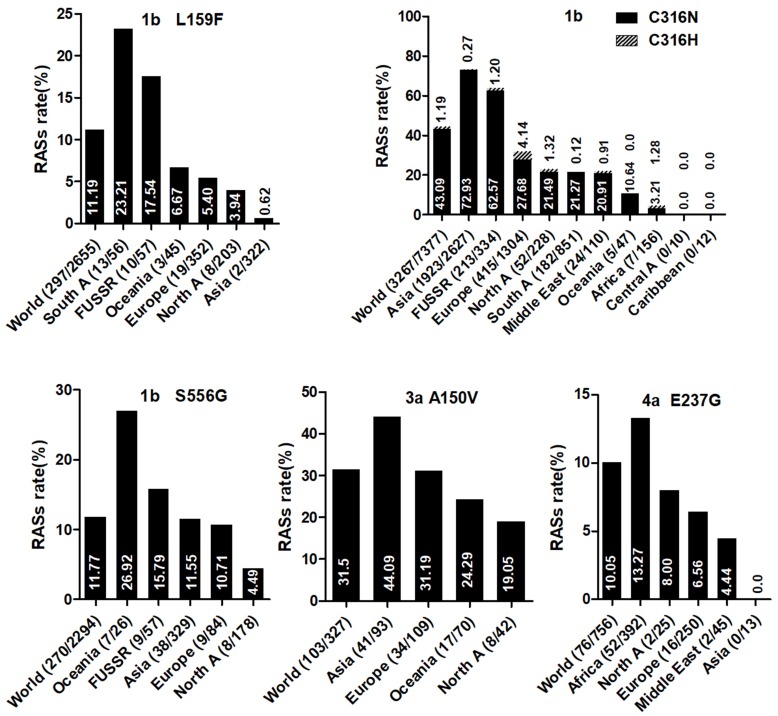
Country of origin information was available for approximately 70% of the analyzed sequences. We classified these sequences into Asia, Europe, North America, Central America, South America, Former USSR, Oceania, Africa, Caribbean, or Middle East clusters according to geographic region definitions in the Los Alamos HCV Database. The majority of RASs in most HCV subtypes were similarly distributed among different geographic regions worldwide. NS3 RASs with distinctly variable prevalence by geographic region including Q8OK in GT1a, V36L in GT1a, S122G in GT1b and so on (Figure 1, P<0.05). Q80K in GT1a was mostly prevalent in North America (54.49%, 679/1246), followed by Europe (22.66%, 246/1090), Asia (6.98%, 3/43), Oceania (6.62%, 9/136), and South America (1.03%, 4/390). NS5A RASs (L28M, R30Q, Q54H, and Y93H in GT1b, L31M in GT2b, and E62L in GT3a) varied significantly in geographic prevalence (Figure 2, all P-values <0.05). L28M, R30Q, and Y93H in GT1b showed the highest prevalence in Asia. NS5B RASs exhibited distinct global distribution patterns are present in Figure 3. C316N/H was found mostly in Asia (73.20%, 1923/2627), followed by in the Former USSR (63.77%, 213/334), Europe (31.82%, 415/1304), North America (22.81%, 52/228), South America (21.39%, 182/851), Oceania (10.64%, 5/47), the Middle East (21.82%, 24/110), Africa (4.49%, 7/156), Central America (0%, 0/10), and the Caribbean (0%, 0/12). In contrast, Asia had the lowest prevalence of L159F in GT1b (0.62%, 2/322), while S556G in GT1b commonly appeared in Oceania (26.92%, 7/26) but infrequently in North America (4.49%, 8/178). A150V in GT3a was most prevalent in Asia (44.09%), followed by Europe (31.19%), Oceania (24.29%), and North America (19.05%).
Figure 1.
The global and regional frequency of naturally occurring NS3 RASs that showed unequal distribution by geographic regions. In each plot, except for the first bar representing the global prevalence, geographic regions were arranged in descending order according to the frequency of RASs. Sequences were clustered into Asia, Europe, North America, Central America, South America, Former USSR, Oceania, Africa, Caribbean or Middle East. In each plot, regions with <10 sequences were not shown. Region definition was according to the Los Alamos HCV Database. “A” in North A, South A and Central A denotes America.
Figure 2.
NS5A RASs rate with significantly different frequencies among different geographic regions. In each plot, the first bar represents the global prevalence and geographic regions were arranged in descending order according to the frequency of RASs. Sequences were clustered into Asia, Europe, North America, Central America, South America, Former USSR, Oceania, Africa, Caribbean or Middle East. In each plot, only regions with at least 10 sequences were shown. Region definition was according to the Los Alamos HCV Database. “A” in North A, South A and Central A denotes America.
Figure 3.
NS5B RASs rate with significantly different frequencies among different geographic regions. In each plot, the first bar represents the global prevalence and geographic regions were arranged in descending order according to the frequency of RASs. Sequences were clustered into Asia, Europe, North America, Central America, South America, Former USSR, Oceania, Africa, Caribbean or Middle East. In each plot, only regions with at least 10 sequences were shown. Region definition was according to the Los Alamos HCV Database. “A” in North A, South A and Central A denotes America.
Naturally occurring combined RASs
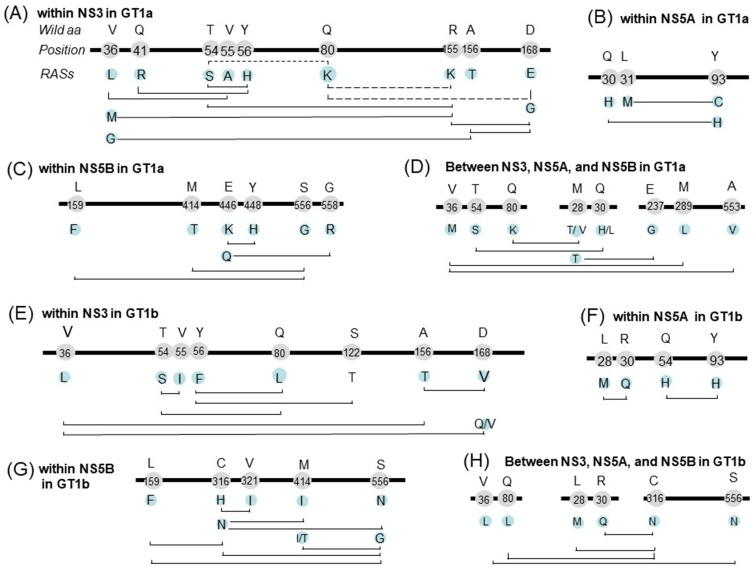
The associations of RASs within and between the HCV NS3, NS5A, and NS5B genes were investigated using a hypergeometric test to detect significantly more or less frequent RAS co-occurrences. This analysis identified pairs of variants that may result in improved or reduced fitness, and RASs were defined as co-occurring or mutually exclusive based on their observed frequencies. Subtypes GT1a and 1b were separately analyzed. For each pair of RASs, only overlapping sequences were used.
Dozens of RAS pairs were identified. In GT1a, RAS combinations within NS3 is shown in Figure 4A. Q80K has three partners (R155K, D168E, and T54S), but the frequencies of all their combinations was significantly lower than expected. Q80K and R155K were respectively observed in 31.87% (2275/7137) and 0.96% (69/7137) of GT1a sequences, but the pair was only found in 0.056% (4/7137) of sequences compared with the expected level (0.308%). Thus, these RASs were considered a mutually exclusive pair. All of these RASs, except Q80K, showed significantly higher co-occurrences with other RASs than expected. For example, R155K tends to be present with V36M, D168G, and T54S. Co-occurring pairs in NS5A included L31M+Y93C and Q30H+Y93H (Figure 4B), and those in NS5B included NI-L159F+NNI-S556G and NNI-M414T+NNI-S556G (Figure 4C). We also identified some co-occurring RASs pairs among NS3, NS5A, and NS5B, including NS3-Q80K+NA5A-M28T/V, NS3-T54S+NS5A-Q30H/L and NS3-V36M+NS5B-A553V and so on (Figure 4D). Two RASs from different regions co-occurred rarely in this study (one or two of the 734 sequences). This means that each of these RASs occurred rarely, but they tend to be co-occurred.
Figure 4.
Co-occurring and mutually exclusive RAS pairs within NS3 (A), NS5A (B) and NS5B (C) and among regions (D) in GT1a; within NS3 (E), NS5A (F) and NS5B (G) and among regions (H) in GT-1b. Bold line represents HCV genome, the figures on the line indicate the location of amino acid, the characters above the location indicate the wild type amino acids, and the characters below the location indicate RASs. The solid, and dashed line connecting two RASs indicate co-occurring RAS pairs (significantly more frequent appearance than expected) and mutually exclusive RAS pairs (significantly infrequent occurrence than expected), respectively. Amino acid residue position is numbered relative to the first amino acid of the NS3, NS5A, or NS5B region.
For GT1b, co-occurring pairs in NS3, NS5A, and NS5B were shown in Figure 4E, F, and G respectively. NS3 RAS pairs included T54S+Q80L, Y56F+S122T and so on. NS5A pairs included Y93H+Q54H and L28M+R30Q. Within NS5B, similar to GT1a, pairs NI-L159F+NNI-S556G and NNI-M414I/T+NNI-S556G were identified. Other pairs included C316H+V321I. 95.4% of sequences with 316H have 321I. Co-occurring pairs between NS3, NS5A, and NS5B, included NS5A-L28M+NS5B-C316N, NS5A-R30Q+NS5B-C316N, and NS3-V36L+NS5B-S556N (Figure 4H).
Discussion
This study investigated naturally occurring RASs among all 86 confirmed HCV subtypes using nucleotide sequences from multiple public databases. We analyzed the frequency and distribution of RASs based on HCV subtype and global geographic regions. In addition, co-occurring RAS and mutually exclusive RAS pairs were identified in subtypes GT1a and 1b within or between the NS3, NS5A, and NS5B genes.
The frequency of Q80K in NS3 varied by both HCV subtype and geographic regions. This RAS was detected in nearly one-third of HCV GT1a sequences and was particularly prevalent in North America, which corroborates findings from previous studies. The NS3 R155K and D168E substitutions, which confer resistance to simeprevir and cross-resistance to other NS3/4A protease inhibitors, appeared in 0.96% and 0.23% of HCV GT1a sequences, respectively. Frequencies of Q80K+R155K and Q80K+D168E were lower than expected, and the pairs appeared to be mutually exclusive. However, these observations contrast with those from another study in which 83% (29/35) of patients infected with HCV GT1a harboring Q80K who experienced virologic failure with simeprevir plus Peg-IFNα/RBV developed virus with a treatment-emergent R155K.41 R155 enables protein conformation favorable for interactions with the quinoline moiety of simeprevir (TMC435), and the salt bridge network between Q80, R155, and D168 within the complex stabilizes this conformation. The R155K mutation results in loss of the salt bridge between residue 155 and Asp168, which leads to reduced simeprevir efficacy.42
Y93H was most prevalent in GT1b (dominant in Asia), and present in 1.35% GT3a and the two 7a and one 7b sequences. Its distribution may be associated with polymorphism of some immune genes. Nguyen et al suggested that the frequency of Y93H in patients who were IFNL3 rs12979860 CC major homozygotes (30%, 3/10) was higher than in the non-CC group in Ireland (11.1%, 4/36),43 and Asian patients also had a high frequency of IFNL3 rs12979860 CC (approximately 90%).44 This association may explain the prevalence of Y93H in Asia. Y93H variants reduce viral sensitivity to ledipasvir in the GT3 HCV subtype.45 The low frequency of Y93H in GT3a is consistent with previous reports, which found only 1 or 2 patients carrying Y93H of among approximately 50 patients at baseline.46–48
The NI-specific RAS S282T, the only RAS to confer in vitro resistance to sofosbuvir, was detected in 27 sequences from GT1a, GT1b, GT2b, GT2h, GT3a, and GT4a. Although this RAS was widely distributed geographically, the most recent notation of this RAS in the searched databases was from 2008. This finding suggests S282T may be deleterious to HCV fitness and could explain why S282T has not been recently identified in samples from clinical trials and has only been found in a few patients with viral relapse in recent years.49–52 The first case involving the S282T variant was reported when viral breakthrough occurred at week 12 in a patient infected with genotype GT3a.49
L159F is not associated with reduced sofosbuvir susceptibility, although this RAS was frequently detected with C316N. The high frequency of this double mutation was reported in untreated Brazilian patients infected with GT1b.53 The combination of L159F with C316N was also frequently found in GT1b-infected patients who failed to respond to sofosbuvir/ribvirin or other sofosbuvir-based regimens.54 Notably, we found a very high prevalence of C316N, but a very low occurrence of L159F, in Asia. As demonstrated in a study of Japanese patients, deep sequencing showed that 30.0% of patients with C316N also carried L159F, indicating that the variant is present but not easily detected due to low abundance.55 S556G significantly co-occurred with C316N in GT1b sequences, which reflects results from a previous study showed this combination after treatment failure with three DAAs (paritaprevir, ombitasvir, and dasabuvir) in GT1b-infected patients.56
Two important points need to be addressed. The first one is whether the observed RASs are the result of natural HCV variation or of transmission from patients who selected RASs during DAA treatment is unclear. This information was not available in overwhelming studies. In clinical practice, it is difficult to determine the source of infection. The second one is about sequences with highly similarity. Although we have excluded all the clones from the same individuals and only kept one sequence from DAA naive patients with multiple visits, we cannot rule out the possibility that there may be some sequences originated from the same individual at different time points but are not specified in NCBI or related publications.
In conclusion, we obtained the knowledge about the geographic and subtype specific prevalence for an updated list of RASs. Our data may be of special relevance for those countries where highly effective antivirals might not be available. Considering the geographic and subtype specific RASs prevalence will help the clinicians to make optimal treatment choices. The RASs pairs both mutually exclusive and co-occurring would benefit anti-HCV drug development. In addition, given that the emergence of RASs is a growing issue in the setting of current treatment with DAAs, the results provide valuable data on the baseline prevalence, which can be used to monitor for increasing antiviral resistance in the future.
Acknowledgments
We are grateful to Dr. Hongmei Mo for the suggestion. This work was supported by the Natural Science Foundation of China [grant number 81301415], Natural Science and Technology Major Project [grant number 2014ZX10002002] and Program for JLU Science and Technology Innovative Research Team [grant number 2017TD-08].
Abbreviations
DAA, direct-acting antiviral; GT, genotype; HCV, hepatitis C virus; NI, nucleos(t)ide inhibitors; NNI, non-nucleoside inhibitors; RAS, resistance-associated substitution; SVR, sustained virologic response.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74–81. doi: 10.1111/j.1478-3231.2008.01934.x [DOI] [PubMed] [Google Scholar]
- 2.Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61(1 Suppl):S58–S68. doi: 10.1016/j.jhep.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683 [DOI] [PubMed] [Google Scholar]
- 4.Guedj J, Dahari H, Rong L, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A. 2013;110(10):3991–3996. doi: 10.1073/pnas.1203110110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103 [DOI] [PubMed] [Google Scholar]
- 6.Smith DB, Bukh J, Kuiken C, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59(1):318–327. doi: 10.1002/hep.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Meer AJ, Veldt BJ, Feld JJ, et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308(24):2584–2593. doi: 10.1001/jama.2012.144878 [DOI] [PubMed] [Google Scholar]
- 8.Hadziyannis SJ, Sette H Jr, Morgan TR, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010 [DOI] [PubMed] [Google Scholar]
- 9.Mangia A, Santoro R, Minerva N, et al. Peginterferon alfa-2b and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2005;352(25):2609–2617. doi: 10.1056/NEJMoa042608 [DOI] [PubMed] [Google Scholar]
- 10.Pang PS, Planet PJ, Glenn JS. The evolution of the major hepatitis C genotypes correlates with clinical response to interferon therapy. PLoS One. 2009;4(8):e6579. doi: 10.1371/journal.pone.0006579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asselah T, Boyer N, Saadoun D, Martinot-Peignoux M, Marcellin P. Direct-acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN-free treatment and future perspectives. Liver Int. 2016;36 Suppl 1:47–57. doi: 10.1111/liv.13027 [DOI] [PubMed] [Google Scholar]
- 12.McCown MF, Rajyaguru S, Kular S, Cammack N, Nájera I. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob Agents Chemother. 2009;53(5):2129–2132. doi: 10.1128/AAC.01598-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cento V, Mirabelli C, Salpini R, et al. HCV genotypes are differently prone to the development of resistance to linear and macrocyclic protease inhibitors. PLoS One. 2012;7(7):e39652. doi: 10.1371/journal.pone.0039652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53(5):1742–1751. doi: 10.1002/hep.24262 [DOI] [PubMed] [Google Scholar]
- 15.McPhee F, Hernandez D, Yu F, et al. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology. 2013;58(3):902–911. doi: 10.1002/hep.26388 [DOI] [PubMed] [Google Scholar]
- 16.Karino Y, Toyota J, Ikeda K, et al. Characterization of virologic escape in hepatitis C virus genotype 1b patients treated with the direct-acting antivirals daclatasvir and asunaprevir. J Hepatol. 2013;58(4):646–654. doi: 10.1016/j.jhep.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 17.Komatsu TE, Boyd S, Sherwat A, et al. Regulatory analysis of effects of hepatitis C virus NS5A polymorphisms on efficacy of elbasvir and grazoprevir. Gastroenterology. 2017;152(3):586–597. doi: 10.1053/j.gastro.2016.10.017 [DOI] [PubMed] [Google Scholar]
- 18.Ji F, Wei B, Yeo YH, et al. Systematic review with meta-analysis: effectiveness and tolerability of interferon-free direct-acting antiviral regimens for chronic hepatitis C genotype 1 in routine clinical practice in Asia. Aliment Pharmacol Ther. 2018;47(5):550–562. doi: 10.1111/apt.14507 [DOI] [PubMed] [Google Scholar]
- 19.Panel AIHG. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015;62(3):932–954. doi: 10.1002/hep.27950 [DOI] [PubMed] [Google Scholar]
- 20.European Association for Study of L. EASL recommendations on treatment of hepatitis C 2015. J Hepatol. 2015;63(1):199–236. doi: 10.1016/j.jhep.2015.03.025 [DOI] [PubMed] [Google Scholar]
- 21.Bartels DJ, Sullivan JC, Zhang EZ, et al. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J Virol. 2013;87(3):1544–1553. doi: 10.1128/JVI.02294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paolucci S, Fiorina L, Mariani B, et al. Naturally occurring resistance mutations to inhibitors of HCV NS5A region and NS5B polymerase in DAA treatment-naive patients. Virol J. 2013;10:355. doi: 10.1186/1743-422X-10-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costantino A, Spada E, Equestre M, et al. Naturally occurring mutations associated with resistance to HCV NS5B polymerase and NS3 protease inhibitors in treatment-naive patients with chronic hepatitis C. Virol J. 2015;12:186. doi: 10.1186/s12985-015-0414-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Rao HY, Xie XW, et al. Direct-acting antiviral agents resistance-associated polymorphisms in chinese treatment-naive patients infected with genotype 1b hepatitis C virus. Chin Med J (Engl). 2015;128(19):2625–2631. doi: 10.4103/0366-6999.166038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Chen ZW, Li H, Ren H, Hu P. Prevalence of hepatitis C virus-resistant association substitutions to direct-acting antiviral agents in treatment-naive hepatitis C genotype 1b-infected patients in western China. Infect Drug Resist. 2017;10:377–392. doi: 10.2147/IDR.S146595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep. 2016;6:20310. doi: 10.1038/srep20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kliemann DA, Tovo CV, Da Veiga AB, de Mattos AA, Wood C. Polymorphisms and resistance mutations of hepatitis C virus on sequences in the European hepatitis C virus database. World J Gastroenterol. 2016;22(40):8910–8917. doi: 10.3748/wjg.v22.i40.8910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patino-Galindo JA, Salvatierra K, Gonzalez-Candelas F, López-Labrador FX. Comprehensive screening for naturally occurring hepatitis C virus resistance to direct-acting antivirals in the NS3, NS5A, and NS5B genes in worldwide isolates of viral genotypes 1 to 6. Antimicrob Agents Chemother. 2016;60(4):2402–2416. doi: 10.1128/AAC.02776-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welzel TM, Bhardwaj N, Hedskog C, et al. Global epidemiology of HCV subtypes and resistance-associated substitutions evaluated by sequencing-based subtype analyses. J Hepatol. 2017;67(2):224–236. doi: 10.1016/j.jhep.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 30.Sorbo MC, Cento V, Di Maio VC, et al. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: update 2018. Drug Resist Updat. 2018;37:17–39. doi: 10.1016/j.drup.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 31.Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol. 2016;64(2):486–504. doi: 10.1016/j.jhep.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky JM. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology. 2016;151(1):70–86. doi: 10.1053/j.gastro.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 33.Ng TI, Tripathi R, Reisch T, et al. In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS3/4A protease inhibitor glecaprevir. Antimicrob Agents Chemother. 2018;62(1):e01620–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wing PAC, Jones M, Cheung M, et al. Amino acid substitutions in genotype 3a hepatitis C virus polymerase protein affect responses to sofosbuvir. Gastroenterology. 2019;157:692–704.e9. doi: 10.1053/j.gastro.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McPhee F, Ueland J, Vellucci V, et al. Impact of preexisting hepatitis C virus genotype 6 NS3, NS5A, and NS5B polymorphisms on the in vitro potency of direct-acting antiviral agents. Antimicrob Agents Chemother. 2019;63(4). doi: 10.1128/AAC.00779-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng G, Tian Y, Doehle B, et al. In vitro antiviral activity and resistance profile characterization of the hepatitis C virus NS5A inhibitor ledipasvir. Antimicrob Agents Chemother. 2016;60(3):1847–1853. doi: 10.1128/AAC.02524-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dvory-Sobol H, Han B, Lu J, et al. In vitro resistance profile of hepatitis C virus NS5A inhibitor velpatasvir in genotypes 1 to 6. J Viral Hepat. 2019;26(8):991–1001. doi: 10.1111/jvh.13116 [DOI] [PubMed] [Google Scholar]
- 38.Han B, Parhy B, Zhou E, et al. In vitro susceptibility of hepatitis C virus genotype 1 through 6 clinical isolates to the pangenotypic NS3/4A inhibitor voxilaprevir. J Clin Microbiol. 2019;57(4). doi: 10.1128/JCM.01844-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano T, Akuta N, Suzuki F, et al. Role of NS5A-L31/Y93 double wild-type in failure of glecaprevir/pibrentasvir double therapy in two patients with a history of direct-acting antiviral agent failure: an ultra-deep sequencing analysis. Intern Med. 2019. doi: 10.2169/internalmedicine.2604-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumada H, Suzuki Y, Ikeda K, et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology. 2014;59(6):2083–2091. doi: 10.1002/hep.27113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington PR, Komatsu TE, Deming DJ, et al. Impact of hepatitis C virus polymorphisms on direct-acting antiviral treatment efficacy: regulatory analyses and perspectives. Hepatology. 2018;67(6):2430–2448. doi: 10.1002/hep.29693 [DOI] [PubMed] [Google Scholar]
- 42.Xue W, Pan D, Yang Y, Liu H, Yao X. Molecular modeling study on the resistance mechanism of HCV NS3/4A serine protease mutants R155K, A156V and D168A to TMC435. Antiviral Res. 2012;93(1):126–137. doi: 10.1016/j.antiviral.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 43.Nguyen LT, Hall N, Sheerin D, Carr M, De Gascun CF. Naturally occurring HCV NS5A/B inhibitor resistance-associated mutations to direct-acting antivirals. Antivir Ther. 2016;21(5):447–453. doi: 10.3851/IMP3025 [DOI] [PubMed] [Google Scholar]
- 44.Wu R, Chi X, Wang X, et al. IFNL4 ss469415590 polymorphism contributes to treatment decisions in patients with chronic hepatitis C virus genotype 1b, but not 2a, infection. Infect Genet Evol. 2016;39:132–140. doi: 10.1016/j.meegid.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 45.Hernandez D, Zhou N, Ueland J, Monikowski A, McPhee F. Natural prevalence of NS5A polymorphisms in subjects infected with hepatitis C virus genotype 3 and their effects on the antiviral activity of NS5A inhibitors. J Clin Virol. 2013;57(1):13–18. doi: 10.1016/j.jcv.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 46.Peiffer KH, Sommer L, Susser S, et al. Interferon lambda 4 genotypes and resistance-associated variants in patients infected with hepatitis C virus genotypes 1 and 3. Hepatology. 2016;63(1):63–73. doi: 10.1002/hep.28255 [DOI] [PubMed] [Google Scholar]
- 47.Leroy V, Angus P, Bronowicki JP, et al. Daclatasvir, sofosbuvir, and ribavirin for hepatitis C virus genotype 3 and advanced liver disease: a randomized phase III study (ALLY-3+). Hepatology. 2016;63(5):1430–1441. doi: 10.1002/hep.28473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartolini B, Giombini E, Taibi C, et al. Characterization of naturally occurring NS5A and NS5B polymorphisms in patients infected with HCV genotype 3a treated with direct-acting antiviral agents. Viruses. 2017;9(8):212. doi: 10.3390/v9080212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker A, Filke S, Lubke N, et al. Detection of a genetic footprint of the sofosbuvir resistance-associated substitution S282T after HCV treatment failure. Virol J. 2017;14(1):106. doi: 10.1186/s12985-017-0779-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svarovskaia ES, Dvory-Sobol H, Parkin N, et al. Infrequent development of resistance in genotype 1–6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis. 2014;59(12):1666–1674. doi: 10.1093/cid/ciu697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donaldson EF, Harrington PR, O’Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61(1):56–65. doi: 10.1002/hep.27375 [DOI] [PubMed] [Google Scholar]
- 52.Hedskog C, Dvory-Sobol H, Gontcharova V, et al. Evolution of the HCV viral population from a patient with S282T detected at relapse after sofosbuvir monotherapy. J Viral Hepat. 2015;22(11):871–881. doi: 10.1111/jvh.12405 [DOI] [PubMed] [Google Scholar]
- 53.Peres-da-Silva A, Brandao-Mello CE, Lampe E. Prevalence of sofosbuvir resistance-associated variants in Brazilian and worldwide NS5B sequences of genotype-1 HCV. Antivir Ther. 2017;22(5):447–451. doi: 10.3851/IMP3131 [DOI] [PubMed] [Google Scholar]
- 54.Di Maio VC, Cento V, Lenci I, et al. Multiclass HCV resistance to direct-acting antiviral failure in real-life patients advocates for tailored second-line therapies. Liver Int. 2017;37(4):514–528. doi: 10.1111/liv.13327 [DOI] [PubMed] [Google Scholar]
- 55.Ito J, Suda G, Yamamoto Y, et al. Prevalence and characteristics of naturally occurring sofosbuvir resistance-associated variants in patients with hepatitis C virus genotype 1b infection. Hepatol Res. 2016;46(13):1294–1303. doi: 10.1111/hepr.12685 [DOI] [PubMed] [Google Scholar]
- 56.Dietz J, Susser S, Vermehren J, et al. Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology. 2018;154(4):976–988.e4. doi: 10.1053/j.gastro.2017.11.007 [DOI] [PubMed] [Google Scholar]