Abstract
Objective
This study investigated whether levels of signaling pathways and inflammatory adipokines in epicardial fat regulate cardiovascular risks in humans and mice.
Methods
Epicardial fat was obtained from the hearts of patients with heart failure requiring coronary artery bypass surgery, and signaling pathways were compared with visceral fat. The genetic profile of epicardial and visceral fat from humans was also compared with genetic profiles of epicardial and visceral fat in obese mice. Left ventricular (LV) fractional shortening was measured in obese mice before and after treatment with inducers of mitochondrial signaling heme oxygenase 1 (HO‐1)‐peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha (PGC1α). An RNA array/heat map on 88 genes that regulate adipose tissue function was used to identify a target gene network.
Results
Human epicardial fat gene profiling showed decreased levels of mitochondrial signaling of HO‐1‐PGC1α and increased levels of the inflammatory adipokine CCN family member 3. Similar observations were seen in epicardial and visceral fat of obese mice. Improvement in LV function was linked to the increase in mitochondrial signaling in epicardial fat of obese mice.
Conclusions
There is a link between cardiac ectopic fat deposition and cardiac function in humans that is similar to that which is described in obese mice. An increase of mitochondrial signaling pathway gene expression in epicardial fat attenuates cardiometabolic dysfunction and LV fractional shortening in obese mice.
Introduction
Obesity is a significant risk factor for heart failure, with a prevalence of 70% in individuals with morbid obesity for 20 years and 90% in those with morbid obesity for 30 years 1. Adipose tissue is an active endocrine organ that secretes multiple signaling molecules, among them leptin, adiponectin, and resistin 2. Pericardial thickness has a direct association with inappropriately high left ventricular (LV) mass and an inverse association with fractional shortening (FS) 3, and it also decreases LV function 2, 4. In contrast, echocardiographic assessment of epicardial fat may be a reliable marker of cardiovascular risk, at least in females 2.
Enlarged white adipose cells secrete several inflammatory cytokines 5, 6. Elevated adipokine CCN family member 3 (CCN3; also known as nephroblastoma overexpressed [NOV]), which is a multifamily protein of the CCN (cellular communication network) family, is elevated in obesity. Elevated NOV levels are associated with inflammation and tissue damage 7. NOV is a matricellular protein that regulates multiple cellular activities, including cell adhesion, migration, proliferation, differentiation, and survival. Induction of NOV increased adipose tissue deposition, enhanced cholesterol, and increased insulin resistance and sleep apnea 8. In particular, epicardial fat is a distinct source of adipokines, reactive oxygen species (ROS), and inflammatory cytokines, and it has been tied to significant cardiac remodeling because of a decrease in the levels of heme oxygenase 1 (HO‐1) 9, 10 and peroxisome proliferator‐activated receptor (PPAR) gamma coactivator 1‐alpha (PGC1α) 11, 12. HO‐1 is regarded as the first line of defense against oxidative stress given that oxidative stress is a strong inducer of HO, which comprises both an inducible and a constitutive form (HO‐1 and HO‐2, respectively) and catalyzes the oxidative degradation of heme into iron, biliverdin, and carbon monoxide (CO). An increase in HO‐1 expression triggers an increase in heme degradation and in the production of CO and bilirubin, an antiapoptotic and an antioxidant respectively, both of which decrease cardiac remodeling 10, 13, 14. An increase in mouse epicardial fat deposition is associated with a decrease of HO‐1 levels in fat and a resultant increase in ROS 9, 11. Importantly, perturbations in the levels of HO‐1 were shown to regulate mitochondrial biogenesis 15 and levels of mitochondria carriers and their function 16.
PGC1α, a regulator of PPAR‐α in brown adipose tissue rich with mitochondria, has been shown to have a critical role in thermogenesis 17, in maintaining mitochondrial biogenesis and function, and in cellular energy metabolism 18. Transgenic mice with mildly elevated levels of muscle PGC1α are resistant to age‐related obesity and diabetes, suggesting that PGC1α stimulates the secretion of factors that affect the function of other tissues 19. Adipose‐specific PGC1α deficiency decreased mitochondrial biogenesis, glucose uptake, and the development of insulin resistance 20. Moreover, HO‐1 expression was mediated via PGC1α activation and, indirectly, it can further increase PGC1α 21. Overexpression of PGC1α in cardiac tissue of mice activates mitochondrial biogenesis and proliferation 22, 23. Conversely, mice lacking PGC1α in adipose tissue and fed a high‐fat diet developed insulin resistance and increased levels of circulating lipids 20. PGC1α along with the transcriptional regulator PR domain containing 16 (PRDM16) are the major facilitators of adipocyte browning and are responsible for thermogenic activation of brown fat 24, 25. In humans, adipose tissue is located beneath the skin (subcutaneous fat), around internal organs (visceral fat), in bone marrow (yellow bone marrow), intramuscularly (muscular system), and in breast tissue. Adipose tissue is found in specific locations; one such area of visceral adiposity is epicardial fat, located around the heart, which is a major determinant of cardiovascular risk.
In this study, we examined signaling pathways in adipocyte markers of epicardial fat from hearts of humans with advanced heart failure and requiring coronary artery bypass surgery and compared them with visceral fat obtained from age‐ and sex‐matched individuals undergoing abdominal surgery. Alterations in levels of inflammatory mediators and cytoprotective/antioxidant autacoids are related to, and appear to be responsible for, the development of heart failure in the obese mouse. Specifically, local adipose tissue appears to have a more pronounced effect on the myocardium than other visceral adipose tissue.
To identify novel relevant epicardial fat adipokines, we combined a genome‐wide and heat map analysis of gene expression in adipocytes that allowed us to identify levels of novel regulatory genes in epicardial and visceral fat. The results indicate that in both humans and mice, HO‐1, PGC1α, and adiponectin gene expression follows the same pattern. HO‐1, PGC1α, and mitochondrial brown fat uncoupling protein 1 (UCP1) correlate positively with adiponectin and each other, suggesting networks for these message expressions. Furthermore, an increase of HO‐1‐PGC1α is associated with a decrease in inflammatory adipokines including NOV and improvement in LV function.
Methods
Animal protocols
All mouse experimental protocols were performed following an Institutional Animal Care and Use Committee of New York Medical College approved protocol in accordance with the Guide for the Care and Use of Laboratory Animals. We used 16‐week‐old male mice homozygous for the diabetes spontaneous mutation (db/db) as a model of obesity‐induced diabetic cardiomyopathy, as these mice develop insulin resistance and hyperglycemia and fail to control gluconeogenesis, leading to peripheral neuropathy and myocardial disease. These mice, on C57BL/6J background (Jackson Laboratories, Bar Harbor, Maine), were divided into two groups with six mice per group following a 16‐week acclimatization period, weighing approximately 54 g at the start of the experiment. The two groups were as follows: (1) control (lean) and (2) group treated with epoxyeicosatrienoic acid (EET) agonist, an inducer of HO‐1, injected intraperitoneally twice per week for 8 weeks at a dose of 1.5 mg/kg of body weight. Echocardiography was performed as previously described 26, 27.
Patient population
Following informed consent, epicardial fat specimens were obtained from 16 sequential patients who were overweight and undergoing coronary artery bypass surgery. Visceral fat specimens were obtained from age‐ and sex‐matched controls of six donors, three females and three males, undergoing abdominal surgery. Written informed consent was received from participants prior to inclusion in the study. The human specimens were obtained at St. Mary's Medical Center in Huntington, West Virginia. The protocol and informed consent were approved by the Marshall University Institutional Review Board.
Human echocardiograms
Echocardiograms were performed on a Philips iE33 machine (Philips Healthcare, Highland Heights, Ohio). All echocardiograms were performed by a standard protocol in a laboratory accredited by the Intersocietal Accreditation Commission on Echocardiography Laboratories. The measurements were performed according to the recommendations set forth by the American Society of Echocardiography 28.
Sample collection in humans
During surgery, samples of epicardial fat and two tubes of blood were obtained, placed on ice, and stored at −80°C. Samples were shipped on ice to New York Medical College for further analysis.
Echocardiogram measurement of FS in mice
Echocardiography was performed using a 12‐MHz probe on isoflurane anesthetized mice. Images of the LV diameter were obtained in M‐mode and used to measure the end‐diastolic and end‐systolic diameters from which the LV FS was calculated 11.
Real‐time quantitative polymerase chain reaction
We pooled the pericardial and visceral fat data from both humans and mice. The correlations between the different gene expressions within the specific participants are presented. Polymerase chain reaction (PCR) array analysis of mRNAs was carried out in randomly selected participants as well as in mice. Total RNA was extracted from epicardial and visceral adipose tissue with TRIzol (Ambion, Austin, Texas). A Biotek plate reader and the Take3 plate (Biotek, Winooski, Vermont) were used to determine RNA at an absorbance of 260 nm. RNA measurements were subsequently evaluated by the A260 to A280 ratio. A High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, Massachusetts) was used to synthesize complementary DNA (cDNA) from total RNA. TaqMan Fast Universal PCR Master Mix (2×) on a 7500 HT Fast Real‐Time PCR System (Applied Biosystems) was used to perform real‐time PCR. Specific TaqMan Gene Expression Assay probes (Thermo Fisher Scientific, Waltham, Massachusetts) for mouse HO‐1, PGC1α, NOV, mitofusin 2 (MFN2), tumor necrosis factor (TNF‐α), IL4, PRDM16, adiponectin, and GAPDH were used as previously described 9, 11.
ScienCell's GeneQuery Human Preadipocyte Cell Biology qPCR Array Kit (catalog number GK103; ScienCell Research Laboratories, Inc., Carlsbad, California) was used as an array to find expression of target genes. GeneQuery qPCR array kits are quantitative PCR ready in a 96‐well plate format, with each well containing one primer set that can specifically recognize and efficiently amplify a target gene's cDNA. Each GeneQuery plate contains eight controls. Five target housekeeping genes (β‐actin, GAPDH, LDHA, NONO, and PPIH) enable normalization of data. The following genes were analyzed in both epicardial and visceral adipose tissue 9, 21: for preadipocyte cell markers, ADIPOQ, LEP, HOXC8/9, TBX1, TMEM26, UCP1, CIDEA, PRDM16, SLC7A10, and P2RX5; for proliferation, CCND1, PROKR1, LYRM1, FTO, SLC2A4, PDGFA, IL4, IGF1, FGF5, MAPK1/3, and TNFSF10; for differentiation and adipogenesis, PPARG, CEBPA/B, KLF2/5/15, SOCS1/3, INSR, DLK1, ADD1, and SREBF1; for extracellular matrix and cytoskeleton, MMP1/2/3, FAP, DPP4, TGFB1, COL1A1/2, and SMAD2/4; for lipid metabolism, FABP5/7, LBP, LPL, PLIN2, ACSL1, and LIPC; for obesity, MC4R, UCP3, ADRB2/3, POMC, FFAR4, and CARTPT; and for liposarcoma, FUS, MDM2, HMGA2, DDIT3, CDK4, EWSR1, DES, and VIM.
RNA microarray
Gene expression is presented as a heat map of log10‐transformed mean values (n = 3) relative to the mean values (n = 16) expressed in the human and mice pericardial versus visceral subgroup. The color code appears as a legend in the upper left‐hand corner of the graph. Values of relative expression ranged from 0.15 to 16.5 or log10‐transformed values of −0.83 to 1.22. Genes are ordered from highest increase in expression to greatest decrease in expression in the high‐fat subgroup. Note that the subgroup of enhanced visceral fat demonstrates marked attenuation of the changes compared with pericardial fat. In the short list, we selected the 10 genes that had the highest increase in expression and the 10 genes that had the greatest decrease in expression in visceral versus pericardial fat. Again, the color code for the log10‐transformed data is shown in the upper left‐hand corner of the graph and shows pericardial fat versus visceral fat changes. The correlation plot shows the Pearson correlation between different gene expressions in the entire set of data (e.g., visceral fat vs. pericardial fat). We noted substantial positive and negative correlations of gene expression. As we performed network analysis, we saw that to a large degree, there was a group of genes (about one‐half) that correlated positively with each other and negatively with the remaining genes (again, about one‐half). This is illustrated by choosing the genes focused on in the short list of top 10 upregulated and top 10 downregulated genes. Very clearly, the top 10 upregulated genes correlated positively with each other (pericardial fat) and the top 10. Details of this method are well described 29.
Statistical analysis
Data analysis was performed using the R programming language (The R Foundation, Vienna, Austria), with network analysis performed on correlation plots using the gplots package (https://cran.r-project.org/web/packages/gplots/index.html). Data values are expressed as means (SE). Individual group means were compared using unpaired t test. The null hypothesis was rejected at P < 0.05.
Results
We enrolled 16 sequential patients undergoing a planned cardiac surgery, including 15 coronary bypass surgeries alone and 1 coronary bypass plus valve replacement. Baseline characteristics are shown in Table 1. The mean BMI of the participants was 28.3 (SD 6.8). The mean age was 65.8 (9.9) years, and 50% were female. Data regarding cardiac risk factors were collected. All participants were hypertensive, and 50% had diabetes. Chronic kidney disease was present in 25%, and 37.5% were smokers.
Table 1.
Visceral fat specimens obtained from age‐ and sex‐matched controls undergoing abdominal surgery
| Age, y | 65.8 ± 9.9 |
| Female | 8/16 (50%) |
| BMI, kg/m2 | 28.3 ± 6.8 |
| Hypertension | 16/16 (100%) |
| Diabetes | 8/16 (50%) |
| Smoking | 6/16 (37.5%) |
| Chronic kidney disease | 4/16 (25%) |
| Total cholesterol, mg/dL | 135.8 ± 36.0 |
| HDL, mg/dL | 43.1 ± 21.4 |
| LDL, mg/dL | 78.9 ± 30.4 |
| Triglycerides, mg/dL | 122.8 ± 24.5 |
| Fasting glucose, mg/dL | 126.3 ± 35.5 |
| Epicardial fat thickness by echo, mm | 6.7 ± 1.8 |
| Ejection fraction, % | 49.7 ± 8.8 |
| FS, % | 23.6 ± 8.1 |
Data given as means ± SD.
HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; FS, fractional shortening.
Laboratory data were available for lipid studies in 14 of 16 participants. The average total cholesterol was 135.8 mg/dL. The average low‐density lipoprotein cholesterol was 78.9 mg/dL, and the mean high‐density lipoprotein cholesterol was 43.1 mg/dL. The average triglyceride level among participants was 122.8 mg/dL. The lipid data may be affected by fluctuations in cholesterol levels during acute events and by treatment with statins. The mean fasting glucose level was 126.3 mg/dL.
Echocardiographic data was obtained prior to surgery in all participants. The average thickness of epicardial fat by echocardiography was 6.7 mm. The mean ejection fraction was 49.7%, with a mean FS of 23.6%.
Human epicardial fat and visceral fat exhibit similar differences in expression levels of PGC1α, HO‐1, adiponectin, UCP1, PRDM16, and MFN2
The mRNA expression levels of PGC1α, HO‐1, and adiponectin in human epicardial fat were significantly (P < 0.05) reduced compared with visceral fat during reverse transcription PCR analysis (Figure 1A‐1C). Similarly, expression levels of uncoupling gene UCP1, thermogenesis gene PRDM16, and mitochondrial dynamics‐related mitofusion gene MFN2 were also significantly (P < 0.05) decreased in epicardial compared with visceral fat (Figure 1D‐1F). Furthermore, solute carrier family 2 member 4 (SLC2A4) expression was decreased in epicardial fat (Figure 1G).
Figure 1.
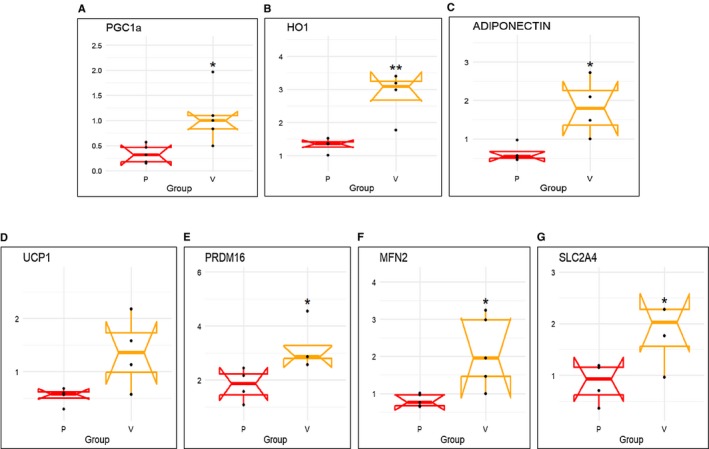
mRNA expression of (A) PGC1α, (B) HO‐1, (C) adiponectin, (D) UCP1, (E) PRDM16, (F) MFN2, and (G) SLC2A4 in human epicardial (P) vs. visceral adipose (V) tissues. Results are means ± SE; n = 4. *P < 0.05 vs. epicardial fat; **P < 0.02 vs. epicardial fat.
Human epicardial fat displays increased expression of adipogenic and inflammatory markers
There was higher mRNA expression of inflammatory adipokine CCN3/NOV in epicardial fat (Figure 2A) compared with visceral fat. Moreover, the mRNA expression of adipogenic CCAAT/enhancer‐binding protein alpha (CEBP‐α) (P < 0.05) was significantly upregulated in human epicardial fat compared with visceral fat (Figure 2B), and expression of inflammatory TNF‐α and IL4 mRNA expression was significantly higher (P < 0.05) in epicardial fat compared with visceral fat (P < 0.05) (Figure 2C‐2D).
Figure 2.
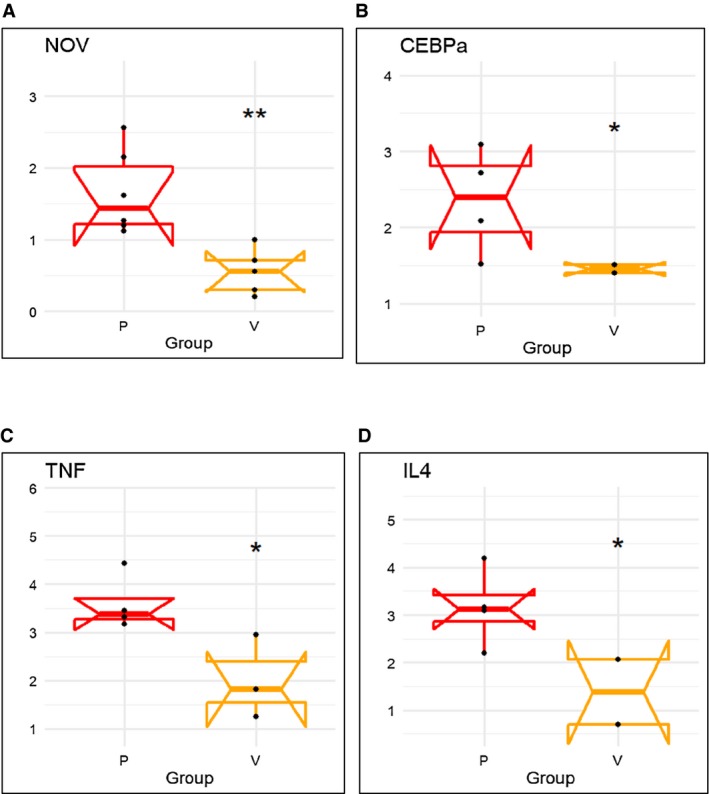
mRNA expression of inflammatory markers (A) NOV, (B) CEBP‐α, (C) TNF‐α, and (D) IL4 in human epicardial (P) vs. visceral adipose (V) tissues. Results are means ± SE; n = 4. *P < 0.05 vs. epicardial fat; **P < 0.02 vs. picardial fat.
RNA array analyses identify changes in correlation coefficients of gene expression in human epicardial and visceral fat
The upper section of Figure 3 shows a color code for the positive correlation coefficients as yellow to red and negative as blue to black. In human genes (Figure 3), it is clear that IL4 correlated with TNF‐α and CEBP‐α and that those latter two correlated very tightly with each other as well as positively with NOV. This group of four correlated negatively with MFN2, adiponectin, UCP1, PRDM16, HO‐1, and SLC2A4 genes, of which HO‐1 and SLC2A4 correlated closely with each other. UCP1 and adiponectin correlated positively with each other, suggesting networks for these message expressions. MFN2 correlated with HO‐1 and SLC2A4 but not as tightly as they correlated with each other. In Figure 3, UCP1 is red but almost as red with adiponectin, and its correlation is less strong with MFN2, almost in the no correlation range. PRDM16 was very tightly connected to HO‐1 and SLC2A4.
Figure 3.
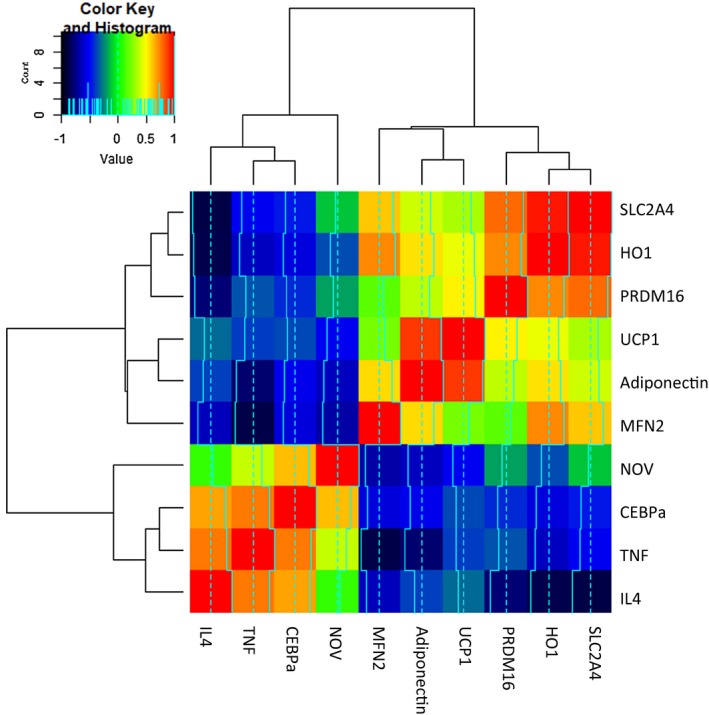
RNA array analyses changes in correlation coefficient gene expression of SLC2C4, HO‐1, PRDM16, UCP1, adiponectin, MFN2, NOV, CEBP‐α, TNF‐α, and IL4 both in human epicardial (P) and visceral fat (V). Data from pericardial and visceral fat were compared using a heat map. Results are means ± SE; n = 4.
HO‐1 and PGC1α in mouse epicardial fat versus visceral fat and LV FS
Decreased levels of PGC1α and HO‐1 were found in mouse epicardial fat compared with visceral fat (P < 0.05; Figure 4A‐4B). The expression of adiponectin mRNA was decreased in epicardial versus visceral fat (P < 0.05) (Figure 4C). Similarly, levels of the mitochondrial uncoupling gene UCP1 and PRDM16, a thermogenic gene, were decreased in epicardial fat compared with the levels found in visceral fat (P < 0.05; Figure 4D‐4E). The decrease in HO‐1 and PGC1α was associated with a decrease in the mitochondrial biogenesis gene MFN2. The levels of MFN2 in epicardial adipose tissue were less than 50% of those in visceral fat (P < 0.05) (Figure 4F). Furthermore, we examined the effect of the increase of HO‐1 and PGC1α levels on cardiac function in mice. Cardiac function, measured by echocardiography and calculation of LV FS, was impaired (P < 0.05 in db/db mice when compared with lean mice). Mice (db/db) were administered an EET agonist, with an inducer of HO‐1‐PGC1α for 8 weeks, followed by measuring FS (as previously described) 10, 11. As depicted in Figure 4G, FS was reduced (P < 0.05) in untreated obese mice, while the treated mice (EET agonist) exhibited improved heart function as measured by increased FS. FS in wild‐type obese mice as measured by M‐mode echocardiography was 55% in lean mice, while the FS in obese mice was reduced to 29%, increasing to 56% after EET agonist treatment (Figure 4G).
Figure 4.
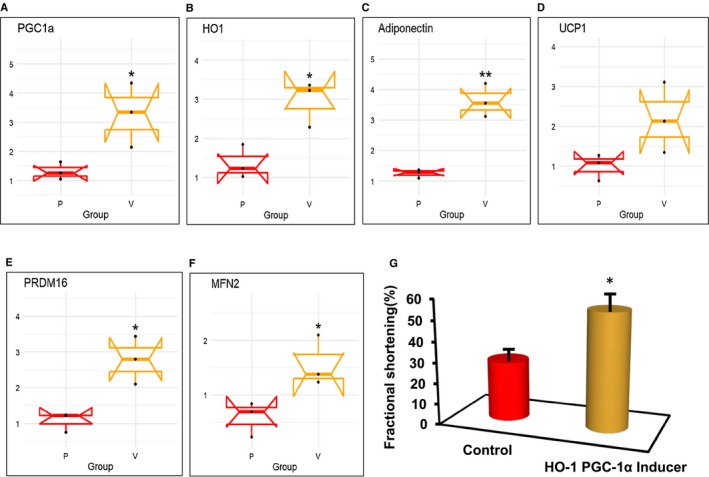
mRNA expression of (A) PGC1α, (B) HO‐1, (C) adiponectin, (D) UCP1, (E) PRDM16, and (F) MFN2 and in mouse epicardial (P) vs. visceral adipose (V) tissues. Results are means ± SE; n = 4. *P < 0.05 vs. epicardial fat; **P < 0.02 vs. epicardial fat. (G) Increase of HO‐1‐PGC1α using EET agonist as an inducer improved FS. Results are means ± SE; n = 5. *P < 0.05 vs. db/db (saline) mice. Echocardiogram measured LV and FS as previously described by Singh et al. 11.
Differential levels of adiponectin and inflammatory adipokines in mouse visceral fat and epicardial fat
Levels of inflammatory adipokine NOV, IL4, and TNF‐α in epicardial fat of obese mice were markedly elevated (P < 0.05) compared with levels found in visceral fat (Figure 5A‐5C).
Figure 5.
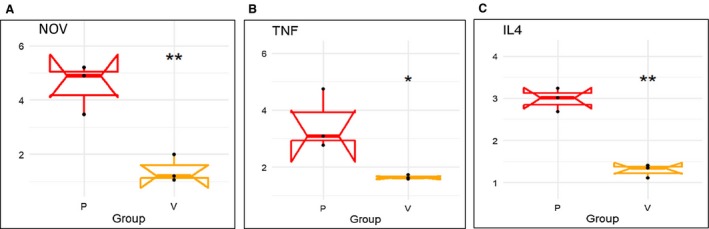
mRNA expression of inflammatory markers (A) NOV, (B) TNF‐α, and (C) IL4 in mouse epicardial (P) vs. visceral adipose (V) tissues. Results are means ± SE; n = 4. *P < 0.05 vs. epicardial fat; **P < 0.02 vs. epicardial fat.
RNA array analyses identify similar changes in correlation coefficients of gene expression in mouse epicardial and visceral fat
Although identifying fewer genes, the mouse heat map demonstrates similarities to the human in that HO‐1, PGC1α, and adiponectin correlated closely with high levels of TNF, NOV, and IL4 (Figure 6). Red represents a complete correlation, while the color orange in this figure represents R = 0.65 to 0.7. Conversely, UCP1 correlated negatively with IL4 and TNF‐α, represented by blue and black. IL4 and MFN2 were negatively correlated (Figure 6).
Figure 6.
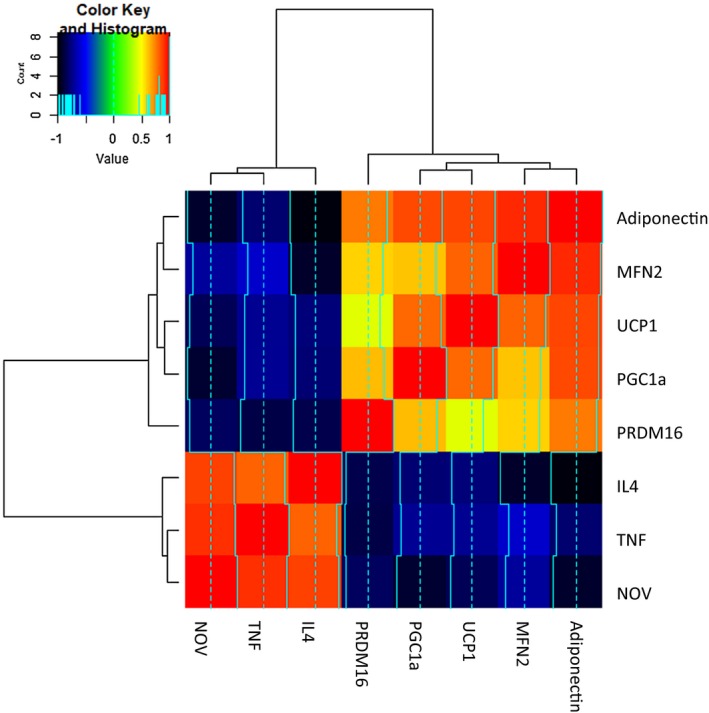
FS in lean and obese mice and RNA array analyses changes in correlation coefficient gene expression of adiponectin, MFN2, UCP1, PGC1α, PRMD16, IL4, TNF, and NOV in epicardial (P) and visceral (V) fat. Data from pericardial and visceral fat were compared using a heat map. Results are means ± SE; n = 4.
Discussion
In the present report, we describe signaling pathways in the epicardial fat of patients with overweight or obesity and coronary disease that correspond to pathways that we have previously described in epicardial fat of an obese mouse model. In addition, we describe adverse signaling expression pathways in the epicardial fat of overweight humans with vascular disease that are significantly more prominent than those found in visceral fat, another observation in concert with that seen in our mouse model 9, 11, 12. This suggests that the accumulation of inflammatory cytokines and chemokines in epicardial fat has a significantly greater toxic effect on the heart than those emanating from visceral fat, both in humans and mice. Additionally, epicardial fat NOV is highly expressed compared with visceral fat. More importantly, we identify PGC1α and HO‐1 as candidate markers for attenuation of LV dysfunction, and, as such, they may be the basis for the development of personalized therapeutic approaches to this problem caused by obesity. The correlations between different gene expressions within the specific participants are presented. For example, SLC2A4 gene expression correlates extremely well with HO‐1 gene expression between visceral and pericardial fat. HO‐1‐PGC1α‐UCP1 correlate positively with adiponectin and each other, suggesting networks for these message expressions. Furthermore, an increase of HO‐1‐PGC1α is associated with a decrease in inflammatory adipokines including NOV and improvement in LV function.
Adipose tissue exerts powerful effects on both vascular and cardiac function 30, 31. In particular, excess epicardial fat is associated with cardiac remodeling and cardiomyopathy 32, 33. Epicardial fat is anatomically adjacent to the epicardium and is not separated by any fascial layer 34. The lack of a significant barrier between epicardial fat and the myocardium allows for the transport of adipokines into the heart with relative ease by several mechanisms, including a concentration gradient. This is suggested by our prior observation of a greater expression of NOV in the epicardial fat than in the myocardium of obese mice 9.
HO‐1 reduces adiposity and vascular dysfunction in obese mice by several mechanisms, among which is the increased expression of adiponectin and an increase in bilirubin and CO, products of HO‐1‐mediated heme degradation, known to attenuate vascular dysfunction in hypertension 35, 36, 37, 38, 39. In the heart, the anti‐inflammatory effects of bilirubin include increasing nitric oxide bioavailability 40. Similarly, increased bilirubin decreases levels of inflammatory cytokines as well as markers of endoplasmic reticulum stress 41. Reduction in HO‐1 expression is associated with an increase in ROS, resulting in organ damage in both humans and mice 42, 43.
We have previously shown that ablation of HO‐1 is associated with a reduction in PGC1α, which, itself, is responsible for a decrease in MFN2 and a resulting increase in white fat over brown‐like fat 6, 44. As noted, HO‐1 expression is also mediated via PGC1α activation and, indirectly, can further increase PGC1α by enhancing EET activity. As PGC1α is downstream of EET, it becomes clear that HO‐1, EET, and PGC1α together constitute a feedback loop that has critical ramifications for cardiac function (Figure 7).
Figure 7.
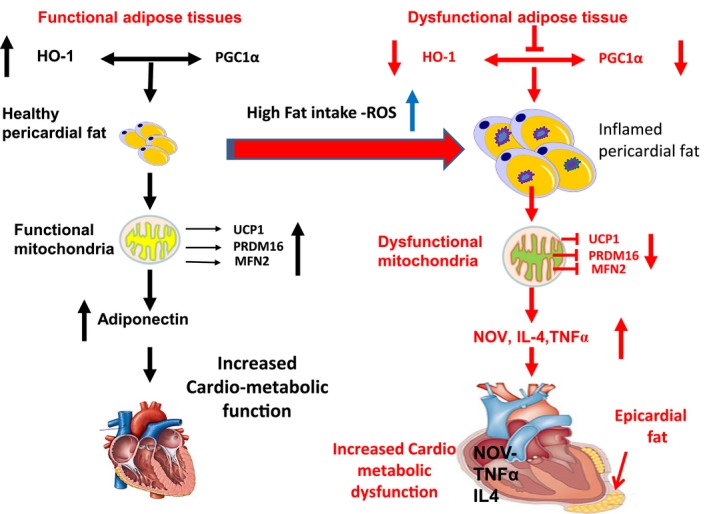
Schematic depiction of epicardial fat of obese mice and humans with coronary disease. There is a significant reduction in HO‐1, PGC1α, adiponectin, MFN2, and PRDM16 when compared with visceral fat of lean mice and age‐ and sex‐matched human controls respectively. This is accompanied by a reciprocal increase in NOV and the inflammatory marker TNF‐α in epicardial fat.
Inhibition of PGC1α is accompanied by a reduction in mitochondrial biogenesis as manifested by a concurrent reduction in MFN2 and thermogenic genes such as PRDM16. This, in turn, results in cardiac remodeling and cardiomyopathy as well as reduced energy expenditure and conversion of brown to white adipose tissue. This process can be reversed in the mouse by induction of the HO‐1‐PGC1α axis. Increased levels of HO‐1 and PGC1α are essential in restoring LV function 9, 10, 11.
The increase of FS in obese mice treated with EET agonist significantly decreased NOV protein expression in epicardial fat, which concurrently upregulated HO‐1‐PGC1α downstream signaling and restored ejection fraction and heart function. The observed reduction of HO‐1 and PGC1α in untreated obese mice was associated with a marked increase in NOV, indicating that inflammatory marker NOV is located downstream of either HO‐1 or PGC1α. Moreover, downregulation of NOV is associated with a reduction of other inflammatory adipokines 9, 11, 12. We have previously demonstrated that NOV is interrelated with HO‐1 and PGC1α in the obese db/db mouse and that it is located downstream of PGC1α in the signaling pathway 9.
In conclusion, we demonstrate in the epicardial fat of humans with coronary disease that there is a significant reduction in HO‐1, PGC1α, adiponectin, MFN2, and PRDM16 when compared with the visceral fat of age‐ and sex‐matched controls. This is accompanied by a reciprocal increase in NOV and TNF‐α in epicardial fat. This latter observation is identical to that reported in the mouse 11 as is the reduction in the activity of HO‐1‐PGC1α. Moreover, reduction in activity of HO‐1‐PGC1α in epicardial fat appears to promote myocardial dysfunction and cardiomyopathy (Figure 4). Pooled epericardial and visceral fat data from both the human and mouse demonstrate the correlations between the different gene expressions that are similar in both humans and mice. This study provides the first evidence for a difference in activity of HO‐1‐PGC1α between epicardial fat and visceral fat and demonstrates for the first time that these signaling pathways previously described in the mouse appear to be active in human epicardial fat as well. Stimulation of HO‐1‐PGC1α in the mouse is associated with significant improvement in the ejection fraction of the hearts of mice with obesity and diabetic cardiomyopathy 9, 11. Hence, the current observations open the way toward additional human studies that may further elucidate these pathways and consequently lead to novel targets for the treatment both of obesity itself and its related cardiomyopathy syndromes.
Acknowledgments
We thank Jennifer Brown for her editorial assistance in preparing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
See Commentary pg. https://doi.org/10.1002/oby.22629
Funding agencies: This work was supported by the National Institutes of Health (grant 1R56HL139561; NGA).
Disclosure: The authors declared no conflict of interest.
Author contributions: Study design: NGA and JIS; data collection: SPS, JIS, ET, GA‐B, and BE; data analysis: NGA, JIS, and SPS; data interpretation: JIS and NGA; literature search: NGA, JAM, SPS, YG, and MG; generation of figures: JIS and SPS; writing of manuscript: NGA, JIS, and JAM.
References
- 1. Alpert MA, Terry BE, Mulekar M, et al. Cardiac morphology and left ventricular function in normotensive morbidly obese patients with and without congestive heart failure, and effect of weight loss. Am J Cardiol 1997;80:736‐740. [DOI] [PubMed] [Google Scholar]
- 2. Fernandez Munoz MJ, Basurto Acevedo L, Cordova Perez N, et al. Epicardial adipose tissue is associated with visceral fat, metabolic syndrome, and insulin resistance in menopausal women. Rev Esp Cardiol (Engl Ed) 2014;67:436‐441. [DOI] [PubMed] [Google Scholar]
- 3. Pucci G, Battista F, de Vuono S, et al. Pericardial fat, insulin resistance, and left ventricular structure and function in morbid obesity. Nutr Metab Cardiovasc Dis 2014;24:440‐446. [DOI] [PubMed] [Google Scholar]
- 4. Rosito GA, Massaro JM, Hoffmann U, et al. Pericardial fat, visceral abdominal fat, cardiovascular disease risk factors, and vascular calcification in a community‐based sample: the Framingham Heart Study. Circulation 2008;117:605‐613. [DOI] [PubMed] [Google Scholar]
- 5. Peterson SJ, Shapiro JI, Thompson E, et al. Oxidized HDL, adipokines, and endothelial dysfunction: a potential biomarker profile for cardiovascular risk in women with obesity. Obesity (Silver Spring) 2019;27:87‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh S, Grant I, Meissner A, Kappas A, Abraham N. Ablation of adipose‐HO‐1 expression increases white fat over beige fat through inhibition of mitochondrial fusion and of PGC1alpha in female mice. Horm Mol Biol Clin Investig 2017;31. doi: 10.1515/hmbci-2017-0027 [DOI] [PubMed] [Google Scholar]
- 7. Pakradouni J, Le GW, Calmel C, et al. Plasma NOV/CCN3 levels are closely associated with obesity in patients with metabolic disorders. PLoS One 2013;8:e66788. doi: 10.1371/journal.pone.0066788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weingarten J, Bellner L, Peterson S, et al. The association of NOV/CCN3 with obstructive sleep apnea (OSA): preliminary evidence of a novel biomarker in OSA. Horm Mol Biol Clin Investig 2017;31. doi: 10.1515/hmbci-2017-0029 [DOI] [PubMed] [Google Scholar]
- 9. Cao J, Singh SP, McClung J, et al. EET intervention on Wnt1, NOV and HO‐1 signaling prevents obesity‐induced cardiomyopathy in obese mice. Am J Physiol Heart Circ Physiol 2017;313:H368‐H380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monu SR, Pesce P, Sodhi K, et al. HO‐1 induction improves the type‐1 cardiorenal syndrome in mice with impaired angiotensin II‐induced lymphocyte activation. Hypertension 2013;62:310‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh SP, McClung JA, Bellner L, et al. CYP‐450 epoxygenase derived epoxyeicosatrienoic acid contribute to reversal of heart failure in obesity‐induced diabetic cardiomyopathy via PGC‐1 alpha activation. Cardiovasc Pharm Open Access 2018;7:233. doi: 10.4172/2329-6607.1000233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schragenheim J, Bellner L, Cao J, et al. EET enhances renal function in obese mice resulting in restoration of HO‐1‐Mfn1/2 signaling, and decrease in hypertension through inhibition of sodium chloride co‐transporter. Prostaglandins Other Lipid Mediat 2018;137:30‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abraham NG, Rezzani R, Rodella L, et al. Overexpression of human heme oxygenase‐1 attenuates endothelial cell sloughing in experimental diabetes. Am J Physiol Heart Circ Physiol 2004;287:H2468‐H2477. [DOI] [PubMed] [Google Scholar]
- 14. Abraham NG, Junge JM, Drummond GS. Translational significance of heme oxygenase in obesity and metabolic syndrome. Trends Pharmacol Sci 2016;37:17‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase‐1 regulates cardiac mitochondrial biogenesis via Nrf2‐mediated transcriptional control of nuclear respiratory factor‐1. Circ Res 2008;103:1232‐1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Noia MA, Van DS, Palmieri F, et al. Heme oxygenase‐1 enhances renal mitochondrial transport carriers and cytochrome C oxidase activity in experimental diabetes. J Biol Chem 2006;281:15687‐15693. [DOI] [PubMed] [Google Scholar]
- 17. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold‐inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998;92:829‐839. [DOI] [PubMed] [Google Scholar]
- 18. Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC‐1. Cell 1999;98:115‐124. [DOI] [PubMed] [Google Scholar]
- 19. Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC‐1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A 2009;106:20405‐20410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Kleiner S, Mepani RJ, Laznik D, et al. Development of insulin resistance in mice lacking PGC‐1alpha in adipose tissues. Proc Natl Acad Sci U S A 2012;109:9635‐9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh SP, Schragenheim J, Cao J, Falck JR, Abraham NG, Bellner L. PGC‐1 alpha regulates HO‐1 expression, mitochondrial dynamics and biogenesis: role of epoxyeicosatrienoic acid. Prostaglandins Other Lipid Mediat 2016;125:8‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arany Z, He H, Lin J, et al. Transcriptional coactivator PGC‐1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab 2005;1:259‐271. [DOI] [PubMed] [Google Scholar]
- 23. Rowe GC, Jiang A, Arany Z. PGC‐1 coactivators in cardiac development and disease. Circ Res 2010;107:825‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008;454:961‐967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hondares E, Rosell M, Diaz‐Delfin J, et al. Peroxisome proliferator‐activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC‐1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J Biol Chem 2011;286:43112‐43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schreyer SA, Wilson DL, LeBoeuf RC. C57BL/6 mice fed high fat diets as models for diabetes‐accelerated atherosclerosis. Atherosclerosis 1998;136:17‐24. [DOI] [PubMed] [Google Scholar]
- 27. Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet‐induced type II diabetes in C57BL/6J mice. Diabetes 1988;37:1163‐1167. [DOI] [PubMed] [Google Scholar]
- 28. Lang RM, Bierig M, Devereux RB, et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography's Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Ecocardiogr 2005;18:1440‐1463. [DOI] [PubMed] [Google Scholar]
- 29. Werner JU, Tödter K, Xu P, et al. Comparison of fatty acid and gene profiles in skeletal muscle in normal and obese C57BL/6J mice before and after blunt muscle injury. Front Physiol 2018;9:19. doi: 10.3389/fphys.2018.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akoumianakis I, Tarun A, Antoniades C. Perivascular adipose tissue as a regulator of vascular disease pathogenesis: identifying novel therapeutic targets. Br J Pharmacol 2017;174:3411‐3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antonopoulos AS, Antoniades C. The role of epicardial adipose tissue in cardiac biology: classic concepts and emerging roles. J Physiol 2017;595:3907‐3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fuster JJ, Ouchi N, Gokce N, Walsh K. Obesity‐induced changes in adipose tissue microenvironment and their impact on cardiovascular disease. Circ Res 2016;118:1786‐1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graner M, Pentikainen MO, Nyman K, et al. Cardiac steatosis in patients with dilated cardiomyopathy. Heart 2014;100:1107‐1112. [DOI] [PubMed] [Google Scholar]
- 34. Iacobellis G, Corradi D, Sharma AM. Epicardial adipose tissue: anatomic, biomolecular and clinical relationships with the heart. Nat Clin Pract Cardiovasc Med 2005;2:536‐543. [DOI] [PubMed] [Google Scholar]
- 35. Hosick PA, Stec DE. Heme oxygenase, a novel target for the treatment of hypertension and obesity? Am J Physiol Regul Integr Comp Physiol 2012;302:R207‐R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawamura K, Ishikawa K, Wada Y, et al. Bilirubin from heme oxygenase‐1 attenuates vascular endothelial activation and dysfunction. Arterioscler Thromb Vasc Biol 2005;25:155‐160. [DOI] [PubMed] [Google Scholar]
- 37. Li M, Kim DH, Tsenovoy PL, et al. Treatment of obese diabetic mice with a heme oxygenase inducer reduces visceral and subcutaneous adiposity, increases adiponectin levels, and improves insulin sensitivity and glucose tolerance. Diabetes 2008;57:1526‐1535. [DOI] [PubMed] [Google Scholar]
- 38. Nicolai A, Li M, Kim DH, et al. Heme oxygenase‐1 induction remodels adipose tissue and improves insulin sensitivity in obesity‐induced diabetic rats. Hypertension 2009;53:508‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Braud L, Pini M, Muchova L, et al. Carbon monoxide‐induced metabolic switch in adipocytes improves insulin resistance in obese mice. JCI Insight 2018;3:e123485. doi: 10.1172/jci.insight.123485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bakrania B, Du Toit EF, Ashton KJ, et al. Hyperbilirubinemia modulates myocardial function, aortic ejection, and ischemic stress resistance in the Gunn rat. Am J Physiol Heart Circ Physiol 2014;307:H1142‐1149. [DOI] [PubMed] [Google Scholar]
- 41. Dong H, Huang H, Yun X, et al. Bilirubin increases insulin sensitivity in leptin‐receptor deficient and diet‐induced obese mice through suppression of ER stress and chronic inflammation. Endocrinology 2014;155:818‐828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohta K, Yachie A, Fujimoto K, et al. Tubular injury as a cardinal pathologic feature in human heme oxygenase‐1 deficiency. Am J Kidney Dis 2000;35:863‐870. [DOI] [PubMed] [Google Scholar]
- 43. Poss KD, Tonegawa S. Reduced stress defense in heme oxygenase 1‐deficient cells. Proc Natl Acad Sci U S A 1997;94:10925‐10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanella L, Sodhi K, Kim DH, et al. Increased heme‐oxygenase 1 expression decreases adipocyte differentiation and lipid accumulation in mesenchymal stem cells via upregulation of the canonical Wnt signaling cascade. Stem Cell Res Ther 2013;4:28. doi: 10.1186/scrt176 [DOI] [PMC free article] [PubMed] [Google Scholar]


