Abstract
Down syndrome (DS), trisomy 21, is marked by intellectual disability and a premature aging profile including degeneration of the basal forebrain cholinergic neuron (BFCN) projection system, similar to what is seen in Alzheimer’s disease (AD). Although data indicate that perinatal maternal choline supplementation (MCS) alters the structure and function of these neurons in the Ts65Dn mouse model of DS and AD (Ts), how MCS affects the molecular profile of vulnerable BFCNs is unknown. We investigated the genetic signature of BFCNs obtained from Ts and disomic (2N) offspring of Ts65Dn dams maintained on a MCS diet (Ts+, 2N+) or a choline-normal diet (ND) from mating until weaning, then maintained on ND until 4.4–7.5 months of age. Brains were then collected and prepared for choline acetyltransferase (ChAT) immunohistochemistry and laser capture microdissection followed by RNA extraction and custom-designed microarray analysis. Findings revealed upregulation of select transcripts in classes of genes related to the cytoskeleton (Tubb4b), AD (Cav1), cell death (Bcl2), presynaptic (Syngr1), immediate early (Fosb, Arc), G protein signaling (Gabarap, Rgs10), and cholinergic neurotransmission (Chrnb3) in Ts compared to 2N mice, which were normalized with MCS. Moreover, significant downregulation was seen in select transcripts associated with the cytoskeleton (Dync1h1), intracellular signaling (Itpka, Gng3, Mlst8), and cell death (Ccng1) in Ts compared to 2N mice that were normalized with MCS. This study provides valuable insight into mechanisms of genotype-dependent differences and the effects of MCS at the molecular level within a key vulnerable cell type in DS and AD.
Keywords: Microarrays, basal forebrain cholinergic neurons, maternal choline supplementation, Down syndrome, laser capture microdissection, Ts65Dn
Introduction
Down syndrome (DS) is a multi-faceted condition caused by triplication of human chromosome 21 (HSA21) that results in altered cardiac, respiratory, endocrine, gastrointestinal, and immunological systems (Bonamico et al., 2001; Cohen, 2006; Dyken, Lin-Dyken, Poulton, Zimmerman, & Sedars, 2003; Fong & Brodeur, 1987; Freeman et al., 2008). In addition to physiological anomalies, individuals with DS present with cognitive deficits in the domains of learning, memory, and language, as well as increased incidence of neuropsychiatric conditions (Alexander et al., 1997; Haxby & Schapiro, 1992; Oliver, Crayton, Holland, Hall, & Bradbury, 1998). While there is significant intra-individual variability in the degree of physiological and cognitive dysfunction, intellectual disability (ID) is considered a hallmark of DS (Epstein, 1995; Maatta, Tervo-Maatta, Taanila, Kaski, & Iivanainen, 2006). Compounding the ID seen in individuals with DS is a premature aging phenotype (Franceschi et al., 2019) that includes a neuropathological profile similar to that seen in Alzheimer’s disease (AD), namely, the deposition of amyloid-beta (Aβ) plaques and tau-containing neurofibrillary tangles (NFTs) by early midlife, accompanied by a clinical presentation of dementia in >50 % of individuals over the age of fifty (Dekker et al., 2018; Hof et al., 1995; Mann, Yates, & Marcyniuk, 1984; Perez et al., 2019; Thase, 1982; Wegiel, Wisniewski, Dziewiatkowski, Popovitch, & Tarnawski, 1996; Wisniewski, Dalton, McLachlan, Wen, & Wisniewski, 1985; Wisniewski, Wisniewski, & Wen, 1985).
In both AD and DS, there is age-associated degeneration of basal forebrain cholinergic neurons (BFCNs) (Casanova, Walker, Whitehouse, & Price, 1985; Coyle, Oster-Granite, Reeves, & Gearhart, 1988; Jorgensen, Brooksbank, & Balazs, 1990; Mann et al., 1984; Mufson, Bothwell, & Kordower, 1989; Sendera et al., 2000; Whitehouse et al., 1983; Yates et al., 1983; Yates, Simpson, Maloney, Gordon, & Reid, 1980) located within the medial septum/vertical limb of the diagonal band (MS/VDB), that project to the hippocampus and play a critical role in memory function (Hasselmo & Sarter, 2011; Mesulam, Mufson, Wainer, & Levey, 1983; Rye, Wainer, Mesulam, Mufson, & Saper, 1984). Dysfunction in this cholinergic projection system is also found in the Ts65Dn mouse model of DS making it a valuable translational model to assess cellular and molecular factors underlying degeneration of BFCNs, as well as possible treatment paradigms (Ash et al., 2014; Berger-Sweeney, 2003; Cataldo et al., 2003; Contestabile, Fila, Bartesaghi, Contestabile, & Ciani, 2006; Cooper et al., 2001; Granholm, Sanders, & Crnic, 2000; Holtzman et al., 1996; Hunter et al., 2003; Kelley et al., 2016; Kelley et al., 2014a; Moon et al., 2010; Seo & Isacson, 2005; Strupp et al., 2016).
A potential therapeutic strategy for BFCN degeneration with translational potential is perinatal maternal choline supplementation (MCS). In the developing fetus, choline is involved in effective neural tube closure, organogenesis, central nervous system cell membrane synthesis, and gene expression related to altering DNA methylation (Blusztajn, Cermak, Holler, & Jackson, 1998; Blusztajn, Slack, & Mellott, 2017; Cooney, Dave, & Wolff, 2002; Fisher, Zeisel, Mar, & Sadler, 2001, 2002; Niculescu, Yamamuro, & Zeisel, 2004; Wurtman, Cansev, Sakamoto, & Ulus, 2009; Zeisel, 2000; Zeisel & Blusztajn, 1994). When supplemented in utero and in early development, choline has both immediate and long-term beneficial effects in healthy disomic rats and mice (Holler, Cermak, & Blusztajn, 1996; Li et al., 2004; Loy, Heyer, Williams, & Meck, 1991; Meck, Smith, & Williams, 1988, 1989; Meck & Williams, 1997, 1999, 2003; Mellott, Williams, Meck, & Blusztajn, 2004; Pyapali, Turner, Williams, Meck, & Swartzwelder, 1998; Sandstrom, Loy, & Williams, 2002; Tees, 1999) and in rodent models of prenatal ethanol exposure, Rett syndrome, and status epilepticus (Holmes et al., 2002; Nag & Berger-Sweeney, 2007; Nag, Mellott, & Berger-Sweeney, 2008; Thomas, Abou, & Dominguez, 2009; Thomas, Biane, O’Bryan, O’Neill, & Dominguez, 2007; Ward, Agarwal, Wang, Berger-Sweeney, & Kolodny, 2008). Previously, we demonstrated that MCS during pregnancy and lactation attenuates cognitive dysfunction and BFCN degeneration in adult Ts65Dn progeny (Ash et al., 2014; Kelley et al., 2016; Kelley et al., 2014a; Moon et al., 2010; Powers et al., 2017; Strupp et al., 2016). Despite these encouraging findings, the molecular and cellular mechanisms associated with MCS-mediated attenuation of BFCN degeneration remains an under investigated area.
To address this, the present study examined the molecular signature of MS/VDB neurons, identified immunohistochemically using the cholinergic neuron marker choline acetyltransferase (ChAT), and acquired by laser capture microdissection (LCM) in Ts65Dn (Ts) and disomic (2N) mice (Alldred et al., 2018; Mufson et al., 1989). Since the MS/VDB region contains a heterogeneous population of neurons, analyses based on whole region homogenates are ill suited for BFCN-specific profile analysis (Ginsberg, Che, Wuu, Counts, & Mufson, 2006; Ginsberg et al., 2011; Mesulam et al., 1983; Rye et al., 1984). LCM provides a rigorous and reproducible method to isolate single cells based on a specific cellular phenotype (Ginsberg et al., 2018; Ginsberg et al., 2017; Ginsberg, Mufson, et al., 2010) and can be combined with custom-designed microarrays containing probes relevant to BFCNs, DS, cognitive dysfunction, and AD that hybridize the cDNA made from LCM BFCN mRNA isolates (Ginsberg & Che, 2014). This approach is highly effective for comparative group analysis and provides valuable insight into the molecular pathophysiology of DS, AD, and MCS treatment.
Methods
Ts65Dn mouse model
The Ts65Dn DS mouse model has segmental trisomy of murine chromosome Mmu16 and a centromeric low-coding region of Mmu17 imposed by radiation-induced reciprocal translocation (Akeson et al., 2001; Dierssen, Herault, & Estivill, 2009; Duchon et al., 2011; Kahlem et al., 2004; Reeves et al., 1995; Sturgeon & Gardiner, 2011). The probable triplicated section of Mmu17 is not coding sequence (CDS) dense and there were no genes from the triplicated segment of Mmu17 on our custom-designed array. For clarity, we denote not triplicated using the notation Mmu16NT and Mmu17NT.
Subjects
Female Ts65Dn and male C57BL/6JEiJ×B6EiC3SnF1/J mice were obtained from Jackson Laboratories (Bar Harbor, Maine, USA) for breeding as described previously (Ash et al., 2014; Kelley et al., 2014a). Use of C57BL/6JEiJ×B6EiC3SnF1/J male breeding pairs provided mixed litters of disomic (2N) and segmentally trisomic (Ts) offspring. The males were used for a series of structure-function investigations (Ash et al., 2014; Powers et al., 2017) accounting for the greater number of female mice in the present study. Genotyping was performed via qRT-PCR from tail snip or ear punch samples sent to Jackson Laboratories. Mice homozygous for the autosomal recessive retinal deterioration Pde6brd1 gene were excluded from analysis (Holtzman et al., 1995; Keeler, 1966). Animals were bred and maintained at Cornell University (Ithaca, NY, USA). All procedures were approved by the Institutional Animal Care and Use Committee of Cornell University, which conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Maternal choline supplementation (MCS)
One-half of the dams received a diet supplemented with choline (5.1 g/kg choline chloride in AIN-76A, Dyets, Inc., Bethlehem, PA, USA) beginning at the time of breeding and continuing until weaning (offspring, 2N+ and Ts+). Non-supplemented dams received a diet controlled for choline content (1.1 g/kg choline chloride in AIN-76A, Dyets, Inc.), referred to as normal diet (offspring, 2N and Ts). Both MCS and normal diet offspring remained with their original dams until weaning. At weaning mice were housed in same-sex, mixed-genotype groups and maintained on normal diet. At all stages, experimenters were blind to genotype. Groups consisted of 10 mice each (Table 1). The amount of choline supplementation is comparable to that used in other studies (Holler et al., 1996; Li et al., 2004; Loy et al., 1991; Meck et al., 1988; Meck & Williams, 1999; Mellott et al., 2004; Pyapali et al., 1998; Sandstrom et al., 2002; Schenk & Brandner, 1995; Tees, 1999), and translates to values within the normal range of human consumption and the recommended range for pregnant and nursing female humans (Detopoulou, Panagiotakos, Antonopoulou, Pitsavos, & Stefanadis, 2008; Sciences & Medicine, 1998).
Table 1.
Subject table
| n† | Age range (mean) | Sex (F:M)‡ | |
|---|---|---|---|
| 2N§ | 10 | 4.5–7.5 (5.9) mos | 8:2 |
| 2N+ | 10 | 4.5–7.5 (5.9) mos | 7:3 |
| Ts | 10 | 4.5–7.1 (6.1) mos | 9:1 |
| Ts+ | 10 | 4.4–7.5 (6.0) mos | 9:1 |
Group sizes;
F, female; M, male;
2N, disomic mice; Ts, Ts65Dn mice;
, maternal choline supplementation;
Tissue preparation
At 4.4–7.5 mos (Table 1) offspring were deeply anesthetized by intraperitoneal injection of a ketamine:xylazine (90mg/kg:9.5mg/kg) solution and transcardially perfused with 4 % paraformaldehyde. Brains were removed from the calvarium and placed in the same fixative for 24 h, stored at 4° C, followed by transfer into a 30 % sucrose solution until sectioning on a freezing sliding microtome in the coronal plane at 40 μm thickness into 6 series (240 μm between sections), and stored at 4° C in a cryoprotectant solution (30 % glycerol, 30 % ethylene glycol, 40 % phosphate buffer) until immunolabeling.
Immunohistochemistry
ChAT immunohistochemistry was performed as described previously (Kelley, Perez, Overk, Wynick, & Mufson, 2011). A full series of free-floating sections from each animal was washed and incubated in sodium metaperiodate to inhibit endogenous peroxidase activity. Next, tissue was washed in TBS containing 0.25 % Triton X-100, and incubated in a blocking solution consisting of 3 % serum in TBS/Triton X-100 to enhance primary antibody penetrance and block nonspecific binding. Tissue was then incubated overnight with the goat polyclonal antibody for ChAT (1:1000; Millipore, Billerica, MA, USA), in a solution of TBS/Triton X-100 with 1 % serum. All washes and incubations were carried out at room temperature on a shaker table.
Following overnight incubation in primary antibody, sections were washed in TBS and incubated for 1 h with a biotinylated secondary antibody (Vector Laboratories, Inc., Burlingame, CA, USA). To amplify the signal, sections were incubated for 1 h in an avidin-biotin-complex solution (Elite kit, Vector Laboratories, Inc.). Before and after chromogen reaction, tissue was washed in a sodium imidazole acetate buffer (0.68 g imidazole, 6.8 g sodium acetate trihydrate per 1 L distilled water, pH 7.4 with glacial acetic acid). ChAT reactivity was visualized with a solution consisting of 0.05 % 3,3’-diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich, St. Louis, MO, USA), 1 % nickel (II) ammonium sulfate hexahydrate and 0.0015 % H2O2 resulting in a black reaction product (Kelley et al., 2014b). Sections were rinsed, mounted on glass slides and dried overnight at room temperature. All procedures were performed blind to genotype and treatment and no gross features disrupted the blinding.
Laser capture microdissection (LCM)
An Arcturus LCM system connected to an inverted microscope with a software-operated stage was used to capture individual ChAT-immunolabeled MS/VDB neurons. The MS/VDB neurons were pooled to ensure that sufficient numbers of ChAT-positive perikarya would be available for analysis. The LCM system captures individual cells through a near-infared laser-charged thermoplastic membrane that adheres tissue excised with an ultraviolet laser. One hundred ChAT-positive MS/VDB neurons per subject were identified with a 1× lens (10× magnification) and cells were outlined with a lasso tool using a 40× lens (magnification 400×) (Mufson et al., 1989). Extracted cholinergic neurons were distributed across the MS/VDB using four slides from a 240 μm series. Throughout the LCM procedure groups were randomized across genotype and treatment.
RNA isolation and cDNA synthesis
RNASE AWAY (Molecular BioProducts, San Diego, CA, USA) was used to clean all surfaces and tools at each step. RNA was isolated using a standard TRIzol-chloroform-isopropanolol protocol. Membranes with LCM isolates were separated from the plastic caps and homogenized by treatment with cold TRIzol Reagent (1 ml, 4° C; Ambion, Carlsbad, CA, USA) at 4° C then put on ice for 5–10 min. For phase separation, chloroform was added to the membrane-TRIzol mixture, vortexed and let sit for 5 min at room temperature. Samples were then centrifuged > 10,000 RPM at 4° C for 20 min. The aqueous RNA phase was extracted and added to the nucleic acid coprecipitant linear acrylamide (Ambion) to aid in recovery and visualization of pellet at later stages. Isopropanolol was added and mixed by inverting, for RNA precipitation. Samples were then centrifuged (12,000 RPM) at 4° C for 20 min resulting in a visible RNA pellet. Liquid phase was disposed and samples were washed with 80 % EtOH and centrifuged at 12,000 RPM at 4° C for 7 min. Pellets were re-suspended in 24 μl diethyl pyrocarbonate (DEPC)-treated H2O (Sigma, St. Louis, MO, USA) and treated with an RNASE inhibitor (0.5 μl; SUPERase-In, Ambion).
Terminal continuation (TC) RNA amplification was employed to make dsRNA using methods optimized previously (Alldred, Che, & Ginsberg, 2009). Briefly, a polyT primer was hybridized to the polyA tail of the mRNA by adding 1 μl polyT primer to 6 μl sample and running one cycle (65° 2 min, 45° 1 min). We performed first strand synthesis by adding Superscript III (Invitrogen, Carlsbad, CA, USA), Master Mix (5×FirstStrand, DTT, 10 mM dNTP Invitrogen), RNase Inhibitor (Ambion), TC primers, in DEPC-treated H2O and running one cycle (50° 60 min, 65° 15 min). Lastly, primers were re-annealed and RNA degraded through adding PCR buffer (Applied Biosystems, Branchburg, New Jersey, USA) and RNaseH (Ambion) in DEPC-treated H2O and running one cycle (37° 30 min, 95° 3 min, 60° 3 min). cDNA was purified using centrifugal filters (Amicon Ultra 0.5mL 30K NMWL cellulose membrane) as per manufacturer’s instructions (Millipore).
Membrane hybridization
Nitrocellulose membranes with embedded probes were used for hybridization of cDNA, as described previously (Alldred et al., 2018). Membranes were stripped in a weak base (0.4N NaOH) in glass tubes placed in a rotating hybridization oven (45 min at 45° C), rinsed (0.2 M Tris pH 7.2, 0.1% SDS (Gibco), 0.1% SSC 20× (Fisher); 1× 1 min, 2× 10 min, 20° C), and incubated in a prehybridization buffer (formamide, Denhardts solution, 20× SSPE (Ambion), 0.1% SDS, 10%, and boiled sheared salmon sperm DNA (Ambion) in DEPC-treated H2O; 4 h, 42° C). Samples were incubated in a cocktail (10× transcription buffer, DEPC-treated H2O, 400nM NaCl, dACGTPs (Ambion), DTT, dUTP, RNAse inhibitor), T7 RNA polymerase (Epicentre Biotechnologies, San Diego, CA, USA), and the isotope P33 (5 h, 37° C). Samples were added to the membranes and incubated for 18 h at 42° C in a rotating hybridization oven. At all stages, a nylon mesh was used to prevent the membranes from sticking to themselves in the hybridization tubes. A total of 12 membranes were used, batches were processed in groups of 6, and systematic stratified sampling across membranes, batches, and groups was carefully mapped and employed. Two to five membranes were run per animal (average of two per animal).
Membrane hybridization was visualized by transferring the radioactive signal to a phosphor screen placed in a cassette (Amersham Biosciences, Little Chalfont, UK) that applied constant and even pressure for 24 h at room temperature. Images were obtained with a Kodak Digital Science Image Station 440CF (Rochester, NY, USA), and light intensity was quantified with ImageQuant software (Amersham Biosciences).
Nomenclature and chromosome loci
All genes represented by the membrane-embedded probes were re-identified using the Mouse Genome Informatics (MGI) online databases accessed September 2018 (Jackson Laboratories, Bar Harbor, ME, USA) (Smith et al., 2018) and represent the most recent nomenclature using genome build GRCm38. Gene functional clustering is based on a combination of MGI description and literature relevant to DS and AD.
Membrane normalization and statistical analysis
Post-normalization, the quotients were averaged across repeat arrays, for each gene, within subject. Membrane probes vary in hybridization strength, and were treated separately with each probe as an independent variable. The arrays are a tool for cross-group comparative analysis, rather than precise recordings of expression levels within a subject. Each membrane included negative control probes placed throughout the coordinates. For each gene the signal intensity ratio was modeled as a function of mouse group, using mixed effects models with random mouse effect to account for the correlation between repeated assays on the same mouse (McCulloch, Searle, & Neuhaus, 2008). Significance was set at p < 0.01, two-sided; false discovery rate (FDR) based on an empirical null distribution due to strong correlation between genes was controlled at level q < 0.1 (Benjamini & Hochberg, 1995; Efron, 2007). There was no association between age and expression levels for any gene (-0.000 < R2 < 0.280).
Nanostring expression verification
For expression verification, NanoString nCounter was performed on select codesets designed in conjunction with NanoString Technologies (Seattle, WA, USA) utilizing 55 genes known to be involved in DS and AD pathology with five internal negative controls and five internal positive control genes. Normalization was performed utilizing the NanoString reference gene normalization protocol (Veldman-Jones et al., 2015). Because of tissue limitation and genes chosen, hippocampal sections were used for verification. This precludes possible confounds from the heterogeneity of MS/VDB neuron subpopulations (Mesulam et al., 1983; Rye et al., 1984). NanoString nCounter statistical analysis was performed on normalized gene expression levels as described previously (Schafer, Dolgalev, Alldred, Heguy, & Ginsberg, 2015). Each gene was modeled as a function of the mouse study group, using mixed effects models with random mouse effect (Alldred, Lee, Petkova, & Ginsberg, 2015a, 2015b; McCulloch et al., 2008), with significance at α = 0.05, two-sided.
Results
Differences in total expression profiles
Custom-designed microarray results revealed significant differences in expression profiles across genotypes and treatments (Figs. 1 and 2A). After normalizing to background levels and calculating within-row z scores, we found that the average gene expression for Ts was increased to 1.48-fold 2N mice levels. In treatment groups, average expression level in 2N+ was increased to 1.05-fold compared to 2N, and Ts+ was increased 5.13-fold relative to Ts levels (Fig. 2A). Since comparison of male and female mice showed male expression levels fell within 1.5 × interquartile range of female expression levels we pooled the data (Tukey, 1977).
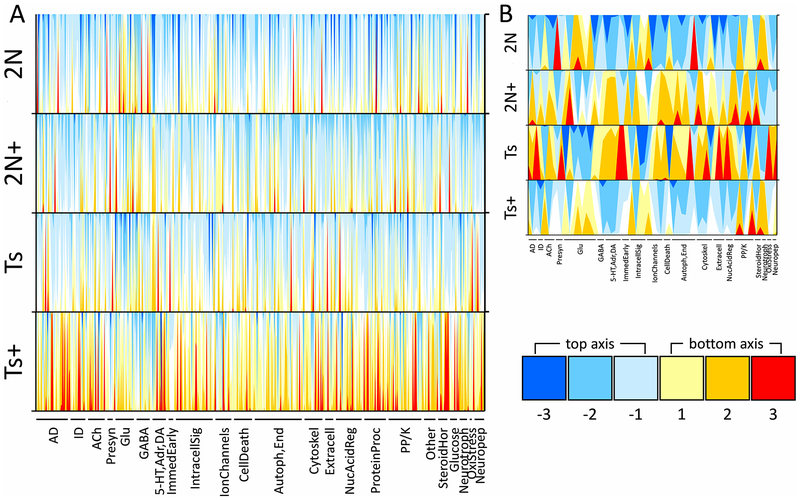
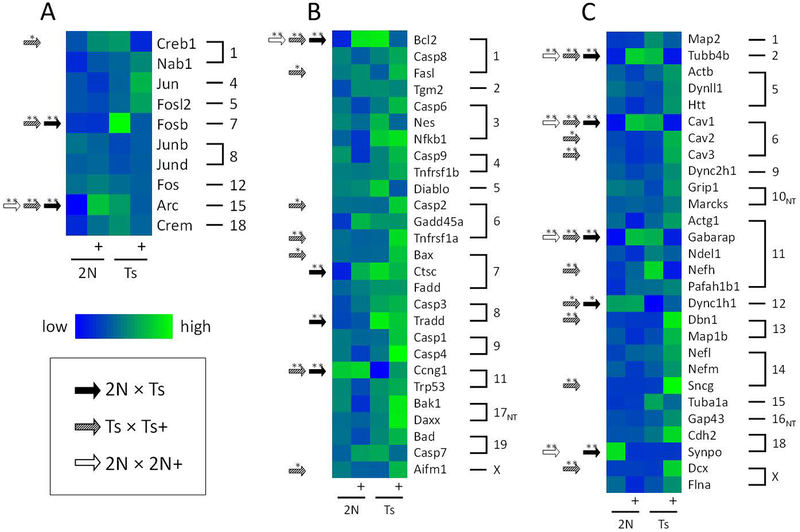
Figure1.
Horizon plots display transcription signature of MS/VDB basal forebrain cholinergic neurons from Ts65Dn (Ts) and disomic (2N) mice exposed to maternal choline supplementation (MCS, +) or normal diet from conception to weaning. A. Two-axis horizon plots show the relative expression level of all genes examined, using normalized row (gene) z-scores across the four groups. Negative values descend from the top abscissa (blues) and positive values ascend from the lower abscissa (reds and yellows) as shown in the legend at the lower right. Horizon plots stack data to provide an illustration for intensity of differences across genotype and treatment groups by overlaying fold-change surface plots and color-coding for overt determination of patterns. Each stacked surface plot (i.e. each color) represents a one-fold change, such that the lighter shades (-1 and 1 in the legend) are nearer to the mean expression which is set at 0, and darker shades descend/ascend accordingly (−2, −3 and 2, 3 in the legend). Use of two x-axes prevents overlap of negative and positive z-scores. Gene classes are listed below the 4 horizon plots and each spike can be thought of as a gene in that class (AD, Alzheimer’s disease; ID, intellectual disability; ACh, acetylcholine; Presyn, presynaptic; Glu, glutamate; GABA, gamma aminobutyric acid; 5-HT,Adr,DA, serotoninergic, adrenergic, and dopaminergic; IntracellSig, intracellular signaling; Autoph,End, autophagy and endosomal; Cytoskel, cytoskeleton; Extracell, extracellular matrix and nontransmitter inter-cellular signaling; NucAcidReg, nucleic acid regulation; ProteinProc, protein processing; PP/K, protein phosphatase and kinase; SteroidHor, steroid hormones; Neurotroph, neurotrophins; OxiStress, oxidative stress; Neuropep, neuropeptides). B. The horizon plot that results when only the genes with significant genotype differences in normal diet conditions (2N × Ts) as well as significant alteration with MCS in Ts mice (Ts+ × Ts) is shown (p < 0.01 using mixed effects model with false discovery rate based on null distribution; 10 mice per group). As opposed to the overall expression differences shown in A, the plots in B shows a clear expression pattern of elevated levels in Ts and 2N+ mice and lower levels in 2N and Ts+ mice, providing an overview of the genotype and treatment interaction effect in this murine model with MCS.
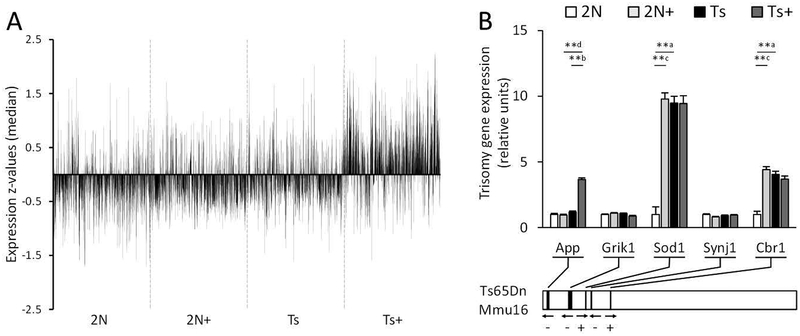
Figure 2.
Total expression and trisomy related gene expression levels of genes on a custom-designed microarray were measured in isolated basal forebrain cholinergic neurons from the MS/VDB. A. Vertical spike plot shows median z-scores per probe from a membrane microarray analysis on cholinergic neurons in the MS/VDB of Ts65Dn mice (Ts) and disomic littermates (2N). From conception to weaning, dams and pups were exposed to either normal diet or maternal choline supplementation (+). There are clear treatment and genotype effects when transcription comparison results are examined as a whole, including an overall increase in expression of Ts+ mice genes compared to the other groups. B. Bar graph shows expression levels of five genes triplicated in the Ts65Dn mouse model. The chromosome segment underneath diagrams to-scale locations of the genes on the chromosome. ** p < 0.005 using a mixed effects model with false discovery rate based on null distribution, n = 10 mice per group; a, Ts × 2N; b, Ts+ × Ts; c, 2N+ × 2N; d, Ts+ × 2N+
Expression of triplicated Mmu16 genes
Several genes triplicated in Ts mice have human orthologs implicated in cognitive dysfuntion and AD. The following five triplicated gene transcripts were on the custom-designed microarray: amyloid beta A4 precursor protein (APP, gene App), glutamate receptor ionotropic kainate 1 (Grik1), superoxide dismutase 1 soluble (Sod1), synaptojanin 1 (Synj1), and carbonyl reductase 1 (Cbr1) (Fig. 2B). Upregulation was found in Sod1 and Cbr1 (Fig. 2B). MCS significantly increased expression of App in Ts65Dn mice, and Sod1 and Cbr1 in 2N mice (Fig. 2B).
Alterations in AD-related transcripts
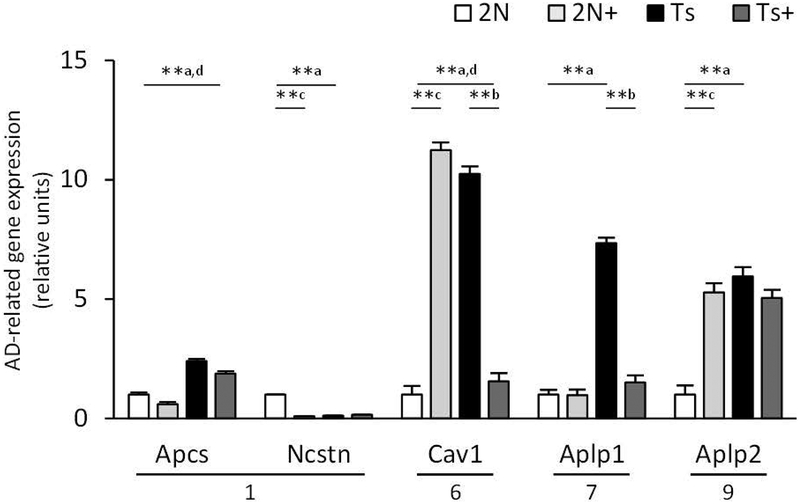
Of the 42 microarray probes representing AD-associated genes the following were differentially regulated across genotypes: caveolin 1/caveolae protein (Cav1), APP-like protein 1 (Aplp1) and 2 (Aplp2), and serum amyloid P-component (Apcs) were significantly upregulated in Ts compared to 2N mice, and nicastrin (Ncstn) was significantly downregulated (Table 2, Fig. 3). Apcs and Cav1 expression was downregulated by MCS in Ts mice, and Aplp1 upregulated (Table 2, Fig. 3). Additionally, MCS significantly increased genes involved in APP processing and transport (presenilin 2, Psen2; low density lipoprotein receptor-related protein 1, Lrp1; membrane metallo endopeptidase, Mme), intracellular signaling (cyclin-dependent kinase 5 regulatory subunit 1 p35, Cdk5r1; APP binding family A member 1, Apba1), AD plaque constituent (beta-2 microglobulin, B2m; synuclein beta, Sncb), as well as perlecan/heparan sulfate proteoglycan 2 (Hspg2), bromodomain PHD finger transcription factor (Bptf), and versican (Vcan), but significantly decreased NEDD8 activating enzyme E1 subunit 1 (Nae1) in Ts mice (Table 2). Expression of Cav1, Aplp2, and microtubule-associated protein tau (Mapt2) was upregulated and Ncstn downregulated by MCS in 2N mice (Table 2).
Table 2.
Comparison of Alzheimer’s disease related gene expression levels between genotypes and treatments
| Gene | Mmu† | 2N+ fold of 2N‡ | Ts fold of 2N | Ts+ fold of 2N | Ts+ fold of Ts |
|---|---|---|---|---|---|
| Cav1 | 6 | 11.24** | 10.25** | 1.56 | 0.15** |
| Aplp1 | 7 | 0.97 | 7.34** | 1.50 | 0.20** |
| Aplp2 | 9 | 5.28** | 5.95** | 5.05** | 0.85 |
| Apcs | 1 | 0.60 | 2.40** | 1.87** | 0.78 |
| Psen2 | 1 | 1.87 | 1.68 | 2.40** | 1.43* |
| Nae1 | 8 | 1.19 | 1.61 | 0.38 | 0.24** |
| Mapt2 | 11 | 5.93** | 1.36 | 1.90 | 1.39 |
| B2m | 2 | 1.50 | 1.35 | 2.14** | 1.59** |
| Sncb | 13 | 1.17 | 1.19 | 2.80** | 2.36** |
| App& | 16 | 0.95 | 1.17 | 3.66** | 3.12** |
| Apba1 | 19 | 0.98 | 1.06 | 2.37** | 2.23** |
| Vcan | 13 | 0.98 | 1.01 | 1.63* | 1.62** |
| Cdk5r1 | 11 | 0.98 | 1.00 | 1.49* | 1.49* |
| Mapt4 | 11 | 1.04 | 0.99 | 1.72** | 1.74** |
| Mapt2n6p | 11 | 0.89 | 0.97 | 4.97** | 5.13** |
| Mapt1n6p | 11 | 0.82 | 0.96 | 1.38 | 1.44* |
| Mme | 3 | 0.92 | 0.95 | 1.46 | 1.54** |
| Lrp1 | 10NT | 0.81 | 0.93 | 1.71** | 1.84** |
| Hspg2 | 4 | 0.90 | 0.82 | 1.36 | 1.66** |
| Bptf | 11 | 0.63 | 0.74 | 2.29** | 3.10** |
| Ncstn | 1 | 0.09** | 0.12** | 0.15** | 1.33 |
Mus musculus chromosome using build Grmc38, NT denotes not triplicated;
2N, disomic mice, Ts, Ts65Dn,
maternal choline supplementation;
, p < 0.005;
, p < 0.01;
gene is triplicated in Ts65Dn mice
Figure 3.
Bar graph shows expression levels of Alzheimer disease (AD) related genes. Ts65Dn (Ts) and disomic (2N) mice were exposed to a maternal choline supplementation (+) or normal diet from conception to weaning and gene expression levels were measured in basal forebrain cholinergic neurons at 4.4–7.5 mos. Multiple AD-associated genes were upregulated in Ts compared with 2N mice including Cav1 and Aplp1, which were downregulated in Ts+ compared with Ts mice. * p < 0.01, ** p < 0.005, mixed effects model with false discovery rate based on null distribution, n = 10 mice per group; a, Ts × 2N; b, Ts+ × Ts; c, 2N+ × 2N; d, Ts+ × 2N+; chromosomes are shown under genes
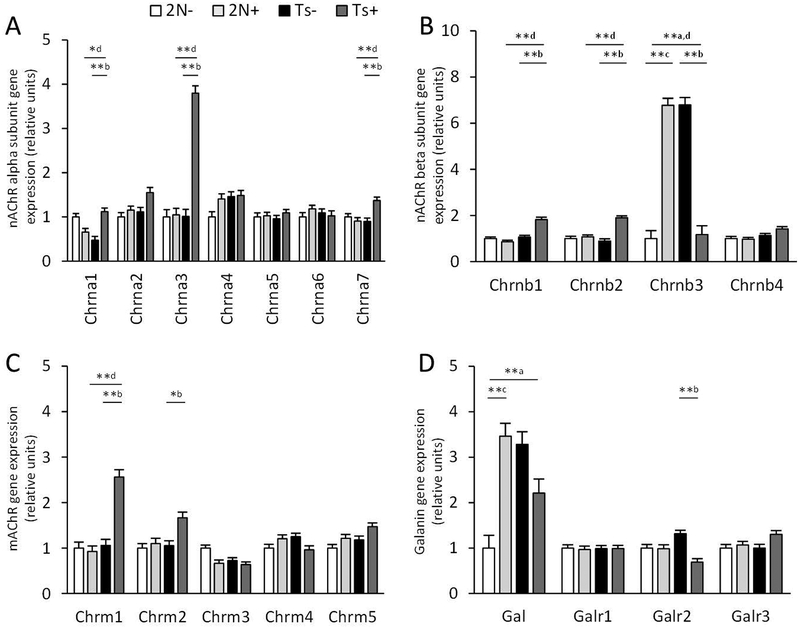
Alterations in cholinergic and galaninergic transcripts
Nicotinic cholinergic receptor (nAChR) beta polypeptide 3 (Chrnb3) transcript levels were upregulated in Ts mice, and MCS normalized Chrnb3 expression to 2N levels (Fig. 4). No statistically significant genotype changes were seen in the family of muscarinic receptors (mAChRs) or cholinergic synthesizing and transport genes between genotypes in the normal diet condition. MCS upregulated expression of nAChR alpha polypeptides 3 (Chrna3) and 7 (Chrna7), beta polypeptides 1 (Chrnb1) and 2 (Chrnb2), and mAChR 1 (Chrm1) and 2 (Chrm2) in Ts mice (Fig. 4). Chrnb3 was also significantly upregulated by MCS in 2N mice (Fig. 4).
Figure 4.
Bar graph shows expression levels of cholinergic receptor (AChR) and galanin (Gal) related genes. Ts65Dn (Ts) and disomic (2N) mice were exposed to a maternal choline supplementation (+) or normal diet from conception to weaning and gene expression levels were measured in basal forebrain cholinergic neurons at 4.4–7.5 mos. A-B. Multiple nicotinic AChR (nAChR) alpha (Chrna, A) and beta (Chrnb, B) subunits were upregulated in Ts+ compared to Ts mice, but only Chrnb3 was elevated in 2N+ compared with 2N mice. C. Muscarinic AChR (mAChR) genes (Chrm) were stably expressed in 2N+ and 2N mice, while Chrm1 and Chrm2 were significantly elevated in Ts+ compared to Ts mice. D. Transcripts for Gal were elevated in Ts compared with 2N mice but not Gal receptor genes (Galr). * p < 0.01, ** p < 0.005, mixed effects model with false discovery rate based on null distribution, n = 10 mice per group; a, Ts × 2N; b, Ts+ × Ts; c, 2N+ × 2N; d, Ts+ × 2N+
Galanin (Gal), a gene encoding the neuropeptide Gal that co-localizes with BFCNs in rodents (Perez, Wynick, Steiner, & Mufson, 2001), was significantly upregulated in Ts mice (Fig. 4). Gal receptor (GalR) transcript levels were similar between genotypes under normal diet conditions. MCS downregulated Galr2 expression in Ts mice, and upregulated expression of Gal in 2N mice.
Immediate-early gene transcripts
Immediate early genes FBJ osteosarcoma oncogene B (Fosb) and activity regulated cytoskeletal-associated protein (Arc) were significantly upregulated in Ts mice. MCS significantly decreased expression of Fosb and Arc as well as cAMP responsive element binding protein 1 (Creb1), although levels of Fosb remained 2.5-fold higher that 2N mice (p < 0.07) and Arc 4.2-fold higher (Table 3, Fig. 5). Arc was significantly upregulated by MCS in 2N mice (Table 3, Fig. 5).
Table 3.
Comparison of intracellular signaling related gene expression levels between genotypes and treatments
| Gene | Mmu† | 2N+ fold of 2N‡ | Ts fold of 2N | Ts+ fold of 2N | Ts+ fold of Ts |
|---|---|---|---|---|---|
| Immediate early genes | |||||
| Fosb | 7 | 0.99 | 10.46** | 2.52 | 0.24** |
| Arc | 15 | 8.26** | 6.50** | 4.15** | 0.64** |
| Creb1 | 1 | 1.17 | 1.12 | 0.84 | 0.75* |
| Second messenger and receptor cascades | |||||
| Rgs4 | 1 | 9.19** | 8.62** | 8.04** | 0.93 |
| Rgs5 | 1 | 9.40** | 8.36** | 7.29** | 0.87 |
| Rgs10 | 7 | 7.85** | 7.62** | 1.19 | 0.16** |
| Nos2 | 11 | 2.52** | 2.65** | 2.42** | 0.91 |
| Gnas | 2 | 1.06 | 1.16 | 8.01** | 6.94** |
| Rgs2 | 1 | 0.99 | 1.09 | 3.27** | 3.00** |
| Gna11 | 10NT | 0.90 | 1.06 | 1.64* | 1.54** |
| Nrg2 | 18 | 0.92 | 0.95 | 1.56** | 1.65** |
| Tsc2# | 17NT | 0.92 | 0.94 | 1.89* | 2.01** |
| Adcy1 | 11 | 0.92 | 0.92 | 1.38 | 1.50** |
| Adcy5 | 16NT | 0.87 | 0.89 | 1.38 | 1.54** |
| Adcy4 | 14 | 0.92 | 0.88 | 1.31 | 1.49* |
| Adcy8 | 15 | 0.85 | 0.86 | 1.94** | 2.25** |
| Itpr1 | 5 | 0.79 | 0.55 | 1.05 | 1.90* |
| Mlst8 | 17NT | 0.81 | 0.19** | 0.82 | 4.34** |
| Gng3 | 19 | 0.85 | 0.18** | 1.09 | 6.04** |
| Itpka | 2 | 0.09** | 0.11** | 1.15 | 10.32** |
Mus musculus chromosome using build Grmc38, NT denotes not triplicated;
2N, disomic mice, Ts, Ts65Dn,
, maternal choline supplementation;
, p < 0.005;
, p < 0.01;
gene is implicated in intellectual disability
Figure 5.
Heatmaps show gene expression levels in basal forebrain cholinergic neurons for select gene classes between disomic (2N) and Ts65Dn (Ts) mice exposed to normal diet or maternal choline supplementation (+). A. Heatmap of immediate early gene expression levels shows significant increases in Arc and Fosb expression levels between Ts and 2N mice (black arrows) and partial normalization with MCS (hatched arrows). Lowest expression levels are in blue (Arc, 2N), and highest in green (Fosb, Ts+). B. Significant differences in expression levels were seen in multiple cell death associated genes between Ts and 2N mice. Partial normalization occurred with MCS for Bcl2 and Ccng in Ts mice, and MCS increased expression of Bcl2 in 2N mice (white arrows). Lowest expression levels are in blue (Ccng1, Ts), and highest in green (Casp4, Ts+). C Expression levels of cytoskeleton proteins associated with intraneuronal structure and transport show significant differences both structural, Tubb4b, and transport, Dync1h1, related genes in Ts compared to 2N mice. MCS resulted in normalizing decreases of Tubb4b, Cav1, and Gabarap expression levels in Ts mice, and increased expression in Cav2, Cav3, Nefh, Dbn1, Sncg, and Dcx in Ts+ compared to Ts mice. Lowest expression levels are in blue (Dync1h1, Ts), and highest in green (Sncg, Ts+). The grouping outline is based on mouse chromosome numbers, listed on the right. *, p < 0.01; **, p < 0.005, mixed effects model with false discovery rate based on null distribution
Intracellular signaling related transcripts
Of the 55 intracellular signaling related transcripts examined, six were significantly increased in Ts compared with 2N mice, including Aplp1 and Aplp2 genes associated with AD (Table 2, Fig. 3), regulator of G-protein signaling 4 (Rgs4), 5 (Rgs5), and 10 (Rgs10), and gas neurotransmitter nitric oxide synthase 2 inducible (Nos2) (Table 3), whereas three intracellular signaling genes were significantly decreased inositol 1,4,5-trisphosphate 3-kinase A (Itpka); and G-protein related MTOR associated protein LST8 homolog, Mlst8; and G-protein gamma 3, Gng3) (Table 3). No genes associated with adenylyl cyclase (AC) signaling on the custom-designed array were significantly changed across genotype under normal diet conditions. MCS normalized expression levels of Rgs10 (5.40-fold decrease) and Itpka, Mlst8, and Gng3 (average 5.90-fold increase) in Ts mice (Table 3). There were twelve genes with equivalent expression levels between 2N and Ts mice but altered by MCS in Ts mice, including the tuberous sclerosis 2 (Tsc2) gene implicated in ID (Table 3), and the AD associated gene Cdk5r1 (Table 2).
Cell death-related transcripts
Of the 27 gene probes on the custom-designed microarray related to cell death signaling, expression levels of B cell leukemia/lymphoma 2 (Bcl2), cathepsin C (Ctsc), and tumor necrosis factor (TNF) receptor superfamily member 1a (TNFRSF1A)-associated via death domain (Tradd) were upregulated in Ts mice, and cyclin G1 (Ccng1) was downregulated (Table 4, Fig. 5B). MCS partially reversed these differences in expression for Bcl2 and Ccng1 in Ts mice, although levels were still significantly different from 2N mice (Table 4, Fig. 5B). MCS significantly increased expression of five genes in Ts65Dn mice: Fas ligand/TNF superfamily member 6 (Fasl), caspase 2 (Casp2), Tnfrsf1a, BCL2-associated X protein (Bax), apoptosis-inducing factor mitochondrion-associated 1 (Aifm1) (Table 4, Fig. 5B). MCS significantly increased expression of Bcl2 in 2N mice (Table 4, Fig. 5B). Aside from Bcl2 in Ts mice, no genes associated with cell death were decreased with MCS in either genotype.
Table 4.
Comparison of cell death related gene expression levels between genotypes and treatments
| Gene | Mmu† | 2N+ fold of 2N‡ | Ts fold of 2N | Ts+ fold of 2N | Ts+ fold of Ts |
|---|---|---|---|---|---|
| Tradd | 8 | 1.17 | 2.96** | 2.30** | 0.78 |
| Bcl2 | 1 | 2.42** | 2.57** | 1.58 | 0.62** |
| Ctsc | 7 | 1.80 | 2.11** | 1.91** | 0.91 |
| Bax | 7 | 1.05 | 0.96 | 1.43 | 1.48* |
| Fasl | 1 | 1.02 | 0.96 | 1.45 | 1.52* |
| Casp2 | 6 | 1.03 | 0.90 | 1.36 | 1.50* |
| Tnfrsf1a | 6 | 0.95 | 0.86 | 1.39 | 1.62** |
| Aifm1 | X | 0.75 | 0.74 | 1.21 | 1.64* |
| Ccng1 | 11 | 1.01 | 0.22** | 0.71** | 3.25** |
Mus musculus chromosome using build Grmc38, NT denotes not triplicated;
2N, disomic mice, Ts, Ts65Dn,
, maternal choline supplementation;
, p < 0.005;
, p < 0.01
Cytoskeletal and presynaptic transcripts
There were 28 probes related to neuronal cytoskeleton and associated structural proteins. Upregulation of tubulin beta 4B class IVB (Tubb4b) and gamma-aminobutyric acid (GABA) receptor associated protein (Gabarap) was found in Ts compared to 2N mice (Table 5; Fig. 5C). Cytoskeletal elements dynein cytoplasmic 1 heavy chain 1 (Dync1h1) and synaptopodin (Synpo) were significantly decreased in Ts compared to 2N mice (Table 5; Fig. 5C). Expression levels of Tubb4b, Gabarap, and Dync1h1 were normalized by MCS. MCS treatment produced an upregulation of caveolin 2 (Cav2), caveolin 3 (Cav3), drebrin 1 (Dbn1), synuclein gamma (Sncg), and doublecortin (Dcx) expression in Ts mice (Fig. 5C) but significantly upregulated Tubb4b and Gabarap, and downregulated Synpo in 2N mice (Table 5; Fig. 5C).
Table 5.
Comparison of cytoskeletal and presynaptic related gene expression levels between genotypes and treatments
| Gene | Mmu† | 2N+ fold of 2N‡ | Ts fold of 2N | Ts+ fold of 2N | Ts+ fold of Ts |
|---|---|---|---|---|---|
| Cytoskeletal | |||||
| Gabarap | 11 | 8.87** | 8.16** | 2.05 | 0.25** |
| Tubb4b | 2 | 3.29** | 3.01** | 0.81 | 0.27** |
| Nefh | 11 | 1.16 | 1.62 | 0.88 | 0.54** |
| Dbn1 | 13 | 1.05 | 1.13 | 2.13** | 1.88** |
| Sncg | 14 | 1.06 | 1.11 | 4.94** | 4.46** |
| Dcx | X | 0.97 | 1.02 | 2.22** | 2.17** |
| Cav2 | 6 | 0.93 | 0.92 | 1.37 | 1.48* |
| Cav3 | 6 | 0.87 | 0.84 | 1.46** | 1.74** |
| Dync1h1 | 12 | 0.86 | 0.37* | 0.77 | 2.08* |
| Synpo | 18 | 0.05** | 0.06** | 0.07** | 1.06 |
| Presynaptic | |||||
| Syngr1 | 15 | 0.89 | 14.64** | 0.99 | 0.07** |
| Snap29 | 8 | 2.21** | 1.13 | 1.38 | 1.22 |
| Bsn | 9 | 0.91 | 1.00 | 0.65 | 0.66* |
| Snca | 6 | 0.26** | 0.33** | 0.61* | 1.87 |
Mus musculus chromosome using build Grmc38, NT denotes not triplicated;
2N, disomic mice, Ts, Ts65Dn,
, maternal choline supplementation;
, p < 0.005;
, p < 0.01
Of the genes related to presynaptic function, upregulation of synaptogyrin 1 (Syngr1) and downregulation of synuclein, alpha (Snca) was found in Ts mice (Table 5). MCS significantly decreased expression of Syngr1 and bassoon (Bsn) in Ts mice; while in 2N mice, MCS upregulated synaptosomal-associated protein 29 (Snap29) and downregulated Snca (Table 5).
Autophagosome and protein degradation transcripts
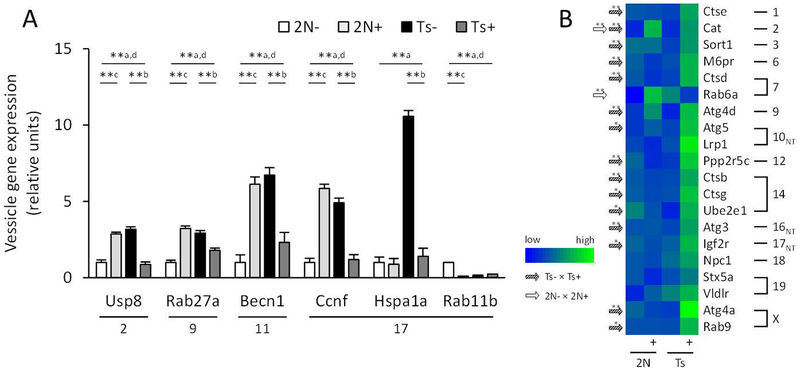
Expression of multiple autophagosome-associated genes was significantly increased in Ts compared with 2N mice including autophagy related beclin 1 (Becn1) and those encoding for proteins involved in the ubiquitin degradation process: cyclin F (Ccnf), heat shock protein 1A (Hspa1a), and ubiquitin specific peptidase 8 (Usp8). Additionally, we found an increase in exosomal member of RAS oncogene family 27a (Rab27a) and a decrease in endosomal Rab11b in Ts compared to 2N mice (Fig. 6A). In Ts mice, MCS decreased expression of Becn1, Ccnf, Hspa1a, Usp8, and Rab27a (Fig. 6A), and in 2N mice of Rab11b (Fig. 6A). In addition MCS increased expression of lysosomal and autophagic cathepsin B (Ctsb), D (Ctsd), E (Ctse), and G (Ctsg), autophagy related 3 (Atg3) and 5 (Atg5), autophagy related cysteine peptidase 4A (Atg4a) and 4D (Atg4d), catalase (Cat), protein phosphatase 2 regulatory subunit B’ gamma (Ppp2r5c), insulin-like growth factor 2 receptor (Igf2r), ubiquitin-conjugated enzyme E2E 1 (Ube2e1), and Rab9, as well as endosomal mannose-6-phosphate receptor (M6pr) and sortilin 1 (Sort1) (Figure 6B).
Figure 6.
Bar graph and heatmap show significant changes in gene expression levels in basal forebrain cholinergic neurons for vesicular endosomal, exosomal, and autophagy-related genes between disomic (2N) and Ts65Dn (Ts) mice exposed to normal diet or maternal choline supplementation (+). A. Bar graph shows increased expression of five genes related to endosomes and autophagy in Ts mice compared to 2N mice and normalization with MCS. ** p < 0.005 using a mixed effects model with false discovery rate based on null distribution, n = 10 mice per group; a, Ts × 2N; b, Ts+ × Ts; c, 2N+ × 2N; d, Ts+ × 2N+ B. Heatmap of autophagy, endosome, and exosome gene expression levels shows significant changes with MCS in Ts (hatched arrows) and 2N (white arrows) mice. Lowest expression levels are in blue (Rab6a, 2N), and highest in green (Atg4a, Ts+).
Transcript expression validation
Validation of select transcripts in MS/VDB BFCNs was performed by NanoString nCounter through hippocampal CA1 subregional analysis (a target for MS/VDB cholinergic projections), not single population analysis, as part of our ongoing studies of the septohippocampal system in Ts and 2N offspring following MCS. NanoString nCounter hippocampal CA1 subregional analysis revealed expression level changes in Synpo (downregulation) and Psen2 (upregulation) concurrent with the findings reported using custom-designed microarrays. Specifically, downregulation of Synpo in Ts compared with 2N mice found via microarray paralleled significant downregulation of CA1 Synpo transcript levels in Ts compared with 2N mice via NanoString nCounter (p < 0.007). Moreover, downregulation of Synpo by MCS in Ts BFCNs mirrored significant downregulation of Synpo in hippocampal CA1 subregional analysis of Ts+ mice (p < 0.01). Further, upregulation of Psen2 by MCS in BFCNs was also validated by NanoString nCounter of Psen2 in hippocampal CA1 subregional analysis of Ts+ compared with Ts mice (p < 0.02). Frozen tissue from the MS/VDB region was not available for transcript expression validation.
Discussion
In DS there is a striking loss of cholinergic neurons located within the basal forebrain (Coyle, Oster-Granite, & Gearhart, 1986; Fodale, Mafrica, Caminiti, & Grasso, 2006; Schliebs & Arendt, 2011), an area that provides the major cholinergic input to the hippocampal component of the medial temporal lobe memory circuit (Berger-Sweeney, 2003; Hampel et al., 2018; Hasselmo & Sarter, 2011; Mesulam et al., 1983; Whitehouse et al., 1983). Recently, studies have demonstrated that dietary MCS during pregnancy and lactation attenuates cognitive dysfunction and BFCN degeneration in adult Ts65Dn progeny (Ash et al., 2014; Kelley et al., 2016; Kelley et al., 2014a; Moon et al., 2010; Powers et al., 2017; Strupp et al., 2016). Despite the importance of the cholinergic basal forebrain projection system in cognitive function, little is known about the molecular pathobiology of BFCNs in DS (Alldred et al., 2018; Alldred et al., 2015a, 2015b; Chrast et al., 2000; Perez et al., 2019). Here, we present single population neuronal expression data showing alterations to the transcriptional signature of MS/VDB cholinergic neurons obtained from offspring of Ts65Dn dams treated with MCS or a choline-controlled diet during gestation and nursing. Overall, we found genotype-dependent alterations in select transcripts from gene ontology categories related to AD, G-protein intracellular signaling, immediate early gene responses, cell death, cytoskeletal structure and transport, autophagy, protein degradation, presynaptic function, neurotransmission, and neuromodulation. Interestingly, MCS corrected aberrant expression of various genes in cytoskeletal, G-protein intracellular signaling, immediate early, and cell death functional pathways.
Consistent with prior reports in humans with DS and in Ts mice, we did not find a gene dosage effect for the triplicated segment in Ts mice (Choi et al., 2009; Gardiner, 2004; Holtzman et al., 1995; Pritchard & Kola, 1999). For example, expression levels for the App gene were comparable between 2N and Ts mice; and MCS had a differential effect, increasing App expression in Ts+, while levels were unchanged in 2N+ mice. Although elevated levels of the protein product APP have been reported in Ts mice, data do not suggest an increase in Aβ processing enzymes or in the deposition of Aβ moieties; moreover, App mRNA has a shorter half-life in Ts than 2N mice (Choi et al., 2009; Holtzman et al., 1996; Hunter et al., 2003; Salehi et al., 2006). In an APP/PS1 AD mouse model, MCS resulted in decreased levels of soluble Aβ40 and Aβ42 peptides and reduced size of amyloid-like plaques in the hippocampus (Mellott et al., 2017), suggesting there may be further downstream effects in addition to the mRNA changes we found.
We previously reported ChAT activity increases in the hippocampus at 15–19 months of age in Ts and Ts+ compared with treatment-matched 2N mice (Kelley et al., 2016), corresponding with reports from independent labs of increases in ChAT enzyme activity at 10–12 months of age in the hippocampus, olfactory bulb, and isocortex of Ts compared with 2N mice (Contestabile et al., 2006; Seo & Isacson, 2005). In the present study, we did not find genotype-specific alterations in cholinergic synthesizing (choline acetyltransferase, Chat), transport (solute carrier family 18 vesicular monoamine member 3, Slc18a3), or degradation (acetylcholinesterase, Ache; butyrylcholinesterase, Bche) transcripts within BFCNs in Ts compared to 2N mice at 4–8 months of age, suggesting enzyme activity changes are either age dependent or occur via a mechanism other than direct mRNA regulation in MS/VDB cholinergic neurons. In Ts offspring exposed to MCS there was no change in Chat, Slc18a3, or Ache, but there was an upregulation in several nAChR and mAChR genes, suggesting alterations in local network signaling may occur at the transcription level, while ACh metabolic factors remain unaltered. Prior work in disomic rats has shown that MCS results in a functional profile of decreased ACh turnover (marked by decreased protein activity levels of ChAT, AChE, and high-affinity choline uptake) along with increased evoked ACh release, in the hippocampus during the first two months of life (Blusztajn et al., 1998; Cermak et al., 1999; Cermak, Holler, Jackson, & Blusztajn, 1998; Meck, Williams, Cermak, & Blusztajn, 2007). Whether the changes observed in disomic rats with MCS persist throughout adult life and whether they are discoverable in BFCNs is unknown (Cermak et al., 1999). We did not find transcript changes in the MS/VDB of 2N mice that mirror activity alterations in the hippocampus of MCS exposed rats, suggesting that if a similar downregulation occurs in the current model, it is not at the transcription level in these neurons. There are conflicting reports on whether the forebrain cholinergic system of individuals with DS is altered from birth (Becker, et al., 1991; Kish, et al., 1989; Casanova, et al., 1985; McGeer, et al., 1985).
Alterations in intracellular signaling transcripts within Ts BFCNs compared with 2N mice may provide insight into the failure of the septohippocampal system to respond appropriately to stimuli in trisomic mice. A prior study found no difference in hippocampal ACh baseline release rates in Ts compared to 2N mice (4 months of age), but a significantly greater release rate in 2N mice during the performance of a hippocampal dependent memory-attention task that was not seen in Ts mice (Chang & Gold, 2008). In the present study, expression of two immediate early genes, Fosb and Arc, were significantly elevated in Ts mice, and significantly decreased with MCS. The genotype-dependent elevation in immediate early genes is reminiscent of previously reported increases in protein levels for proto-oncogene c-Fos (FOS), phosphorylated FOS (pFOS), and ARC in the hippocampus of 7-month-old Tc1 mice, a murine model of DS with a transchromosomic copy of HSA21 (Ahmed et al., 2013). The fact that we did not find Ts-specific elevations in Fos, fos-like antigen 2 (Fosl2), or other immediate early genes suggests a specific dysregulation of Arc and Fosb transcription within BFCNs in the Ts65Dn mouse. Of note, the protein encoded by Arc interacts with mRNA or the lipid bilayer and is involved in long-term depression at the synapse whereas the other immediate early genes examined function through polymerase and transcription factor binding, related to nuclear regulatory roles prior to mRNA turnover (Barylko et al., 2018; Dynes & Steward, 2012; Minatohara, Akiyoshi, & Okuno, 2015).
The synaptic-related markers, Snca and Syngr1, were significantly altered in Ts mice and normalized with MCS. Functionally, Snca encodes alpha synuclein, a protein that is involved in presynaptic signaling and vesicle trafficking, and an effector in the pathogenesis of an autosomal dominant form of Parkinson disease (PD), as well as a component of Lewy bodies in sporadic PD and amyloid beta plaques in AD (Kessler et al., 2018; Mukaetova-Ladinska et al., 2000; Xu & Pu, 2016). Syngr1 encodes synaptogyrin 1, a protein involved in vesicle trafficking and synaptic plasticity (Belfort & Kandror, 2003; Janz et al., 1999). The ability of MCS to normalize expression of these synaptic genes may partially underlie the prolonged benefits of MCS on septohippocampal-dependent memory tests seen at 6–12 months in Ts mice (Ash et al., 2014; Kelley et al., 2016; Strupp et al., 2016). Of note, alpha synuclein and synaptogyrin are not involved in vesicle docking or release and genotype dependent differences represent a problem with vesicle maintenance and synaptic plasticity rather than neurotransmitter exocytosis (Ben Gedalya et al., 2009; Janz et al., 1999; Vargas et al., 2017).
Analysis of cell stress and apoptosis related transcripts revealed an increase in the expression of Bcl2, Ctsc, and Tradd in Ts compared with 2N mice. Bcl2 encodes an outer mitochondrial membrane protein that can regulate apoptosis by controlling permeability, while Ctsc encodes the protein cathepsin C, which is involved in peptidase regulation and proteolysis associated with the apoptotic process. By contrast, Tradd is involved in extrinsic apoptotic signaling as a cytoplasmic effector for death domain-containing receptors. Functionally, an upregulation of Tradd, suggests that the TNF or Fas signaling pathway is activated, however we did not find alterations in downstream effectors such as Casp3, Casp7 or Bad. We also found a significant decrease between Ts and 2N mice for Ccng1 expression, which encodes cyclin G1, a negative regulator of apoptosis. MCS normalized expression of Bcl2 and Ccng1 without significantly altering Ctsc or Tradd expression levels in Ts mice. Alterations in these cell survival genes suggest either an ongoing neurodegenerative stress response or a system that remains in an immature state. MCS exposure in Ts mice increased expression of genes involved in cytochrome C mediated stress responses and apoptosis including pro-apoptotic Bax and Casp2. However, there were no alterations in downstream effectors, suggesting that while a stress response may be ongoing, apoptotic processes are quiescent in MCS exposed Ts mice.
Although prior studies in this mouse model have focused on increases in protein markers for early endosomes, such as Rab5b, as indicative of neuronal transport dysfunction (Cataldo et al., 2003; Delcroix et al., 2004; Ginsberg, Alldred, et al., 2010; Salehi et al., 2006), we did not find any genotype specific changes in this class of genes. In the present study, we found increased expression of genes coding for protein products involved in the ubiquitin degradation system (Usp8, Ccnf, Hspa1a) and autophagy (Becn1), and decreased expression of Rab11b, protein product involved in receptor recycling, in Ts compared to 2N mice. Hspa1a is elevated in response to oxidative stress and misfolded proteins, and protein expression is increased in the deposits of AD and other neurodegenerative diseases (Hauser et al., 2005; Leak, 2014; Leverenz, Miller, Dobie, Peskind, & Raskind, 2001). Notably we found that MCS corrected elevated levels of Hspa1a in Ts while not altering expression levels in 2N mice.
Interestingly, there was a significant increase in gene expression of the neuropeptide Gal in Ts mice compared with 2N mice. Gal and GalRs co-localize with BFCNs during development and into adulthood in rodents (Chan-Palay, 1988; Melander et al., 1985; Mufson, Cochran, Benzing, & Kordower, 1993; Perez et al., 2001; Vogels, Renkawek, Broere, ter Laak, & van Workum, 1989) and BFCN degeneration is associated with an increase in Gal and GalRs as well as hypertrophy of galaninergic fibers (Beal, MacGarvey, & Swartz, 1990; Chan-Palay, 1988; Counts, He, Che, Ginsberg, & Mufson, 2009; Kelley et al., 2011; Mufson, Counts, Perez, & Binder, 2005; Sendera et al., 2000). Functionally, Gal may be neuroprotective by preventing Aβ-induced cell death or by decreasing cleavage of caspase 3 and caspase 9 in AD (Ding, MacTavish, Kar, & Jhamandas, 2006; Elliott-Hunt et al., 2011; Vogels et al., 1989). Whether the increase in Gal expression is triggered by an extracellular or inter-cellular event, or is a developmentally related transcriptional bystander effect remains to be investigated. While Gal expression was not significantly altered with MCS in TS mice, we found the expression Galr2 was significantly decreased with MCS in Ts mice. Prior studies have highlighted GalR2 agonists as providing the same neuroprotection as Gal (Ding et al., 2006; Sipkova et al., 2017), and based on its role in disease pathology, this can be a general indicator of the health of the MS/VDB system (Beal et al., 1990).
In the present study, we observed comparable Cdk5 expression levels between Ts and 2N animals, but a paradoxical increase in MCS treated Ts mice. Cdk5, which encodes for a serine/threonine kinase involved in cytoskeletal dynamics, plays a role brain development (Yoo and Lubec, 2001) and in the formation of tau pathology in AD and neurodegeneration in other diseases (Bu, Li, Davies, & Vincent, 2002; Imahori & Uchida, 1997; Kimura, Ishiguro, & Hisanaga, 2014; Nguyen, Lariviere, & Julien, 2001; Patrick et al., 1999). A prior study revealed no change in hippocampal CDK5 levels at 7 months in the Tc1 mouse model of DS (Ahmed et al., 2013). On the other hand, CDK5 protein levels were increased in the hippocampus at 4 months of age, with no change in functional markers p25 and p35 in Ts mice (Cruz & Tsai, 2004; Pollonini et al., 2008; Yoo & Lubec, 2001). Interestingly, it has been reported that the amount of p25 (marker of CDK5 activity and neurotoxicity) in the frontal cortex of patients with AD or DS is lower than in controls (Yoo and Lubec, 2001). Although the sample size was small, the study raises questions as to the exact role that CDK5 and its regulatory subunits play in DS pathobiology.
Potential limitations
In the present study, we relied on mRNA extraction from fixed mouse brains to examine relative gene expression levels across groups. This type of single-time-point analysis assumes the sample is representative of a steady-state measurement. Based on prior studies, the average half-life of mRNA is estimated at 10 hours, and transcripts that have a high production and high decay rate (< 2 h half-life) may be functionally different from longer-lived transcripts but are expected to maintain a steady state expression level (Lam et al., 2001; Yang et al., 2003). With this in mind, we minimized stress by deeply anesthetizing each animal prior to entering the perfusion suite, and circadian rhythm interactions by conducting randomized perfusions across treatment groups on the same day within a 5-hour window. With regard to the latter, we did not find significant differences between groups in protein phosphatase 1 catalytic subunit genes (Ppp1ca, Ppp1cb, and Ppp1cc), which play a role in circadian regulation and photoperiod entrainment. However, we cannot rule out effects due to intrinsic factors such as possible polymorphisms in select genes that affect turnover rates (Puga et al., 2005), which are unexplored in this mouse model, and the custom-designed array platform is not designed to detect.
It is important to bear in mind that the Ts65Dn murine model is representative of a specific pathology found in DS and AD, notably age-related degeneration of the septohippocampal cholinergic system. This model lacks the classic neuropathological hallmarks of aging DS and AD, namely amyloid plaques and neurofibrillary tangles. Moreover, the model does not recreate the full genetic condition of human trisomy 21; several genes triplicated in humans with DS are not present in the Ts65Dn model and there are triplicated genes in Ts65Dn mice that are not trisomic in human DS (Sturgeon & Gardiner, 2011). These considerations should be kept in mind when extrapolating the present findings to other species and treatment models. Despite this caveat, the Ts65Dn mouse does provide a unique model with reproducibility for the study of the cellular and molecular pathobiology that trisomy exerts upon the septohippocampal memory circuit. Expression profiles reported herein implicate key signaling pathways beyond canonical AD-related genes in the pathogenesis of MS/VDB neurons in DS and AD. The datasets derived using a vulnerable-cell specific population provide tangible targets related to mechanistic alterations and functional consequences of septohippocampal degeneration. These investigations begin the process of defining which transcript (and ultimately encoded proteins) changes are primary and which are secondary within BFCNs in response to MCS in Ts65Dn mice, which has direct implications for translation to human disease.
Acknowledgements:
Support by NIH grants HD057564 (BJS, EJM, SDG), PO1AG014449 (EJM, SDG), AG043375 (EJM & SDG), AG107617 (SDG); and the Alzheimer’s Association, IIRG-12-237253 (SDG).
Footnotes
Conflict of Interest Statement
The authors have no conflicts of interest that might be perceived as influencing their objectivity.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Ahmed MM, Dhanasekaran AR, Tong S, Wiseman FK, Fisher EM, Tybulewicz VL, & Gardiner KJ (2013). Protein profiles in Tc1 mice implicate novel pathway perturbations in the Down syndrome brain. Hum Mol Genet, 22(9), 1709–1724. doi: 10.1093/hmg/ddt017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeson EC, Lambert JP, Narayanswami S, Gardiner K, Bechtel LJ, & Davisson MT (2001). Ts65Dn -- localization of the translocation breakpoint and trisomic gene content in a mouse model for Down syndrome. Cytogenet Cell Genet, 93(3–4), 270–276. doi: 10.1159/000056997 [DOI] [PubMed] [Google Scholar]
- Alexander GE, Saunders AM, Szczepanik J, Strassburger TL, Pietrini P, Dani A, … Schapiro MB (1997). Relation of age and apolipoprotein E to cognitive function in Down syndrome adults. Neuroreport, 8(8), 1835–1840. [DOI] [PubMed] [Google Scholar]
- Alldred MJ, Chao HM, Lee SH, Beilin J, Powers BE, Petkova E, … Ginsberg SD (2018). CA1 pyramidal neuron gene expression mosaics in the Ts65Dn murine model of Down syndrome and Alzheimer’s disease following maternal choline supplementation. Hippocampus, 28(4), 251–268. doi: 10.1002/hipo.22832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, Che S, & Ginsberg SD (2009). Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods, 177(2), 381–385. doi: 10.1016/j.jneumeth.2008.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, Lee SH, Petkova E, & Ginsberg SD (2015a). Expression profile analysis of hippocampal CA1 pyramidal neurons in aged Ts65Dn mice, a model of Down syndrome (DS) and Alzheimer’s disease (AD). Brain Struct Funct, 220(5), 2983–2996. doi: 10.1007/s00429-014-0839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, Lee SH, Petkova E, & Ginsberg SD (2015b). Expression profile analysis of vulnerable CA1 pyramidal neurons in young-Middle-Aged Ts65Dn mice. J Comp Neurol, 523(1), 61–74. doi: 10.1002/cne.23663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash JA, Velazquez R, Kelley CM, Powers BE, Ginsberg SD, Mufson EJ, & Strupp BJ (2014). Maternal choline supplementation improves spatial mapping and increases basal forebrain cholinergic neuron number and size in aged Ts65Dn mice. Neurobiol Dis, 70, 32–42. doi: 10.1016/j.nbd.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko B, Wilkerson JR, Cavalier SH, Binns DD, James NG, Jameson DM, … Albanesi JP (2018). Palmitoylation and Membrane Binding of Arc/Arg3.1: A Potential Role in Synaptic Depression. Biochemistry, 57(5), 520–524. doi: 10.1021/acs.biochem.7b00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, MacGarvey U, & Swartz KJ (1990). Galanin immunoreactivity is increased in the nucleus basalis of Meynert in Alzheimer’s disease. Ann Neurol, 28(2), 157–161. doi: 10.1002/ana.410280207 [DOI] [PubMed] [Google Scholar]
- Belfort GM, & Kandror KV (2003). Cellugyrin and synaptogyrin facilitate targeting of synaptophysin to a ubiquitous synaptic vesicle-sized compartment in PC12 cells. J Biol Chem, 278(48), 47971–47978. doi: 10.1074/jbc.M304174200 [DOI] [PubMed] [Google Scholar]
- Ben Gedalya T, Loeb V, Israeli E, Altschuler Y, Selkoe DJ, & Sharon R (2009). Alpha-synuclein and polyunsaturated fatty acids promote clathrin-mediated endocytosis and synaptic vesicle recycling. Traffic, 10(2), 218–234. doi: 10.1111/j.1600-0854.2008.00853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 12. [Google Scholar]
- Berger-Sweeney J (2003). The cholinergic basal forebrain system during development and its influence on cognitive processes: important questions and potential answers. Neurosci Biobehav Rev, 27(4), 401–411. [DOI] [PubMed] [Google Scholar]
- Blusztajn JK, Cermak JM, Holler T, & Jackson DA (1998). Imprinting of hippocampal metabolism of choline by its availability during gestation: implications for cholinergic neurotransmission. J Physiol Paris, 92(3–4), 199–203. [DOI] [PubMed] [Google Scholar]
- Blusztajn JK, Slack BE, & Mellott TJ (2017). Neuroprotective Actions of Dietary Choline. Nutrients, 9(8). doi: 10.3390/nu9080815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonamico M, Mariani P, Danesi HM, Crisogianni M, Failla P, Gemme G, … Medical Genetic, G. (2001). Prevalence and clinical picture of celiac disease in italian down syndrome patients: a multicenter study. J Pediatr Gastroenterol Nutr, 33(2), 139–143. [DOI] [PubMed] [Google Scholar]
- Bu B, Li J, Davies P, & Vincent I (2002). Deregulation of cdk5, hyperphosphorylation, and cytoskeletal pathology in the Niemann-Pick type C murine model. J Neurosci, 22(15), 6515–6525. doi:20026692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Walker LC, Whitehouse PJ, & Price DL (1985). Abnormalities of the nucleus basalis in Down’s syndrome. Ann Neurol, 18(3), 310–313. doi: 10.1002/ana.410180306 [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, Villar A, … Nixon RA (2003). App gene dosage modulates endosomal abnormalities of Alzheimer’s disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci, 23(17), 6788–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak JM, Blusztajn JK, Meck WH, Williams CL, Fitzgerald CM, Rosene DL, & Loy R (1999). Prenatal availability of choline alters the development of acetylcholinesterase in the rat hippocampus. Dev Neurosci, 21(2), 94–104. doi: 10.1159/000017371 [DOI] [PubMed] [Google Scholar]
- Cermak JM, Holler T, Jackson DA, & Blusztajn JK (1998). Prenatal availability of choline modifies development of the hippocampal cholinergic system. FASEB J, 12(3), 349–357. [DOI] [PubMed] [Google Scholar]
- Chan-Palay V (1988). Galanin hyperinnervates surviving neurons of the human basal nucleus of Meynert in dementias of Alzheimer’s and Parkinson’s disease: a hypothesis for the role of galanin in accentuating cholinergic dysfunction in dementia. J Comp Neurol, 273(4), 543–557. doi: 10.1002/cne.902730409 [DOI] [PubMed] [Google Scholar]
- Chang Q, & Gold PE (2008). Age-related changes in memory and in acetylcholine functions in the hippocampus in the Ts65Dn mouse, a model of Down syndrome. Neurobiol Learn Mem, 89(2), 167–177. doi: 10.1016/j.nlm.2007.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Berger JD, Mazzella MJ, Morales-Corraliza J, Cataldo AM, Nixon RA, … Mathews PM (2009). Age-dependent dysregulation of brain amyloid precursor protein in the Ts65Dn Down syndrome mouse model. J Neurochem, 110(6), 1818–1827. doi: 10.1111/j.1471-4159.2009.06277.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, … Antonarakis SE (2000). The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res, 10(12), 2006–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen WI (2006). Current dilemmas in Down syndrome clinical care: celiac disease, thyroid disorders, and atlanto-axial instability. Am J Med Genet C Semin Med Genet, 142C(3), 141–148. doi: 10.1002/ajmg.c.30102 [DOI] [PubMed] [Google Scholar]
- Contestabile A, Fila T, Bartesaghi R, Contestabile A, & Ciani E (2006). Choline acetyltransferase activity at different ages in brain of Ts65Dn mice, an animal model for Down’s syndrome and related neurodegenerative diseases. J Neurochem, 97(2), 515–526. doi: 10.1111/j.1471-4159.2006.03769.x [DOI] [PubMed] [Google Scholar]
- Cooney CA, Dave AA, & Wolff GL (2002). Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr, 132(8 Suppl), 2393S–2400S. doi: 10.1093/jn/132.8.2393S [DOI] [PubMed] [Google Scholar]
- Cooper JD, Salehi A, Delcroix JD, Howe CL, Belichenko PV, Chua-Couzens J, … Mobley WC (2001). Failed retrograde transport of NGF in a mouse model of Down’s syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc Natl Acad Sci U S A, 98(18), 10439–10444. doi: 10.1073/pnas.181219298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts SE, He B, Che S, Ginsberg SD, & Mufson EJ (2009). Galanin fiber hyperinnervation preserves neuroprotective gene expression in cholinergic basal forebrain neurons in Alzheimer’s disease. J Alzheimers Dis, 18(4), 885–896. doi: 10.3233/JAD-2009-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Oster-Granite ML, & Gearhart JD (1986). The neurobiologic consequences of Down syndrome. Brain Res Bull, 16(6), 773–787. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Oster-Granite ML, Reeves RH, & Gearhart JD (1988). Down syndrome, Alzheimer’s disease and the trisomy 16 mouse. Trends Neurosci, 11(9), 390–394. [DOI] [PubMed] [Google Scholar]
- Cruz JC, & Tsai LH (2004). Cdk5 deregulation in the pathogenesis of Alzheimer’s disease. Trends Mol Med, 10(9), 452–458. doi: 10.1016/j.molmed.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Dekker AD, Sacco S, Carfi A, Benejam B, Vermeiren Y, Beugelsdijk G, … De Deyn PP (2018). The Behavioral and Psychological Symptoms of Dementia in Down Syndrome (BPSD-DS) Scale: Comprehensive Assessment of Psychopathology in Down Syndrome. J Alzheimers Dis, 63(2), 797–819. doi: 10.3233/JAD-170920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcroix JD, Valletta J, Wu C, Howe CL, Lai CF, Cooper JD, … Mobley WC (2004). Trafficking the NGF signal: implications for normal and degenerating neurons. Prog Brain Res, 146, 3–23. [DOI] [PubMed] [Google Scholar]
- Detopoulou P, Panagiotakos DB, Antonopoulou S, Pitsavos C, & Stefanadis C (2008). Dietary choline and betaine intakes in relation to concentrations of inflammatory markers in healthy adults: the ATTICA study. Am J Clin Nutr, 87(2), 424–430. doi: 10.1093/ajcn/87.2.424 [DOI] [PubMed] [Google Scholar]
- Dierssen M, Herault Y, & Estivill X (2009). Aneuploidy: from a physiological mechanism of variance to Down syndrome. Physiol Rev, 89(3), 887–920. doi: 10.1152/physrev.00032.2007 [DOI] [PubMed] [Google Scholar]
- Ding X, MacTavish D, Kar S, & Jhamandas JH (2006). Galanin attenuates beta-amyloid (Abeta) toxicity in rat cholinergic basal forebrain neurons. Neurobiol Dis, 21(2), 413–420. doi: 10.1016/j.nbd.2005.08.016 [DOI] [PubMed] [Google Scholar]
- Duchon A, Raveau M, Chevalier C, Nalesso V, Sharp AJ, & Herault Y (2011). Identification of the translocation breakpoints in the Ts65Dn and Ts1Cje mouse lines: relevance for modeling Down syndrome. Mamm Genome, 22(11–12), 674–684. doi: 10.1007/s00335-011-9356-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyken ME, Lin-Dyken DC, Poulton S, Zimmerman MB, & Sedars E (2003). Prospective polysomnographic analysis of obstructive sleep apnea in down syndrome. Arch Pediatr Adolesc Med, 157(7), 655–660. doi: 10.1001/archpedi.157.7.655 [DOI] [PubMed] [Google Scholar]
- Dynes JL, & Steward O (2012). Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol, 520(14), 3105–3119. doi: 10.1002/cne.23073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B (2007). SIZE, POWER AND FALSE DISCOVERY RATES. The Annals of Statistics, 35(4), 27. doi: 10.1214/009053606000001460 [DOI] [Google Scholar]
- Elliott-Hunt CR, Holmes FE, Hartley DM, Perez S, Mufson EJ, & Wynick D (2011). Endogenous galanin protects mouse hippocampal neurons against amyloid toxicity in vitro via activation of galanin receptor-2. J Alzheimers Dis, 25(3), 455–462. doi: 10.3233/JAD-2011-110011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ (1995). Epilogue: toward the twenty-first century with Down syndrome--a personal view of how far we have come and of how far we can reasonably expect to go. Prog Clin Biol Res, 393, 241–246. [PubMed] [Google Scholar]
- Fisher MC, Zeisel SH, Mar MH, & Sadler TW (2001). Inhibitors of choline uptake and metabolism cause developmental abnormalities in neurulating mouse embryos. Teratology, 64(2), 114–122. doi: 10.1002/tera.1053 [DOI] [PubMed] [Google Scholar]
- Fisher MC, Zeisel SH, Mar MH, & Sadler TW (2002). Perturbations in choline metabolism cause neural tube defects in mouse embryos in vitro. FASEB J, 16(6), 619–621. [DOI] [PubMed] [Google Scholar]
- Fodale V, Mafrica F, Caminiti V, & Grasso G (2006). The cholinergic system in Down’s syndrome. J Intellect Disabil, 10(3), 261–274. doi: 10.1177/1744629506067615 [DOI] [PubMed] [Google Scholar]
- Fong CT, & Brodeur GM (1987). Down’s syndrome and leukemia: epidemiology, genetics, cytogenetics and mechanisms of leukemogenesis. Cancer Genet Cytogenet, 28(1), 55–76. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Gensous N, Bacalini MG, Conte M, & Salvioli S (2019). Accelerated bio-cognitive aging in Down syndrome: State of the art and possible deceleration strategies. Aging Cell, e12903. doi: 10.1111/acel.12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SB, Bean LH, Allen EG, Tinker SW, Locke AE, Druschel C, … Sherman SL (2008). Ethnicity, sex, and the incidence of congenital heart defects: a report from the National Down Syndrome Project. Genet Med, 10(3), 173–180. doi: 10.1097/GIM.0b013e3181634867 [DOI] [PubMed] [Google Scholar]
- Gardiner K (2004). Gene-dosage effects in Down syndrome and trisomic mouse models. Genome Biol, 5(10), 244. doi: 10.1186/gb-2004-5-10-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, … Che S (2010). Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry, 68(10), 885–893. doi: 10.1016/j.biopsych.2010.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Gunnam SM, Schiroli C, Lee SH, Morgello S, & Fischer T (2018). Expression profiling suggests microglial impairment in human immunodeficiency virus neuropathogenesis. Ann Neurol, 83(2), 406–417. doi: 10.1002/ana.25160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, & Che S (2014). Methods and compositions for amplification and detection of microRNAs (miRNAs) and noncoding RNAs (ncRNAs) using the signature sequence amplification method (SSAM). Recent Adv DNA Gene Seq, 8(1), 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Wuu J, Counts SE, & Mufson EJ (2006). Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem, 97(2), 475–487. doi: 10.1111/j.1471-4159.2006.03764.x [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Malek-Ahmadi MH, Alldred MJ, Che S, Elarova I, Chen Y, … Mufson EJ (2017). Selective decline of neurotrophin and neurotrophin receptor genes within CA1 pyramidal neurons and hippocampus proper: Correlation with cognitive performance and neuropathology in mild cognitive impairment and Alzheimer’s disease. Hippocampus. doi: 10.1002/hipo.22802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, & Che S (2011). Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Chem Neuroanat, 42(2), 102–110. doi: 10.1016/j.jchemneu.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Counts SE, Wuu J, Alldred MJ, Nixon RA, & Che S (2010). Regional selectivity of rab5 and rab7 protein upregulation in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis, 22(2), 631–639. doi: 10.3233/JAD-2010-101080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granholm AC, Sanders LA, & Crnic LS (2000). Loss of cholinergic phenotype in basal forebrain coincides with cognitive decline in a mouse model of Down’s syndrome. Exp Neurol, 161(2), 647–663. doi: 10.1006/exnr.1999.7289 [DOI] [PubMed] [Google Scholar]
- Hampel H, Mesulam MM, Cuello AC, Farlow MR, Giacobini E, Grossberg GT, … Khachaturian ZS (2018). The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain, 141(7), 1917–1933. doi: 10.1093/brain/awy132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmo ME, & Sarter M (2011). Modes and models of forebrain cholinergic neuromodulation of cognition. Neuropsychopharmacology, 36(1), 52–73. doi: 10.1038/npp.2010.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MA, Li YJ, Xu H, Noureddine MA, Shao YS, Gullans SR, … Vance JM (2005). Expression profiling of substantia nigra in Parkinson disease, progressive supranuclear palsy, and frontotemporal dementia with parkinsonism. Arch Neurol, 62(6), 917–921. doi: 10.1001/archneur.62.6.917 [DOI] [PubMed] [Google Scholar]
- Haxby JV, & Schapiro MB (1992). Longitudinal study of neuropsychological function in older adults with Down syndrome. Prog Clin Biol Res, 379, 35–50. [PubMed] [Google Scholar]
- Hof PR, Bouras C, Perl DP, Sparks DL, Mehta N, & Morrison JH (1995). Age-related distribution of neuropathologic changes in the cerebral cortex of patients with Down’s syndrome. Quantitative regional analysis and comparison with Alzheimer’s disease. Arch Neurol, 52(4), 379–391. [DOI] [PubMed] [Google Scholar]
- Holler T, Cermak JM, & Blusztajn JK (1996). Dietary choline supplementation in pregnant rats increases hippocampal phospholipase D activity of the offspring. FASEB J, 10(14), 1653–1659. [DOI] [PubMed] [Google Scholar]
- Holmes GL, Yang Y, Liu Z, Cermak JM, Sarkisian MR, Stafstrom CE, … Blusztajn JK (2002). Seizure-induced memory impairment is reduced by choline supplementation before or after status epilepticus. Epilepsy Res, 48(1–2), 3–13. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Kilbridge J, Chen KS, Rabin J, Luche R, Carlson E, … Mobley WC (1995). Preliminary characterization of the central nervous system in partial trisomy 16 mice. Prog Clin Biol Res, 393, 227–240. [PubMed] [Google Scholar]
- Holtzman DM, Santucci D, Kilbridge J, Chua-Couzens J, Fontana DJ, Daniels SE, … Mobley WC (1996). Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc Natl Acad Sci U S A, 93(23), 13333–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CL, Isacson O, Nelson M, Bimonte-Nelson H, Seo H, Lin L, … Granholm AC (2003). Regional alterations in amyloid precursor protein and nerve growth factor across age in a mouse model of Down’s syndrome. Neurosci Res, 45(4), 437–445. [DOI] [PubMed] [Google Scholar]
- Imahori K, & Uchida T (1997). Physiology and pathology of tau protein kinases in relation to Alzheimer’s disease. J Biochem, 121(2), 179–188. [PubMed] [Google Scholar]
- Janz R, Sudhof TC, Hammer RE, Unni V, Siegelbaum SA, & Bolshakov VY (1999). Essential roles in synaptic plasticity for synaptogyrin I and synaptophysin I. Neuron, 24(3), 687–700. [DOI] [PubMed] [Google Scholar]
- Jorgensen OS, Brooksbank BW, & Balazs R (1990). Neuronal plasticity and astrocytic reaction in Down syndrome and Alzheimer disease. J Neurol Sci, 98(1), 63–79. [DOI] [PubMed] [Google Scholar]
- Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, … Yaspo ML (2004). Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res, 14(7), 1258–1267. doi: 10.1101/gr.1951304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler C (1966). Retinal degeneration in the mouse is rodless retina. J Hered, 57(2), 47–50. [DOI] [PubMed] [Google Scholar]
- Kelley CM, Ash JA, Powers BE, Velazquez R, Alldred MJ, Ikonomovic MD, … Mufson EJ (2016). Effects of Maternal Choline Supplementation on the Septohippocampal Cholinergic System in the Ts65Dn Mouse Model of Down Syndrome. Curr Alzheimer Res, 13(1), 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CM, Perez SE, Overk CR, Wynick D, & Mufson EJ (2011). Effect of neocortical and hippocampal amyloid deposition upon galaninergic and cholinergic neurites in AbetaPPswe/PS1DeltaE9 mice. J Alzheimers Dis, 25(3), 491–504. doi: 10.3233/JAD-2011-102097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CM, Powers BE, Velazquez R, Ash JA, Ginsberg SD, Strupp BJ, & Mufson EJ (2014a). Maternal choline supplementation differentially alters the basal forebrain cholinergic system of young-adult Ts65Dn and disomic mice. J Comp Neurol, 522(6), 1390–1410. doi: 10.1002/cne.23492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CM, Powers BE, Velazquez R, Ash JA, Ginsberg SD, Strupp BJ, & Mufson EJ (2014b). Sex differences in the cholinergic basal forebrain in the Ts65Dn mouse model of Down syndrome and Alzheimer’s disease. Brain Pathol, 24(1), 33–44. doi: 10.1111/bpa.12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C, Atasu B, Hanagasi H, Simon-Sanchez J, Hauser AK, Pak M, … Lohmann E (2018). Role of LRRK2 and SNCA in autosomal dominant Parkinson’s disease in Turkey. Parkinsonism Relat Disord, 48, 34–39. doi: 10.1016/j.parkreldis.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Kimura T, Ishiguro K, & Hisanaga S (2014). Physiological and pathological phosphorylation of tau by Cdk5. Front Mol Neurosci, 7, 65. doi: 10.3389/fnmol.2014.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, … Staudt LM (2001). Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol, 2(10), RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak RK (2014). Heat shock proteins in neurodegenerative disorders and aging. J Cell Commun Signal, 8(4), 293–310. doi: 10.1007/s12079-014-0243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz JB, Miller MA, Dobie DJ, Peskind ER, & Raskind MA (2001). Increased alpha 2-adrenergic receptor binding in locus coeruleus projection areas in dementia with Lewy bodies. Neurobiol Aging, 22(4), 555–561. [DOI] [PubMed] [Google Scholar]
- Li Q, Guo-Ross S, Lewis DV, Turner D, White AM, Wilson WA, & Swartzwelder HS (2004). Dietary prenatal choline supplementation alters postnatal hippocampal structure and function. J Neurophysiol, 91(4), 1545–1555. doi: 10.1152/jn.00785.2003 [DOI] [PubMed] [Google Scholar]
- Loy R, Heyer D, Williams CL, & Meck WH (1991). Choline-induced spatial memory facilitation correlates with altered distribution and morphology of septal neurons. Adv Exp Med Biol, 295, 373–382. [DOI] [PubMed] [Google Scholar]
- Maatta T, Tervo-Maatta T, Taanila A, Kaski M, & Iivanainen M (2006). Mental health, behaviour and intellectual abilities of people with Down syndrome. Downs Syndr Res Pract, 11(1), 37–43. [DOI] [PubMed] [Google Scholar]
- Mann DM, Yates PO, & Marcyniuk B (1984). Alzheimer’s presenile dementia, senile dementia of Alzheimer type and Down’s syndrome in middle age form an age related continuum of pathological changes. Neuropathol Appl Neurobiol, 10(3), 185–207. [DOI] [PubMed] [Google Scholar]
- McCulloch CE, Searle SR, & Neuhaus JM (2008). Generalized, linear, and mixed models (2nd ed.). Hoboken, N.J.: Wiley. [Google Scholar]
- Meck WH, Smith RA, & Williams CL (1988). Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev Psychobiol, 21(4), 339–353. doi: 10.1002/dev.420210405 [DOI] [PubMed] [Google Scholar]
- Meck WH, Smith RA, & Williams CL (1989). Organizational changes in cholinergic activity and enhanced visuospatial memory as a function of choline administered prenatally or postnatally or both. Behav Neurosci, 103(6), 1234–1241. [DOI] [PubMed] [Google Scholar]
- Meck WH, & Williams CL (1997). Characterization of the facilitative effects of perinatal choline supplementation on timing and temporal memory. Neuroreport, 8(13), 2831–2835. [DOI] [PubMed] [Google Scholar]
- Meck WH, & Williams CL (1999). Choline supplementation during prenatal development reduces proactive interference in spatial memory. Brain Res Dev Brain Res, 118(1–2), 51–59. [DOI] [PubMed] [Google Scholar]
- Meck WH, & Williams CL (2003). Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci Biobehav Rev, 27(4), 385–399. [DOI] [PubMed] [Google Scholar]
- Meck WH, Williams CL, Cermak JM, & Blusztajn JK (2007). Developmental periods of choline sensitivity provide an ontogenetic mechanism for regulating memory capacity and age-related dementia. Front Integr Neurosci, 1, 7. doi: 10.3389/neuro.07.007.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander T, Staines WA, Hokfelt T, Rokaeus A, Eckenstein F, Salvaterra PM, & Wainer BH (1985). Galanin-like immunoreactivity in cholinergic neurons of the septum-basal forebrain complex projecting to the hippocampus of the rat. Brain Res, 360(1–2), 130–138. [DOI] [PubMed] [Google Scholar]
- Mellott TJ, Huleatt OM, Shade BN, Pender SM, Liu YB, Slack BE, & Blusztajn JK (2017). Correction: Perinatal Choline Supplementation Reduces Amyloidosis and Increases Choline Acetyltransferase Expression in the Hippocampus of the APPswePS1dE9 Alzheimer’s Disease Model Mice. PLoS One, 12(3), e0174875. doi: 10.1371/journal.pone.0174875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellott TJ, Williams CL, Meck WH, & Blusztajn JK (2004). Prenatal choline supplementation advances hippocampal development and enhances MAPK and CREB activation. FASEB J, 18(3), 545–547. doi: 10.1096/fj.03-0877fje [DOI] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Wainer BH, & Levey AI (1983). Central cholinergic pathways in the rat: an overview based on an alternative nomenclature (Ch1-Ch6). Neuroscience, 10(4), 1185–1201. [DOI] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, & Okuno H (2015). Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front Mol Neurosci, 8, 78. doi: 10.3389/fnmol.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Chen M, Gandhy SU, Strawderman M, Levitsky DA, Maclean KN, & Strupp BJ (2010). Perinatal choline supplementation improves cognitive functioning and emotion regulation in the Ts65Dn mouse model of Down syndrome. Behav Neurosci, 124(3), 346–361. doi: 10.1037/a0019590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, Bothwell M, & Kordower JH (1989). Loss of nerve growth factor receptor-containing neurons in Alzheimer’s disease: a quantitative analysis across subregions of the basal forebrain. Exp Neurol, 105(3), 221–232. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, Cochran E, Benzing W, & Kordower JH (1993). Galaninergic innervation of the cholinergic vertical limb of the diagonal band (Ch2) and bed nucleus of the stria terminalis in aging, Alzheimer’s disease and Down’s syndrome. Dementia, 4(5), 237–250. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, Counts SE, Perez SE, & Binder L (2005). Galanin plasticity in the cholinergic basal forebrain in Alzheimer’s disease and transgenic mice. Neuropeptides, 39(3), 233–237. doi: 10.1016/j.npep.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Hurt J, Jakes R, Xuereb J, Honer WG, & Wischik CM (2000). Alpha-synuclein inclusions in Alzheimer and Lewy body diseases. J Neuropathol Exp Neurol, 59(5), 408–417. [DOI] [PubMed] [Google Scholar]
- Nag N, & Berger-Sweeney JE (2007). Postnatal dietary choline supplementation alters behavior in a mouse model of Rett syndrome. Neurobiol Dis, 26(2), 473–480. doi: 10.1016/j.nbd.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Nag N, Mellott TJ, & Berger-Sweeney JE (2008). Effects of postnatal dietary choline supplementation on motor regional brain volume and growth factor expression in a mouse model of Rett syndrome. Brain Res, 1237, 101–109. doi: 10.1016/j.brainres.2008.08.042 [DOI] [PubMed] [Google Scholar]
- Nguyen MD, Lariviere RC, & Julien JP (2001). Deregulation of Cdk5 in a mouse model of ALS: toxicity alleviated by perikaryal neurofilament inclusions. Neuron, 30(1), 135–147. [DOI] [PubMed] [Google Scholar]
- Niculescu MD, Yamamuro Y, & Zeisel SH (2004). Choline availability modulates human neuroblastoma cell proliferation and alters the methylation of the promoter region of the cyclin-dependent kinase inhibitor 3 gene. J Neurochem, 89(5), 1252–1259. doi: 10.1111/j.1471-4159.2004.02414.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C, Crayton L, Holland A, Hall S, & Bradbury J (1998). A four year prospective study of age-related cognitive change in adults with Down’s syndrome. Psychol Med, 28(6), 1365–1377. [DOI] [PubMed] [Google Scholar]
- Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, & Tsai LH (1999). Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature, 402(6762), 615–622. doi: 10.1038/45159 [DOI] [PubMed] [Google Scholar]
- Perez SE, Miguel JC, He B, Malek-Ahmadi M, Abrahamson EE, Ikonomovic MD, … Mufson EJ (2019). Frontal cortex and striatal cellular and molecular pathobiology in individuals with Down syndrome with and without dementia. Acta Neuropathol. doi: 10.1007/s00401-019-01965-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Wynick D, Steiner RA, & Mufson EJ (2001). Distribution of galaninergic immunoreactivity in the brain of the mouse. J Comp Neurol, 434(2), 158–185. [DOI] [PubMed] [Google Scholar]
- Pollonini G, Gao V, Rabe A, Palminiello S, Albertini G, & Alberini CM (2008). Abnormal expression of synaptic proteins and neurotrophin-3 in the Down syndrome mouse model Ts65Dn. Neuroscience, 156(1), 99–106. doi: 10.1016/j.neuroscience.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers BE, Kelley CM, Velazquez R, Ash JA, Strawderman MS, Alldred MJ, … Strupp BJ (2017). Maternal choline supplementation in a mouse model of Down syndrome: Effects on attention and nucleus basalis/substantia innominata neuron morphology in adult offspring. Neuroscience, 340, 501–514. doi: 10.1016/j.neuroscience.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard MA, & Kola I (1999). The “gene dosage effect” hypothesis versus the “amplified developmental instability” hypothesis in Down syndrome. J Neural Transm Suppl, 57, 293–303. [PubMed] [Google Scholar]
- Puga I, Lainez B, Fernandez-Real JM, Buxade M, Broch M, Vendrell J, & Espel E (2005). A polymorphism in the 3’ untranslated region of the gene for tumor necrosis factor receptor 2 modulates reporter gene expression. Endocrinology, 146(5), 2210–2220. doi: 10.1210/en.2004-1366 [DOI] [PubMed] [Google Scholar]
- Pyapali GK, Turner DA, Williams CL, Meck WH, & Swartzwelder HS (1998). Prenatal dietary choline supplementation decreases the threshold for induction of long-term potentiation in young adult rats. J Neurophysiol, 79(4), 1790–1796. doi: 10.1152/jn.1998.79.4.1790 [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, … Davisson MT (1995). A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet, 11(2), 177–184. doi: 10.1038/ng1095-177 [DOI] [PubMed] [Google Scholar]
- Rye DB, Wainer BH, Mesulam MM, Mufson EJ, & Saper CB (1984). Cortical projections arising from the basal forebrain: a study of cholinergic and noncholinergic components employing combined retrograde tracing and immunohistochemical localization of choline acetyltransferase. Neuroscience, 13(3), 627–643. [DOI] [PubMed] [Google Scholar]
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, … Mobley WC (2006). Increased App expression in a mouse model of Down’s syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron, 51(1), 29–42. doi: 10.1016/j.neuron.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Sandstrom NJ, Loy R, & Williams CL (2002). Prenatal choline supplementation increases NGF levels in the hippocampus and frontal cortex of young and adult rats. Brain Res, 947(1), 9–16. [DOI] [PubMed] [Google Scholar]
- Schafer MJ, Dolgalev I, Alldred MJ, Heguy A, & Ginsberg SD (2015). Calorie Restriction Suppresses Age-Dependent Hippocampal Transcriptional Signatures. PLoS One, 10(7), e0133923. doi: 10.1371/journal.pone.0133923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk F, & Brandner K (1995). Indirect effects of peri- and postnatal choline treatment on place-learning abilities in rat. Psychobiology, 23(4), 12. doi: 10.3758/BF03333077 [DOI] [Google Scholar]
- Schliebs R, & Arendt T (2011). The cholinergic system in aging and neuronal degeneration. Behav Brain Res, 221(2), 555–563. doi: 10.1016/j.bbr.2010.11.058 [DOI] [PubMed] [Google Scholar]
- Sciences N. A. o., & Medicine I. o. (1998) Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. Washington (DC). [PubMed] [Google Scholar]
- Sendera TJ, Ma SY, Jaffar S, Kozlowski PB, Kordower JH, Mawal Y, … Mufson EJ (2000). Reduction in TrkA-immunoreactive neurons is not associated with an overexpression of galaninergic fibers within the nucleus basalis in Down’s syndrome. J Neurochem, 74(3), 1185–1196. [DOI] [PubMed] [Google Scholar]
- Seo H, & Isacson O (2005). Abnormal APP, cholinergic and cognitive function in Ts65Dn Down’s model mice. Exp Neurol, 193(2), 469–480. doi: 10.1016/j.expneurol.2004.11.017 [DOI] [PubMed] [Google Scholar]
- Sipkova J, Sida P, Kasprikova N, Kramarikova I, Hynie S, & Klenerova V (2017). Effect of Stress on the Expression of Galanin Receptors in Rat Heart. Folia Biol (Praha), 63(3), 98–104. [PubMed] [Google Scholar]
- Smith CL, Blake JA, Kadin JA, Richardson JE, Bult CJ, & Mouse Genome Database G (2018). Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res, 46(D1), D836–D842. doi: 10.1093/nar/gkx1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strupp BJ, Powers BE, Velazquez R, Ash JA, Kelley CM, Alldred MJ, … Ginsberg SD (2016). Maternal Choline Supplementation: A Potential Prenatal Treatment for Down Syndrome and Alzheimer’s Disease. Curr Alzheimer Res, 13(1), 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon X, & Gardiner KJ (2011). Transcript catalogs of human chromosome 21 and orthologous chimpanzee and mouse regions. Mamm Genome, 22(5–6), 261–271. doi: 10.1007/s00335-011-9321-y [DOI] [PubMed] [Google Scholar]
- Tees RC (1999). The influences of rearing environment and neonatal choline dietary supplementation on spatial learning and memory in adult rats. Behav Brain Res, 105(2), 173–188. [DOI] [PubMed] [Google Scholar]
- Thase ME (1982). Longevity and mortality in Down’s syndrome. J Ment Defic Res, 26(Pt 3), 177–192. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Abou EJ, & Dominguez HD (2009). Prenatal choline supplementation mitigates the adverse effects of prenatal alcohol exposure on development in rats. Neurotoxicol Teratol, 31(5), 303–311. doi: 10.1016/j.ntt.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Biane JS, O’Bryan KA, O’Neill TM, & Dominguez HD (2007). Choline supplementation following third-trimester-equivalent alcohol exposure attenuates behavioral alterations in rats. Behav Neurosci, 121(1), 120–130. doi: 10.1037/0735-7044.121.1.120 [DOI] [PubMed] [Google Scholar]
- Tukey JW (1977). Exploratory data analysis. Reading, MA: Addison-Wesley. [Google Scholar]
- Vargas KJ, Schrod N, Davis T, Fernandez-Busnadiego R, Taguchi YV, Laugks U, … Chandra SS (2017). Synucleins Have Multiple Effects on Presynaptic Architecture. Cell Rep, 18(1), 161–173. doi: 10.1016/j.celrep.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, … Marshall G (2015). Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res, 75(13), 2587–2593. doi: 10.1158/0008-5472.CAN-15-0262 [DOI] [PubMed] [Google Scholar]
- Vogels OJ, Renkawek K, Broere CA, ter Laak HJ, & van Workum F (1989). Galanin-like immunoreactivity within Ch2 neurons in the vertical limb of the diagonal band of Broca in aging and Alzheimer’s disease. Acta Neuropathol, 78(1), 90–95. [DOI] [PubMed] [Google Scholar]
- Ward BC, Agarwal S, Wang K, Berger-Sweeney J, & Kolodny NH (2008). Longitudinal brain MRI study in a mouse model of Rett Syndrome and the effects of choline. Neurobiol Dis, 31(1), 110–119. doi: 10.1016/j.nbd.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Wegiel J, Wisniewski HM, Dziewiatkowski J, Popovitch ER, & Tarnawski M (1996). Differential susceptibility to neurofibrillary pathology among patients with Down syndrome. Dementia, 7(3), 135–141. [DOI] [PubMed] [Google Scholar]
- Whitehouse PJ, Struble RG, Hedreen JC, Clark AW, White CL, Parhad IM, & Price DL (1983). Neuroanatomical evidence for a cholinergic deficit in Alzheimer’s disease. Psychopharmacol Bull, 19(3), 437–440. [PubMed] [Google Scholar]
- Wisniewski KE, Dalton AJ, McLachlan C, Wen GY, & Wisniewski HM (1985). Alzheimer’s disease in Down’s syndrome: clinicopathologic studies. Neurology, 35(7), 957–961. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Wisniewski HM, & Wen GY (1985). Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Ann Neurol, 17(3), 278–282. doi: 10.1002/ana.410170310 [DOI] [PubMed] [Google Scholar]
- Wurtman RJ, Cansev M, Sakamoto T, & Ulus IH (2009). Administration of docosahexaenoic acid, uridine and choline increases levels of synaptic membranes and dendritic spines in rodent brain. World Rev Nutr Diet, 99, 71–96. doi: 10.1159/000192998 [DOI] [PubMed] [Google Scholar]
- Xu L, & Pu J (2016). Alpha-Synuclein in Parkinson’s Disease: From Pathogenetic Dysfunction to Potential Clinical Application. Parkinsons Dis, 2016, 1720621. doi: 10.1155/2016/1720621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, & Darnell JE Jr. (2003). Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res, 13(8), 1863–1872. doi: 10.1101/gr.1272403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates CM, Simpson J, Gordon A, Maloney AF, Allison Y, Ritchie IM, & Urquhart A (1983). Catecholamines and cholinergic enzymes in pre-senile and senile Alzheimer-type dementia and Down’s syndrome. Brain Res, 280(1), 119–126. [DOI] [PubMed] [Google Scholar]
- Yates CM, Simpson J, Maloney AF, Gordon A, & Reid AH (1980). Alzheimer-like cholinergic deficiency in Down syndrome. Lancet, 2(8201), 979. [DOI] [PubMed] [Google Scholar]
- Yoo BC, & Lubec G (2001). p25 protein in neurodegeneration. Nature, 411(6839), 763–764; discussion 764–765. doi: 10.1038/35081146 [DOI] [PubMed] [Google Scholar]
- Zeisel SH (2000). Choline: an essential nutrient for humans. Nutrition, 16(7–8), 669–671. [DOI] [PubMed] [Google Scholar]
- Zeisel SH, & Blusztajn JK (1994). Choline and human nutrition. Annu Rev Nutr, 14, 269–296. doi: 10.1146/annurev.nu.14.070194.001413 [DOI] [PubMed] [Google Scholar]