Abstract
Organisms that reproduce sexually utilize a specialized form of cell division called meiosis to reduce their chromosome number by half to generate haploid gametes. Meiosis in females is especially error prone and this vulnerability has a profound impact on human health: it is estimated that 10–25% of human embryos are chromosomally abnormal, and the vast majority of these defects arise from problems with the female reproductive cells (oocytes). Here we highlight recent studies that explore how these important cells divide. Although we focus on work in the model organism C. elegans, we also discuss complementary studies in other organisms that together provide new insights into this crucial form of cell division.
INTRODUCTION
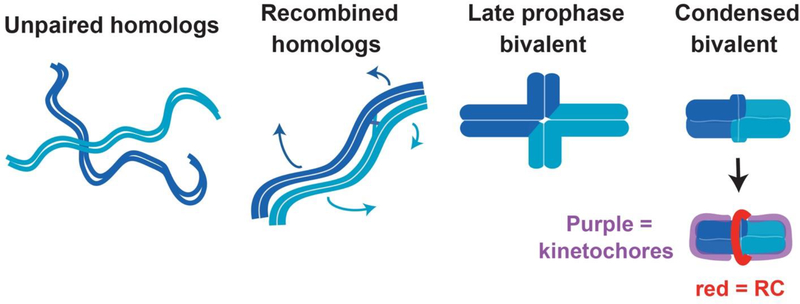
Oocytes have several features that differentiate them from mitotically-dividing cells and therefore necessitate the use of unique mechanisms. First, meiotic cells undergo a specialized cell division program with one round of DNA replication followed by two rounds of division to halve their chromosome number. Segregation during the first meiotic division depends on recombination (crossing over) between paternally- and maternally-derived homologous chromosomes. In C. elegans, there is only one crossover per homolog pair that is typically formed off-center, leading to the formation of cruciform bivalents in Meiosis I (MI) with long and short arms (reviewed in [1]) (Figure 1). At Anaphase I, sister chromatid cohesion is released along the short-arm axis of the bivalent, allowing the crossover to be resolved and homologous chromosomes to segregate away from one another. This is followed by a second division, where sister chromatids separate, resulting in haploid gametes. Execution of this complex set of chromosomal events requires mechanisms to precisely pattern meiotic chromosomes such that they can align on the spindle and be faithfully segregated during each division.
Figure 1. Chromosome organization during C. elegans meiosis.
During Meiosis I, homologous partner chromosomes (depicted in light and dark blue) pair and recombine to form bivalents. In C. elegans, there is one crossover per homolog pair that tends to form off-center; the recombined chromosomes then reorganize around this crossover (depicted with blue arrows), resulting in cruciform bivalents with long and short arms. These bivalents then condense further prior to the meiotic divisions, such that the short arms are largely indistinguishable. Kinetochore proteins form cup-like structures (purple) that surround the two ends of the bivalent, and a multi-protein ring complex (RC; red) forms around the short arm axis.
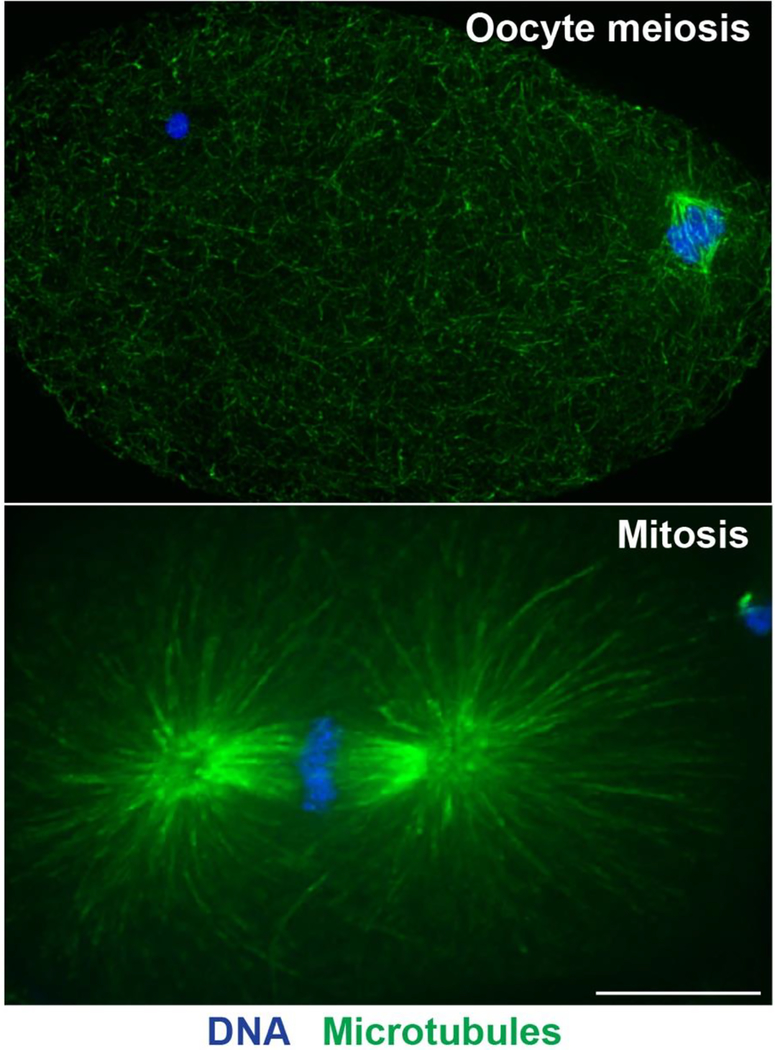
Another distinguishing feature of oocytes of many species is that they lack centriole- containing centrosomes, which nucleate microtubules and act as structural cues to define and organize the spindle poles during mitosis and male meiosis (thus, oocyte spindles are “acentriolar”). Consequently, oocyte spindles assemble using a different pathway and are morphologically distinct from spindles containing centrosomes; acentriolar spindles are smaller and lack astral microtubules at the poles (Figure 2). How these spindles form and then subsequently mediate chromosome segregation are important questions.
Figure 2. Comparison of spindle morphology with and without centrosomes.
Shown are C. elegans spindles in oocyte meiosis (top) compared to the mitotic one-cell stage embryo (bottom); microtubules are in green and DNA in blue. Acentriolar oocyte spindles are much smaller and lack astral microtubules at the poles. Bar = 10μm.
The model organism C. elegans has emerged as a powerful system to address these questions. These worms are transparent and the oocyte meiotic divisions are rapid, allowing visualization in live, intact animals. Moreover, they are amenable to a wide variety of experimental manipulations, facilitating combined genetic, genomic, and cytological approaches. Recent work in this system coupled with complementary work in other organisms has deepened our understanding of how acentriolar oocyte spindles form and how chromosomes congress and segregate on these spindles.
Acentriolar spindle assembly and organization
Recent work has shed light on some of the mechanisms by which oocytes organize microtubules into a bipolar spindle in the absence of centrosomes. One major pathway was discovered through studies of mouse oocytes. In this system, small microtubule asters called microtubule organizing centers (MTOCs) start out dispersed in the cytoplasm and then cluster together near the chromosomes and reorganize into a bipolar spindle, suggesting that self- organization of these structures drives acentriolar spindle assembly [2]. In contrast, live imaging of human oocytes has demonstrated that spindle assembly proceeds without MTOCs, demonstrating the existence of other mechanisms [3]. Interestingly, work in C. elegans oocytes has demonstrated that its pathway of spindle assembly looks similar to human [4], suggesting that it could be a powerful model for uncovering these mechanisms.
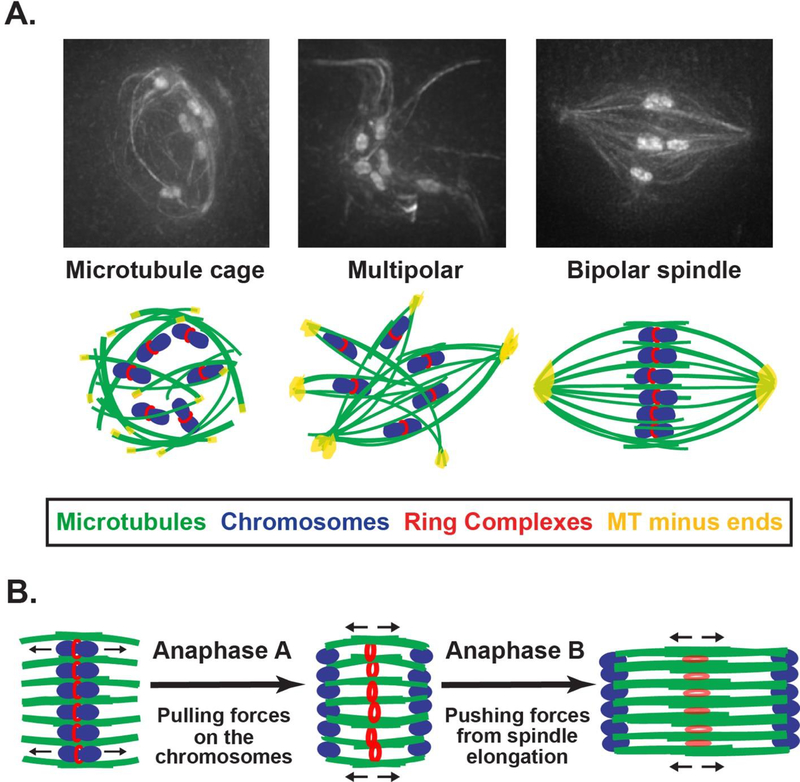
As the meiotic divisions are initiated in C. elegans, a diffuse haze of tubulin initially appears within the disassembling nucleus as it begins to break down [5]. Then, microtubules nucleate and assemble into a cage-like structure located inside the disassembling envelope; the circular shape of this array is thought to arise from constraints on the microtubules by the nuclear envelope remnants. Subsequently, microtubules are reorganized such that the minus ends are sorted to the periphery of the structure, and then these ends are organized into multiple nascent poles that coalesce to form the bipolar spindle (Figure 3A) [4]. These steps then repeat during Meiosis II, although the cage-like structure does not form since there is no disassembling nuclear envelope; instead microtubules appear to nucleate in the vicinity of the chromosomes, suggesting that there may be different mechanisms for microtubule formation in MI and MII [4].
Figure 3. Models for spindle assembly and chromosome segregation during oocyte meiosis.
A.) Shown are oocytes expressing GFP::tubulin and GFP::histone (to mark microtubules and chromosomes, respectively), at the major stages of acentriolar spindle formation (top), adapted from [4]. Corresponding cartoons are shown below each image. Microtubules first form a cage- like array inside the disassembling nuclear envelope. The minus ends are then sorted to the periphery of the array where they are organized into nascent poles that coalesce until bipolarity is achieved.
B.) Models for chromosome segregation. During Anaphase A, chromosomes are subjected to pulling forces (arrows in cartoon on the left), facilitating poleward movement along laterally- associated microtubule bundles. RCs (red) are removed from chromosomes and remain intact in the center of the spindle, wedging open the microtubule bundles and therefore creating wide microtubule “channels”. In Anaphase B, the spindle elongates from the center (arrows in the middle/right cartoons), driving chromosomes further apart. During this stage the RCs elongate and disassemble, so the microtubule bundles are no longer wedged open and move closer together, causing the center of the spindle to narrow.
Currently, it is not known how microtubules are initially nucleated in the early stages of spindle assembly. γ-tubulin is present in vicinity of the disassembling nuclear envelope, but depletion of this protein does not lead to obvious spindle defects [6, 7]. Moreover, the Ran pathway, which has been shown to be important for spindle assembly in mouse, Drosophila, and human oocytes (reviewed in [8]) has been reported to be dispensable for chromosome segregation in C. elegans oocytes [9], although a detailed characterization has not been performed. Given this gap in knowledge, uncovering factors facilitating microtubule nucleation during the two meiotic divisions is an important area of future study.
In contrast, some factors mediating later steps of acentriolar spindle assembly have been uncovered. KLP-18 (a kinesin-12 family motor) and MESP-1 (an auxiliary protein) are required for sorting microtubule minus ends to the periphery of the assembling spindle [4, 10–12]. Moreover, several factors have been implicated in focusing spindle poles, including the MEI-1/2 microtubule severing complex (katanin) [13] [14], the microtubule minus-end binding protein ASPM-1 [12], and dynein [15, 16]. Finally, the kinesin-13 family member MCAK (KLP-7) is required for proper pole organization and for regulating microtubule length [5, 17–19]. Investigating how these factors collectively work to promote both initial pole formation and also the coalescence of multiple poles into a bipolar spindle will be important to understand these critical aspects of acentriolar spindle assembly.
Another key question is how microtubules within the bipolar spindle are organized into a functional array that can drive chromosome congression and segregation. In some species (e.g., Xenopus and Drosophila), acentriolar spindles lack long microtubules that extend from the poles to the chromosomes and instead are comprised of short microtubules organized into a tiled array [20–23]. This is also likely in C. elegans oocytes, as a partial electron microscopy reconstruction of a metaphase spindle revealed the presence of numerous short microtubules [24]. Upon depletion of MEI-1/2, the spindle was comprised of fewer, longer microtubules, suggesting that katanin’s severing activity produces short microtubules that can be arranged by other factors into a bipolar spindle of the correct length [24]. The highly homologous minus-end-directed kinesins KLP-15 and KLP-16 appear to be required for this function; upon depletion of these proteins, microtubules cannot reach the multipolar stage and instead collapse into a dense array of short microtubules. Thus, these factors may organize short microtubules into longer bundles that can facilitate chromosome dynamics [25].
Chromosome congression is mediated by lateral microtubule interactions
C. elegans meiotic chromosomes have a number of unique features that facilitate their congression and segregation. First, C. elegans chromosomes are holocentric, which means that kinetochore proteins load along the entire chromosome and therefore appear to cup each half of the bivalent in Meiosis I (Figure 1) [26]. Interestingly, while in spermatocytes these cup-like kinetochores form end-on microtubule attachments, in oocytes microtubules instead run along the sides of the bivalents and appear to predominantly form lateral associations [11, 19]. However, despite the lack of end-on attachments, depletion of kinetochore components causes defects in chromosome orientation on oocyte spindles, suggesting that kinetochores help align the bivalents within the lateral bundles [27].
In the absence of canonical kinetochore attachments, chromosome congression relies on a protein complex that forms a ring around the center of each bivalent (or around the sister chromatid interface in Meiosis ||) [11], called the ring complex, or “RC”. This complex has been shown to exhibit an unusual behavior in oocytes experimentally arrested in metaphase, with the complex stretching away from the chromosomes towards microtubule plus ends, suggesting that it can generate plus-end-directed forces [15]. One component of the RC that could provide this activity is the kinesin-4 family member KLP-19, which has been proposed to “walk” chromosomes along the lateral bundles to the center of the spindle [11]. However, it is possible that other RC components could also provide plus-end forces that promote congression.
In addition to KLP-19, the RCs are comprised of many other conserved cell division proteins including the kinase BUB-1, the CENP-F homologs HCP-1/2, the CLASP homolog CLS- 2 [27], MCAK [17, 18], condensin I component CAPG-1 [28], and the Chromosomal Passenger Complex (CPC) [11], which contains AIR-2/Aurora B kinase. RC assembly occurs during early prometaphase, concurrently with nuclear envelope breakdown. The CPC first rearranges from what appears to be a linear localization along the short arm axis [29], to a ring-like structure encircling this region [11]. Then, other proteins are progressively recruited, with the CPC required for the targeting of all other known components [11, 27, 30, 31]. Additionally, the RC appears to be organized in layers, with AIR-2 close to the DNA, BUB-1 and KLP-19 in a middle layer, and HCP-1/2 and CLS-2 on the outside [27], suggesting that the RC is structured as layers of subcomplexes. The small ubiquitin-like modifier SUMO plays an important role in RC assembly [31]. SUMO and its conjugating enzymes UBC-9 and GEI-17 (E2 and E3 enzymes, respectively) localize to the RC, and GEI-17 depletion prevents the targeting of most other RC components, causing chromosome congression defects. Moreover, multiple RC components have been shown to be SUMOylated either in vitro or in vivo while others contain SUMO interaction motifs (SIMs) [31]. Thus, a network of SUMO-SIM interactions appears to drive the assembly of the RC during prometaphase, ultimately building a structure that can mediate chromosome congression.
Multiple mechanisms coordinate to drive chromosome segregation
Interestingly, depletion of kinetochore components does not slow chromosome movement during anaphase, demonstrating that chromosome segregation is also driven by a non-canonical mechanism [27]. At the metaphase-to-anaphase transition, the protease separase relocalizes from the kinetochores to the RCs, where it is thought to cleave cohesin on the short arm axis of each bivalent [15]. This release of cohesion enables the chromosomes to separate and is also coordinated with the removal of the RCs from the chromosomes [15, 27]. Concomitantly, the spindle shortens and the spindle poles broaden [32, 33]. Then, chromosomes move on this shortened spindle towards the poles, representing Anaphase A-like poleward movement. Subsequently, the spindle elongates in a process analogous to Anaphase B, driving the chromosomes further apart [34]. The mechanisms driving chromosome segregation during these two phases of anaphase have recently been the subject of much interest.
One idea is that chromosome movement is driven by a population of microtubules that polymerizes between the separating chromosomes and pushes on their inside surfaces to drive them apart [27, 35]. However, a number of studies have provided evidence that microtubule pushing cannot be the only force mediating segregation and is unlikely to operate as proposed. Specifically, analysis of Anaphase A spindle organization using both light [15, 34] and electron [19] microscopy failed to reveal a population of microtubules contacting the inside surfaces of chromosomes; instead microtubules were shown to run along the sides of separating chromosomes, forming “channels” that the chromosomes reside in as they move towards the poles (Figure 3B). Thus, this type of pushing mechanism is unlikely to operate during Anaphase A. Alternatively, since the RCs are removed from chromosomes as they separate [27] and remain within the channels [15], another model is that removal of the plus-end forces generated by the RCs enables minus-end-directed poleward movement along the laterally-associated microtubule bundles; this would represent a “pulling” rather than a “pushing” force. However, it is not known what generates this force. Although dynein inhibition causes lagging chromosomes and was thus proposed to facilitate poleward movement [15], a number of more recent studies have demonstrated that dynein inhibition does not alter chromosome segregation rates, calling this idea into question [34, 35]. However, since full dynein inhibition causes spindle defects [15, 16], it is unclear whether the reported depletion/inhibition conditions fully inactivated dynein function, so this question has not been conclusively resolved. Identifying the factors generating poleward forces during Anaphase A is therefore an important area of future study.
Once chromosomes reach the poles, the spindle elongates in Anaphase B (Figure 3B) [34]. Unlike Anaphase A, it is possible that this phase of segregation could be driven by pushing forces coming from the center of the spindle; in this view, chromosomes that have already reached the spindle poles are pushed further apart as the spindle lengthens from the middle due to microtubule polymerization [19, 34]. This idea is supported by laser ablation experiments, where severing microtubules between separating chromosomes during Anaphase B was shown to halt chromosome movement [35]. The mechanisms driving this spindle elongation are not completely understood, but they have been shown to rely on the doublecortin homolog ZYG-8 [34]. Moreover, the anaphase spindle is stabilized during the elongation phase by complementary mechanisms involving the microtubule crosslinking protein SPD-1 (PRC1) and the minus-end-directed kinesins KLP-15 and KLP-16 [25].
During Anaphase B, the RCs begin to disassemble, the channels become less apparent, and the center of the spindle narrows (Figure 3B). Different components leave the RCs at different times, and as Anaphase B proceeds, the RCs appear to lose structural integrity, first flattening before they disappear [15, 36]. RC disassembly has been shown to be dependent on the SUMO protease ULP-1, suggesting that removal of SUMO from an RC component (or components) is required to disassemble the structures [36]. Interestingly, RC disassembly is delayed following a variety of experimental perturbations that cause chromosome segregation errors, suggesting that the disassembly process is regulated [37]. Under these conditions, RCs remain intact and the channels remain wide as the spindles elongate during Anaphase B. This suggests that the channel narrowing that normally occurs during Anaphase B is not an active part of the mechanism that drives spindle elongation and chromosome segregation. Instead this change in spindle morphology may simply be a consequence of RC disassembly; as the RCs break down, the microtubule bundles that form the channels are not held apart and therefore move closer together.
One important outstanding question is the relative importance of Anaphase A and B mechanisms to chromosome segregation. Since the spindle shortens at the metaphase to anaphase transition, Anaphase A normally occurs over a short distance, and thus the majority of chromosome movement occurs during Anaphase B [34]. This suggests that during wild type anaphase, this second phase of the segregation process may be more critical. However, in katanin mutants where the spindles do not significantly shorten, chromosomes are still able to move poleward [6], suggesting that Anaphase A mechanisms are capable of mediating segregation over greater distances. Moreover, it is possible that the reason oocytes delay RC disassembly under error conditions is to keep Anaphase A mechanisms active throughout anaphase (i.e., facilitating chromosome-to-pole movement through wide channels as the spindle elongates) [37]; if this conjecture is correct, it would imply that there is an advantage to having these mechanisms active. Finally, a recent study demonstrated that when chromosomes lag in the center of the spindle during the Anaphase B phase of segregation, they appear stretched and elongated, suggesting that they are subjected to pulling rather than pushing forces [38]. This result could either suggest that Anaphase B is not solely driven by pushing forces, or it could be another example of a condition in which Anaphase A “pulling” mechanisms remain active throughout anaphase. Regardless of which of these interpretations is correct, these findings reaffirm that anaphase is not solely driven by pushing forces and therefore it will be important to investigate how the different forces operating on chromosomes are generated and coordinated.
Complementary work in other systems
Altogether, important questions still remain about how acentriolar spindles form and mediate chromosome congression and segregation, and it will be important to test the models generated using C. elegans in other organisms. Notably, in both mouse and Drosophila oocytes, end-on kinetochore attachments are suppressed until after bipolar spindles assemble, suggesting a role for other types of chromosome-microtubule interactions prior to this stage [39–41]. Thus, findings generated in C. elegans, which do not have canonical kinetochore attachments, could potentially inform future studies in these organisms.
Conversely, discoveries in other organisms are also generating new hypotheses that in the future can be tested in C. elegans. Notably, recent studies in Drosophila have provided insights into how microtubules are nucleated in oocytes. It has been shown that augmin, which participates in microtubule nucleation in mitosis by recruiting γ-tubulin onto spindle microtubules [42–44], is not required for bulk nucleation in oocytes [45, 46]. However, careful analysis has demonstrated that a stable population of augmin at the spindle poles promotes full microtubule assembly [46]. Additionally, a complementary nucleation pathway has been identified, where the kinesin-6 family motor Subito recruits the γ-tubulin complex to the spindle equator; this Subito-γ- tubulin interaction is suppressed away from chromosomes [47]. Thus, spatial regulation of multiple microtubule nucleation pathways promotes acentriolar spindle assembly in Drosophila.
Additionally, there have also been recent discoveries about spindle organization in Drosophila oocytes. It has been demonstrated that the activities of multiple families of kinesin motors (kinesin-5, kinesin-6, kinesin-12, and kinesin-14) are coordinated to promote spindle symmetry, and that disrupting this balance causes asymmetric spindles with misaligned chromosomes [48]. Moreover, there have been new insights into the spatial regulation of one of these motors. It was discovered that 14-3-3 proteins interact with Ncd (kinesin-14), and that this interaction prevents Ncd from binding to microtubules. However, this interaction is antagonized by phosphorylation of Ncd by Aurora B, thus enabling Ncd to bind microtubules and promote spindle assembly in the vicinity of chromosomes [49].
There has also been rapid progress in understanding mammalian meiosis in recent years, including studies of both mouse and human oocytes. These discoveries have been highlighted in a number of recent reviews, focusing on topics such as the regulation of the meiotic divisions [50], the assembly and positioning of the meiotic spindle [51, 52], chromosome segregation [53], meiotic drive [54], and causes of aneuploidy [55]. Altogether, it is clear that oocyte meiosis is becoming a topic of much interest, suggesting that we may soon begin to unlock the mysteries of how these important cells divide.
ACKNOWLEDGMENTS
We are grateful to members of the Wignall lab, the WiLa ICB, and the fantastic meiosis community for valuable discussions over the years that have shaped our thinking. S.M.W. was supported by NIH R01GM124354.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE
The authors declare no conflicts of interest.
REFERENCES
Highlighted papers:
* Of interest
** Of outstanding interest
- 1.Schvarzstein M, Wignall SM, and Villeneuve AM (2010). Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev 24, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuh M, and Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- 3.Holubcova Z, Blayney M, Elder K, and Schuh M (2015). Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff ID, Tran MV, Mullen TJ, Villeneuve AM, and Wignall SM (2016). Assembly of C. elegans acentrosomal spindles occurs without evident MTOCs and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol Biol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gigant E, Stefanutti M, Laband K, Gluszek-Kustusz A, Edwards F, Lacroix B, Maton G, Canman JC, Welburn JP, and Dumont J (2017). Inhibition of ectopic microtubule assembly by the kinesin-13 KLP-7MCAK prevents chromosome segregation and cytokinesis defects in oocytes. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNally K, Audhya A, Oegema K, and McNally FJ (2006). Katanin controls mitotic and meiotic spindle length. J Cell Biol 175, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobinnec Y, Fukuda M, and Nishida E (2000). Identification and characterization of Caenorhabditis elegans gamma-tubulin in dividing cells and differentiated tissues. J Cell Sci 113 Pt 21, 3747–3759. [DOI] [PubMed] [Google Scholar]
- 8.Cavazza T, and Vernos I (2015). The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front Cell Dev Biol 3, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Askjaer P, Galy V, Hannak E, and Mattaj IW (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell 13, 4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segbert C, Barkus R, Powers J, Strome S, Saxton WM, and Bossinger O (2003). KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol Biol Cell 14, 4458–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wignall SM, and Villeneuve AM (2009). Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nature cell biology 11, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly AA, Osterberg V, Christensen S, Price M, Lu C, Chicas-Cruz K, Lockery S, Mains PE, and Bowerman B (2014). Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol Biol Cell 25, 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srayko M, Buster DW, Bazirgan OA, McNally FJ, and Mains PE (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 14.McNally K, Berg E, Cortes DB, Hernandez V, Mains PE, and McNally FJ (2014). Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol Biol Cell 25, 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, and Wignall SM (2015). Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HY, Mains PE, and McNally FJ (2005). Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol 169, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly AA, Sugioka K, Chuang CH, Lowry JB, and Bowerman B (2015). KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J Cell Biol 210, 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han X, Adames K, Sykes EM, and Srayko M (2015). The KLP-7 Residue S546 Is a Putative Aurora Kinase Site Required for Microtubule Regulation at the Centrosome in C. elegans. PLoS One 10, e0132593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redemann S, Lantzsch I, Lindow N, Prohaska S, Srayko M, and Muller-Reichert T (2018). A Switch in Microtubule Orientation during C. elegans Meiosis. Curr Biol 28, 2991–2997 e2992. [DOI] [PubMed] [Google Scholar]; ** This study uses careful EM reconstructions to assess spindle morphology during different stages of anaphase, demonstrating that chromosomes segregate through microtubule-free channels in Anaphase A, followed by spindle elongation and channel narrowing in Anaphase B.
- 20.Burbank KS, Groen AC, Perlman ZE, Fisher DS, and Mitchison TJ (2006). A new method reveals microtubule minus ends throughout the meiotic spindle. J Cell Biol 175, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brugues J, Nuzzo V, Mazur E, and Needleman DJ (2012). Nucleation and transport organize microtubules in metaphase spindles. Cell 149, 554–564. [DOI] [PubMed] [Google Scholar]
- 22.Skold HN, Komma DJ, and Endow SA (2005). Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J Cell Sci 118, 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang ZY, Hallen MA, and Endow SA (2009). Mature Drosophila meiosis I spindles comprise microtubules of mixed polarity. Curr Biol 19, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srayko M, O’Toole E T, Hyman AA, and Muller-Reichert T (2006). Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol 16, 1944–1949. [DOI] [PubMed] [Google Scholar]
- 25.Mullen TJ, and Wignall SM (2017). Interplay between microtubule bundling and sorting factors ensures acentriolar spindle stability during C. elegans oocyte meiosis. PLoS Genet 13, e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper shows that the minus-end-directed kinesins KLP-15 and KLP-16 are required for microtubule bundling and organization during acentriolar spindle assembly, and also uncovers a role for the microtubule crosslinking protein SPD-1 (PRC1) in bundling anaphase spindle microtubules, thus revealing complementary mechanisms that promote spindle stability and facilitate chromosome segregation.
- 26.Monen J, Maddox PS, Hyndman F, Oegema K, and Desai A (2005). Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol 7, 1248–1255. [DOI] [PubMed] [Google Scholar]
- 27.Dumont J, Oegema K, and Desai A (2010). A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nature cell biology 12, 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. (2009). Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Carvalho CE, Zaaijer S, Smolikov S, Gu Y, Schumacher JM, and Colaiacovo MP (2008). LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev 22, 2869–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collette KS, Petty EL, Golenberg N, Bembenek JN, and Csankovszki G (2011). Different roles for Aurora B in condensin targeting during mitosis and meiosis. J Cell Sci 124, 3684–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelisch F, Tammsalu T, Wang B, Jaffray EG, Gartner A, and Hay RT (2017). A SUMO-Dependent Protein Network Regulates Chromosome Congression during Oocyte Meiosis. Mol Cell 65, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper shows that RC assembly is dependent on components of the SUMO pathway, and also demonstrates that some RC proteins are SUMOylated in vitro and in vivo, while others have SUMO interaction motifs (SIMs). These findings suggest that a SUMO-SIM network drives RC assembly and thus promotes chromosome congression.
- 32.Albertson DG, and Thomson JN (1993). Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res 1, 15–26. [DOI] [PubMed] [Google Scholar]
- 33.Yang HY, McNally K, and McNally FJ (2003). MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev Biol 260, 245–259. [DOI] [PubMed] [Google Scholar]
- 34.McNally KP, Panzica MT, Kim T, Cortes DB, and McNally FJ (2016). A Novel Chromosome Segregation Mechanism During Female Meiosis. Mol Biol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laband K, Le Borgne R, Edwards F, Stefanutti M, Canman JC, Verbavatz JM, and Dumont J (2017). Chromosome segregation occurs by microtubule pushing in oocytes. Nat Commun 8, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This work shows that chromosome movement during Anaphase B is inhibited by either severing microtubules between separating chromosomes or by depleting the CLASP homolog CLS-2, suggesting that this phase of anaphase is driven by polymerization of microtubules in the center of the spindle, leading to spindle elongation.
- 36.Davis-Roca AC, Divekar NS, Ng RK, and Wignall SM (2018). Dynamic SUMO remodeling drives a series of critical events during the meiotic divisions in Caenorhabditis elegans. PLoS Genet 14, e1007626. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper characterizes the process of RC disassembly in anaphase, demonstrating that it is a progressive process where components are removed from the structures at different times, and revealing that RC stability is regulated by a balance between SUMO conjugating and deconjugating activity.
- 37.Davis-Roca AC, Muscat CC, and Wignall SM (2017). Caenorhabditis elegans oocytes detect meiotic errors in the absence of canonical end-on kinetochore attachments. J Cell Biol 216, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This work demonstrates that C. elegans oocytes delay anaphase RC disassembly in the presence of a variety of meiotic defects, demonstrating that errors can be detected in these cells and revealing a mechanism that regulates anaphase progression. When RC disassembly is delayed, microtubule channels remain wide as the spindle elongates in Anaphase B, suggesting that channel narrowing is a consequence of RC disassembly and is not an active part of the mechanism that drives spindle elongation.
- 38.Vargas E, McNally K, Friedman JA, Cortes DB, Wang DY, Korf IF, and McNally FJ (2017). Autosomal Trisomy and Triploidy Are Corrected During Female Meiosis in Caenorhabditis elegans. Genetics 207, 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunet S, Maria AS, Guillaud P, Dujardin D, Kubiak JZ, and Maro B (1999). Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol 146, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gluszek AA, Cullen CF, Li W, Battaglia RA, Radford SJ, Costa MF, McKim KS, Goshima G, and Ohkura H (2015). The microtubule catastrophe promoter Sentin delays stable kinetochore-microtubule attachment in oocytes. J Cell Biol 211, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radford SJ, Hoang TL, Gluszek AA, Ohkura H, and McKim KS (2015). Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Drosophila Oocytes. PLoS Genet 11, e1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goshima G, Mayer M, Zhang N, Stuurman N, and Vale RD (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petry S, Groen AC, Ishihara K, Mitchison TJ, and Vale RD (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, Obuse C, and Goshima G (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci U S A 106, 6998–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meireles AM, Fisher KH, Colombie N, Wakefield JG, and Ohkura H (2009). Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colombie N, Gluszek AA, Meireles AM, and Ohkura H (2013). Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet 9, e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rome P, and Ohkura H (2018). A novel microtubule nucleation pathway for meiotic spindle assembly in oocytes. J Cell Biol 217, 3431–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radford SJ, Go AM, and McKim KS (2017). Cooperation Between Kinesin Motors Promotes Spindle Symmetry and Chromosome Organization in Oocytes. Genetics 205, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaven R, Bastos RN, Spanos C, Rome P, Cullen CF, Rappsilber J, Giet R, Goshima G, and Ohkura H (2017). 14-3-3 regulation of Ncd reveals a new mechanism for targeting proteins to the spindle in oocytes. J Cell Biol 216, 3029–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper provides insight into the spatial regulation of the motor Ncd in Drosophila oocytes, uncovering a mechanism by which the activity of proteins are regulated to promote spindle assembly around chromosomes.
- 50.Sanders JR, and Jones KT (2018). Regulation of the meiotic divisions of mammalian oocytes and eggs. Biochemical Society transactions 46, 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mogessie B, Scheffler K, and Schuh M (2018). Assembly and Positioning of the Oocyte Meiotic Spindle. Annu Rev Cell Dev Biol 34, 381–403. [DOI] [PubMed] [Google Scholar]
- 52.Gruss OJ (2018). Animal Female Meiosis: The Challenges of Eliminating Centrosomes. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mihajlovic AI, and FitzHarris G (2018). Segregating Chromosomes in the Mammalian Oocyte. Curr Biol 28, R895–R907. [DOI] [PubMed] [Google Scholar]
- 54.Lampson MA, and Black BE (2017). Cellular and Molecular Mechanisms of Centromere Drive. Cold Spring Harb Symp Quant Biol 82, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webster A, and Schuh M (2017). Mechanisms of Aneuploidy in Human Eggs. Trends Cell Biol 27, 55–68. [DOI] [PubMed] [Google Scholar]