Abstract
Malaria continues to be a global health burden, threatening over 40% of the world’s population. Drug resistance in Plasmodium falciparum, the etiological agent of the majority of human malaria cases, is compromising elimination efforts. New approaches to treating drug-resistant malaria benefit from defining resistance liabilities of known antimalarial agents and compounds in development and defining genetic changes that mediate loss of parasite susceptibility. Here, we present protocols for in vitro selection of drug-resistant parasites and for site-directed gene editing of candidate resistance mediators to test for causality.
Keywords: Malaria, Plasmodium falciparum, Drug resistance, Selections, Gene editing, ZFN, CRISPR/Cas9
1. Introduction
In 2017, there were an estimated 219 million reported cases of malaria, resulting in 435,000 deaths, the majority due to Plasmodium falciparum [1]. The World Health Organization recommends artemisinin-based combination therapies (ACTs) as first-line treatments for malaria [1]. Alarmingly, resistance has emerged to artemisinin—the cornerstone of ACTs [2–5]—as well as to several partner drugs and earlier first-line therapies. Detailed analyses of genetic determinants of antimalarial resistance benefit molecular surveillance campaigns to monitor the appearance and spread of drug resistance and inform therapeutic strategies to overcome resistance. Important insights in drug development are also gained by subjecting compounds in the antimalarial pipeline to in vitro drug selections, in order to assess the resistance liabilities and search for their resistance mechanisms that in some instances correspond to the actual drug targets.
To simulate the evolution of parasites that happens in clinical settings, several groups including ours have pressured parasites with sublethal concentrations of drugs, enriching for parasites that have mutated as a means to survive drug pressure [6–8]. In vitro drug resistance selections can be performed in a single-step, stepwise, or pulse manner. Briefly, single-step selections involve large numbers of parasites pressured at one drug concentration, often 3–5× IC50 values, where the IC50 is calculated as the concentration that produces half-maximal growth inhibition of parasites exposed to drug for 48 or 72 hr. The benefit of this method is that drug-resistant parasites are obtained relatively quickly, typically within 1–2 months. However, this requires handling large amounts of culture (often multiple flasks of 100–200 mL each) and also necessitates large quantities of red blood cells and culture media. Stepwise selections are easier to handle, as these can be performed using parasite cultures of only 5 mL. Parasites are incubated with drug media starting at 1× IC50, and the concentration of drug exposure is slowly increased over time. Parasites must be monitored carefully, and this selection method can result in a lengthy selection period (requiring several months of continuous culture). In situations where drug-resistant parasites cannot be obtained with either of the methods described, an alternative method is to briefly pulse parasites with drug. Here, parasites are exposed to drug for 1–2 days, then drug is washed off, and parasites are allowed to recover. To obtain drug-resistant parasites that stably maintain their phenotype, we prefer single-step selections.
Once drug-resistant mutants are obtained, these are typically cloned by limiting dilution, and resistance is verified by observing a shift in IC50 values. Genetic mutations are then identified by whole-genome sequencing or, in the case of known candidates, Sanger sequencing. To confirm that these mutations confer drug resistance, the mutation is reintroduced into drug-sensitive parasites, or the mutation in drug-resistant parasites is reverted to the wild-type sequence to confer drug sensitivity. Genetic manipulation of P. falciparum has historically been challenging. With the newfound ability to precisely introduce double-strand breaks in the genome, as opposed to earlier methods of single or double crossover-based recombination methods [9, 10], researchers no longer need to wait for stochastic breaks in the DNA in order to achieve homologous recombination. Site-directed gene editing via zinc-finger nucleases (ZFN) or clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated gene 9 (Cas9) has revolutionized our ability to efficiently edit parasite genomes [11–13]. While ZFNs were developed first for Plasmodium, CRISPR/Cas9 has become a more widely used tool and thus is first discussed below.
The CRISPR/Cas9 system widely used in gene-editing technologies relies on the type II endonuclease Cas9 [14–16], which cleaves double-stranded DNA three nucleotides upstream of the protospacer-adjacent motif (PAM) via its RuvC and HNH nuclease domains. Cas9 is directed to cleavage sites by complementary base pairing with the guide RNA, which is composed of two distinct components: the CRISPR RNA (crRNA) and the trans-activating CRISPR RNA (tracrRNA). In a landmark discovery, Jinek et al. demonstrated that a single-guide RNA (sgRNA) can be engineered to act in place of the crRNA and tracrRNA [14]. Thus, to mediate a particular site-directed break, one only needs to replace the sgRNA targeting sequence in vectors that express sgRNA and Cas9. A break in the integrity of the genome is potentially catastrophic and must be repaired. P. falciparum do not possess components of the nonhomologous end-joining (NHEJ) pathway. Instead, these parasites rely primarily on homologous recombination to repair DNA breaks [17, 18]. Thus, P. falciparum gene editing can be mediated by supplying three elements: (1) a Cas9 endonuclease, (2) a sgRNA, and (3) a donor template with homology regions upstream and downstream of the cleavage site. An important consideration is that donor templates must include silent mutations at the Cas9 recognition sites to prevent cleavage of either the supplied donor template or the template that has been integrated into the genome. Nucleotide mutations within the PAM or in the seed region, which is the 12 nucleotide sequence directly upstream of the PAM motif, are most effective in disrupting Cas9 binding [14]. The ability to direct double-stranded breaks rather than waiting for a break to occur stochastically has dramatically reduced the amount of time needed to genetically manipulate parasites. In addition, the relatively low cost of performing CRISPR/Cas9 has led to its adoption in many laboratories.
The pioneering studies of CRISPR/Cas9 in P. falciparum, published within months of each other in Nature Biotechnology and Nature Methods, employed two-plasmid approaches to express Cas9, sgRNA, and a donor template [12, 13]. At the same time, studies in P. yoelii employed a one-plasmid approach [19]. These studies utilize Streptococcus pyogenes Cas9 (SpCas9), which has a PAM recognition motif of 5′-NGG-3′downstream of the protospacer element [14]. Ghorbal et al. delivered SpCas9 on the pUF1 plasmid, which also contains a yeast dihydroorotate dehydrogenase (ydhodh) expression cassette that confers resistance to PfDHODH inhibitors such as DSM1 [20]. The second plasmid, pL7, delivered a sgRNA expressed under the P. falciparum U6 snRNA polymerase III promoter and a donor template. pL7 also expresses human dihydrofolate reductase (hdhfr) and yfcu (yeast cytosine deaminase and uridyl phosphoribosyl transferase), allowing positive selection with WR99210 and negative selection with 5-fluorocytosine, respectively [21, 22]. Gene knockouts were successfully obtained with transfection of pL7 as an intact or a linearized plasmid. Using CRISPR/Cas9, Ghorbal et al. engineered gene knockouts and marker-free point mutations. Successful editing was observed within 3 weeks of transfection of ring-stage parasites and in as little as 8 days with nucleofection of schizont stages [13].
Because the P. falciparum U6 snRNA polymerase III promoter had not been well defined, Wagner et al. chose instead to use promoter and terminator sequences from T7 RNA polymerase to express sgRNA. pCas9-sgRNA-T, which contains a blasticidin S-deaminase (bsd) selectable marker, was used to deliver SpCas9 and the sgRNA [12]. The second plasmid, pT7-RNAP-HR, delivers T7 RNA polymerase and the donor template. This group selected only for the Cas9-expressing plasmid and obtained successful marker-free gene disruption within 4–6 weeks.
For similar reasons, Marcus Lee (group leader at the Wellcome Sanger Institute in Hinxton, UK, formerly in the Fidock lab at Columbia University in New York, USA) also decided on a two-plasmid approach in which sgRNA transcription was driven by a T7 RNA polymerase promoter sequence. The first plasmid, named pDC2-Cas9-sgRNA-hdhfr, delivers SpCas9 driven by the calmodulin promoter and a sgRNA driven by the T7 RNA polymerase promoter, and also contains the selectable marker hdhfr. The second plasmid, pDC2-T7Pol-donor-bsd, expresses a calmodulin-driven T7 RNA polymerase and a bsd selectable marker. These plasmids have been used to mediate point mutations in three genes, and successful editing was observed within 3–4 weeks [23–25]. By replacing the T7 promoter with a U6 snRNA polymerase III promoter (pDC2-cam-Cas9-U6-sgRNA-hdhfr), he obviated the need to express T7 RNA polymerase, and thus the second plasmid was simplified to only express the donor template and bsd (pDC2-donor-bsd). Using these plasmids, successful editing was obtained within 2–8 weeks of transfection [6, 24].
An alternative plasmid-free approach is to deliver SpCas9 protein, a dual guide RNA (dgRNA) comprising of both tracrRNA and crRNA, and a single-strand template DNA for repair. Parasites were selected using a compound that confers resistance to parasites that incorporate the mutation of interest, and editing was obtained within 4–5 weeks post-electroporation [26].
Multiple groups have used original or modified pUF1 and pL7 plasmids [13] to introduce point mutations [27], generate reporter lines [28], tag proteins of interest [29, 30], or produce gene knockouts [31, 32]. Importantly, this technology can now be used to demonstrate the nonessentiality of conserved genetic elements [33] and genes [30, 34–36], a subset of which had earlier been proposed to be essential due to previous unsuccessful knockout attempts. Variations of these plasmids have been used to achieve particular goals. In a conditional knockdown study, three plasmids were used, one each for SpCas9, sgRNA, and the donor template, respectively [36]. The pDC2-cam-Cas9-U6-sgRNA-hdhfr vector [6, 24] was modified to pDC2-Cas9-hDHFRyFCU, in which hDHFR is fused with yFCU and used in combination with a markerless plasmid encoding the dimerization-competent Cre (DiCre) recombinase and flanked with sequences to insert the recombinase into p230p or pfs47. Marker-free parasite clones were successfully obtained 7 weeks post-transfection in this study, thereby establishing the benefit of using CRISPR/Cas9 to rapidly generate parasite lines expressing dimerization-competent Cre (DiCre)-recombinase [37]. P. falciparum parasites constitutively expressing Cas9 have also been generated. These parasites, when paired with a tetracycline-controlled conditional knockdown system, have been used to investigate the essentiality of several genes [38–40].
In the rodent malaria parasite P. yoelii, CRISPR/Cas9 has been used for gene deletion, gene insertion, and the introduction of point mutations [19]. Here, SpCas9, sgRNA, and the donor template were delivered on a single plasmid. sgRNA was driven by a P. yoelii U6 snRNA promoter, while SpCas9 was driven by the P. berghei eef1α promoter. Also encoded in this pYC vector was the donor template that included regions for homologous recombination and a hdhfr cassette that mediate resistance to the selection agent pyrimethamine. P. yoelii transfected with these constructs were then intravenously injected into mice for parasite propagation, and pyrimethamine was supplied in the drinking water 1 day after parasite injection. Edited parasites were observed in as little as 5 days postinjection [19]. This strategy was also used to investigate the functions of the PyApiP2 gene family using knockout studies, in which 12 out of 24 genes were successfully targeted [41]. Zhang et al. then modified pYC such that hdhfr was replaced with a hdhfryfcu cassette to allow positive and subsequent negative selection, which as described earlier permits sequential genetic modification of parasites. This strategy was used to tag an ookinete surface protein and to confirm using knockout studies that genes shown to be involved in ookinete motility in P. berghei have similar functions in P. yoelii [42].
Precise gene editing can also be mediated by ZFNs, whose use in P. falciparum predated CRISPR/Cas9. This technology relies on a pair of zinc fingers that each recognize unique 15–18 nucleotide DNA sequences located several nucleotides apart on either strand of the DNA helix. Each zinc-finger sequence is fused to separate halves of the FokI restriction endonuclease, such that the enzyme can refold as an obligate heterodimer upon binding of the two DNA-binding zinc-finger proteins. This results in the introduction of a double-stranded break [43, 44]. ZFN-mediated gene editing in P falciparum has proven to be efficient [11, 45–47] but requires custom design of ZFNs and can be cost-prohibitive.
Here we describe a method for single-step in vitro resistance selections, followed by gene editing via a two-plasmid U6 expression-based CRISPR/Cas9 strategy or ZFNs in P. falciparum.
2. Materials
2.1. Parasite Cultures
Dd2, NF54, 3D7, Cam3.II, and V1/S parasite lines, available from the Malaria Research and Reference Reagent Resource Center (MR4, Manassas, VA—http://www.malaria.mr4.org), are routinely used for in vitro drug resistance selections and gene editing. The procedures described here are not restricted to these particular parasites.
RPMI 1640 with l-glutamine (Invitrogen, Carlsbad, CA), 25 mM N-2-hydroxyethylpiperazine-N′ −2-ethanesulfonic (HEPES; CalBiochem, San Diego, CA), 50 mg/L hypoxanthine (Sigma-Aldrich, St. Louis, MO), 0.25% sodium bicarbonate (NaHCO3; Sigma-Aldrich) and 0.01 mg/mL gentamicin (Invitrogen) medium is supplemented with 0.5% Albumax II (Invitrogen) to constitute complete medium. Store at 4 °C. Media used in this protocol are always complete media.
Human red blood cells (RBCs) are obtained from Interstate Blood Bank (Memphis, TN). RBC must be fresh and no more than 2 weeks old when used.
2.2. In Vitro Drug Resistance Selections
Drug compound to be examined.
SYBR Green I (Invitrogen by Thermo Fisher Scientific, Waltham, MA), supplied at 10,000× in DMSO. It detects parasite DNA and is used at 1×. Store at −20 °C, avoid repeat freeze-thaw cycles, and keep away from light.
Mitotracker Deep Red FM (Molecular Probes, Eugene, OR) supplied at 50 μg and reconstituted in DMSO to 500 μM. It detects mitochondria-active parasites and is used at 100 nM. Store at −20 °C, avoid repeat freeze-thaw cycles, and keep away from light.
BD Accuri C6 Plus Flow Cytometer to measure drug inhibition of parasite growth (see Note 1).
Softwares FlowJo (Becton Dickinson) and Prism (GraphPad).
2.3. Site-Specific Editing Using CRISPR/Cas9 or ZFN
Plasmids for CRISPR/Cas9-mediated editing express Cas9, sgRNA sequence that targets the endonuclease to its cleavage site, and a donor template containing silent mutations at the Cas9 binding site.
Plasmids for ZFN-mediated editing express a pair of ZFNs (custom-ordered from Sigma-Aldrich, St. Louis, MO) and a donor repair template that contains silent mutations at the ZFN binding site. Several customized ZFNs, specific for pfcrt, eef1a, pfmdr1, k13, lipoic acid ligase (lipB), elongase1, or pfs47, have already been published or obtained by the Fidock lab and are available upon request to that lab.
For both types of site-directed gene editing, we use a two-plasmid approach in which one plasmid encodes a bsd selectable marker, while the other encodes a hdhfr expression cassette, thus allowing plasmid selection using blasticidin and the antifolate drug WR99210 [21, 24, 25, 48].
2.4. Parasite Transfection and Selection of Recombinant Lines
Cytomix (1×): 120 mM KC1, 0.2 mM CaCl2, 2 mM EGTA, 10 mM MgCl2, 25 mM HEPES, 5 mM K2HPO4, 5 mM KH2PO4; pH 7.6. Store at room temperature.
An electroporator, e.g., Gene Pulser XCell (BioRad, Hercules, CA) and 0.2-cm cuvettes (BioRad).
Blasticidin HCl (Invitrogen) is dissolved in tissue culture-grade water at 10 mg/mL, sterile-filtered, and stored in aliquots at −80 °C. Parasites are exposed to a concentration of 2.5 μg/mL.
WR99210 (Jacobus Pharmaceuticals, Princeton, NJ; molecular weight 394.35) is dissolved in tissue culture-grade water, and working stocks of sterile-filtered, 25 μM WR99210 are stored in aliquots at −80 °C. Parasites are exposed to a concentration of 2.5 nM.
3. Methods
3.1. In Vitro Drug Resistance Selections
Thaw parasite of choice, and grow under standard hypoxic conditions (5% O2, 5% CO2, 90% N2; [48]). Recently cloned parasites are preferred to minimize genetic drift.
Determine the concentration at which drug compound inhibits 50% growth (IC50). Parasites predominantly in the ring stages (see Note 2) seeded at 0.2% parasitemia and 1% hematocrit in a 96-well plate are exposed to serially increasing concentrations of drug compound. After incubation for 72 h at 37 °C in a hypoxic environment (5% O2, 5% CO2, 90% N2), samples are resuspended, and 5 μL of each well is removed and mixed with 40 μL of stain (1× SYBR Green I and 100 nM MitoTracker Deep Red, diluted in 1× phosphate-buffered saline (PBS)). Assays are read on a BD Accuri C6 Flow Cytometer and analyzed using FlowJo®. IC50 values are determined by nonlinear regression analysis using the Prism software. At least three biological repeats are performed to accurately determine IC50 values [25, 49].
For single-step selections, large numbers of parasites (2 × 109) are pressured at 3–5× IC50, and the experiment is performed in triplicate. Grow parasites to the appropriate volume, ensuring that cultures are always healthy and maintained at a parasitemia between 1% and 5%. Prior to beginning the selection, it is preferable to start with a culture with lower volume and higher parasitemia rather than higher volume and lower parasitemia (even though this gives the same number of parasites) to ensure that the parasites are actively growing. A 100 mL culture at 5% parasitemia and 4% hematocrit corresponds to 2 × 109 parasites (see Note 3). Since this experiment is performed in triplicate, you will need >300 mL of culture in total before starting. These cultures will then be diluted from 5% to ~2–2.5%, so each of the triplicate flasks will eventually hold ~200–250 mL. When beginning the selections, cultures should be predominantly ring stages. If culturing in multiple vessels, combine all cultures together. Remove 10 mL cultures to freeze in Glycerolyte 57 (Fenwal) as a preselection comparative line. Add the appropriate amount of drug (5× IC50) and RBC to achieve ~2–2.5% parasitemia and 4% hematocrit (see Note 4). Divide into three flasks, and incubate at 37 °C in hypoxic conditions.
Early the next day, remove the supernatant and supply cultures with fresh medium containing the selection concentration of drug. Parasitemia is monitored by Giemsa staining a thin blood smear. Continue this procedure for 6 days, ensuring that the parasitemia is sufficiently reduced. By day 6, parasitemia should be close to zero.
On day 7, refresh drug medium and add fresh red blood cells, equivalent to an additional 0.5% hematocrit.
Refresh drug medium every other day. Once parasites are no longer detected, monitoring can be reduced from every other day to once a week.
On day 14, and every week thereafter, fresh RBC is added, and total culture volume is reduced. The new total culture volume will be 75% of the current total volume. To achieve this, culture medium is aspirated, and a thin blood smear is made to check parasitemia. Measure or estimate the remaining volume and discard 50% of this culture. Add fresh RBC to achieve 4% hematocrit for 1/4 of the total new volume. Add fresh drug media to achieve the new reduced total volume (see Note 5 for sample calculation). Culture volume is repeatedly diminished in this way every 7 days until cultures are 50 mL, a volume that we find easy to handle and maintain. At this point, when it’s time to refresh RBC, 1/4 of resuspended culture is removed, 1 mL of fresh RBC (from the 50% hematocrit stock) is added, and volume is brought up to 50 mL with fresh drug media. Throughout this process, continue to refresh drug medium every 2–3 days (three times per week is a useful schedule).
The experiment is carried out to until parasites are visually detected by microscopy. Cultures are discarded if no parasites are visible by day 60 (see Note 6).
3.2. Site-Specific Editing Using CRISPR/Cas9: Design of Plasmids
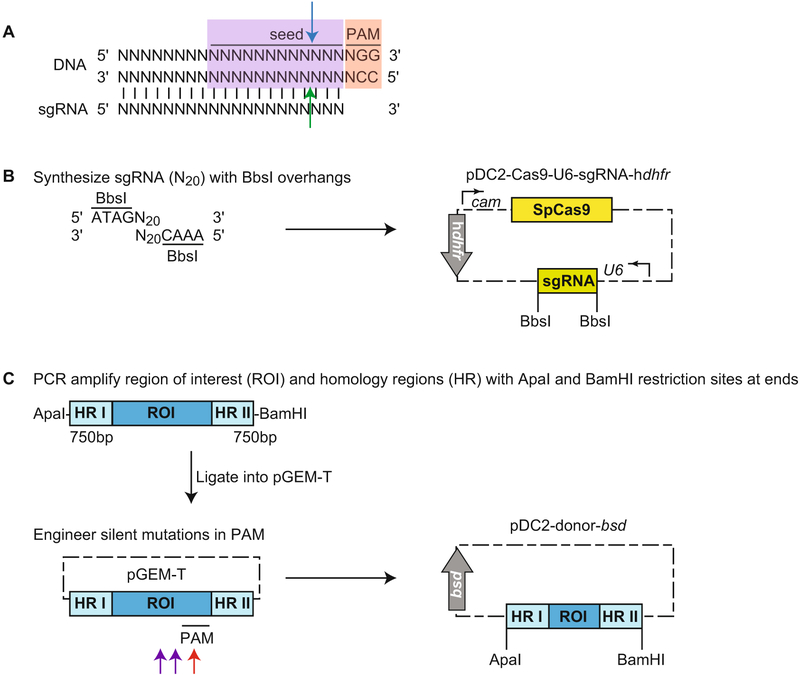
This strategy outlines a two-plasmid approach using the U6 promoter to drive sgRNA expression. The first plasmid encodes a P falciparum calmodulin-driven SpCas9, a P. falciparum U6 promoter-driven sgRNA, and a hdhfr selectable marker. The second plasmid encodes the region to be replaced, and homology arms on either side, plus a bsd selectable marker (Fig. 1) [6].
Identify suitable guide RNAs that will bind to 20 nucleotides upstream of a 5′-NGG-3′ PAM motif (see Note 7). The study by Ghorbal et al. demonstrated that unlike its counterpart in mammalian cells, P. falciparum RNA polymerase III does not have a strict requirement for a G nucleotide at the 5′ end to initiate transcription. PAMs and sgRNA can be on either strand of DNA. The cleavage site, which occurs three nucleotides upstream of the PAM sequence, should be as close as possible to sequences to be modified to facilitate efficacy, although we have had success with cleavage sites up to 750 bp away from the modified site [25]. In addition, ensure that the sgRNA binds to a sequence that is unique within the genome. This can be done using a variety of tools, including https://chopchop.rc.fas.harvard.edu/, E-CRISP, or downloading Protospacer Workbench [50] from http://www.protospacer.com/. Select the best two guides by proximity to site of interest or the Doench score. Be sure to select guide sequences with G or C nucleotides interspersed along the sequence, and avoid sequences that contain long tracts of only A or T nucleotides.
Custom synthesize these sequences with BbsI restriction site overhangs, which allows insertion behind the U6 promoter in pDC2-Cas9-U6-sgRNA-hdhfr [6]. To do so, order phos-phorylated primers with 5′-ATAGN20 as the forward primer and 3′-AAACN20 as the reverse primer, where N20 is the sgRNA identified above. Note that only the protospacer, and not the PAM sequences, are included in these N20. Anneal primer pairs to generate dsDNA, and insert into BbsI-digested pDC2-Cas9-U6-sgRNA-hdhfr (Fig. 1a, b).
Select a donor that contains the region of interest, with homology arms on either side of ~750 bp each (see Note 8). PCR amplify this region using primers with ApaI and BamHI restriction sites at the 5′ and 3′ ends, respectively. Clone into pGEM-T for further modification of nucleotides at sites of interest to achieve desired mutations/insertions/deletions. Mutate at least one nucleotide at the PAM site (Fig. 1a, red box, c, red arrow) or two nucleotides in the seed region (the first 12 nucleotides upstream of the PAM site) closest to the PAM (Fig. 1a, purple box, c, purple arrows) to prevent Cas9-mediated cleavage of the donor plasmid or integrated DNA. Sequence both strands of the donor template in full to avoid plasmids with unwanted mutations. Insert fragment into ApaI-and BamHI-digested pDC2-donor-bsd plasmid [6] (Fig. 1c).
Fig. 1.
CRISPR/Cas9-mediated editing using a two-plasmid approach. (a) This diagram shows the sgRNA binding via complementary base pairing to the 20 nucleotides of DNA upstream of the PAM sequence (red box). When custom ordering primers to engineer double-stranded sgRNAs, do not include the PAM sequence. When brought to the DNA sequence by sgRNA, Cas9 mediates a blunt double-stranded break via its RuvH nuclease domain, which cleaves three nucleotides upstream of the PAM in the sequence that is complementary to the recognized DNA strand (blue arrow), and its HNH nuclease domain, which cleaves the strand that is recognized (green arrow). The seed region (purple box) is the 12 nucleotides immediately upstream of the PAM. Mutations within the PAM will be most effective in blocking Cas9 recognition and binding to DNA (and thus cleavage). If it is not possible to introduce silent mutations within the PAM, mutate two nucleotides within the seed region, preferably close to the PAM. (b) pDC2-Cas9-U6-sgRNA-hdhfr delivers SpCas9 and a U6-driven sgRNA and contains a hdhfr selectable marker. Custom synthesize the sgRNA with BbsI overhangs, and then insert into BbsI-digested pDC2-Cas9-hdhfr to generate pDC2-Cas9-U6-sgRNA-hdhfr. (c) pDC2-donor-bsd delivers the donor repair template and contains a bsd selectable marker. PCR amplify a fragment containing the region of interest (ROI) flanked by ~750 bp homology region (HR), with ApaI and BamHI restriction sites at the ends. Clone the fragment into pGEM-T to engineer silent mutations at the Cas9 binding site. Ideally, one nucleotide within the PAM is mutated (red arrow). If this is not possible, two nucleotides within the seed region and close to the PAM site are mutated (purple arrows). Insert this fragment into ApaI-and BamHI-digested pDC2-dsrfto generate pDC2-donor-bsd
This general protocol can also be followed if using other pairs of plasmids. For cloning strategies using pUF1-Cas9 and pL7 plasmids [13] (see Note 9). For cloning into pCas9-sgRNA-T and pT7-RNAP-HR plasmids [12] (see Note 10). To clone into pDC2-Cas9-sgRNA-hdhfr and pDC2-T7 pol-donor-bsd plasmids [24, 25] (see Note 11). Lastly, a one-plasmid approach can also be employed to deliver SpCas9, sgRNA, and a donor template (see Note 12).
3.3. Site-Specific Editing Using ZFN: Design of Plasmids
Pairs of ZFNs can be ordered through Sigma-Aldrich (CompoZr ZFNs, Product No. CSTZFN-1KT). Specify the target region of interest, and ensure that the cleavage site is close to the site of the desired modification. Select the two most active pairs based on the yeast proxy cleavage assays, results of which are provided by the company.
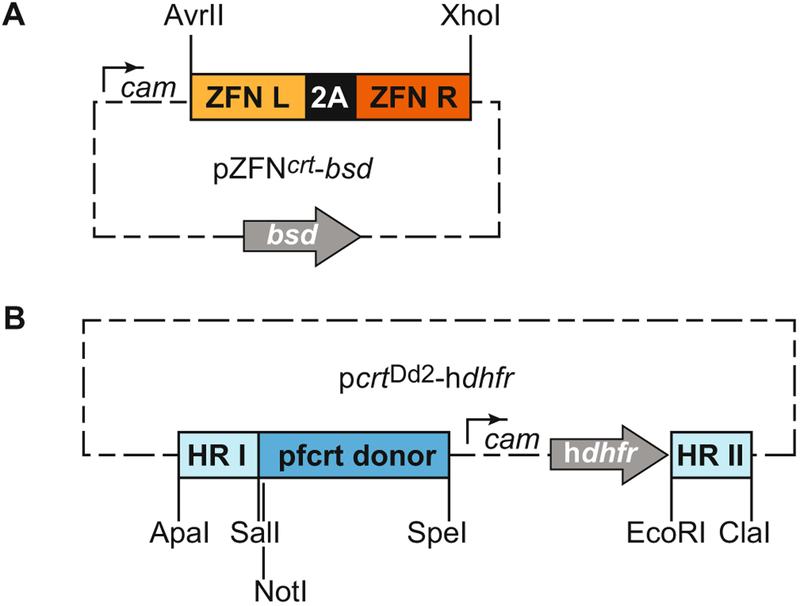
The ZFN pair is cloned into a pDC2 vector expressing a bsd selectable marker (pZFN-bsd), downstream of a calmodulin promoter and upstream of a hsp86 3′ untranslated region (UTR) [11]. The ZFN described here can be excised with AvrII and XhoI restriction enzymes to be replaced with custom ZFNs of interest that have been digested with NheI and XhoI (Fig. 2a). The ZFN pair is expressed from the same promoter and is separated by a “2A ribosome skip peptide” sequence that yields two separate proteins translated from one combined transcript.
Select a donor that contains the region of interest, with homology arms on either side of about 750 bp each (see Note 8). Amplify this region by PCR, cloning into pGEM-T for further modifications including gene mutations and silent binding site mutations. To prevent FokI-mediated cleavage of the donor plasmid or the integrated plasmid, engineer silent mutations at the binding sites. This is then cloned into a pDC2 vector encoding the hdhfr cassette [11] (Fig. 2b).
Fig. 2.
ZFN-mediated editing using a two-plasmid approach. (a) pZFNcrt-bsd delivers two zinc-finger sequences fused to separate halves of the FokI restriction endonuclease and contains a bsd selectable cassette. This particular pair of ZFNs can be excised with AvrII and XhoI restriction enzymes to be replaced with custom ZFNs digested with NheI and XhoI. (b) pcrtDd2-hdhfr delivers the donor repair template. This vector and the restriction enzyme sites that can be used to clone in donor regions are shown as described in [11]. Note that donor templates can be made on any standard P. falciparum expression plasmid
3.4. Parasite Transfection and Selection of Recombinant Lines
50 μg of each plasmid is required for transfection. In the case of ZFN, these are the plasmids encoding ZFN and that of the donor. In the case of CRISPR/Cas9, these plasmids encode Cas9 and sgRNA and the donor. When preparing the plasmids, directly resuspend these plasmids in 1× Cytomix.
When ready to transfect, parasites should be at ~5% parasitemia and consist predominantly of ring stages. 2.5 mL of culture should be used per transfection. Parasite cultures are harvested, pelleted at 500 × g for 3 min, supernatant removed, and resuspended in an equal volume of 1 × Cytomix. This resuspension is then spun again at 500 × g for 3 min and resuspended in 300–400 μL of 1× Cytomix. Total volume of parasites + plasmids should be ~450 μL, so add 1× Cytomix according to the volume of plasmid DNA. Be sure to keep a stock of frozen parasites made the day of electroporation to have an appropriate control for future comparisons with transfected parasites.
Put the plasmids in a 1.5 mL centrifuge tube. Mix in the resuspended parasite culture. Transfer to an electroporation cuvette.
Prepare one well (per transfection) of a six-well plate with 4 mL media containing 4% hematocrit. Aliquot 1 mL media per transfection into a conical tube.
Settings for electroporation of infected red blood cells are voltage of 0.31 kV and capacitance of 950 μF. Electric resistance is infinite, and cuvette gap is 2 mm.
Immediately after electroporation, add 1 mL of complete media to the cuvette.
Transfer the contents of the cuvette to the six-well plate, and place at 37 °C. Once the RBC have settled, tilt the plate at a 45° angle, remove the supernatant, and add fresh culture medium. Incubate at 37 °C.
The next day, transfected parasites are exposed to culture medium with 2.5 nM WR99210 and 2.5 μg/mL blasticidin for 6 days. 1.5 nM WR99210 is sufficient if parasites express wild-type P. falciparum dhfr. If preferred, drug selection can be kept on until edited parasites are detected. Parasitemia is monitored to ensure it does not exceed 10%. Parasites should be undetectable 6–7 days post-transfection. Evidence of abundant gametocytes on day 6 is a sign of culture stress and reduces the likelihood of successful transfections.
When parasites are detectable by microscopy, usually 14–21 days post-electroporation, culture them to ~5% parasitemia, harvest the gDNA, and verify by PCR whether parasites now harbor the desired changes. It is important to verify that binding site mutations are present to be certain that mutations were incorporated by CRISPR/Cas9- and/or ZFN-mediated gene editing. CRISPR/Cas9-mediated editing often edits close to 100% and may obviate further parasite cloning. If mixed cultures prevail, clone out bulk cultures by limiting dilution to obtain pure populations.
4. Notes
Parasitemia can also be assayed via other means, e.g., hypoxanthine incorporation, determination of SYBR Green I staining (only) with a plate reader, or DAPI (Molecular Probes, Eugene, OR)-stained parasites on a high-content imager such as the Operetta CLS (Perkin Elmer, Waltham, MA). We note that parasite inhibition assays with only SYBR Green I may yield skewed IC50 values [49].
We have found that there is generally no necessity to tightly synchronize parasites. A culture with 70–90% rings produces good IC50 growth curves. If the drug compound is particularly sensitive to parasite stage, parasites can be synchronized using 5% sorbitol [51].
Assuming one parasite per infected red blood cell (iRBC), parasite numbers can be approximated by the following formula: iRBC/mL = (1 × 1010) × (% parasitemia) × (% hematocrit).
It is imperative to start with no more than 3% parasitemia; we find 2% parasitemia is ideal. Cultures with a high starting parasitemia will crash before the drug has a chance to take effect. Slow-acting drugs, or drugs that act on different stages of the lifecycle, may allow parasitemia to increase before killing.
When reducing culture volumes, if at day 14 the current flask volume is 200 mL, then the new total volume will be 150 mL. If after aspirating the supernatant you are left with 100 mL, discard 50 mL. To this, add 3 mL fresh RBC (washed and stored at 50% hematocrit). The calculation for this is that 1/4 of 150 mL is 37.5 mL, and to achieve a 4% hematocrit, it takes 1.5 mL packed RBC (at 100% hematocrit). Since our RBC is washed and stored at 50% hematocrit, we will need to add 3 mL of washed RBC.
If selection with 5× IC50 yields no parasite-positive flasks after 60 days, the experiment can be repeated at a lower drug concentration (e.g., 3× IC50). Alternatively, stepwise selections where a single 6-well culture is pressured at 1× IC50 can also be performed. A yet milder selection strategy is pulsing parasites with drug for a day, then removing drug and allowing them to recover, and repeating the process until resistant parasites emerge.
The method described is for Streptococcus pyogenes Cas9 (SpCas9)-mediated editing, which is most commonly used in P. falciparum and was used in pioneering studies [12, 13, 19]. SpCas9 recognizes the PAM motif 5′-NGG-3′ [14, 15]. If space on a vector is limiting, the smaller Staphylococcus aureus Cas9 (SaCas9) is an attractive alternative. SaCas9 recognizes the motif S’-NNGRRT-S’ where R represents A or G [52].
Longer homology arms lead to higher recombination efficiency. If the region of interest to be inserted is large, this will generate large plasmids that might be unstable and scramble in Escherichia coli or provide very low yields. In this case, homology arms can be reduced to as little as 250 bp.
The strategy described in Ghorbal et al. [13] employs a two-plasmid approach and a U6-expressed sgRNA. No modification is necessary to pUF1-Cas9, which delivers SpCas9 and has a ydhodh selection marker. pL7 delivers U6-expressed sgRNA, donor template, and hdhfr and yfcu selectable markers. The donor template is constructed by PCR amplification of the region of interest and homology arms on either side from parasite of interest and subsequent modification at the PAM or seed regions (Fig. 1a). Alternatively, the donor is custom synthesized. In both instances, the donor template is cloned into pL7 using AflII/SpeI and NcoI/EcoRI restriction sites. The pL7 containing donor template is then digested with BtGZI, and sgRNA is inserted using the In-Fusion HD Cloning Kit. Parasites harboring pUF1-Cas9 are selected with DSM1, and parasites transformed with pL7 are selected with WR99210, which can be followed with 5-fluorocytosine for negative selection [13].
The strategy described in Wagner et al. [12] employs a two-plasmid approach and a T7 polymerase-expressed sgRNA. PCR amplify the region of interest and left and right homology arms, introduce silent mutations in the PAM or seed regions (Fig. 1a), and then assemble into SalI-digested pT7-RNAP in a Gibson reaction to produce pT7-RNAP-HR. This plasmid also delivers T7 RNA polymerase. To insert a T7 polymerase-driven sgRNA into pCas9, order custom primers to synthesize the double-stranded sgRNA, and then PCR amplify this product using the T7 promoter adaptor and T7 terminator adaptor primers as described in [12]. This is then inserted into pCas9 using SalI restriction sites to generate pCas9-sgRNA-T. This plasmid will deliver SpCas9 and sgRNA and has a bsd selectable marker. Transfections should be selected with bsd.
The strategy described in Ng et al. [25] employs a two-plasmid approach and a T7 polymerase-expressed sgRNA. Custom synthesize sgRNA with BbsI overhangs, which allows insertion behind the T7 promoter in pDC2-Cas9-hdhfr to produce pDC2-Cas9-sgRNA-hdhfr. This plasmid delivers SpCas9, a T7 promoter-driven sgRNA, and a hdhfr selectable marker. To generate the donor plasmid, PCR amplify the region of interest with SacI and AatII restriction sites at the 5′ and 3′ ends, respectively. Introduce silent mutations at the PAM or seed regions (Fig. 1a), and then insert fragment into SacI- and AatII-digested pDC2-T7pol-bsd to generate pDC2-T7pol-donor-bsd. This plasmid expresses a T7 RNA polymerase and a bsd selectable marker. Select transfections with WR99210 and bsd [25].
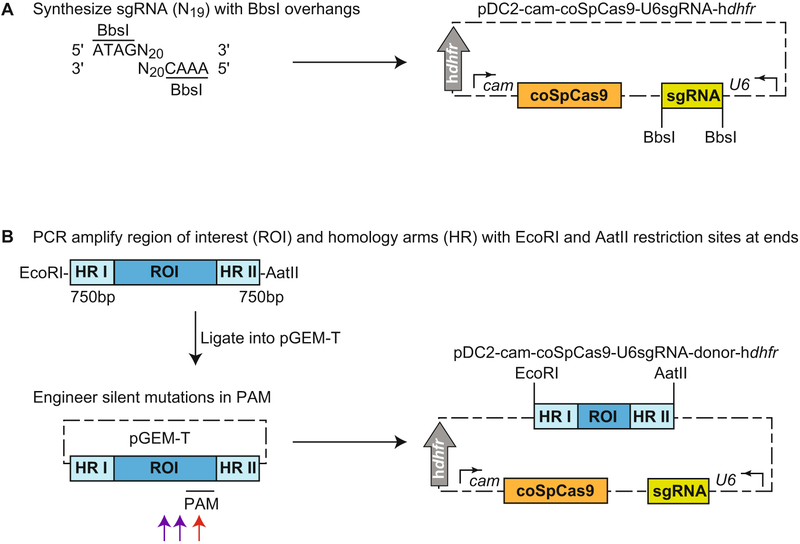
A one-plasmid approach for P. falciparum CRISPR/Cas9 editing has also been developed by Marcus Lee (Fig. 3). pDC2-cam-coSpCas9-U6sgRNA-donor-hdhfr delivers SpCas9 codon-optimized for P. falciparum (coSpCas9), U6-expressed sgRNA, a donor template, and contains a hdhfr selectable marker. Insert sgRNA generated using the methods described in Subheading3.2 into a BbsI-digested pDC2-cam-coSpCas9-U6-hdhfr to generate pDC2-cam-coSpCas9-U6sgRNA-hdhfr (Fig. 3a). PCR amplify the region of interest and homology arms with EcoRI and AatII restriction sites at the 5′ and 3′ ends, respectively, and engineer silent mutations in the PAM or seed regions (Fig. 1a). Then insert this fragment into EcoRI- and AatII-digested pDC2-cam-coSpCas9-U6sgRNA-hdhfr to generate pDC2-cam-coSpCas9-U6sgRNA-donor-hdhfr (Fig. 3b). Select transfections with WR99210.
Fig. 3.
CRISPR/Cas9-mediated editing using a one-plasmid approach. The final vector, pDC2-cam-coSpCas9-U6sgRNA-donor-hdhfr, delivers SpCas9 codon-optimized for P. falciparum, a U6-driven sgRNA, and the donor template and contains a hdhfr selectable marker, (a) Custom synthesize sgRNA with BbsI overhangs, and then insert into BbsI-digested pDC2-cam-coSpCas9-U6-hdhfr to generate pDC2-cam-coSpCas9-U6sgRNA-hdhfr. (b) PCR amplify a fragment containing the region of interest (ROI) flanked by ~750 bp homology region (HR), with EcoRI and AatII restriction sites at the ends. Clone the fragment into pGEM-T to engineer silent mutations within the PAM or seed region. Ideally, one nucleotide at the PAM site is mutated (red arrow). If this is not possible, two nucleotides in the seed region and close to the PAM are mutated (purple arrows). Insert this fragment into EcoRI- and AatII-digested pDC2-cam-coSpCas9-U6sgRNA-hdhfr to generate pDC2-cam-coSpCas9-U6sgRNA-donor-hdhfr
Acknowledgments
We thank Marcus Lee for his development of several T7- and U6-based editing systems that are discussed herein.
References
- 1.World Health Organization (2018) World malaria report 2018. https://www.who.int/malaria/publications/world-malaria-report-2018/report/en
- 2.Dondorp AM, Nosten F, Yi P et al. (2009) Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley EA, Dhorda M, Fairhurst RM et al. (2014) Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takala-Harrison S, Jacob CG, Arze C et al. (2015) Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211:670–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tun KM, Imwong M, Lwin KM et al. (2015) Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis 15:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonoiki E, Ng CL, Lee MC et al. (2017) A potent antimalarial benzoxaborole targets a Plasmodium falciparum cleavage and polyadenylation specificity factor homologue. Nat Commun 8:14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanaerschot M, Lucantoni L, Tao L et al. (2017) Hexahydroquinolines are antimalarial candidates with potent blood stage and transmission-blocking activity. Nat Microbiol 2:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowell AN, Istvan ES, Lukens AK et al. (2018) Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidhu AB, Verdier-Pinard D, Fidock DA (2002) Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Koning-Ward TF, Gilson PR, Crabb BS (2015) Advances in molecular genetic systems in malaria. Nat Rev Microbiol 13:373–387 [DOI] [PubMed] [Google Scholar]
- 11.Straimer J, Lee MC, Lee AH et al. (2012) Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat Methods 9:993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner JC, Platt RJ, Goldfless SJ et al. (2014) Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods 11:915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghorbal M, Gorman M, Macpherson CR et al. (2014) Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821 [DOI] [PubMed] [Google Scholar]
- 14.Jinek M, Chylinski K, Fonfara I et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P, Yang L, Esvelt KM et al. (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AH, Symington LS, Fidock DA (2014) DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol Mol Biol Rev 78:469–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkman LA, Lawrence EA, Deitsch KW (2014) Malaria parasites utilize both homologous recombination and alternative end joining pathways to maintain genome integrity. Nucleic Acids Res 42:370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Xiao B, Jiang Y et al. (2014) Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5: e01414–e01414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganesan SM, Morrisey JM, Ke H et al. (2011) Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Mol Biochem Parasitol 177:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fidock DA, Wellems TE (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A 94:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duraisingh MT, Triglia T, Cowman AF (2002) Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol 32:81–89 [DOI] [PubMed] [Google Scholar]
- 23.LaMonte G, Lim MY, Wree M et al. (2016) Mutations in the Plasmodium falciparum cyclic amine resistance locus (PfCARL) confer multidrug resistance. MBio 7:e00696–e00616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim MY, LaMonte G, Lee MC et al. (2016) UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat Microbiol 1:16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng CL, Siciliano G, Lee MC et al. (2016) CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol Microbiol 101:381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford ED, Quan J, Horst JA et al. (2017) Plasmid-free CRISPR/Cas9 genome editing in Plasmodium falciparum confirms mutations conferring resistance to the dihydroisoquinolone clinical candidate SJ733. PLoS One 12: e0178163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bansal A, Ojo KK, Mu J et al. (2016) Reduced activity of mutant calcium-dependent protein kinase 1 is compensated in Plasmodium falciparum through the action of protein kinase G. MBio 7:e02011–e02016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogollon CM, van Pul FJ, Imai T et al. (2016) Rapid generation of marker-free P. falciparum fluorescent reporter lines using modified CRISPR/Cas9 constructs and selection protocol. PLoS One 11:e0168362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuang D, Qiao J, Li Z et al. (2017) Tagging to endogenous genes of Plasmodium falciparum using CRISPR/Cas9. Parasit Vectors 10:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miliu A, Lebrun M, Braun-Breton C et al. (2017) Shelph2, a bacterial-like phosphatase of the malaria parasite Plasmodium falciparum, is dispensable during asexual blood stage. PLoS One 12:e0187073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nacer A, Claes A, Roberts A et al. (2015) Discovery of a novel and conserved Plasmodium falciparum exported protein that is important for adhesion of PfEMP1 at the surface of infected erythrocytes. Cell Microbiol 17:1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bansal A, Molina-Cruz A, Brzostowski J et al. (2018) PfCDPK1 is critical for malaria parasite gametogenesis and mosquito infection. Proc Natl Acad Sci U S A 115:774–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryant JM, Regnault C, Scheidig-Benatar C et al. (2017) CRISPR/Cas9 genome editing reveals that the intron is not essential for var2csa gene activation or silencing in Plasmodium falciparum. MBio 8:e00729–e00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wezena CA, Alisch R, Golzmann A et al. (2017) The cytosolic glyoxalases of Plasmodium falciparum are dispensable during asexual blood-stage development. Microb Cell 5:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bansal A, Molina-Cruz A, Brzostowski J et al. (2017) Plasmodium falciparum calcium-dependent protein kinase 2 is critical for male gametocyte exflagellation but not essential for asexual proliferation. MBio 8:e01656–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobb DW, Florentin A, Fierro MA et al. (2017) The exported chaperone PfHsp70x is dispensable for the Plasmodium falciparum intraerythrocytic life cycle. mSphere 2:e00363–e00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knuepfer E, Napiorkowska M, van Ooij C et al. (2017) Generating conditional gene knockouts in Plasmodium - a toolkit to produce stable DiCre recombinase-expressing parasite lines using CRISPR/Cas9. Sci Rep 7:3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walczak M, Ganesan SM, Niles JC et al. (2018) ATG8 is essential specifically for an autophagy-independent function in apicoplast biogenesis in blood-stage malaria parasites. MBio 9: e02021–e02017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasamu AS, Glushakova S, Russo I et al. (2017) Plasmepsins IX and X are essential and draggable mediators of malaria parasite egress and invasion. Science 358:518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidik SM, Huet D, Ganesan SM et al. (2016) A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166:1423–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Li Z, Cui H et al. (2017) Systematic CRISPR-Cas9-mediated modifications of Plasmodium yoelii ApiAP2 genes reveal functional insights into parasite development. MBio 8: e01986–e01917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Gao H, Yang Z et al. (2017) CRISPR/Cas9 mediated sequential editing of genes critical for ookinete motility in Plasmodium yoelii. Mol Biochem Parasitol 212:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyon Y, Vo TD, Mendel MC et al. (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8:74–79 [DOI] [PubMed] [Google Scholar]
- 44.Miller JC, Holmes MC, Wang J et al. (2007) An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25:778–785 [DOI] [PubMed] [Google Scholar]
- 45.McNamara CW, Lee MC, Lim CS et al. (2013) Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504:248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straimer J, Gnadig NF, Witkowski B et al. (2015) K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabryszewski SJ, Modchang C, Musset L et al. (2016) Combinatorial genetic modeling of pfcrt-mediated drug resistance evolution in Plasmodium falciparum. Mol Biol Evol 33:1554–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fidock DA, Nomura T, Wellems TE (1998) Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol 54:1140–1147 [DOI] [PubMed] [Google Scholar]
- 49.Ekland EH, Schneider J, Fidock DA (2011) Identifying apicoplast-targeting antimalarials using high-throughput compatible approaches. FASEB J 25:3583–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacPherson CR, Scherf A (2015) Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat Biotechnol 33:805–806 [DOI] [PubMed] [Google Scholar]
- 51.Lambros C, Vanderberg JP (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65:418–420 [PubMed] [Google Scholar]
- 52.Ran FA, Cong L, Yan WX et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]