Abstract
microRNAs (miRNAs) are considered as master regulators of biological processes. Dysregulation of miRNA expression has been implicated in many human diseases. Driven by the key biological roles and the therapeutic potential, developing methods for miRNA regulation has become an intense research area. Due to favorable pharmacological properties, small molecule-based miRNA inhibition emerges as a promising strategy and significant progresses have been made. However, it remains challenging to regulate miRNA using small molecules because of the inherent difficulty in RNA targeting and inhibition. Herein we outline the workflow of generating bifunctional small molecule inhibitors blocking miRNA biogenesis through proximity-enabled inactivation of Dicer, an enzyme required for the processing of precursor miRNA (pre-miRNA) into mature miRNA. By conjugating a weak Dicer inhibitor with a pre- miRNA binder, the inhibitor can be delivered to the Dicer processing site associated with the targeted pre-miRNA, and as a result inhibiting Dicer-mediated pre-miRNA processing. This protocol can be applicable in producing bifunctional inhibitors for different miRNAs.
Keywords: miRNA inhibition, Dicer, RNA binder, RNase inhibitor
1. Introduction
microRNAs (miRNAs) are small non-coding RNAs that regulate gene expression post-transcriptionally. By binding to the target mRNAs through complimentary base pairing, miRNAs can either induce mRNA degradation or repress translation. It is estimated that more than 60% of mammalian genes, associated with various biological processes, are modulated by miRNAs1–2. Dysregulation of miRNAs has been associated with many diseases3. Therefore, miRNAs have been considered as a new and important therapeutic target. Significant efforts have been dedicated in developing miRNA regulation methods. Several exiting progresses in this regard have been made so far. For example, antisense oligonucleotides (ASOs) has been applied for blocking the function of miRNA4, and has been in clinical trials5–6. However, ASOs suffer from low cellular uptake efficiency, endosome trapping, inherent off-targeting, and poor pharmacokinetic profiles7. Alternatively, developing small molecules for miRNA regulation has recently emerged as a promising method that can address issues faced by ASOs.
The biogenesis of miRNA starts with the transcription of the miRNA gene in the nucleus. The primary transcript (pri-miRNA) produced is processed by ribonuclease (RNase) Drosha to give a stem-loop structure, precursor miRNA (pre-miRNA), which is exported to the cytoplasm. The pre-miRNA is then cleaved by RNase Dicer to produce miRNA duplex. One strand of the duplex (mature miRNA) is loaded onto the Argonaute (AGO) protein to form the miRNA-induced silencing complex (miRISC). miRISC uses mature miRNA as a guide to target mRNAs and achieve the regulation in expression8.
Small molecules disrupting any step in the processing pathway may block the production of miRNA. For example, miRNA inhibitors have been reported to repress miRNA gene transcription9–10, pri/pre- miRNA processing or AGO protein loading11–12. A lot of recent studies focus on the identification of small molecules targeting pri/pre-miRNAs. Those molecules are expected to interrupt the interaction between pri/pre-miRNA with Drosha/Dicer, thus resulting in the inhibition of miRNA production. Many of those inhibitors were identified through library screening, including affinity-based13–21, in vitro22–26 and cellular27 activity-based screening. In addition, computational approaches were also used to discover miRNA inhibitors13, 28–31. Alternatively, miRNA inhibitors can be obtained by modifying known RNA binders. For example, additional RNA binding moiety can be conjugated onto RNA binder to achieve multivalent recognition and increase the binding affinity or specificity to the target pri/pre-miRNA23, 32–35.Conjugation of an RNA cleaving structure onto pri-miRNA binder can result in selectively degradation of the target pri-miRNA36. The degradation of pri-miRNA could also be achieved using RNase recruiting molecule made by conjugation of a pri-miRNA binder with an RNase recruitment moiety37. These studies demonstrated the potential of regulating miRNA by modifying current RNA binders to enhance their activities.
2. Description of the method
We present a method to generate bifunctional small molecules inhibiting miRNA maturation by targeting the enzymatic activity of selected Dicer molecules. Dicer is well known for its function in cleaving long double stranded RNAs into small RNAs, including miRNA and small interfering RNAs (siRNAs). Increasing evidence shows that the function of Dicer is not limited to RNA cleavage. It also regulates other cellular processes38. Thus, enzymatic inhibition of Dicer does not only disrupt pre-miRNA processing, but also interferes other cellular processes associated with Dicer. To achieve selective inhibition in the processing of a targeted pre-miRNA, we developed a proximity-enabled Dicer inactivation strategy that has been proven to be feasible in generating miR-21 inhibitors39–40. In this approach, bifunctional molecules are designed to deliver a weak Dicer inhibitor to the cleavage site of the Dicer molecule associated with the targeted pre-miRNA (Fig. 1). The bifunctional molecules consist of a pre-miRNA binding unit and a Dicer inhibiting unit. A low affinity Dicer inhibiting unit is selected so that it cannot affect the pan-cellular function of Dicer when presented at low concentrations. However, the recognition of a target pre-miRNA by the RNA binding unit in the bifunctional molecule brings the conjugated Dicer inhibitor unit into the close proximity to the pre-miRNA cleavage site of the bound Dicer molecule, thus causing the blockage of Dicer activity and inhibiting the production of the target miRNA. The identification of strong and selective pre-miRNA binders has been challenging. However, such pre-miRNA binders do not always give potent miRNA inhibitors with satisfactory biological activities. The bifunctional strategy provides an alternative inhibitory mechanism that decouples the binding and inhibition. As Dicer-mediated pre-miRNA cleavage is a general process involved in miRNA biogenesis, we expect that this bi-functional Dicer inhibition approach should be applicable in generating inhibitors for different miRNAs when appropriate pre-miRNA binding units for individual pre-miRNAs are identified. We describe below the protocols to develop such bifunctional molecules for miRNA regulation using miR-21 as an example.
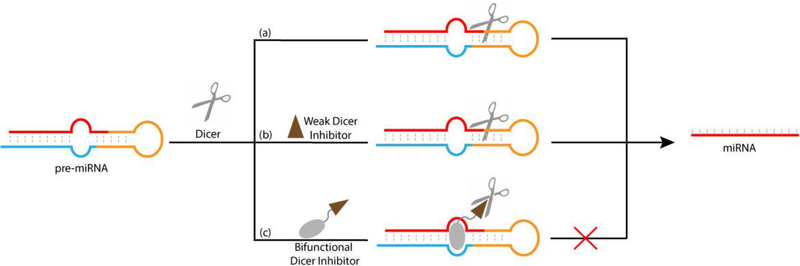
Fig. 1.
An illustration of the approach: (a) miRNA is produced from pre-miRNA by Dicer processing; (b) Weak Dicer inhibitor does not affect the activity of Dicer; (c) Bifunctional molecule inhibits the enzymatic activity of Dicer complexed with pre-miRNA.
3. Protocols
The development of bifunctional molecules involves four steps: (i) the identification and validation of pre-miRNA binding units by fluorescence polarization (FP)-based assays, (ii) the selection and synthesis of Dicer inhibiting unit, (iii) the conjugation of the two functional units through click chemistry, and (iv) the evaluation of miRNA inhibition activity and selectivity. These protocols provide the general procedures that can be followed to develop and test new bifunctional molecules for inhibiting the maturation of miRNAs not necessarily limited to the miR-21 used as an example here.
3.1. Materials and equipment
Chemical reagents of molecular biology grade were used for the screening assay and purchased from Sigma-Aldrich unless specified. All solutions and buffers were prepared with RNase free water. A morpholino-based ASO, Anti-miR-21 A (Sequence: AGTCAACATCAGTCTGATAAGCTAC), was purchased from Gene Tools. A phosphorothioate-based ASO, Anti-miR-21 B (Sequence: CAACATCAGTCTGATAAGCTAC), was obtained from Integrated DNA Technologies. pCAGGS-Flag-hsDicer plasmid DNA was obtained from Addgene (plasmid # 41584; http://n2t.net/addgene:41584; RRID:Addgene_41584)41. pCMV-miR21 was obtained from Addgene (plasmid # 20381 ; http://n2t.net/addgene:20381 ; RRID:Addgene_20381)42. MSCV-miR-34a was obtained from Addgene (plasmid # 63932 ; http://n2t.net/addgene:63932 ; RRID:Addgene_63932)43. The FP data were collected with a microplate reader (SpectraMax i3X, Molecular Devices) equipped with a fluorescence polarization detection cartridge. A default instrument setting was used except Z height (3 mm), and integration time (100 ms). Phosphor imaging was carried out using an exposure cassette (GE Healthcare Bio-Sciences AB) and Personal Molecular Imager (Bio-Rad). Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR) was performed on CFX96 Real Time PCR System (Bio-Rad).
3.2. Identification and validation of pre-miRNA binding unit
The RNA binding unit can be chosen either from known pre-miRNA binders or from library screening. Many screening methods have been developed for identifying pre-miRNA ligands44. We introduced a FP based displacement screening assay (Fig. 2). In this assay, a fluorescently labeled pre-miRNA binder serves as a reference compound. When it forms a complex with the target pre-miRNA, the degree of FP increases due to a reduction in the rate of fluorophore tumbling. When screening a compound library, any compound with a stronger pre-miRNA binding affinity will displace the reference compound from the complex, resulting in a decreased polarity of the fluorescence light. Thus, a strong pre-miRNA binder can be identified by a reduced FP signal in the assay.
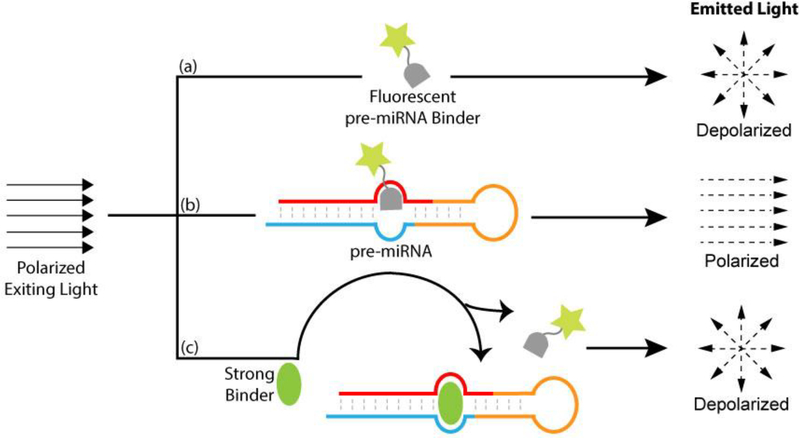
Fig. 2.
An illustration of the FP-based screening and binding assay. Upon excitation by polarized light, (a) a free fluorescent pre-miRNA binder emits depolarized light; (b) when complexed to pre-miRNA, it gives polarized light due to reduced tumbling; (c) a strong pre-miRNA binder replaces the fluorescent binder from the complex, resulting in decreased polarity of the light emitted.
3.2.1. Preparation of pre-miRNA
To prepare pre-miR-21 for screening, the full-length pre-miR-21 was prepared enzymatically by in vitro transcription using T7 RNA polymerase (Ambion). The sequence of pre-miR-21 was obtained from miRbase (http://www.mirbase.org/)45. The DNA template was obtained by primer extension using Taq polymerase (Ambion). The forward primer contains a T7 promoter sequence (GAAATTAATACGACTCACTATAGG) followed by the first 46 nucleotides of pre-miR-21 (TGTCGGGTAGCTTATCAGACTGATGTTGACTGTTGAATCTCATGGC). The reverse complimentary sequence of the last 48 nucleotides of pre-miR-21 was used as the sequence of reverse primer (TGTCAGACAGCCCATCGACTGGTGTTGCCATGAGATTCAACAGTCAAC). The two primers have 22 nucleotide overlapping. The in vitro transcription reaction was carried out following vendor’s protocol and followed by RQ1 DNase (Promega) treatment to digest the template DNA. The reaction was then purified by phenol:chloroform extraction and ethanol precipitation. The RNA was dissolved in water and stored at - 20 °C. It was allowed to refold as follows before use: RNA was heated to 94 °C for 2 min and then cooled to 4 °C at a rate of 1 °C/s using a thermal cycler (S1000, Bio-Rad).
3.2.2. Preparing the reference compound for screening
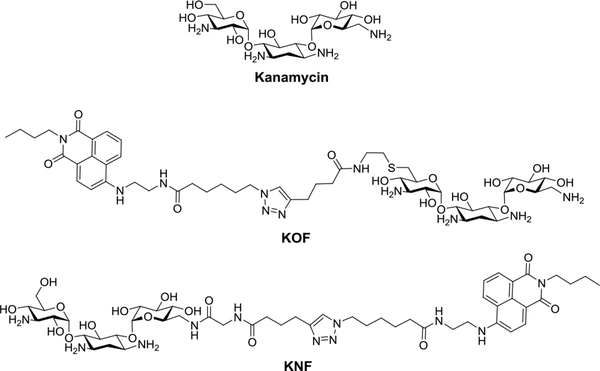
In the FP-based screening assay, a reference compound can be produced by labeling a known binder for the pre-miRNA of interest with a fluorophore. It is possible that the modification may disrupt the binding of the compound to the RNA depending on where the modification occurs. As a result, different labeling sites on the RNA binder may have to be tested and the binding of the resulting fluorescently labeled reference compound to the target pre-miRNA needs to be validated. For example, a known pre- miR-21 binder, kanamycin, was conjugated with a fluorophore at 2 different sites (Fig. 3)39. After testing the binding to pre-miR-21, only one of the 2 resulting compounds, KOF, retained the binding affinity to pre-miR-2139 as determined by the FP-based binding assay described in Section 3.2.4.
Fig. 3.
The structures of kanamycin and its fluorophore-tagged derivatives.
3.2.3. The FP screening assay
For optimal screening result, the concentration of the reference compound to be used in the assay has to be determined first. It should be less than dissociate constant (Kd) and the concentration of pre- miRNA, so that a greater assay window could be created. To get good signal-to-noise ratio, it is recommended to use a concentration that gives a read of more than 10,000 counts in fluorescence intensity mode. One can determine the optimal concentration by making a serial dilution of the reference compound and examining the signal generated. After identifying optimal working concentration of the reference compound KOF in the assay, to identify pre-miR-21 binder, 30 nM of KOF was incubated with pre-miR-21 (1 μM) and individual library compound (3 μM) in cacodylate buffer (pH 7.4, 10 mM, 0.01% Triton X-100) at room temperature for 1 h. The solution was then distributed into black 96-well plates (100 μL/well) for FP data collection. Control experiments, containing KOF with or without pre-miR-21, were also included. The experiment was performed in triplicates. The hits were selected by examining the decrease of average FP relative to that of KOF with pre-miR-21. The larger amount of FP signal decrease indicates the better displacement of the reference compound and the stronger the binder is. For example, neomycin (Fig. 4) was identified as a hit that reduced FP signal to a level that matches that of free KOF, suggesting a complete displacement of KOF from the pre-miR-21.
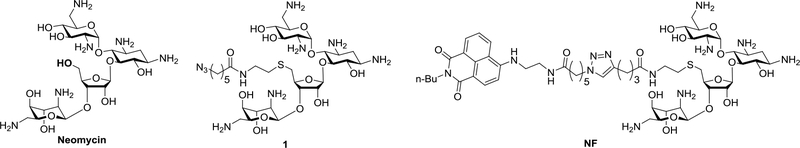
Fig. 4.
Structures of Neomycin and its derivatives.
3.2.4. Hit validation
A validation analysis is required to confirm the binding of the hits from the screening to the target pre- miRNA. Many biochemical methods have been used for studying the interaction between a small molecule and an RNA46. We used an FP-based binding assay adapted from the literature47 for this purpose. Neomycin, the best hit from the above screening for pre-miR-21, was labeled with a fluorophore via the primary hydroxyl site (NF, Fig. 4)39 and subsequently used in the FP assay upon pre- miR-21 titration. 30 nM of NF was mixed with various concentrations of pre-miR-21 (3 nM to 4 μM) in cacodylate buffer (pH 7.4, 10 mM, 0.01% Triton X-100) and incubated at room temperature for 1 h. The solution was then distributed into black 96-well plates (100 μL/well) for FP data collection. The experiment was performed in triplicates. The Kd was obtained by plotting the mean of FP versus pre- miR-21 concentration using Equation (1), where P0 is the polarization of NF alone, P is the measured polarization at each pre-miR-21 concentration [RNA]total, ΔP is the total change of polarization upon saturation, [F]total is the total concentration of NF.
| Equation (1) |
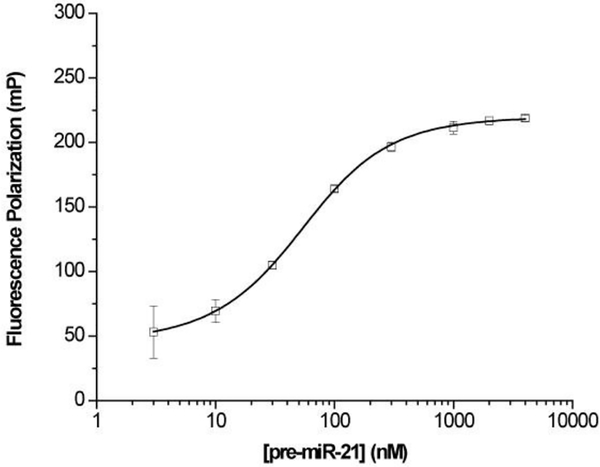
A typical saturation binding curve of NF to pre-miR-21 was shown in Fig. 539. A Kd of 38 nM was determined from the binding data, which confirms the binding of neomycin to pre-miR-21. In addition, the strong binding of NF to pre-miR-21 indicates that the modification through the primary hydroxyl site did not disrupt the binding of neomycin to pre-miR-21. As mentioned above, structural modification may disrupt the binding of a binder to the target RNA. This could lead to false negative results in the hit validation stage. Therefore, the site of fluorescent labeling on the hit molecules in the validation assays should be carefully considered and tested in individual cases. It is possible that multiple labeling sites have to be tested.
Fig. 5.
Fluorescence polarization of NF in presence of pre-miR-21.
3.2.5. Generating the clickable version of pre-miRNA binding unit
The validation analysis can provide valuable information regarding the optimal modification site on the hit compound to be used to conjugate the Dicer inhibiting unit without interrupting pre-miRNA binding. We applied click chemistry for such conjugation to generate bifunctional molecules. For this purpose, an azide-appended neomycin (Compound 1, Fig. 4) modified via the primary hydroxyl site as suggested from the studies in section 3.2.4, was synthesized39. This azido pre-miRNA binding unit was used in section 3.4 to synthesize bifunctional molecules.
3.3. Selection and synthesis of the Dicer inhibiting unit
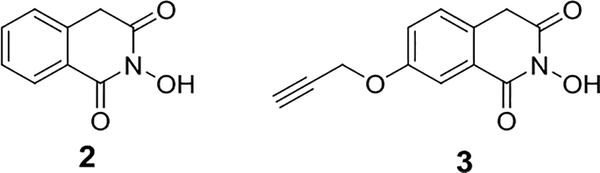
Fluorescence-based high-throughput screening assay has been developed for discovering general Dicer inhibitor48, but no small-molecule inhibitor has been reported so far to our knowledge. As a member of the RNase III family, Dicer cleaves RNA in a mechanism similar as other RNases in the family do49. Many small-molecule RNase inhibitors have been reported that could potentially be tested for Dicer inhibition50–52. We found that an influenza endonuclease, 2-hydroxyisoquinoline-1,3(2H,4H)-dione (Compound 2, Fig. 6), inhibits the activity of Dicer weakly39. To produce a clickable version of the Dicer inhibition unit to conjugate to the pre-miRNA binding unit, a weak Dicer inhibitor such as 2 has to be functionalized with an alkyne group through a linker. As the pharmacophore of 2 consists of the three oxygen atoms considered to be critical for the inhibitory activity50, we introduced a propargyl group away from these critical functionalities and obtained Compound 339 to be coupled to the azido neomycin 1. To identify the resulting bifunctional molecule with optimal activity, it is recommended to generate a set of bifunctional molecules with linkers of different lengths to connect the Dicer inhibition and pre- miRNA binding units for testing. This can be implemented by synthesizing a series of Compound 3 variants with different lengths of linkers between the alkyne and Dicer inhibitor39 for the following click coupling in section 3.4. We expect that the same Dicer inhibition unit described here can be used in generating new Dicer inactivating bifunctional molecules for other miRNAs when the pre-miRNA binding unit and the linker are optimized for targeting other miRNAs. Compound 3 is stable under neutral condition except that the enolization of the carbonyl group in position 3 may occur very slowly. It is worth noting that, however, we found it can decompose in the aqueous solution at low pH. Under the common purification protocol using high performance liquid chromatography (HPLC) with a typical 0.1% trifluoroacetic acid (TFA) elution solution, the eluted fractions have to be kept on ice or frozen and the solvents should be removed as soon as possible at low temperature to prevent Compound 3 from rapid decomposing.
Fig. 6.
Structures of Dicer inhibitors.
3.4. Generating bifunctional molecules
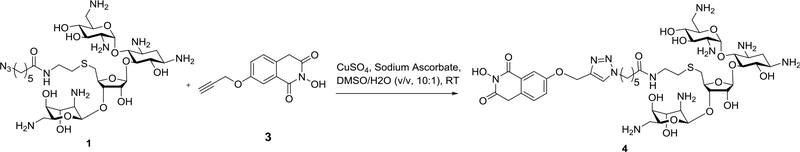
The conjugation of the two functional units was accomplished using Copper-catalyzed click chemistry under standard reaction condition (Fig. 7). To a mixture of the alkyne-modified Dicer inhibiting unit 3 (100 mM) and azido-modified RNA binder 1 (100 mM) in DMSO (200 μL) was added aqueous solutions of CuSO4 (0.1 eq, 10 μL) and sodium ascorbate (0.2 eq, 10 μL). The mixture was stirred at room temperature. The reaction usually completes within 2 h. The product Compound 4 was purified by HPLC using C-18 column eluted with H2O/CH3CN containing 0.1% TFA. To avoid the decomposition of the bifunctional molecules under acidic condition as mentioned in Section 3.3, the fractions were kept frozen during the purification process. The solvents were removed by freeze drying. The compounds can be stably stored at −80 °C as solid or as stock solution in water.
Fig. 7.
Synthesis of the bifunctional conjugates via click chemistry.
3.5. Evaluation of the in vitro miRNA inhibition activity
To evaluate the activity of bifunctional molecules in blocking pre-miRNA processing, a reconstituted Dicer-mediated in vitro pre-miRNA cleavage assay using recombinant Dicer protein and 32P labeled pre- miRNA was carried out as described below.
3.5.1. Dicer enzyme preparation
Dicer enzyme expressed in insect cells is commercially available (Genlantis) and can be used directly in the activity assays. However, we found its activity for pre-miRNA processing could be inconsistent and vary from batch to batch. Alternatively, the recombinant FLAG-tagged Dicer can be expressed and purified in mammalian cells using plasmid DNA pCAGGS-Flag-hsDicer. Human embryonic kidney (HEK) 293T cells were used to express Dicer protein due to their high transfection efficiency and high expression level. To express recombinant Dicer, HEK293T cells were cultured in DMEM (Gibco) supplemented with 10% FBS and 2 mM GlutaMAX (Life Technologies) at 37 °C in a humidified incubator containing 5% CO2. No antibiotics were added in the cell culture. 6 × 106 cells were plated in a 15-cm dish and grown for 16 h to around 60% confluency. 9 μg of the plasmid DNA was used to transfect the cells with Lipo3000 transfection reagent (Invitrogen) per the manufacture’s protocol. The medium was replaced every day. The cells were washed on dish with PBS (2 × 15 mL) after a 3-day incubation. They were then kept at −80 °C for overnight and thaw on ice. 10 mL of ice-cold PBS were added into the dish. The cells were gently scratched off the dish and transferred into a tube for centrifugation at 4 °C and 600 g for 10 min. After removing the supernatant, the cells were then lysed with 1 mL of ice-cold lysis buffer (Tris 50 mM, NaCl 150 mM, Triton X-100 1%, SDS 0.1%, pH 7.5) containing the cocktail protease inhibitors (ThermoFisher) using a Branson 2510 sonicator at 4 °C (20 × 10 s, at intervals of 20 s). The lysate was centrifuged at 4 °C and 21130 g, for 10 min. The clear supernatant was carefully taken out and incubated with 60 μL 50% slurry of Anti-FLAG M2 affinity beads (Sigma-Aldrich) pre-washed with ice-cold buffer (3 × 1 mL) (Tris 50 mM, NaCl 150 mM, NP-40 0.005%, pH 7.5). It was gently agitated at 4 °C for 16 h and then spun down at 845 g for 2 min. The supernatant was carefully removed and the beads were washed with the same ice-cold washing buffer (2 × 1 mL). The beads with Dicer were finally suspended in a mixture of H2O and glycerol (25 μL/25 μL) and stored at −20 °C. The Dicer on-beads was used for the following assay directly. To minimize the effects of batch-to-batch variation in Dicer purification and activity, it is recommended to quantify the activity of Dicer after each preparation. This is done by a serial dilution of the Dicer/beads from each batch of preparation to cleave 32P labeled pre- miR-21 as described below in Section 3.5.2. The amount of Dicer/beads that was able to cleave pre-miR- 21 (~ 20 ng) completely into mature miR-21 within 5 h can be quantified and defined as 1 unit.
3.5.2. In vitro Dicer inhibition assay
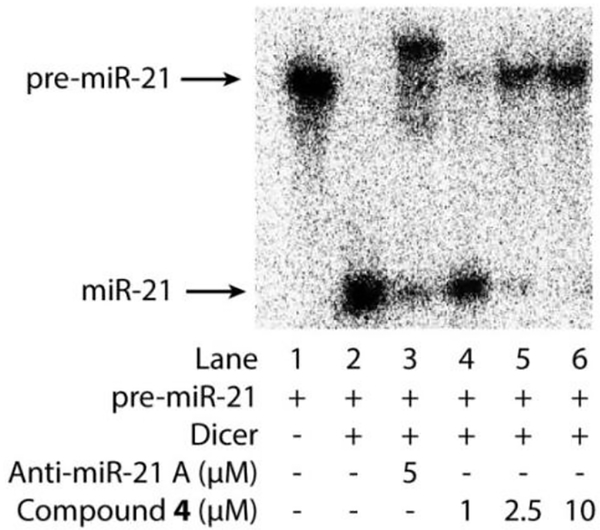
This activity assay allows the evaluation of the activity of bifunctional molecules in inhibiting Dicer- mediated cleavage of pre-miRNA (~70 nucleotides) into mature mi NA (~22 nucleotides) by visualizing and quantifying the relative amounts of the precursor versus the cleaved product on gel under each treatment condition. To carry out the assay for miR-21, 32P labeled pre-miR-21 was in vitro transcribed as described in Section 3.2.1 except that [alpha-P32] uridine 5’-triphosphate (UTP) (Perkinelmer) was added following vendor’s protocol. The transcription reaction product was purified with NucAway spin columns (Invitrogen). The RNA prepared was stored at −20 °C and refolded as described in Section 3.2.1 before use. In the activity assay, the Dicer-mediated reaction was prepared by mixing refolded32 -labeled pre-mi −21 (1 μL, ~20 ng) with recombinant Dicer (1 μL of Genlantis Dicer or 1 unit of home made Dicer on beads) in the presence or absence of varying concentrations of bifunctional molecules (diluted from stock solution in water) in buffer (HEPES 24 mM, NaCl 200 mM, EDTA 0.04 mM, MgCl2 2.5 mM, ATP 1 mM, pH 7.5) to a total volume of 10 μL. The experimental condition with pre-miR-21 only (without Dicer and compound) was used as a control. The mixtures were gently agitated at 37 °C for 5 h. The reactions were stopped by adding equal volume of Gel Loading Buffer II (ThermoFisher Scientific) and incubating at 95 °C for 5 min. The reaction products, pre-miR-21 and the processed short mature miR-21, were resolved by 15% denaturing polyacrylamide gel and analyzed by phosphor imaging (Fig. 8). The activity of bifunctional miR-21 inhibitors can be assessed by their ability to prevent Dicer from cutting the pre-miR-21 into mature miR-21. As shown in Fig. 8, Dicer efficiently cleaved pre-miR-21 into mature miR-21 (Lane 2). Anti-miR-21 A, a morpholino-based ASO designed to block miR-21 maturation, was used as a positive control. It did inhibit pre-miR-21 cleavage significantly at 5 μM (Lane 3). Importantly, the bifunctional Compound 4 inhibited miR-21 production in a dose-dependent manner (Lane 4–6). Quantitative analysis of the inhibition rate could be obtained by quantifying corresponding band intensities of pre-miR-21 and mature miR-21 with Quantity One software (Bio-rad). The selectivity of the conjugate can be evaluated by repeating this Dicer inhibition assay to test and compare its inhibition for other pre-miRNAs40.
Fig. 8.
Electrophoresis gel analysis of 32P labeled pre-miR-21 cleavage mediated by Dicer in the presence of tested compounds.
3.6. miRNA inhibition activity in cell
The activity of small molecule in cell can be evaluated by measuring the level of target miRNA in treated cells. Among all the methodologies, RT-qPCR is considered as the gold standard and widely used for miRNA detection53. Based on this technique, multiple types of miRNA assay are now commercial available. To test miR-21 inhibition activity in cells, HEK293T cells overexpressing pre-miR-21 were treated with tested compound and the miR-21 levels under different treatment conditions were analyzed by RT-qPCR.
3.6.1. Cell culture and transfection
HEK293T cells were cultured as described in Section 3.5.1. They were plated in 24-well plates at 2.0 × 105 cells/well. After overnight incubation, the cells grow to around 70% cell confluency. They were then transfected with plasmid DNA pCMV-miR21 or MSCV-miR-34a using Lipo2000 transfection reagent (Invitrogen). Typically, a mixture of DNA (200 ng), Lipofectamine 2000 (1 μL) and tested compound were incubated in 50 μL of Opti-MEM reduced serum medium (Thermo Fisher Scientific) at room temperature for 5 min. The mixture was then transferred into individual well containing the cells and 500 μL of fresh replaced medium. Cells were harvested for RNA extraction after 22 h of incubation.
3.6.2. RNA extraction and RT-qPCR
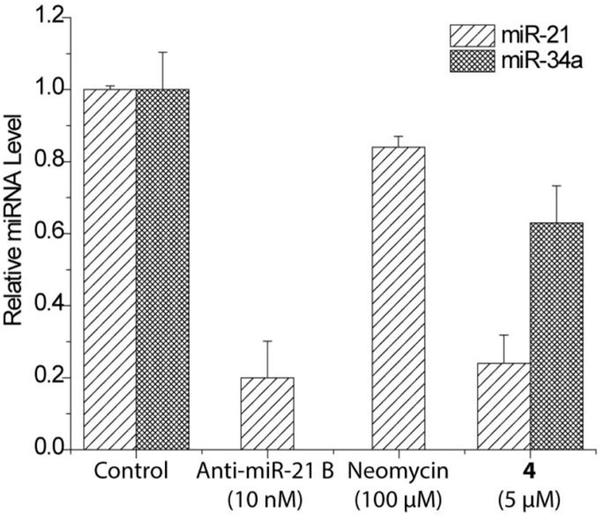
Total RNA was extracted with miRNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. Reverse transcription reaction was performed using a Taqman microRNA RT Kit (Applied Biosystems) to generate cDNA. 10 ng of the total RNA was used in each reverse transcription reaction (15 μL). qPCR reaction was then completed with Taqman Universal PCR Master Mix (Applied Biosystems) and TaqMan miRNA assay (Thermo Fisher Scientific) following the manufacture’s protocol. 1 μL of the cDNA was used in each qPCR reaction (20 μL). The triplicate threshold cycles (Ct) obtained for each treatment were used to determine the relative levels of miRNA normalized to U6 small nuclear RNA using the 2-ΔΔCt method.54 The assay was performed three times independently. As shown in Fig. 9, compound 4 reduced miR-21 by 76% at 5 μM when compared to non-treated condition39. However, no inhibitory activity was observed with neomycin even up to 100 μM. To evaluate the selectivity of the bifunctional conjugate, the level changes of other miRNAs in the cells upon treatment can be analyzed and compared to the change in miR-21. For this purpose, miR-34a was randomly chosen for the test and was found to be also inhibited by 4, however with a much lower potency compared to the inhibition of miR-2139. Thus, although 4 exhibited certain degree of selectivity, it is not specific for inhibiting miR-21 maturation. This is likely resulted from the promiscuous RNA recognition of neomycin. We believe that the selectivity of bifunctional conjugates can be improved by incorporating more specific RNA recognition moieties identified from screening or other approaches.
Fig. 9.
RT-qPCR analysis of miRNA level in HEK293T cells.
4. Conclusion
Bifunctional miRNA inhibitors can be generated by conjugating an RNase inhibitor with a pre-miRNA binder and tested for inhibitory activity following the described method using the bifunctional miR-21 inhibitor as an example. This strategy should be applicable in making new inhibitors for miR-21 and other miRNAs. In contrast to other small-molecule miRNA inhibitor, this approach focuses on targeted enzymatic inactivation of Dicer. The potency and selectivity of the bifunctional molecules rely extensively on the property of the RNA binding unit, which calls for the identification of RNA binder with high affinity and specificity. It is not uncommon that some pre-miRNA binders identified by affinity- based screening are not able to inhibit Dicer processing. The bifunctional strategy described here offers a new way to transform non-inhibitory RNA binders into active RNA inhibitors and open new avenues for miRNA inhibition.
Highlights.
A proximity-enabled miRNA inhibition approach was reported.
The approach relies on the enzymatic inactivation of Dicer by bifunctional small molecules.
miR-21 inhibitor was successfully developed using this method.
This approach could be applied to generate bifunctional inhibitors for other miRNAs.
Acknowledgements
This research was supported by NIH R21CA202831 (F.-S.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Friedman RC; Farh KK-H; Burge CB; Bartel DP, Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009, 19 (1), 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ameres SL; Zamore PD, Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol 2013, 14 (8), 475–488. [DOI] [PubMed] [Google Scholar]
- (3).Li Y; Kowdley KV, MicroRNAs in Common Human Diseases. Genomics, Proteomics Bioinf 2012, 10 (5), 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Stenvang J; Petri A; Lindow M; Obad S; Kauppinen S, Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3 (1), 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Janssen HLA; Reesink HW; Lawitz EJ; Zeuzem S; Rodriguez-Torres M; Patel K; van der Meer AJ; Patick AK; Chen A; Zhou Y; Persson R; King BD; Kauppinen S; Levin AA; Hodges MR, Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med 2013, 368 (18), 1685–1694. [DOI] [PubMed] [Google Scholar]
- (6).van Zandwijk N; Pavlakis N; Kao S; Clarke S; Lee A; Brahmbhatt H; Macdiarmid J; Pattison S; Leslie F; Huynh Y; Linton A; Reid G, P1.02MesomiR 1: A Phase I study of TargomiRs in patients with refractory malignant pleural mesothelioma (MPM) and lung cancer (NSCLC). Ann. Oncol 2015, 26 (suppl_2), ii16–ii16. [Google Scholar]
- (7).Juliano RL; Ming X; Nakagawa O, The Chemistry and Biology of Oligonucleotide Conjugates. Acc. Chem. Res 2012, 45 (7), 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ha M; Kim VN, Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol 2014, 15, 509–524. [DOI] [PubMed] [Google Scholar]
- (9).Gumireddy K; Young DD; Xiong X; Hogenesch JB; Huang QH; Deiters A, Small- molecule inhibitors of microRNA miR-21 function. Angew. Chem. Int. Ed 2008, 47 (39), 7482–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Young DD; Connelly CM; Grohmann C; Deiters A, Small Molecule Modifiers of MicroRNA miR-122 Function for the Treatment of Hepatitis C Virus Infection and Hepatocellular Carcinoma. J. Am. Chem. Soc 2010, 132 (23), 7976–7981. [DOI] [PubMed] [Google Scholar]
- (11).Watashi K; Yeung ML; Starost MF; Hosmane RS; Jeang KT, Identification of Small Molecules That Suppress MicroRNA Function and Reverse Tumorigenesis. J. Biol. Chem 2010, 285 (32), 24707–24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tan GS; Chiu C-H; Garchow BG; Metzler D; Diamond SL; Kiriakidou M, Small Molecule Inhibition of RISC Loading. ACS Chem. Biol 2012, 7 (2), 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Velagapudi SP; Disney MD, Two-dimensional combinatorial screening enables the bottom- up design of a microRNA-10b inhibitor. Chem. Commun 2014, 50 (23), 3027–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Diaz JP; Chirayil R; Chirayil S; Tom M; Head KJ; Luebke KJ, Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA 2014, 20 (4), 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chirayil S; Chirayil R; Luebke KJ, Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res 2009, 37 (16), 5486–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Fukuzumi T; Murata A; Aikawa H; Harada Y; Nakatani K, Exploratory Study on the RNA- Binding Structural Motifs by Library Screening Targeting pre-miRNA-29 a. Chemistry – A European Journal 2015, 21 (47), 16859–16867. [DOI] [PubMed] [Google Scholar]
- (17).Pai J; Hyun S; Hyun JY; Park SH; Kim WJ; Bae SH; Kim NK; Yu J; Shin I, Screening of Pre-miRNA-155 Binding Peptides for Apoptosis inducing Activity Using Peptide Microarrays. J. Am. Chem. Soc 2016, 138 (3), 857–867. [DOI] [PubMed] [Google Scholar]
- (18).Shortridge MD; Walker MJ; Pavelitz T; Chen Y; Yang W; Varani G, A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chem. Biol 2017, 12 (6), 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Connelly CM; Boer RE; Moon MH; Gareiss P; Schneekloth JS, Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. ACS Chem. Biol 2017, 12 (2), 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Maiti M; Nauwelaerts K; Herdewijn P, Pre-microRNA binding aminoglycosides and antitumor drugs as inhibitors of Dicer catalyzed microRNA processing. Bioorg. Med. Chem. Lett 2012, 22 (4), 1709–1711. [DOI] [PubMed] [Google Scholar]
- (21).Yan H; Zhou M; Bhattarai U; Song Y; Zheng M; Cai J; Liang F-S, Cyclic Peptidomimetics as Inhibitor for miR-155 Biogenesis. Mol. Pharmaceutics 2019, 16 (2), 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Klemm CM; Berthelmann A; Neubacher S; Arenz C, Short and Efficient Synthesis of Alkyne- Modified Amino Glycoside Building Blocks. Eur. J. Org. Chem 2009, 2009 (17), 2788–2794. [Google Scholar]
- (23).Vo DD; Staedel C; Zehnacker L; Benhida R; Darfeuille F; Duca M, Targeting the Production of Oncogenic MicroRNAs with Multimodal Synthetic Small Molecules. ACS Chem. Biol 2014, 9 (3), 711–721. [DOI] [PubMed] [Google Scholar]
- (24).Bose D; Jayaraj GG; Kumar S; Maiti S, A Molecular-Beacon-Based Screen for Small Molecule Inhibitors of miRNA Maturation. ACS Chem. Biol 2013, 8 (5), 930–938. [DOI] [PubMed] [Google Scholar]
- (25).Lorenz DA; Kaur T; Kerk SA; Gallagher EE; Sandoval J; Garner AL, Expansion of cat- ELCCA for the Discovery of Small Molecule Inhibitors of the Pre-let-7–Lin28 RNA–Protein Interaction. ACS Medicinal Chemistry Letters 2018, 9 (6), 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lorenz DA; Song JM; Garner AL, High-Throughput Platform Assay Technology for the Discovery of pre-microRNA-Selective Small Molecule Probes. Bioconjugate Chem 2015, 26 (1), 19–23. [DOI] [PubMed] [Google Scholar]
- (27).Bose D; Jayaraj G; Suryawanshi H; Agarwala P; Pore SK; Banerjee R; Maiti S, The Tuberculosis Drug Streptomycin as a Potential Cancer Therapeutic: Inhibition of miR-21 Function by Directly Targeting Its Precursor. Angew. Chem. Int. Ed 2012, 51 (4), 1019–1023. [DOI] [PubMed] [Google Scholar]
- (28).Costales MG; Haga CL; Velagapudi SP; Childs-Disney JL; Phinney DG; Disney MD, Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit. J. Am. Chem. Soc 2017, 139 (9), 3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Velagapudi SP; Gallo SM; Disney MD, Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol 2014, 10 (4), 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Childs-Disney JL; Disney MD, Small Molecule Targeting of a MicroRNA Associated with Hepatocellular Carcinoma. ACS Chem. Biol 2016, 11 (2), 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Haga CL; Velagapudi SP; Strivelli JR; Yang W-Y; Disney MD; Phinney DG, Small Molecule Inhibition of miR-544 Biogenesis Disrupts Adaptive Responses to Hypoxia by Modulating ATM- mTOR Signaling. ACS Chem. Biol 2015, 10 (10), 2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Vo DD; Tran TPA; Staedel C; Benhida R; Darfeuille F; Di Giorgio A; Duca M, Oncogenic MicroRNAs Biogenesis as a Drug Target: Structure-Activity Relationship Studies on New Aminoglycoside Conjugates. Chem. Eur. J 2016, 22 (15), 5350–5362. [DOI] [PubMed] [Google Scholar]
- (33).Nahar S; Ranjan N; Ray A; Arya DP; Maiti S, Potent inhibition of miR-27a by neomycin- bisbenzimidazole conjugates. Chem. Sci 2015, 6 (10), 5837–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Murata A; Otabe T; Zhang J; Nakatani K, BzDANP, a Small-Molecule Modulator of Pre-miR- 29a Maturation by Dicer. ACS Chem. Biol 2016, 11 (10), 2790–2796. [DOI] [PubMed] [Google Scholar]
- (35).Watkins D; Jiang L; Nahar S; Maiti S; Arya DP, A pH Sensitive High-Throughput Assay for miRNA Binding of a Peptide-Aminoglycoside (PA) Library. PLOS ONE 2015, 10 (12), e0144251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li Y; Disney MD, Precise Small Molecule Degradation of a Noncoding RNA Identifies Cellular Binding Sites and Modulates an Oncogenic Phenotype. ACS Chem. Biol 2018, 13 (11), 3065–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Costales MG; Matsumoto Y; Velagapudi SP; Disney MD, Small Molecule Targeted Recruitment of a Nuclease to RNA. J. Am. Chem. Soc 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Song M-S; Rossi JJ, Molecular mechanisms of Dicer: endonuclease and enzymatic activity. The Biochemical journal 2017, 474 (10), 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yan H; Bhattarai U; Guo Z-F; Liang F-S, Regulating miRNA-21 Biogenesis By Bifunctional Small Molecules. J. Am. Chem. Soc 2017, 139 (14), 4987–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yan H; Bhattarai U; Song Y; Liang F-S, Design, synthesis and activity of light deactivatable microRNA inhibitor. Bioorg. Chem 2018, 80, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gurtan AM; Lu V; Bhutkar A; Sharp PA, In vivo structure–function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 2012, 18 (6), 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).ZENG Y; CULLEN BR, Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9 (1), 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Okada N; Lin C-P; Ribeiro MC; Biton A; Lai G; He X; Bu P; Vogel H; Jablons DM; Keller AC; Wilkinson JE; He B; Speed TP; He L, A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 2014, 28 (5), 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lorenz DA; Garner AL, Approaches for the Discovery of Small Molecule Ligands Targeting microRNAs In RNA Therapeutics, Garner AL, Ed. Springer International Publishing: Cham, 2018; pp 79–110. [Google Scholar]
- (45).Kozomara A; Griffiths-Jones S, miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014, 42 (D1), D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Blakeley BD; DePorter SM; Mohan U; Burai R; Tolbert BS; McNaughton BR, Methods for identifying and characterizing interactions involving RNA. Tetrahedron 2012, 68 (43), 8837–8855. [Google Scholar]
- (47).Luedtke NW; Liu Q; Tor Y, NA-Ligand Interactions: Affinity and Specificity of Aminoglycoside Dimers and Acridine Conjugates to the HIV-1 Rev Response Element. Biochemistry 2003, 42 (39), 11391–11403. [DOI] [PubMed] [Google Scholar]
- (48).Podolska K; Sedlak D; Bartunek P; Svoboda P, Fluorescence-Based High-Throughput Screening of Dicer Cleavage Activity. Journal of Biomolecular Screening 2014, 19 (3), 417–426. [DOI] [PubMed] [Google Scholar]
- (49).MacRae IJ; Zhou KH; Li F; Repic A; Brooks AN; Cande WZ; Adams PD; Doudna JA, Structural basis for double-stranded RNA processing by dicer. Science 2006, 311 (5758), 195–198. [DOI] [PubMed] [Google Scholar]
- (50).Parkes KEB; Ermert P; Fassler J; Ives J; Martin JA; Merrett JH; Obrecht D; Williams G; Klumpp K, Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem 2003, 46 (7), 1153–1164. [DOI] [PubMed] [Google Scholar]
- (51).Billamboz M; Bailly F; Barreca ML; De Luca L; Mouscadet JF; Calmels C; Andreola ML; Witvrouw M; Christ F; Debyser Z; Cotelle P, Design, Synthesis, and Biological Evaluation of a Series of 2-Hydroxyisoquinoline-1,3(2H,4H)-diones as Dual Inhibitors of Human Immunodeficiency Virus Type 1 Integrase and the Reverse Transcriptase RNase H Domain. J. Med. Chem 2008, 51 (24), 7717–7730. [DOI] [PubMed] [Google Scholar]
- (52).Tang J; Vernekar SKV; Chen Y-L; Miller L; Huber AD; Myshakina N; Sarafianos SG; Parniak MA; Wang Z, Synthesis, biological evaluation and molecular modeling of 2- Hydroxyisoquinoline-1,3-dione analogues as inhibitors of HIV reverse transcriptase associated ribonuclease H and polymerase. European Journal of Medicinal Chemistry 2017, 133, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Schmittgen TD; Lee EJ; Jiang J; Sarkar A; Yang L; Elton TS; Chen C, Real-time PCR quantification of precursor and mature microRNA. Methods 2008, 44 (1), 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Livak KJ; Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25 (4), 402–408. [DOI] [PubMed] [Google Scholar]