Abstract
The kinetochore drives chromosome segregation at cell division. It acts as a physical link between chromosomes and dynamic microtubules, and as a signaling hub detecting and processing microtubule attachments to control anaphase onset. The mammalian kinetochore is a large macromolecular machine that forms a dynamic interface with the many microtubules that it binds. While we know most of the kinetochore’s component parts, how they work together to give rise to its robust functions remains poorly understood. Here we highlight recent findings that shed light on this question, driven by an expanding physical and molecular toolbox. We present emerging principles that underlie the kinetochore’s robust microtubule grip, such as redundancy, specialization, and dynamicity, and present signal processing principles that connect this microtubule grip to robust computation. Throughout, we identify open questions, and define simple engineering concepts that provide insight into kinetochore function.
Introduction
The kinetochore is the macromolecular machine that connects chromosomes to dynamic spindle microtubules at cell division. For genomic information to be accurately segregated and preserved as cells divide, the attachment of chromosomes to the spindle must be both robust and correct. As such, the kinetochore plays key physical and signaling roles: it must grip spindle microtubules, and must – as a ‘computer’ – process attachment information to signal when anaphase can and cannot begin (‘spindle assembly checkpoint’ (SAC)).
The mammalian kinetochore is built from ~100 protein species present in many copies in a well-defined stoichiometry, and it binds the 15–25 microtubules that make up the kinetochore-fiber (k-fiber) [1,2] (Fig. 1A). We now have a near complete parts list for the mammalian kinetochore, and there are significant efforts to map the stoichiometry, structure and biochemistry of this macromolecular machine [2–4]. Yet, how this machine’s mechanical and computational functions emerge from its component parts has long remained a frontier because of the system’s complexity and since mammalian kinetochores and k-fibers cannot yet be reconstituted in vitro. Expanding physical and molecular toolboxes in cells are now helping us address this question.
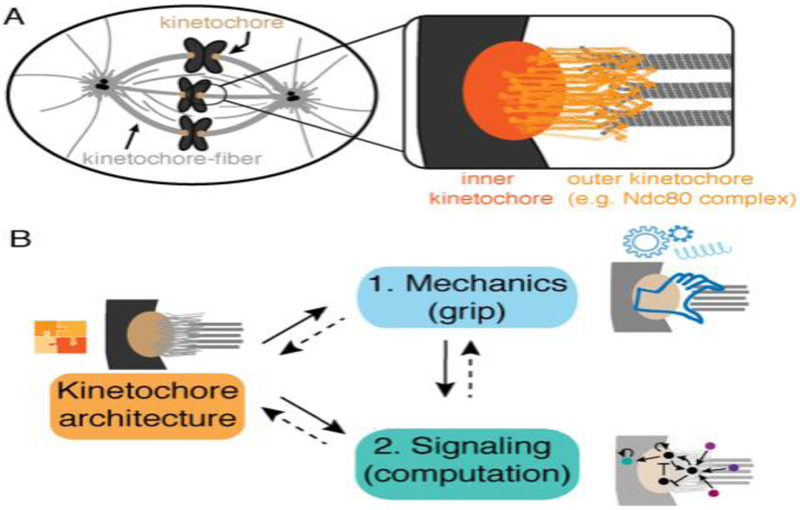
Figure 1. The mammalian kinetochore-microtubule interface.
A) Mammalian kinetochores (brown) connect chromosomes (dark gray) to kinetochore-fibers (“k-fibers”, light gray), bundles of many microtubules. In the inset, the outer kinetochore (light orange, e.g. Ndc80 complexes depicted) is a lawn of proteins that form the microtubule-interacting interface. The inner kinetochore (dark orange) links the outer kinetochore to centromeric chromatin. B) Kinetochore architecture, mechanics and signaling nodes feedback on each other to give rise to robust kinetochore function. In this review, we highlight (solid arrows) how the specific molecular interactions and architecture (orange puzzle pieces) gives rise to robust grip (section (1), blue), and how these in turn give rise to robust computation (section (2), green).
In this review, we focus on how the mammalian kinetochore’s individual parts together, as an ensemble, give rise to some of its key mechanical and signaling functions as it binds the k-fiber’s many microtubules (Fig. 1B). In two parts, we review recent work addressing two questions: (1) How do kinetochores grip multiple microtubules to maintain attachments that are mechanically robust, and yet flexible enough to allow microtubule to grow and shrink and detach to correct errors? (2) How do kinetochores integrate attachment information across multiple microtubules to compute a single output decision and allow anaphase entry? Many of the same proteins are involved in the kinetochore’s microtubule attachment and decision-making, and attachment serves as an input signal and signaling platform for decision-making. As such, we review them together, highlighting engineering principles that generate robust kinetochore function.
(1). Mechanics: Principles of Robust Grip
The inner kinetochore assembles on centromeric chromatin while the outer kinetochore forms the microtubule binding and signaling platform [5,6]. During microtubule attachment, outer kinetochore modules undergo structural rearrangements thought to aid in efficient spindle assembly [7–10]. Kinetochores face a challenging task: if they bind too tightly to microtubules, they avoid detachment but may disrupt microtubule dynamics (growth and shrinkage) or stabilize incorrect attachments, yet if they bind too loosely, they may not be able to correctly move chromosomes into each daughter cell. The kinetochore-microtubule interface must therefore be tuned to achieve robust and dynamic, not just strong, binding. Diverse architectural features of this interface are well-suited to facilitate robust grip (Fig. 2): many outer kinetochore proteins work together to bind many microtubules in a k-fiber (redundancy), a diversity of kinetochore proteins likely contribute to load-bearing (specialization), the mechanics of the interface can be regulated to adjust grip as needed (tunability) and k-fiber microtubules can grow and shrink (dynamicity), generating force to move chromosomes and allowing the spindle to remodel itself. We provide key examples of these features below, focusing on recent work.
Figure 2. Principles of robust grip at the mammalian kinetochore-microtubule interface.
A) Redundancy occurs at multiple levels. For example, many microtubules in the kinetochore-fiber (top) bind a single mammalian kinetochore, and many kinetochore protein copies bind a single microtubule (bottom). B) Kinetochore specialization in binding growing (left) versus shrinking (right) microtubules can occur via differential engagement of kinetochore proteins (top) or via differential localization or regulation of proteins (bottom). C) Tunability of the kinetochore-microtubule interface occurs via diverse regulators including kinases and phosphatases, plus-tip proteins and the binding of different kinetochore proteins over time during mitosis, acting on either kinetochore proteins or k-fiber microtubules. Inset shows examples of three different facets of kinetochore-microtubule interactions that can be tuned by the cell. D) Dynamicity at the kinetochore-microtubule interface occurs from microtubule growth and shrinkage, and it is modulated by a complex network of positive and negative regulators that tune microtubule dynamics and thereby kinetochore velocity. Inset shows potential models for how force at this interface affects microtubule growth or shrinkage velocity, i.e. the force-velocity relationship of this interface.
Redundancy
Redundancy is a hallmark of the mammalian kinetochore-microtubule interface. We use ‘redundancy’ to emphasize the mutiplicity of components, where each and every copy may not be necessary. The kinetochore’s main load-bearing microtubule binding unit, the Ndc80 complex [11,12], has ~250 copies per kinetochore (Fig. 2A) [13], of which only a low fraction (~30%) are engaged with microtubules [14] at any given time during metaphase. Redundancy at the level of kinetochore protein structure (e.g. a single protein may have multiple binding surfaces [15]) or sub-complex architecture (e.g. the multivalent arrangement of couplers [16]) may also be critical for robust tracking of dynamic microtubules by providing multiple contact points. At the kinetochore-microtubule interface, redundancy also occurs at the level of many microtubules (Fig. 2A). Many microtubules compose the mature k-fiber, exceeding the minimum number required for SAC satisfaction [17,18] and the estimated number needed to generate force to move a chromosome [19,20]. The high number of bound microtubules may instead ensure robust segregation by ensuring that turnover of k-fiber microtubules or error correction activities – both essential to function – do not fully disconnect a kinetochore from the spindle [17]. Further, having many redundant kinetochore coupling points may provide more sites for cellular regulation to tune microtubule affinity [21]. A better understanding of the mechanisms setting how many microtubules make a k-fiber and how a k-fiber is assembled [David et al., bioRxiv doi: 10.1101/501445] should provide insight into how the cell regulates the stoichiometry of the kinetochore-microtubule interface and its implications for kinetochore function.
Specialization
Not only do kinetochore proteins play highly specialized biochemical roles, they also play specialized mechanical roles in microtubule binding (Fig. 2B). Some of these proteins may be regulated to correct errors or stabilize proper attachments as mitosis progresses. Defining the specific mechanical functions and relative contributions of different proteins to the mechanics of the kinetochore-microtubule interface will be an important step to understanding kinetochore structure-function. Several proteins act in kinetochore-microtubule attachment in addition to the Ndc80 complex. For example, proteins such as the Ska complex [22–25], Cdt1 [26] and astrin-SKAP [27] have been proposed to act as additional couplers between kinetochores and microtubules, perhaps as “lock-down” factors [23]. Notably, many of these modules’ grip is regulated by the same set of kinases and phosphatases (tunability), which may ensure that whole kinetochore mechanics can be tuned as one ensemble (Fig. 2C) [26,28–30]. Specialization in grip may also arise from the same outer kinetochore protein complexes engaging differently either in structure or in number with growing versus shrinking microtubules (Fig. 2B) [14,31]. Determining which protein modules map to sites of active (energy consuming) and passive (non-energy consuming, e.g. frictional) force generation at the kinetochore-microtubule interface will be an important step forward [31,32]. Just as ascribing specific biochemical functions to kinetochore proteins has helped us understand the mechanisms of kinetochore signaling, mapping specific mechanical functions to diverse proteins will help us understand the underlying engineering principles that drive the kinetochore’s robust grip.
Dynamicity
Robust grip is not only determined by kinetochore composition and architecture, but also by the k-fiber’s dynamicity (Fig. 2D). K-fiber shrinkage powers chromosome movement [20] and k-fiber growth allows movement of the paired sister kinetochore. Thus it is key for the cell to limit growth and shrinkage velocities [33,34], to ensure that shrinking microtubules can move chromosomes on relevant timescales, and that kinetochores can keep track of microtubule ends without losing grip. Plus-end dynamics of individual microtubules within the kinetochore must also be coordinated [35]. An ensemble of regulatory proteins limits the dynamic range of microtubule growth and shrinkage, and imbalance of these regulators can lead to mitotic errors [36,37]. Further, just as microtubule dynamics can generate mechanical force, they are also regulated by force. The ability of force (in a given regime) to stabilize attachments has been directly shown and mapped at the reconstituted budding yeast kinetochore [38] and at grasshopper kinetochores inside cells [19,39], but we still lack a direct and quantitative understanding at the mammalian kinetochore. In principle, the ability of microtubule dynamics to respond to force is well suited to help dissipate force across the spindle, providing ‘slack’ in the system [40]. Flux of k-fiber microtubules towards spindle poles may play a similar role [40–42], and could also enhance kinetochore binding by biasing k-fibers towards a growing state where kinetochores may have specialized engagement [14]. In addition to microtubule end dynamics, the lifetime of components of the kinetochore-microtubule interface must also drive the interface’s mechanical function [43]. For example, the engagement between individual Ndc80 complexes and the microtubule is highly transient, which may allow rapid interface dynamics, while the longer lifetime of k-fiber microtubules is well-suited to ensure that correct microtubule attachments are stable.
Mechanical Insights Across Species
We have much to learn about kinetochore structure-function relationships from studying both differences and similarities between kinetochores across the tree of life [44–46]. For example, studying kinetochores of different sizes [47] or that bind different numbers of microtubules can illuminate diverse strategies that have emerged under different evolutionary constraints to accomplish the same task of chromosome segregation. Much of our understanding of kinetochore mechanics has come from studying species with point (monocentric) kinetochores, yet many species have holocentric kinetochores which face different challenges for grip and coordination of dynamics of microtubules that are spatially distant. Comparing the mechanical function of diverse kinetochore architectures may reveal new principles governing the mechanics of the kinetochore-microtubule interface.
(2). Computation: Principles of Robust Decision-Making
The underlying principles that facilitate robust kinetochore grip directly constrain the inputs that the SAC detects and integrates to make signaling decisions. Indeed, the kinetochore uses the same outer kinetochore module (KMN network) to perform its mechanical and computational functions [48], in both cases in the context of a highly redundant, complex, and dynamic system. The kinetochore accepts input signals from its local environment and produces an output signal that ultimately controls global cell cycle progression (Fig. 3). The inputs that the SAC has been proposed to detect (Ndc80-microtubule binding and tension across the kinetochore) and the output it produces (Mad1 level at that kinetochore) are highly dynamic and variable in magnitude [1,2,49]. Mapping how an input of a certain magnitude is converted into an output is critical to understanding how the kinetochore “computes”. How do we go from mapping the physical architecture of kinetochore-microtubule attachments (the “hardware”) [3] to understanding the network architecture that describes the complex SAC signaling cascade that emerges (the “software”) [50]? Here, we discuss recent advances in our understanding of kinetochore computation. These include which inputs kinetochores detect, how they detect and integrate them across the entire kinetochore structure, and how the structure of the kinetochore-microtubule interface and behavior of the SAC lead to robust chromosome segregation.
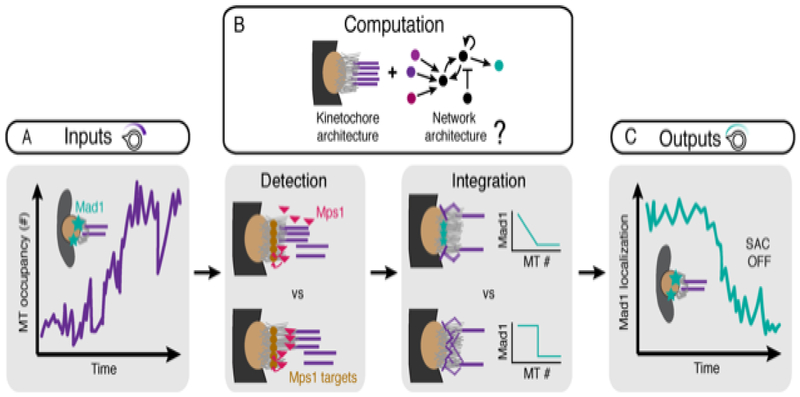
Figure 3. Robust computation at the mammalian kinetochore-microtubule interface.
The kinetochore is the platform for spindle assembly checkpoint signaling in which (A) microtubule occupancy inputs are (B) detected (left) and integrated (right) to compute a (C) checkpoint output, namely the level of Mad1 localization and generation of diffusible inhibitors that prevent anaphase onset. As microtubule occupancy (purple, input) rises during k-fiber formation, Mad1 begins to leave the kinetochore (turquoise, output). Microtubule input detection could, for example, occur through a competition model where microtubules occupy Mps1 binding sites (top), or through a displacement model where microtubule binding distances kinases from substrates (bottom). The kinetochore must integrate signals from detected microtubules to reach an output decision: it could do this linearly, where information from each microtubule is processed independently, or non-linearly where information from different microtubules is processed together to give rise to a threshold number of microtubules for checkpoint satisfaction.
Identifying relevant SAC inputs
A necessary step to understanding kinetochore SAC signal processing is to identify which element of microtubule attachment is the key SAC input signal (Fig. 3A). Two such elements have been proposed: microtubule binding [52,53] and tension generated by dynamic microtubules, either between two kinetochores [54] or across an individual kinetochore [55,56]. Microtubule binding is necessary to turn off the SAC [57], but whether it is sufficient is not yet clear. We now know that tension between kinetochores is not necessary to turn off the SAC [18,57,58]. The role of tension across an individual kinetochore has been harder to probe since pushing forces on chromosomes arms can, in principle, even produce tension on mono-attached or unpaired kinetochores. Tension could deform the kinetochore, and trigger biochemical events, and kinetochore deformations sometimes correlate with a “SAC off” state [54,55]. However, whether these deformations are causal for turning off the SAC is far from clear [59], and metaphase-level deformations that are being measured are not required to turn off the SAC [57,58]. Further, both tension and microtubule binding could result in kinetochore reorganization that is functionally relevant yet undetectable with current deformation mapping approaches. Independent of what exact element of microtubule attachment is the key SAC input signal, this input signal is expected to scale with the number of attached microtubules (“occupancy”).
Signal detection and integration
In order for the kinetochore to convert microtubule attachment into an output signal (ultimately Mad1 loss), it must both detect microtubule attachment signals and integrate them across its structure (Fig. 3B). There are two recent, non-mutually exclusive detection models: Ndc80-microtubule binding may physically displace SAC kinase Mps1 from its targets within the kinetochore [60] or microtubules may physically occupy Mps1-kinetochore binding sites and prevent Mps1 recruitment [61,62] (Fig. 3B). Kinetochore-microtubule binding may also lead to the recruitment of phosphatases that oppose Mps1 [63]. How each kinetochore integrates these local changes in SAC activity at individual microtubule attachment subunits into a global “whole kinetochore” response is less well understood.
Functionally, the degree to which the kinetochore integrates attachment information dictates the quantitative relationship between the input attachment level and output Mad1 kinetochore level (Fig. 3B,C). A highly-integrated checkpoint response could produce a switch-like “digital” input-output relationship, while a response where attachment subunits function independently to trigger an output could produce a linear “analog” input-output relationship (Fig. 3B). Other parameters of this relationship (e.g. threshold microtubule occupancy) also reflect key properties of the SAC signaling network. Currently, our knowledge of how kinetochores count the number of attached microtubules is incomplete. Recent work has demonstrated that ~50% of a metaphase k-fiber is sufficient for Mad1 loss [17,18, Etemad et al., bioRxiv doi: 10.1101/272407], and that the threshold occupancy for Mad1 loss may in fact be lower [64, Kuhn et al., bioRxiv doi: 10.1101/263471]. Feedback loops between attachment, kinases, and phosphatases are likely key for signal integration by amplifying decreases in Mps1 activity upon attachment into large changes in kinetochore phosphorylation state and Mad1 localization [50,65]. This network may help cells to sensitively and rapidly mount a SAC response.
Interplay between kinetochore-microtubule interface mechanics and robust signaling
The properties of the kinetochore-microtubule interface that set robust grip are well suited to maintaining a robust SAC response. First, due to the intrinsic redundancy of the k-fiber, the loss of a single attached microtubule in a metaphase k-fiber is expected to have little overall effect on checkpoint signaling. Absence of redundancy, for example in budding yeast which bind one microtubule per kinetochore [66], results in higher sensitivity to attachment loss. Therefore, species with low redundancy may avoid mitotic delays by biorienting chromosomes early in mitosis [67,68] and rapidly turning the SAC off [69]. Additionally, the tunability and dynamic nature of kinetochoremicrotubule grip may help avoid false positives (e.g. attachments from an incorrect pole that lose Mad1) early in mitosis by destabilizing incorrect attachments, and help avoid false negatives (correct attachments that do not lose Mad1) late in mitosis by preferentially stabilizing correct attachments using specialized proteins.
Cellular-scale implications of kinetochore decision-making
While Mad1 loss occurs independently at each kinetochore, the decision to satisfy the SAC and progress to anaphase is computed over the whole cell. This signal integration is critical to ensure timely and accurate mitosis. The cell requires a balance between a permissive Mad1 loss response that could lead erroneous attachments to allow anaphase (false positives), and a stringent Mad1 loss response that could lead noisy yet correct attachments to prevent anaphase (false negatives) – especially given the dynamicity of kinetochore-microtubule attachments. Because the cell is sensitive to small changes in Mad1 levels at kinetochores [49,71,72], and the number of microtubules in metaphase k-fibers is variable [1], it could, in principle, be highly susceptible to false negatives. However, by removing Mad1 at occupancy levels below the lowest numbers of microtubules typically found in a metaphase k-fiber [17,18,64], the cell can avoid reactivating the SAC upon transient drops in microtubule occupancy during metaphase, avoiding unnecessarily mitotic delays. Conversely, mechanisms exist to prevent cells from immediately responding to false positives: these include cytoplasm-wide SAC activation in early mitosis that is independent of kinetochore attachments [73], and the amplification of kinetochore-generated Mad1 signals in the cytoplasm [49,70,71,72,74].
Computational Insights Across Species
Looking forward, it will be important to understand what gives rise to the SAC’s quantitative input-output relationship and how it may vary in different cell types or across evolution over diverse kinetochore architectures [46]. For example, whether a holocentric C. elegans kinetochore can – or should – remove Mad1 across its entire 2–4 μm structure in response to weak and localized microtubule attachment is not known. Comparison of diverse kinetochores across species may illuminate different strategies for signal detection and integration and provide key insight into the evolution of the SAC signaling network.
Conclusions
How the mammalian kinetochore’s parts list gives rise to its emergent mechanical and computation functions remains a frontier. Still, recent findings building on classic work suggest that key insights are within reach. Principles that underlie the kinetochore’s robust function are emerging: from the redundancy, specialization, and dynamicity that drive robust microtubule attachment, to simple signal processing features such as nonlinearity, sensitivity, and a built-in delay that are well-suited for robust decision-making. Looking forward, approaches and thinking from engineering and systems biology will be key to defining these emergent “whole kinetochore” properties, and for uncovering their molecular bases and functions. Many of these properties remain poorly understood: for example, why do many species’ kinetochores bind so many more microtubules than are mechanically required for moving chromosomes? Budding yeast divide successfully with just one kinetochoremicrotubule, fission yeast kinetochores bind four microtubules, while mammalian kinetochores bind 15–25 microtubules [1]. To define and probe the kinetochore’s emergent properties, we will need tools to physically and molecularly perturb the kinetochore with a new level of control, and to read out quantitative responses: for example, approaches to externally control force and dynamics and to quantitatively rewire kinetochore composition (e.g. with optogenetics [75]) will be critical. Ultimately, the conceptual and experimental integration of kinetochore architecture, mechanics, and signal processing (Fig. 1B) will be essential to understanding how robust chromosome segregation emerges from the kinetochore’s many parts.
Acknowledgements:
We thank Ted Salmon and many colleagues for stimulating discussions, and apologize for not acknowledging all relevant work due to space constraints. We thank the Dumont Lab for critical reading of the manuscript. This work was supported by NIH DP2GM119177 (S.D), NSF CAREER 1554139 (S.D), the NSF Center for Cellular Construction DBI-1548297 (A.F.L, S.D.), a NSF Graduate Research Fellowship (A.F.L., J. K.), UCSF Moritz-Heyman Discovery Fellowships (A.F.L., J. K.), and a UCSF Lloyd Kozloff Fellowship (A.F.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL: Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol 1997, 137:1567–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED: Vertebrate kinetochore protein architecture: Protein copy number. J Cell Biol 2010, 189:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A, Desai A: A Molecular View of Kinetochore Assembly and Function. Biology 2017, 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, et al. : Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENPOPQUR and NDC80. Mol Cell 2018, 71:923–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monda JK, Cheeseman IM: The kinetochore-microtubule interface at a glance. J Cell Sci 2018, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valverde R, Ingram J, Harrison SCSC, Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E, et al. : Conserved Tetramer Junction in the Kinetochore Ndc80 Complex. Cell Rep 2016, 17:1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, Mcewen BF, Mogilner A, Khodjakov A: Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol 2015, 17:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connell CB, Khodjakov A, McEwen BF: Kinetochore flexibility: creating a dynamic chromosome-spindle interface. Curr Opin Cell Biol 2012, 24:40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CA, McAinsh AD, Burroughs NJ: Human kinetochores are swivel joints that mediate microtubule attachments. Elife 2016, 5:3985–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wynne DJ, Funabiki H: Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. 2015, 210:899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A: The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. Cell 2006, 127:983–997. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED: Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006, 127:969–982. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, Badger BL, Salmon ED: A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun 2015, 6:8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo TY, Choi J-M, Conway W, Yu C-H, Pappu RV, Needleman DJ: Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. Elife 2018, 7:e36392. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The authors use fluorescence lifetime imaging microscopy and Förster resonance energy transfer to quantitatively measure the average fraction of Ndc80 bound to microtubules in live human cells under different scenarios. This works provides a mechanistic basis for how tension stabilizes microtubule attachments.
- 15.Monda JK, Whitney IP, Tarasovetc EV., Wilson-Kubalek E, Milligan RA, Grishchuk EL, Cheeseman IM: Microtubule Tip Tracking by the Spindle and Kinetochore Protein Ska1 Requires Diverse Tubulin-Interacting Surfaces. Curr Biol 2017, 27:3666–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkov VA, PJ Huis in ‘t Veld, Dogterom M, Musacchio A: Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. Elife 2018, 7:e36764. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This works uses in vitro assays to study kinetochore structure-function from the bottom up by reconstituting controlled assemblies of Ndc80 particles. The authors show that load-bearing and tip-tracking of single microtubules require two or more Ndc80 complexes. This work provides key insight into how the number of couplers regulates the mechanical behavior of these assemblies in vitro.
- 17.Dudka D, Noatynska A, Smith CA, Liaudet N, McAinsh AD, Meraldi P: Complete microtubule–kinetochore occupancy favours the segregation of merotelic attachments. Nat Commun 2018, 9:2042. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study uses a microtubule-targeting agent to reduce kinetochore-microtubule occupancy by 35%. Kinetochores in this condition were able to satisfy the SAC, indicating that full metaphase microtubule occupancy is not required for mitotic progression.
- 18.Kuhn J, Dumont S: Spindle assembly checkpoint satisfaction occurs via endon but not lateral attachments under tension. J Cell Biol 2017, 216:1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicklas RB: Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol 1983, 97:542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR: Force production by disassembling microtubules. Nature 2005, 438:384–8. [DOI] [PubMed] [Google Scholar]
- 21.Zaytsev AV, Sundin LJR, DeLuca KF, Grishchuk EL, DeLuca JG: Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol 2014, 206:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN: The human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. PNAS. 2018, 115:2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auckland P, Clarke NI, Royle SJ, McAinsh AD: Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol 2017, 216:1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study demonstrates that the Ska complex aids in mechanical coupling at the kinetochore-microtubule interface in mammalian cells since upon Ska depletion there is an increase in kinetochore detachment events on depolymerizing microtubules. This provides key insight into the mechanical contributions of different outer kinetochore proteins to overall kinetochore grip.
- 24.Cheerambathur Dhanya K., Bram Prevo NH Lindsay Lewellyn KDC, Karen Oegema AD: Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex. Dev Cell 2017, 41:424–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janczyk Paweł Ł., Skorupka Katarzyna A., Tooley John G., … TW, Pornillos PTS Owen: Mechanism of Ska Recruitment by Ndc80 Complexes to Kinetochores. Dev Cell 2017, 41:438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal S, Smith KP, Zhou Y, Suzuki A, McKenney RJ, Varma D: Cdt1 stabilizes kinetochore–microtubule attachments via an Aurora B kinase–dependent mechanism. 2018, 217:3446–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kern DM, Monda JK, Su K-C, Wilson-Kubalek EM, Cheeseman IM, Monda JK, Su K-C, Wilson-Kubalek EM, Cheeseman IM: Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. Elife 2017, 6:e26866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLuca KF, Lens SMA, DeLuca JG: Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci 2011, 124:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan YW, Jeyaprakash AA, Nigg EA., Santamaria A: Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol 2012, 196:563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLuca KF, Meppelink A, Broad AJ, Mick JE, Peersen OB, Pektas S, Lens SMA, DeLuca JG: Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J Cell Biol 2018, 217:163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long AF, Udy DB, Dumont S, Dumont Correspondence S, Dumont S: Hec1 Tail Phosphorylation Differentially Regulates Mammalian Kinetochore Coupling to Polymerizing and Depolymerizing Microtubules. Curr Biol 2017, 27:1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dumont S, Salmon ED, Mitchison TJ: Deformations within moving kinetochores reveal different sites of active and passive force generation. Science 2012, 337:355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grishchuk EL: Biophysics of Microtubule End Coupling at the Kinetochore In Centromeres and Kinetochores. Edited by Black BE. Springer, Cham; 2017:397–428. [DOI] [PubMed] [Google Scholar]
- 34.Betterton MD, McIntosh JR: Regulation of chromosome speeds in mitosis. Cell Mol Bioeng 2013, 6:418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armond JW, Vladimirou E, Erent M, McAinsh AD, Burroughs NJ: Probing microtubule polymerisation state at single kinetochores during metaphase chromosome motion. J Cell Sci 2015, 128:1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhoum SF, Compton DA.: Kinetochores and disease: Keeping microtubule dynamics in check! Curr Opin Cell Biol 2012, 24:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakhoum SF, Thompson SL, Manning AL, Compton DA: Genome stability is ensured by temporal control of kinetochore–microtubule dynamics. Nat Cell Biol 2009, 11:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S: Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468:576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King JM, Nicklas RB: Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci 2000, 113:3815–3823. [DOI] [PubMed] [Google Scholar]
- 40.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED: Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol 2003, 162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H: Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J Cell Biol 2009, 186:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganem NJ, Compton DA: Functional Roles of Poleward Microtubule Flux During Mitosis. Cell Cycle 2006, 5:481–485. [DOI] [PubMed] [Google Scholar]
- 43.Dorn JF, Maddox PS: Kinetochore dynamics: How protein dynamics affect chromosome segregation. Curr Opin Cell Biol 2012, 24:57–63. [DOI] [PubMed] [Google Scholar]
- 44.Drinnenberg IA, Henikoff S, Malik HS, Akiyoshi B: Evolutionary Turnover of Kinetochore Proteins: A Ship of Theseus? Trends Cell Biol 2016, 26:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drinnenberg IA, Akiyoshi B: Evolutionary Lessons from Species with Unique Kinetochores In Centromeres and Kinetochores. Edited by Black BE. Springer, Cham; 2017:111–138. [DOI] [PubMed] [Google Scholar]
- 46.Je Van Hooff J, Tromer E, van Wijk LM, Snel B, Kops GJ: Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep 2017, 18:1559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drpic D, Almeida AC, Aguiar P, Renda F, Damas J, Lewin HA, Larkin DM, Khodjakov A, Maiato H: Chromosome Segregation Is Biased by Kinetochore Size. Curr Biol 2018, 28:1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study harnesses the low chromosome number of Indian muntjac cells to demonstrate that variability in kinetochore size critically affects the timing and fidelity of chromosome segregation. The authors show that the kinetochore’s microtubule binding capacity scales with kinetochore size, and that error formation leading to chromosome missegregation occurs more frequently for large kinetochores.
- 48.London N, Biggins S: Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Biol 2014, 15:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collin P, Nashchekina O, Walker R, Pines J: The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol 2013, 15:1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saurin AT: Kinase and Phosphatase Cross-Talk at the Kinetochore. Front Cell Dev Biol 2018, 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waters JC, Chen RH, Murray AW, Salmon ED: Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 1998, 141:1181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connell CB, Lončarek J, Hergert P, Kourtidis A, Conklin DS, Khodjakov A, Loncarek J, Hergert P, Kourtidis A, Conklin DS, et al. : The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol 2008, 183:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janssen LME, Averink TV, Blomen VA, Brummelkamp TR, Medema RH, Raaijmakers JA: Loss of Kif18A Results in Spindle Assembly Checkpoint Activation at Microtubule-Attached Kinetochores. Curr Biol 2018, 28:2685–2696. [DOI] [PubMed] [Google Scholar]
- 54.Uchida KSK, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T: Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 2009, 184:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maresca TJ, Salmon ED: Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol 2009, 184:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Etemad B, Kops GJ: Attachment issues: kinetochore transformations and spindle checkpoint silencing. Curr Opin Cell Biol 2016, 39:101–108. [DOI] [PubMed] [Google Scholar]
- 57.Tauchman EC, Boehm FJ, DeLuca JG: Stable kinetochore–microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun 2015, 6:10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Etemad B, Kuijt TEF, Kops GJPL: Kinetochore–microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun 2015, 6:8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magidson V, He J, Ault JG, O’Connell CB, Yang N, Tikhonenko I, McEwen BF, Sui H, Khodjakov A: Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol 2016, 212:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aravamudhan P, Goldfarb AA, Joglekar AP: The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol 2015, 17:868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiruma Y, Sacristan C, Pachis ST, Adamopoulos A, Kuijt T, Ubbink M, von Castelmur E, Perrakis A, Kops GJPL: Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 2015, 348:1264–1267. [DOI] [PubMed] [Google Scholar]
- 62.Ji Z, Gao H, Yu H: Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 2015, 348:1260–1264. [DOI] [PubMed] [Google Scholar]
- 63.Sivakumar S, Janczyk PŁ, Qu Q, Brautigam CA, Stukenberg PT, Yu H, Gorbsky GJ: The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. Elife 2016, 5:e12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sikirzhytski V, Renda F, Tikhonenko I, Magidson V, McEwen BF, Khodjakov A: Microtubules assemble near most kinetochores during early prometaphase in human cells. J Cell Biol 2018, 217:2647–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using correlative light electron microscopy, this paper shows that many k-fibers assemble close to kinetochores at early prometaphase in mammalian cells. Some prometaphase kinetochores were found attached to very few microtubules and without detectable Mad2, indicating that low microtubule occupancy can prevent SAC activation.
- 65.Funabiki H, Wynne DJ: Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma 2013, 122:135–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterson JB, Ris H: Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci 1976, 22:219–242. [DOI] [PubMed] [Google Scholar]
- 67.He X, Asthana S, Sorger PK: Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000, 101:763–75. [DOI] [PubMed] [Google Scholar]
- 68.Goshima G, Yanagida M: Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 2000, 100:619–33. [DOI] [PubMed] [Google Scholar]
- 69.Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO: Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J Cell Biol 2014, 207:23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C, Whitney IP, Banerjee A, Sacristan C, Sekhri P, Kern DM, Fontan A, Kops GJPL, Tyson JJ, Cheeseman IM, et al. : Ectopic Activation of the Spindle Assembly Checkpoint Signaling Cascade Reveals Its Biochemical Design. Curr Biol 2019, 29:104–119. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Using a synthetic kinetochore-independent SAC activator, the authors establish the dose-response relationship between upstream SAC activators and the cytosolic target of SAC inhibition (APC activity). This work shows that when most kinetochores are attached, the remaining unattached kinetochores are capable of producing disproportionately high levels of SAC inhibitory signal.
- 71.Dick AE, Gerlich DW: Kinetic framework of spindle assembly checkpoint signalling. Nat Cell Biol 2013, 15:1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinrich S, Geissen E-M, Kamenz J, Trautmann S, Widmer C, Drewe P, Knop M, Radde N, Hasenauer J, Hauf S: Determinants of robustness in spindle assembly checkpoint signalling. Nat Cell Biol 2013, 15:1328–1339. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV: Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 2014, 156:1017–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rieder CL, Cole RW, Khodjakov A, Sluder G: The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 1995, 130:941–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Aonbangkhen C, Tarasovetc EV, Ballister ER, Chenoweth DM, Lampson MA: Optogenetic control of kinetochore function. Nat Chem Biol 2017, 13:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]