To the Editor:
CENP-A is a histone H3 variant required for accurate segregation of chromosomes during mitosis. Over the past 20 years, workers have unveiled several structural and mechanical features encoded within CENP-A nucleosomes1–7 that enable the nucleosomes to serve as an epigenetic platform for centromere assembly. In a recent study, Miell et al.8 present evidence that, when measured by atomic force microscopy (AFM), in vitro–reconstituted octamers containing CENP-A have reduced heights compared to those of nucleosome octamers containing H3. These data led the authors to propose that previous AFM images—in which native CENP-A nucleosomes purified from Drosophila or human cells4–6 were shown to possess shorter heights relative to those of native H3 nucleosomes—could have resulted from octameric nucleosomes. The authors propose that CENP-A octamers might be inherently smaller, owing to CENP-A2– histone H42 compaction2,3 within the core of the octamer. The data by Miell et al.8 were surprising. They contradict seminal AFM analysis of in vitro–reconstituted human CENP-A octameric nucleosomes, for which ~580 nm3 octameric volumes were carefully measured over a decade ago9. They also contradict the recently solved crystal structure of the CENP-A octameric nucleosome10 in which, with the exceptions of looser entry and exit DNA, subtle alterations in loop 1 and the unstructured C-terminal six amino acids, a near-perfect atomic correspondence exists between the cores of CENP-A and H3 octameric nucleosomes in vitro. It is puzzling that subtle differences between CENP-A and H3 octameric nucleosomes10 could translate into differences in height8 that would be detected by AFM performed under native conditions.
To examine whether CENP-A octameric nucleosomes are indeed smaller than are H3 nucleosomes in vitro, we obtained recombinant human CENP-A, yeast CENP-ACSE4 and canonical H3 octamers from four independent laboratories, including the source used by Miell et al.8, and reconstituted these histones in equimolar amounts (Supplementary Note and Supplementary Fig. 1) onto centromeric α-satellite–containing or ‘Widom 601’ sequence–containing plasmids, using standardized salt dialysis protocols widely accepted in the chromatin field11,12. We confirmed the quality of the resulting reconstitutions by native PAGE gels (Supplementary Fig. 2), which showed that CENP-A– and H3-containing mono-, di-, tri- and tetranucleosomes released by light micrococcal nuclease (MNase) digestion of the reconstituted plasmids migrated equivalently, results consistent with their equivalent molecular masses and with previously published data. We next applied AFM analyses4–6,13, following generally accepted quality controls, to obtain nucleosomal dimensions of the reconstituted plasmids.
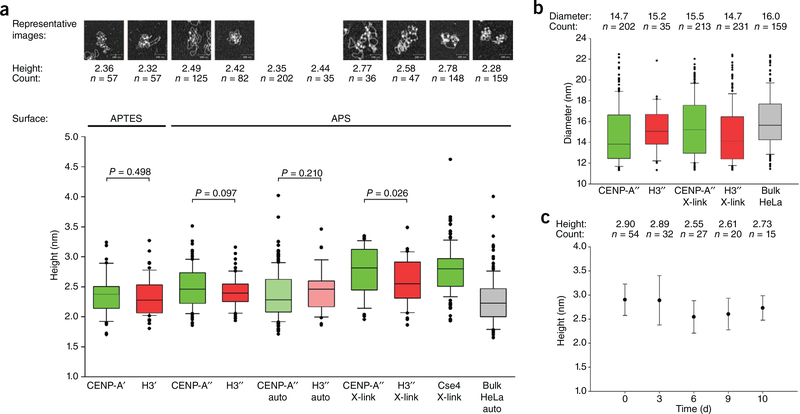
In contrast to the results reported by Miell et al.8, in vitro–reconstituted recombinant CENP-A octameric nucleosomes and H3 octameric nucleosomes, when measured in parallel by AFM, presented no significant differences in height (2.49 ± 0.03 nm versus 2.42 ± 0.03 nm, respectively; Fig. 1a) or diameter (14.69 ± 0.19 nm versus 15.22 ± 0.38 nm, respectively; Fig. 1b). We also considered potential effects of automated analysis (2.35 ± 0.03 nm versus 2.44 ± 0.06 nm, respectively; Fig. 1a), cross-linking (2.77 ± 0.07 nm versus 2.58 ± 0.05 nm, respectively; Fig. 1a) and differences in AFM surfaces (comparison of (3-aminopropyl) triethoxysilane (APTES) and 1-(3-aminopropyl) silatrane (APS); Fig. 1a). None of these treatments resulted in changes to CENP-A octameric heights relative to those of H3 octamers by AFM. We also tested CENP-ACSE4 octamers reconstituted on α-satellite plasmids, noting no significant difference relative to recombinant H3 nucleosomes (2.78 ± 0.03 nm versus 2.42 ± 0.03 nm, respectively; Fig. 1a). Finally, we considered the effect of storage on CENP-ACSE4 octameric nucleosomes (0 d, 2.90 ± 0.04 nm, 3 d, 2.89 ± 0.09 nm; 6 d, 2.55 ± 0.07 nm; 9 d, 2.61 ± 0.07 nm; and 10 d, 2.73 ± 0.07 nm; Fig. 1c), observing that CENP-ACSE4 heights remained relatively constant over a 10-d period of storage. Thus, we conclude that CENP-A and CENPACSE4 do not confer a reduction of height to nucleosome octamers when measured by AFM.
Figure 1.
AFM analysis of recombinant CENP-A and H3 octameric nucleosomes shows no appreciable difference in size. (a) Box plot representing AFM height measurements of reconstituted recombinant CENP-A (green) and H3 (red) nucleosomes and extracted HeLa nucleosomes (gray). Auto, automated analysis; X-link, samples cross-linked with 0.01% glutaraldehyde; single apostrophe, histones from J. Ottesen’s laboratory; double apostrophe, histones from A. Straight’s laboratory. P values from Kolmogorov–Smirnov test for each pair are indicated. The bottom of the box indicates the 25th percentile, the top of the box indicates the 75th percentile, and the horizontal line inside the box indicates the median. The bottom and top whiskers indicate 10th and 90th percentile, respectively, and the black dots indicate outlying values. (b) Box plot of AFM diameter measurements of nucleosomes represented in a. (c) AFM height measurements of the same sample over time. Results from Cse4 octamers cross-linked with 0.01% glutaraldehyde, deposited on APS surface and scanned over the course of 10 d are shown. Error bars, s.d. Raw data are summarized in Supplementary Data Set 1.
Our data are consistent with the crystal structure of the octameric CENP-A nucleosome10 and with previous AFM analyses of either recombinant CENP-A octamers reconstituted by chaperones9 or native CENP-A purified from human cells and reconstituted with histones H2A, H2B and H4 by salt dialysis on α-satellite DNA14. The data are also consistent with the native PAGE analysis of in vitro– reconstituted CENP-A and H3 nucleosomes (Supplementary Fig. 2).
We were unable to determine the causes for the discrepancy of the results from Miell et al.8, compared to the predicted dimensions for octameric CENP-A nucleosomes10, previously published in vitro results9,14 or results presented here. It is possible that subtle experimental variations could result, for example, in incomplete in vitro reconstitutions (causing the formation of homotypic tetramers lacking H2A–H2B dimers) or that differential hydration levels in the samples during AFM analysis could potentially influence the results. However, in our hands, experimental variability in AFM measurements is low, as evidenced by similar results for native or recombinant CENP-A reconstituted nucleosomes obtained over the course of 2 years. AFM measurements are powerful but sensitive to environmental conditions, and they require that control and experimental samples be treated gently and identically and measured in parallel13.
Our data support the notion that features of CENP-A chromatin that make it an unique epigenetic signature in vivo are unlikely to arise merely from the structure of the octameric core of the CENP-A nucleosome but rather, as previously suggested4–7,10,15–22, are caused by other factors, such as chaperones, binding partners, chromatin remodelers, histone modifications and the three-dimensional folded state of the chromatin fiber.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Straight (Stanford University) and J. Ottesen (Ohio State University) for the gift of recombinant human CENP-A and H3 octamers; R. Allshire (University of Edinburgh) for protocols; S. Henikoff (Fred Hutchinson Cancer Research Center) and J. Gerton (Stowers Institute for Medical Research) for the gift of recombinant yeast CSE4 octamers; C. Wu (US National Institutes of Health and Janelia Farm, Howard Hughes Medical Institute) for the gift of the Widom 601 plasmid; R. Gill and M. Bui for technical assistance; T. Misteli, G. Hager and S. Grewal for critical comments and K. Takeyasu for discussions on the AFM analysis presented in ref. 9.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper (doi:10.1038/nsmb.2742).
References
- 1.Sullivan KF, Hechenberger M & Masri KJ Cell Biol. 127, 581–592 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black BE et al. Nature 430, 578–582 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Sekulic N, Bassett EA, Rogers DJ & Black BE Nature 467, 347–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalal Y, Wang H, Lindsay S & Henikoff S PLoS Biol. 5, e218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitriadis EK, Weber C, Gill RK, Diekmann S & Dalal Y Proc. Natl. Acad. Sci. USA 107, 20317–20322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui M et al. Cell 150, 317–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuyama T & Henikoff S Cell 138, 104–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miell MD et al. Nat. Struct. Mol. Biol 20, 763–765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoda K et al. Proc. Natl. Acad. Sci. USA 97, 7266–7271 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachiwana H et al. Nature 476, 232–235 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Dalal Y, Fleury TJ, Cioffi A & Stein A Nucleic Acids Res. 33, 934–945 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noll M, Zimmer S, Engel A & Dubochet J Nucleic Acids Res. 8, 21–42 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walkiewicz MP, Bui M, Quenet D & Dalal Y Methods Mol. Biol. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bui M, Walkiewicz MP, Dimitriadis EK & Dalal Y Nucleus 4, 37–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guse A, Carroll CW, Moree B, Fuller CJ & Straight AF Nature 477, 354–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H et al. Science 340, 1110–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fachinetti D et al. Nat. Cell Biol 15, 1056–1066 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perpelescu M, Nozaki N, Obuse C, Yang H & Yoda KJ Cell Biol. 185, 397–407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloom K & Yeh E Curr. Biol 20, R1040–R1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribeiro SA et al. Proc. Natl. Acad. Sci. USA 107, 10484–10489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filipescu D, Szenker E & Almouzni G Trends Genet. (2013). [DOI] [PubMed] [Google Scholar]
- 22.Gascoigne KE et al. Cell 145, 410–422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger K, Rechsteiner TJ & Richmond TJ Methods Mol. Biol 119, 1–16 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Lyubchenko YL, Gall AA & Shlyakhtenko LS Methods Mol. Biol 148, 569–578 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.