Abstract
Fluorescent indicator displacement (FID) assays are an advantageous approach to convert receptors into optical sensors that can detect binding of various ligands. In particular, the identification of ligands that bind to RNA receptors has become of increasing interest as the roles of RNA in cellular processes and disease pathogenesis continue to be discovered. Small molecules have been validated as tools to elucidate unknown RNA functions, underscoring the critical need to rapidly identify and quantitatively characterize RNA:small molecule interactions for the development of chemical probes. The successful application of FID assays to evaluate interactions between diverse RNA receptors and small molecules has been facilitated by the characterization of distinct fluorescent indicators that reversibly bind RNA and modulate the fluorescence signal. The utility of RNA-based FID assays to both academia and industry has been demonstrated through numerous uses in high-throughput screening efforts, structure-activity relationship studies, and in vitro target engagement studies. Furthermore, the development, optimization, and validation of a variety of RNA-based FID assays has led to general guidelines that can be utilized for facile implementation of the method with new or underexplored RNA receptors. Altogether, the use of RNA-based FID assays as a general analysis tool has provided valuable insights into small molecule affinity and selectivity, furthering the fundamental understanding of RNA:small molecule recognition. In this review, we will summarize efforts to employ FID assays using RNA receptors and describe the significant contributions of the method towards the development of chemical probes to reveal unknown RNA functions.
1. Introduction
Ligand-receptor interactions play a crucial role in the regulation of healthy and disease-related processes and can be harnessed to study and/or target these processes. A fundamental understanding of ligand-receptor interactions is thus important for many areas of basic research and drug discovery; however, specific interactions can be difficult to study in a complex cellular environment. These challenges are particularly relevant for RNA receptors, despite increased interest in small molecule:RNA targeting. One method that has facilitated the detection of ligand-receptor interactions in vitro is the indicator displacement assay (IDA) [1]. In an IDA, a receptor is first bound by an indicator whose optical properties are modulated upon reversible and non-covalent binding (Fig. 1). Then, addition of the ligand, defined as a competitive analyte, can result in displacement of the indicator from the receptor and modulate the optical signal. Pioneered by the Isagawa laboratory [2] and later popularized by the Anslyn laboratory [1, 3], IDAs were first employed as a technique to convert synthetic receptors into optical chemosensors to detect binding and quantify levels of biological ions. The application of IDAs has now been extended to study the recognition of diverse analytes, including small molecules [4–8], anions [9], cations [10], metals [11], metabolite analogues [12], and amino acids [13], using a wide range of synthetic as well as biologically-relevant receptors. Furthermore, IDAs offer a major advantage as a signaling method by avoiding covalent attachment between the indicator and the receptor, which allows for multiple indicators to be potentially used with the same receptor and makes the assay easily adaptable to different receptors and platforms. Fluorescent indicator displacement (FID) assays, which utilize an indicator whose fluorescence properties are modulated upon binding to a receptor, offer additional advantages. FID assays allow for sensitive and quantitative detection, are relatively inexpensive to perform due to convenient availability of a fluorimeter or plate reader, and can be easily formatted to a high-throughput screening (HTS) platform.
Fig. 1.
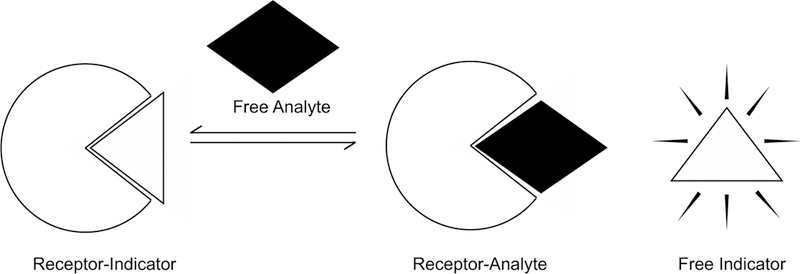
Schematic of an indicator displacement assay. In this example, optical signal intensity is low when the indicator is bound to the receptor and high when the indicator is free in solution.
In this review, we will focus on FID assays that employ RNA as a receptor to detect the binding of small molecule analytes (Fig. 2). RNA, particularly non-coding (ncRNAs), are a newly discovered class of biomolecules implicated in important biological processes such as gene expression and cellular differentiation [14]. The observed dysregulation of ncRNAs in numerous diseases, including cancers, neurodegenerative disorders, and viral infections, has sparked interest within the scientific community to therapeutically target ncRNAs and elucidate their roles in disease states [15]. Small molecules have proven to be a viable avenue to study disease-driving ncRNAs and are advantageous as chemical probes given their tunability and favorable pharmacological properties [16]. Promisingly, the use of FID assays either as a primary high-throughput screen or as a complementary method to validate target engagement in vitro has already greatly aided the development of RNA bioactive chemical probes [17–20].
Fig. 2.
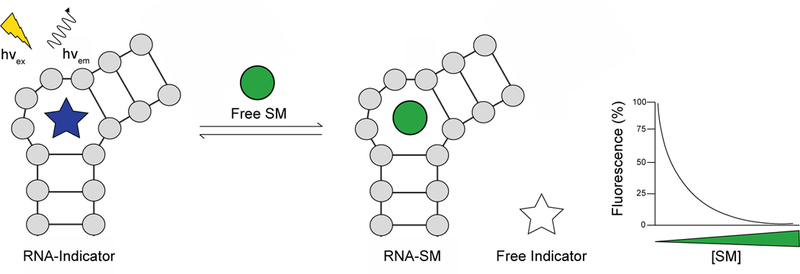
Fluorescent indicator displacement assay to detect RNA:small molecule interactions. In this example, fluorescence is observed when the indicator is bound to the RNA receptor. As the indicator is increasingly displaced by a small molecule (SM) analyte, the fluorescence decreases. Conversely, displacement of an indicator whose fluorescence is quenched when bound to the RNA receptor will result in an increase in fluorescence.
FID assays offer a promising complementary approach to current methods used to detect RNA:small molecule interactions, which have been reviewed previously [21–23]. In comparison to FID assays, other widespread fluorescence-based methods such as Förster resonance energy transfer (FRET), fluorescence polarization (FP), and site-specific labeling typically require incorporation of a fluorescent handle onto the RNA and/or small molecule. FID assays also avoid immobilization of the RNA and/or small molecule, which is necessary when utilizing techniques such as surface plasmon resonance (SPR), second harmonic generation, and small molecule microarrays. Furthermore, FID assays are high-throughput and require small quantities of material in comparison to other traditional methods, including thermal denaturation, isothermal titration calorimetry (ITC), and nuclear magnetic resonance (NMR) spectroscopy. While FID assays allow for native RNA structures and diverse small molecules to be rapidly evaluated in solution, obtainable data is limited to binding constants whereas other techniques enable measurement of stoichiometry, thermodynamic binding parameters, and/or kinetic parameters. Additionally, attention must be given when setting up and interpreting results of an FID assay to ensure a high quality and reliable screening method.
Herein we will highlight numerous examples, through the end of 2018, where implementation of RNA-centric FID assays aided the discovery and furthered understanding of small molecule interactions with RNA. In the following section (Section 2), we aim to survey the indicators that have been successfully studied with biologically relevant RNA receptors. We will introduce both synthetic and commercially available indicators, summarizing the known fluorescence and RNA-binding properties of each indicator and the validation of their use through displacement by diverse small molecule analytes. Additionally, we will draw attention to indicators whose fluorescence and/or RNA-binding properties have been tuned by either synthetic derivatization or through labeling of non-fluorescent ligands, thereby expanding the toolbox of fluorescent probes. In Section 3, we exemplify the utility of FID assays to rapidly detect small molecule interactions with RNA and characterize affinity as well as selectivity of RNA-binding small molecules. We will highlight the extensive use of FID assays in academic and industrial laboratories as primary screens for HTS campaigns and as a complementary approach for small molecule structure-activity relationship (SAR) and target validation studies. Lastly, in Section 4, we aim to accelerate the implementation of RNA-based FID assays by providing a resource to guide the selection of indicators, optimization of assay parameters, and interpretation of displacement data. Altogether, we hope to illustrate the versatility of FID assays to evaluate diverse small molecules and RNAs as well as to highlight the valuable contributions of the method towards fundamental understanding of RNA:small molecule recognition and identifying small molecule probes to study the role of RNA in biological processes and disease.
2. Indicators and RNA Receptors
A wide variety of synthetic and commercially available RNA-binding ligands with varying fluorescence properties and affinities have been utilized as indicators in FID assays. In addition, labeling of non-fluorescent small molecules or peptides with a fluorophore has expanded the repertoire of RNA-binding indicators. The fluorescence properties of indicators can be fine-tuned by derivatization, and, in general, emission intensity is measured through standard fluorescence, FP, and/or FRET using a plate reader or fluorimeter. Furthermore, dual functionality has been observed for several indicators that also inhibit RNA-associated processes in vitro [24]. In this section, we will review the fluorescence and RNA-binding properties of both small molecule-and peptide-based fluorescent indicators (Table 1) as well as relevant applications. We caution the reader in direct comparisons of reported binding affinities, such as those between indicators and RNA receptors, as measurements are highly dependent on the selected buffer conditions [25].
Table 1.
Summary of RNA-Binding Indicators Used in FID Assays.
| Indicator | Excitation (nm) | Emission (nm) | Fluorescence Upon Displacement | RNA Receptor(s) | Reference(s) |
|---|---|---|---|---|---|
| TO | 512 | 533 | turn-OFF | Stem, pre-miR-23b, aptamers (tobramycin, theophylline, GTP, ATP, cAMP, and in vitro selected) | [26–29] |
| TO-PRO-1 | 515 | 531 | turn-OFF | A-site (E. coli), HIV-1 TAR, helix 31 of 16S rRNA, helix 69 of 23S rRNA (E. coli), r(GGGGCC)8, TSL2, 6-nt hairpins, 3×2-nt internal loops, 4×4-nt internal loops, stems | [6, 18, 19, 30–32] |
| SYBR Green I | 497 | 520 | turn-OFF | Theophylline aptamer | [28] |
| SYBR Green II | 497 | 520 | turn-OFF | 6-nt hairpins | [31] |
| X2S | 370 | 450 | turn-ON | RRE-IIB, dsRNA, 1-nt bulges, 6-nt hairpins | [7, 31, 33] |
| X2SS | 424 | 496 | turn-ON | RRE-IIB, dsRNA, pre-miRNAs (21–1, 29a, 30d-1, 92a-1, 122–1, 132–1, 132–2, 326–2, 433–1, 1297–2) | [33, 34] |
| X2SdiMe | 370 | 451 | turn-ON | RRE-IIB, dsRNA, pre-miRNAs (29a, 122) | [33, 35] |
| PYT | 340 | 400 | turn-ON | RRE-IIB, in vitro selected aptamers | [36, 37] |
| F-neo | 490 | 517 | turn-ON | A-site (E. coli), ES7 (C. albicans), pre-miRNA 504, mature miRNAs (7a-5p, 9–5p, 10b-5p, 15a-5p, 19a-5p, 19b-2–5p, 22–5p, 29a-3p, 31–5p, 34a-5p, 126–3p, 130a-5p, 132–5p, 135–5p, 142–5p, 155–5p, 221–5p, 335–5p, 380–5p, 504) | [8, 38, 39] |
| CRT | 553 | 577 | - | In vitro selected aptamers | [40] |
| CRP | 553 | 577 | - | 16S rRNA decoding region | [36] |
| FMN | 455 | 525 | turn-ON | FMN riboswitch (E. coli) | [17] |
| 2-aminopurine | 300 | 370 | turn-ON | Adenine riboswitch (B. subtilis), C74U guanine riboswitch (B. subtilis) | [41, 42] |
| SAM-C8-Cy5 | 649 | 666 | - | SAM-I riboswitches (B. subtilis, P. irgensii, T. tengcongensis) | [12] |
| Ethidium Bromide | 520 | 600 | turn-OFF | dsRNA, bulge (1-, 2-, and 3-nt), RRE, HIV-1 TAR | [43–46] |
| Triazole Orange | 501 | 530 | turn-OFF | Telomeric RNA G-quadruplex (H. sapiens) | [47] |
| ATMND-C2-NH2 | 357 | 406 | turn-ON | A-site (E. coli) | [48] |
| Proflavine | 445 | 505 | - | RRE-IIB | [49, 50] |
| ICR 191 | 415 | 490 | turn-ON | RRE-IIB, HIV-1 TAR | [51, 52] |
| 5-FAM-Tat(49–57)-TAMRA | 500 | 577 | turn-OFF | HIV-1 TAR, HIV-2 TAR, A-site (E. coli), RRE-IIB | [53-55] |
| Rev(34–50)-Fl | 490 | 525 | - | RRE constructs (28-, 47-, and 67-nt) | [36, 56, 57] |
- = indicator monitored through fluorescence polarization
2.1. Small Molecule Indicators
2.1.1. Cyanine Dyes
Cyanine dyes consist of two nitrogen-containing heterocyclic rings linked by a polymethine bridge (Fig. 3) and have been used widely to stain cellular organelles, nucleic acids, and other biological macromolecules [58]. In aqueous solutions, free cyanine dye molecules exhibit little to no fluorescence as a result of torsional motion in the polymethine bridge, leading to non-radiative decay of the excited state. Upon binding nucleic acids, torsional motion of the conjugated dye is restricted and decay of the excited state leads to a 100-to 1,000-fold enhancement in fluorescence [59]. The spectroscopic properties of cyanine dyes can be tuned by varying the length of the polymethine chain, altering the identity of the heteroatoms, and adding substituents to either heterocycle. Numerous cyanine dye derivatives have been synthesized and characterized as well as commercialized, providing fluorescent probes that can be monitored from the visible to near-infrared spectral regions [59]. Aside from cellular staining, several cyanine dyes have been evaluated against biologically-relevant RNAs and are commonly utilized as competitive indicators in FID assays [6, 18, 19, 26–32].
Fig. 3.
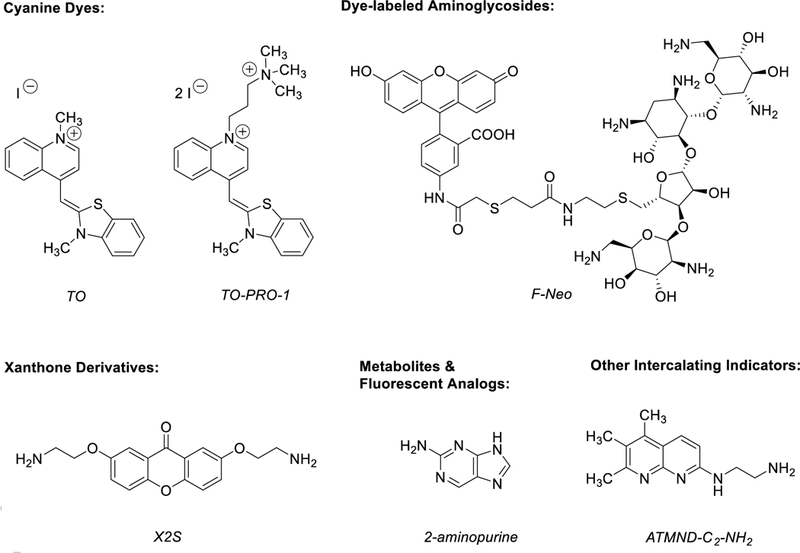
Representative structures of RNA-binding small molecule indicators.
Thiazole orange (TO) is a commercially available cyanine dye first used as an RNA stain for reticulocyte analysis [60] that has since been assessed as an indicator with diverse DNA structures [4, 5] and RNA stem-loop structures, particularly aptamers [26–29]. TO consists of an N-methylquinolinium linked by a single methine to a 3-methylbenzothiazoline and has excitation and emission maximums beyond 500 nm, which is advantageous to limit interference with intrinsic fluorescence of small molecules [61]. Studies undertaken by the Stojanovic and Datta laboratories have demonstrated the promising use of TO as an indicator to detect selective ligand interactions with several RNA aptamers, including tobramycin, guanosine triphosphate (GTP), adenosine triphosphate (ATP), 3’−5’ cyclic adenosine monophosphate, and theophylline aptamers, binding the structures with moderate micromolar affinity (Kd = > 10 µM) [27, 28]. While TO is suspected to bind multiple sites of several RNA aptamers, a significant decrease in TO fluorescence (40–90%) was still observed upon addition of the native ligand [27, 28]. Importantly, RNA aptamers bound by TO were shown to preserve selectivity for the native ligand as little to no TO displacement was observed in the presence of structurally similar negative controls [27–29]. These studies highlight the potential use of TO to characterize the selectivity of RNA aptamers and the chemical space of aptamer-binding ligands.
TO-PRO-1 is a commercially available dye that has been utilized as a general indicator to rapidly screen small molecules against multiple RNAs, allowing for selectivity trends of ligands to be readily assessed. TO-PRO-1 is structurally similar to TO, differing only in charge as a result of a quaternary ammonium substituent on the quinolone ring. Maximum excitation and emission wavelengths of TO-PRO-1 are similar to TO; however, the binding affinity of TO-PRO-1 to double-stranded (ds) DNA has been observed to be 10-fold higher, likely due to increased electrostatic interactions [62]. TO-PRO-1 was first utilized in an FID assay by Asare-Okai and Chow, who characterized and validated the indicator displacement assay using constructs of two well-studied RNAs, namely, bacterial ribosomal A-site and human immunodeficiency virus type 1 (HIV-1) Transactivation Response (TAR) element [6]. Using electrospray ionization mass spectrometry (ESI-MS), displacement of TO-PRO-1 from each RNA by a known ligand was confirmed and correlated with the reduction in the emission intensity of the indicator. TO-PRO-1 was observed to bind A-site RNA with 1:1 stoichiometry and based on the known binding site of each positive control ligand, was thought to bind both RNAs near or adjacent to secondary structures. Furthermore, the authors applied the TO-PRO-1 FID assay to screen aminoglycosides and peptides against two additional bacterial RNA targets, helix 31 of the 16S ribosomal RNA (rRNA) and helix 69 of the 23S rRNA. Other notable examples of TO-PRO-1 applications are from the Disney laboratory where the indicator has been utilized in FID assays as a primary screen to identify RNA-binding compounds that selectively interact with diverse secondary structures and sequences, including stems [32], r(GGGGCC) expansions [18], and randomized RNA libraries comprised of 6-nucleotide (nt) hairpin loops, 3×2-nt symmetrical internal loops, or 4×4-nt asymmetrical internal loops [31]. Notably, TO-PRO-1 binds RNAs that contain unpaired regions with higher affinity (~0.5 µM) as compared to stems (~5 µM) [31, 32]. Furthermore, TO-PRO-1 has been used by Garcia-Lopez, Scapozza, and coworkers to detect selective small molecule interactions with splicing modifiers such as the cis-regulatory RNA element TSL2, a stem-loop RNA structure that overlaps with the 5’ splice site of exon 7 (E7) in the copy of the survival motor neuron 1 gene (SMN2) and regulates E7 inclusion in alternatively spliced transcripts [19]. The TO-PRO-1 FID assay served as a primary screen to evaluate an RNA-focused small molecule library against TSL2 and nucleic acid controls, aiding in the identification of the first bioactive TSL2 structure-stabilizing ligands that promote E7 inclusion and revert spinal muscular atrophy (SMA) molecular phenotypes. Altogether, the examples highlighted demonstrate the versatility of TO-PRO-1 to quickly screen small molecules against diverse and/or underexplored RNAs such as splice sites and identify lead compounds and potential therapeutics.
Both TO and TO-PRO-1 have been used to label non-fluorescent small molecules to generate sequence specific and highly competitive indicators, respectively [29, 30]. For example, the Stojanovic laboratory linked TO to both guanosine monophosphate (GMP) and adenosine monophosphate (AMP) as a strategy to improve the specificity of TO and provide a fluorescent probe that could be used to selectively monitor interactions with corresponding RNA aptamers [29]. Upon binding the respective aptamers, the fluorescence of both TO conjugates increased by more than 100-fold. Both TO conjugates were selective for their corresponding aptamers as the emission intensity was lower or negligible in the presence of other aptamers, mutants, or unrelated nucleic acids. The apparent binding affinity of TO-GMP to GTP aptamers was comparable to that of GMP, while positive cooperativity was observed for TO-AMP binding to ATP aptamers as the affinity increased by several-fold compared to AMP. Titration of the native ligands led to almost complete displacement of each TO conjugate and the selectivity of the aptamer was conserved as other nucleotides were unable to displace TO-AMP or TO-GMP. Similarly, the Disney laboratory linked TO-PRO-1 to an ellipticine derivative (1a) known to bind to the hairpin structure of r(GGGGCC) expansions (Kd = 9.7 µM) and inhibit repeat-associated non-ATG (RAN) translation (IC50 = 11 µM), preventing the expression of toxic proteins associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [18, 30]. The indicator (1a-TOQ) exhibited a 3-fold increase in emission intensity upon binding r(GGGGCC)8 (Kd = 110 nM) and afforded a dose-dependent decrease in fluorescence upon titration of 1a against the r(GGGGCC)8:1a-TOQ complex. To aid the identification of potent inhibitors of ALS/FTD-associated RAN translation, 1a-TOQ was utilized in an FID assay to screen a focused library of 40 compounds that were chemically similar to 1a. Several compounds were observed to decrease 1a-TOQ fluorescence by more than 50% and trends in the chemical features that enhanced recognition of r(GGGGCC)8 were revealed. Ultimately, a single compound (4) was found to be a more potent inhibitor of ALS/FTD-associated RAN translation (IC50 = 1.6 µM) and to bind cellular r(GGGGCC) expansions as confirmed by direct imaging through linking of 4 to TO-PRO-1. Overall, the ability to tune the affinity and/or selectivity of TO and TO-PRO-1 by incorporation of non-fluorescent RNA-binding ligands leverages the utility of indicators to monitor interactions with specific RNAs and to identify highly competitive ligands.
There are several cyanine dyes that have yet to be fully characterized and evaluated as RNA-binding indicators for FID assays. For example, YOYO-1 is a dimeric cyanine dye that consists of two molecules of YO-PRO-1, which is structurally similar to TO-PRO-1 except the indicator contains a benzoxazole ring in place of the benzothiazole ring [62]. YOYO-1 and other commercially available dimeric dyes such as TOTO-1 (dimer of TO) bind nucleic acids with higher affinity compared to their monomer counterparts [59], providing fluorescent probes to identify high affinity RNA:small molecule interactions. Additional monomeric cyanine dyes, including SYBR Green I and II, are commercially available and preliminarily have been evaluated against the theophylline RNA aptamer and a randomized 6-nt hairpin library, respectively [28, 31]. The diverse collection of cyanine dyes that can be readily accessed commercially or fine-tuned synthetically highlights the feasibility to characterize and incorporate these dyes as indicators for a given RNA target.
2.1.2. Xanthone Derivatives
Xanthone is a fused tricyclic scaffold comprised of two benzene rings separated by a 4-tetrahydropyranone (Fig. 3) that is found in a variety of biologically active compounds, including antineoplastics, antimalarials, and antimicrobials [63]. Initial photophysical and spectroscopic studies of xanthone with DNA revealed photoreactivity between the excited state of xanthone and DNA that could be diminished by the addition of electron-donating amino groups to the scaffold [64]. The RNA-binding properties of amino-substituted xanthone derivatives have been extensively studied by the Nakatani laboratory, who used these synthetic analogues as indicators to evaluate small molecules against therapeutically relevant RNAs [7, 24, 33–35]. The fundamental studies, several of which are highlighted below, have provided valuable insights towards the utility of amino-substituted xanthones to develop high quality FID assays and in general, RNA-binding chemical probes.
The Nakatani laboratory first utilized a novel 2,7-Bis(2-aminoethoxy)xanthen-9-one derivative (X2S) as a fluorescent indicator and monitored binding of the indicator to different RNAs, including dsRNA, 1-nt bulges, and the IIB domain of HIV-1 Rev Response element (RRE-IIB) [7]. When bound to RNA, the emission of X2S fluorescence was diminished but recovered upon displacement of the indicator from the RNA in aqueous solutions. Quenching of X2S fluorescence was observed to be more efficient with 1-nt bulges than dsRNA, suggesting that the indicator binds favorably to unpaired regions of RNA. Additionally, binding of X2S was characterized with a truncated sequence of RRE-IIB that retains the critical binding site of Rev peptide necessary for HIV-1 replication [65]. The RRE-IIB construct consisted of a 4-nt hairpin and multiple loops, including a 2×3-nt internal loop and a single adenine bulge. At a 1:2 ratio of RRE-IIB to X2S, the fluorescence of the indicator was ~90% quenched when compared to X2S alone. Measurable fluorescence quenching was also observed with control RNAs that either lacked the hairpin or stem-loops, respectively, suggesting that X2S binds to RRE-IIB at multiple sites. Notably, the authors highlighted that titration of a truncated Rev peptide known to bind near the purine-rich internal loop of RRE-IIB against a preformed X2S:RRE-IIB complex resulted in a rapid increase in fluorescence, suggesting that Rev binds near an X2S-occupied binding site [66]. In comparison, titration of neomycin B led to a slow but steady increase in fluorescence intensity, consistent with binding at a distal site as characterized in prior work by NMR spectroscopy [67]. To validate the X2S FID assay, the LOPAC1280 library was screened to identify small molecules that bind to RRE-IIB [7]. A portion of the library (59/1280 compounds) was removed prior to screening due to fluorescence overlap between the small molecules and X2S. Similar interference was also noted by Tran and Disney when monitoring X2S emission upon incubation with small molecules alone or in conjunction with a randomized 6-nt hairpin RNA library [31], highlighting a potential limitation of the indicator.
To limit interference with small molecule fluorescence, the Nakatani laboratory synthesized and characterized a series of xanthone and thioxanthone derivatives that emit at longer wavelengths compared to X2S [33]. Modifications to X2S included i) increasing the length of the alkyl chain of the aminoethoxy moieties, ii) adding methyl substituents to either the alkyl chain or nitrogen atoms, and iii) replacing the xanthone core with thioxanthone. While slight red-shifts in maximum absorption (370–377 nm) and emission (450–464 nm) wavelengths were observed for xanthone derivatives with increased alkyl chain length and addition of methyl substituents, considerable red-shifts were observed for thioxanthone (X2SS) and its N,N-dimethyl derivative (λEx/λEm = 424/495 nm). A secondary objective of the SAR study was to improve the selectivity of the indicator for internal loops over stems to prevent non-specific binding of the indicator and to potentially allow for site-specific displacement to be monitored. Dissociation constants of X2S and thioxanthone derivatives were determined using an alternative binding model that assumes independent, stoichiometric, and simultaneous binding of the indicator to both the stem and loop regions of RRE-IIB. The ratio of the two dissociation constants (Kd(stem)/Kd(loop)) yielded a selectivity index that described the binding preference of the indicator for either stem or loop structures. X2S derivatives with a propyl chain, butyl chain, or N,N-dimethyl substitution were observed to be largely selective for loops. Only the N,N-dimethyl derivative (X2SdiMe) had a weaker binding affinity for the stem (Kd = 4.3 µM) in comparison to X2S (Kd = 1.7 µM), and it retained about one third affinity of X2S for the loop (Kd = 170 nM). For thioxanthone derivatives, titration data did not fit well with the model and suggested multiple binding events of the indicator to the loop region. Overall, the authors reasoned that the red-shifted emission wavelength of the thioxanthone derivative (X2SS) and the reduced stem affinity of the N,N-dimethyl xanthone derivative (X2SdiMe) make the indicators favorable for use in displacement assays. Indeed, the Nakatani laboratory has demonstrated the utility of these fine-tuned indicators to screen small molecules against pre-micro RNAs (pre-miRNAs) and revealed a dual application of X2SS as an inhibitor of Dicer-mediated processing [24, 34, 35].
2.1.3. Metabolites and Fluorescent Analogs
Metabolites are essential small molecules that are required for cellular processes and can be sensed by regulatory RNA elements such as riboswitches [68]. Riboswitches are typically located at the 5’ untranslated region of bacterial mRNAs and regulate gene expression via metabolite-induced pathways [69]. Upon binding of a cognate metabolite, a riboswitch and its expression platform undergo a conformational change, resulting in modulation of protein synthesis [70]. Inhibition of metabolite binding by competitive small molecules has been validated as a therapeutic strategy to halt gene expression in and prevent growth of bacterial pathogens [17, 71, 72].
Several riboswitch-sensing metabolites are intrinsically fluorescent, providing indicators that allow for direct competition between the native ligand and the small molecule ligand to be monitored. For example, flavin mononucleotide (FMN) is a fluorescent metabolite involved in the riboflavin biosynthesis pathway that binds its native riboswitch with high affinity (Kd = 1.2 nM) [17, 73]. The fluorescence of FMN is quenched upon binding the riboswitch and was utilized by Howe, Roemer, and coworkers to validate the RNA target of a lead small molecule identified in a bacterial phenotypic screen. Displacement of FMN by ribocil-B, one of the most selective RNA bioactives to date, demonstrated competitive and direct ligand binding to the riboswitch and highlighted the utility of RNA-centric FID assays for industrial use in target validation studies [17].
Some riboswitches have been found to accommodate and respond to chemically similar metabolites and/or analogs [74], allowing for the design of fluorescent derivatives. For example, the metabolite adenine and its fluorescent structural isomer, 2-aminopurine (2-AP) (Fig. 3), have been shown to bind with similar affinity to both the adenine riboswitch and a mutant guanine riboswitch (GRA) that has a C74U point mutation (0.47 and 0.25 µM, respectively) [41, 75]. 2-AP strongly emits at 370 nm and can be selectively monitored in the presence of naturally occurring nucleobases [76]. Similar to FMN, the fluorescence of 2-AP is quenched upon binding to RNA via base-stacking interactions. Utilizing 2-AP, Daldrop, Brenk, and coworkers developed an FID assay to experimentally characterize the binding of select small molecules that were virtually screened for binding to the GRA riboswitch [42]. The authors highlighted that the implementation of an FID assay greatly reduced the amount of RNA needed (~99.5%) in comparison to ITC, a commonly used method to assess small molecule binding to riboswitches. Furthermore, 2-AP has been incorporated into other metabolites such as S-Adenosyl-L-methionine (SAM) to provide a fluorescent handle [77]. However, because the 2-AP SAM analog lacks the 6-amino group known to be important for SAM-I riboswitch binding affinity, Hickey and Hammond pursued a SAM analog functionalized at the 3’-hydroxyl of the ribose moiety with a Cy5-labeled linker and developed a competitive FP assay that could be applied in a high-throughput format to screen small molecules against multiple species of SAM-I riboswitches [12]. While riboswitches are particularly well suited for the development of fluorescent analogs, the resulting indicators are likely limited for use with a single riboswitch or class.
2.1.4. Other Intercalating Indicators
Several aromatic-enriched small molecules whose fluorescence is modulated upon binding RNA predominantly through pi-pi stacking interactions have been utilized in FID assays. For example, ethidium bromide (EtBr) is a non-specific ligand known to intercalate into RNA at multiple sites and has been used in FID assays with multiple RNA bulges, a 66-nt construct of RRE (RRE66), and HIV-1 TAR [43, 44, 46, 78]. Furthermore, ICR 191, a commonly used acridine mutagen, has been validated as an indicator with RRE-IIB as emission intensity is recovered upon displacement by Rev peptide or known RRE-binding ligands [51]. Similarly, proflavine is an acridine derivative known to bind RRE with high affinity and 2:1 stoichiometry, competitively inhibiting interactions with Rev peptide [49]. To identify additional competitive small molecules, proflavine was utilized to develop an FID assay that monitors FP at the emission wavelength of the indicator in the presence of an RRE construct that is immobilized to graphene oxide as a means to maximize the change in molecular weight, and thus significantly decrease the FP signal upon displacement of proflavine [50]. Next, triazole orange (TriO) contains a naphthalene core and was first identified to bind a DNA G-quadruplex structure in the promoter region of the alpha subunits of the hypoxia-inducible factor-1 transcription factor and selectively inhibit the growth of cancerous renal and osteosarcoma cell lines [79]. In addition, TriO has been applied as an indicator to characterize the binding of the anticancer agent chelerythrine to biologically relevant RNA G-quadruplexes [47]. Recently, Sato, Nishizawa, and coworkers disclosed an aminoethyl substituted naphthyridine derivative (ATMND-C2-NH2) (Fig. 3) that bound bacterial A-site RNA (E. coli) with higher affinity (0.44 µM) than any other reported non-aminoglycoside ligand [48]. When bound to A-site RNA, the fluorescence of ATMND-C2-NH2 is quenched with higher efficiency (> 95%) compared to other fluorescent indicators that have been used with A-site RNA, particularly fluorophore-labeled aminoglycosides (~65% quenching efficiency) [8, 80]. The authors demonstrated the utility of ATMND-C2-NH2 as an indicator and validated its use by screening several known ribosome-targeting antibiotics. Lastly, bisbenzimidazoles such as Hoechst dyes have preliminarily been evaluated as indicators with DNA aptamers [28] and are known RNA-binding ligands [81, 82], providing a potential new class of intercalating-based indicators to be explored in FID assays against RNA.
2.1.5. Dye-labeled Aminoglycosides
Aminoglycosides are polycationic polysaccharides comprised of a 2-deoxystreptamine core substituted with 1,3-diamino functionality and three to four hydroxyl groups that can provide a linkage to aminosugars (Fig. 3) [83]. Aminoglycosides are known to bind RNA, particularly at the A-site of the bacterial ribosome, and function as bactericidal agents towards both Gram-positive and Gram-negative strains [84–86]. Furthermore, aminoglycosides have been shown to bind many other functional RNAs via electrostatic and hydrogen bonding interactions between the amino and hydroxyl functionalities of aminoglycosides and the bases and/or backbone of RNA [36, 87–90]. The capability of aminoglycosides to interact with a number of diverse RNAs highlights the potential of these molecules to serve as general indicators for competition-based studies. While aminoglycosides are non-fluorescent, the synthetic incorporation of a dye moiety has allowed RNA:aminoglycoside complexes to be detected and monitored upon addition of a competitive small molecule [8, 36, 37, 39, 40].
Pyrene-labeled fluorescent derivatives of tobramycin (PYT) have been synthesized and utilized by the Rando laboratory to quantitatively measure RNA:aminoglycoside interactions [36, 37]. Pyrene was selected as the fluorescent moiety because the dye is known to interact with RNA and its emission intensity is quenched upon binding. The dye moiety was preferentially incorporated at the 6’-amino group of tobramycin and observed to enhance the binding affinity of the aminoglycoside to an RNA aptamer selected from a diverse pool by 200-fold [37]. Additionally, PYT analogs have been evaluated with RRE-IIB, yielding nanomolar dissociation constants that are ~10–20-fold higher compared to unlabeled tombramycin [36]. Using gentamicin C and neomycin B as positive controls, displacement of PYT was observed, while no significant change in fluorescence was observed with the negative controls glucosamine and erthyromicin, validating the use of PYT as an indicator [37]. However, because the pyrene moiety of PYT itself can bind RNA, the indicator has been observed to bind non-specifically, which can limit its utility. Incomplete recovery of fluorescence intensity as a result of multiple binding events can hamper affinity and stoichiometry measurements for competing small molecules as the contribution from each binding site to the change in fluorescence quenching cannot be determined.
To mitigate non-specific binding, fluorescent tags whose signal is not dependent on interactions with RNA bases can be incorporated into aminoglycosides. For example, the Arya laboratory has utilized a fluorescein-labeled derivative of neomycin (F-neo) for HTS of small molecules against a truncated, 27-nt construct of A-site RNA (E. coli) [8]. Fluorescein was incorporated via a thiourea linkage at the C5 of the ribose ring of neomycin, a position that is not predicted to be involved in binding. The fluorescence intensity of F-neo is dependent on the protonation state of fluorescein where the dicarboxylate anion absorbs strongly at 485 nm and the monocarboxylate anion absorbs weakly, leading to different emission intensities. In solution (pH = 7.0), fluorescein is a dianionic species and emits strongly at 517 nm. Upon F-neo binding to RNA, the dianionic species is destabilized by the negative electrostatic potential of the RNA backbone, raising the pKa of fluorescein and shifting the equilibrium towards the non-fluorescent monoanionic species. Molecular modeling studies with the A-site RNA construct suggested that F-neo binds the major groove with the fluorescein moiety positioned adjacent to the phosphates on the edge of the groove. Scatchard analysis of an A-site titration with F-neo revealed that the indicator binds at a single site with a dissociation constant of 43 nM, consistent with other literature reports for unlabeled neomycin. To validate F-neo as a reliable indicator, aminoglycosides known to bind the A-site construct were screened, and the determined relative binding affinities were observed to be similar to those obtained by other methods. The Arya laboratory has since extended the application of F-neo to rapidly screen peptidic-aminosugars against multiple pre-and mature microRNAs, demonstrating the versatility of the indicator with small RNAs [39]. Furthermore, F-neo has also been used to explore the therapeutic potential of targeting eukaryotic ribosomes such as expansion segment 7 (ES7, 209 nts) of the pathogenic fungi Candida albicans [38]. Interestingly, F-neo was found to bind C. albicans ES7 with an estimated 13:1 stoichiometry, nanomolar affinity (Kd = 2.5 nM), and high selectivity (~2,000-fold) compared to human ES7. Screening of a peptidic-aminosugar library (215 compounds) using F-neo resulted in the identification of ~60 compounds that bound ES7 with higher affinity than neomycin, highlighting the successful application of FID assays to study ligand binding to RNAs of larger size.
Aside from measuring fluorescence quenching, the binding of fluorescently-labeled aminoglycosides to RNA has also been monitored by FP [8, 36, 40]. Labeling of tobramycin with 5-carboxytetramethylrhodamine, a farther red-shifted dye compared to fluorescein, provided an indicator (CRT) that bound an in vitro selected RNA aptamer with 1:1 stoichiometry and ~10-fold higher affinity (Kd = 12.3 nM) compared to its fluorescein derivative [40]. However, CRT was found to bind more weakly than unlabeled tobramycin (Kd = 0.77 nM), highlighting a potential drawback of incorporating fluorophores as the dye molecule may interfere with native interactions. In comparison, a 5-carboxytetramethylrhodamine-labeled derivative of paromomycin (CRP) was observed to bind a construct of the 16S rRNA decoding region with ~10-fold higher affinity (Kd = 165 nM) than the unlabeled ligand, demonstrating the importance of evaluating the impact of dye incorporation for each small molecule with every RNA [36].
2.2. Dye-Labeled Peptide Indicators
2.2.1. Tat
Tat is a transcriptional activator protein encoded by HIV-1 [91]. The 86-residue protein activates transcription by binding to the bulge and upper stem region of HIV-1 TAR, leading to increased stabilization and processivity of RNA polymerases [92]. Tat activity is essential for viral replication and therefore, inhibition of the TAR:Tat interaction has been widely viewed as a therapeutic strategy. Tat recognition of TAR has been characterized through the evaluation of peptide fragments, which revealed that a basic arginine-rich subdomain of Tat is critical for binding [93]. Peptide fragments that contain the conserved, positively charged stretch of basic residues have been found to bind multiple RNA bulges in vitro, including the 2-nt bulge motif within the TAR element of human immunodeficiency virus type 2 (HIV-2) [94, 95]. Furthermore, labeling of Tat peptide with a fluorophore(s) has provided a general fluorescence-based indicator that has been utilized to detect small molecule interactions with RNA bulges and related structures [53–55, 96, 97].
For example, Masumoto, Hamasaki, and coworkers employed a fluorescent Tat-derived peptide as an indicator for HTS of small molecules against HIV-1 TAR [53]. Utilizing intramolecular FRET, the 16-residue peptide was labeled with 5-carboxyfluorescein (5-FAM) and tetramethylrhodamine (TAMRA) at the N-terminus and C-terminus, respectively (Fig. 4). Binding of Tat peptide to HIV-1 TAR facilitated FRET, allowing for excitation of 5-FAM and emission detection of TAMRA. The truncated peptide was comprised of the basic domain residues 49–57 of Tat with additional alanine residues added to both termini to prevent unfavorable interactions between the fluorophores and RNA. Additionally, a cysteine residue was introduced into the C-terminus to facilitate incorporation of TAMRA. Titration of HIV-1 TAR against the fluorescently-labeled peptide resulted in a 2.7-fold increase in fluorescence with Kd = 286 nM, assuming 1:1 stoichiometry. Consequently, a decrease in fluorescence was observed upon titration of unlabeled Tat, affording a dissociation constant of 70 nM. Overall, this study demonstrated for the first time the utility of a fluorescent Tat peptide displacement assay to characterize ligand binding interactions with HIV-1 TAR.
Fig. 4.
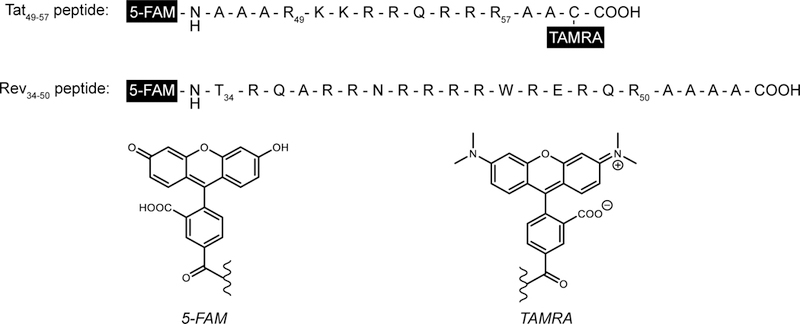
Sequence of FRET pair labeled Tat peptide and fluorophore labeled Rev peptide. Structures of fluorophores 5-FAM and TAMRA used at the N and C-termini shown. Amino acid abbreviations: A = Ala, R = Arg, Q = Gly, N = Asp, T = Thr, W = Trp, E = Glu, C = Cys.
The high affinity of Tat peptide for HIV-1 TAR and literature precedence of Tat peptide to bind other RNAs in vitro [94] inspired Patwardhan, Hargrove, and coworkers to extend the application of the fluorescent peptide displacement assay and screen small molecules against four different RNA targets [54]. Using more stringent buffer conditions, the affinity of the 5-FAM/TAMRA-labeled Tat peptide was first re-examined to bulge RNAs HIV-1 TAR and HIV-2 TAR, then assessed for internal loops, including bacterial ribosomal A-site RNA (E. coli) and RRE-IIB. RNA titrations against the fluorescent peptide yielded low-to-mid nanomolar dissociation values for all RNAs, confirming the capability of Tat to strongly interact with different RNA secondary structures and to be used as a general indicator to screen small molecules. Furthermore, the HTS potential of the Tat peptide displacement assay was demonstrated with three sets of small molecules: (1) aminoglycosides, (2) an in-house developed amiloride derivative library, and (3) a representative set of other known RNA-binding compounds from the literature. Small molecules were screened at two concentrations and compounds displaying >25% displacement at either concentration were designated as hits. Statistical analyses, including agglomerative hierarchical clustering and principal component analysis, revealed patterns within the displacement data and relationships between small molecule structure, RNA binding affinity and selectivity. Overall, this study highlighted the utility of the fluorescent Tat peptide to provide a convenient method to simultaneously evaluate small molecule binding affinity and selectivity for RNA.
2.2.2. Rev
Rev is a ~13 kDa viral regulatory protein that binds to HIV-1 RRE and facilitates the export of unspliced genomic viral RNA from the nucleus into the cytoplasm, where the RNA is translated into viral proteins [98]. In the absence of Rev binding, the expression of late phase HIV-1 structural proteins is halted and the replication of new viral particles is inhibited [65]. Similarly, disruption of the RRE:Rev complex by small molecules has been shown to inhibit HIV-1 replication and proven to be a promising therapeutic strategy towards the treatment of HIV-1 [99, 100]. Minimal RNA and protein substrates required to form the RRE:Rev complex have been determined, providing a manageable model system to monitor in vitro binding of small molecules [101]. Furthermore, the addition of a fluorescent tag to a 17-amino acid Rev peptide fragment (Rev34–50) has yielded a useful indicator that can be utilized to monitor peptide displacement upon small molecule binding [36, 56, 100, 102].
Using a fluorescein-labeled Rev peptide (Rev(34–50)-Fl) (Fig. 4), Wang, Hamasaki, and Rando developed a peptide displacement assay that monitors FP at the emission wavelength of fluorescein in the presence of RRE and competing analytes [36]. In solution, the extent of FP or anisotropy value of Rev(34–50)-Fl is inversely proportional to its tumbling rate that is in part, dependent on the size and shape of Rev(34–50)-Fl. Upon titration with RRE, the anisotropy value of Rev(34–50)-Fl increased, indicating complex formation. In this study, a 47-nt RRE construct known to bind both Rev protein and fragments was used, though FP measurements have also been performed with a longer 67-nt RRE construct [57]. Titration of unlabeled Rev(34–50) against the pre-formed RRE:Rev(34–50)-Fl complex led to displacement of Rev(34–50)-Fl as detected by a decrease in anisotropy, resulting in a dissociation constant of 1.41 nM. Displacement of Rev(34–50)-Fl by small molecules was evaluated for known RRE-binding ligands, neomycin B and tobramycin, and three tobramycin derivatives, revealing that pyrene and arginyl substituted aminoglycosides bind RRE with higher affinity than unmodified ligands. More recently, the peptide displacement assay has been used to screen against commercially-available small molecule libraries, leading to the discovery of new RNA-binding ligands for RRE, including clomiphene, cyproheptadine, and benfluron, that inhibit RRE:Rev complex formation in vitro and interfere with the gene expression processes of HIV-1 in cells [56, 100].
The versatility of the Rev(34–50)-Fl indicator was also shown by Luedtke and Tor, who developed a solid-phase peptide displacement assay and demonstrated its utility to identify selective small molecules that disrupt the RRE:Rev complex [102]. In the assay, a biotinylated 67-nt RRE fragment was first immobilized to streptavidin-modified insoluble agarose beads. Then, Rev(34–50)-Fl was added and the amount of fluorescent peptide bound to RRE was calculated by removing the supernatant and comparing the fluorescence intensity to a calibration curve of Rev(34–50)-Fl concentration as a function of fluorescence. Similar to other literature reports, the dissociation constant for Rev(34–50)-Fl with immobilized RRE was determined to be 2.5 nM, suggesting that the incorporation of a fluorescent tag and immobilization of RNA had no significant effect on the native interaction. The effectiveness of the Rev(34–50)-Fl displacement assay was demonstrated using known RRE-binding aminoglycosides and polycyclic aromatic amidines. Displacement of Rev(34–50)-Fl was monitored as a function of small molecule concentration by collecting the supernatant and comparing its fluorescence intensity to that of the calibration curve. The concentration of small molecule required to displace half of Rev(34–50)-Fl (IC50) was determined and reproducible trends in RRE affinity were observed when compared to values obtained by alternative methods, including FP. However, there were some discrepancies between IC50 values determined by solid-phase displacement in comparison to fluorescence anisotropy, thought to be a result of small molecule fluorescence quenching or interference with fluorescence anisotropy measurements. Indeed, the solid-phase peptide displacement assay can potentially circumvent fluorescence quenching or interference as the supernatant that contains excess small molecule and displaced Rev(34–50)-Fl can be removed, allowing for quantification of Rev(34–50)-Fl that remains on the solid support. Another advantage of the solid-phase Rev peptide displacement assay is the ability to simultaneously assess small molecule binding and selectivity to RRE. Competitor nucleic acids, including transfer RNA (tRNA) and dsDNA, as well as proteins can be added to increase the stringency of the assay and scavenge non-selective small molecules. Under these conditions, only highly selective small molecules will significantly displace Rev(34–50)-Fl and lead to a fluorescence signal in solution. Altogether, these studies highlight the utility of monitoring displacement of a fluorescently-labeled viral peptide, which has aided the identification of high affinity and bioactive compounds that inhibit Rev function, a mechanism of action that is different than currently marketed antiretroviral agents.
3. Example Applications of FID Assays
The ability to use FID assays in a high-throughput format makes the method favorable for screening large small molecule libraries against multiple RNA targets and aiding the rapid identification of RNA:small molecule interactions. For HTS, FID assays have been used as a primary screen, monitoring the percentage of indicator displacement at a single concentration of small molecule in a 384-well plate format. Single-point measurements often provide a “yes or no” response with the percentage of indicator displacement at a specific small molecule concentration denoting whether or not a small molecule binds a target(s). The Disney laboratory and others have utilized single-point measurements to rapidly evaluate small molecule libraries against multiple RNA targets to identify selective RNA-binding compounds [7, 19, 31, 32, 34, 35, 54, 56, 97, 100]. Several notable small molecule libraries that have been screened by an FID assay include the LOPAC library (1,280 compounds) [7, 31], the MyriaScreen chemical library (10,000 compounds, Sigma-Aldrich) [56], a drug-like small molecule library housed at the University of Michigan (103,498 compounds) [97], and two chemical libraries curated from the Open Innovation Center for Drug Discovery (now Drug Discovery Initiative) at the University of Tokyo consisting of 9,600 and 41,119 compounds, respectively [34, 35]. Importantly, Fukuzumi, Nakatani, and coworkers noted that the entire screening of the 41,119 compound library at 10 µM against three RNA targets by a single person took about 60 days, highlighting the applicability to use FID assays for exploratory study of RNA-binding small molecules in an academic setting [35].
FID assays are also useful to further characterize small molecule affinity and selectivity for an RNA target. Often, binding activity is reported as a competitive displacement value (i.e. CD50) or the concentration of small molecule at which 50% displacement of the indicator is observed. Because FID assays are competition-based, the CD50 value of a small molecule can be higher than the value of the dissociation constant measured in the absence of competitor. Dissociation constants can be calculated in FID assays; however, the binding affinity and stoichiometry of the indicator to the receptor must be known [103]. While more time and material consuming than a single-point measurement, determination of CD50 or Kd values for small molecules can be useful to rank and compare compounds, providing a more descriptive and quantitative measure than percentage of indicator displacement alone to elucidate chemical properties that enhance RNA binding affinity and selectivity. There are a number of fundamental studies where incorporation of an FID assay helped to identify and characterize structure-activity relationships of RNA-binding small molecules [43, 55, 104]. In one example, the Hergenrother laboratory utilized an EtBr displacement assay to characterize the binding of a synthetic library of spirocyclic compounds to 1-, 2-, and 3-nt RNA bulges as well as to dsRNA, the majority of which had binding constants > 20 µM for all RNAs evaluated. However, there were several compounds with binding constants < 20 µM for bulge RNAs only and notably, a few that displayed differential binding based on bulge size, revealing important chemical modifications that improved small molecule affinity for RNA bulges and dictated selectivity for specific bulge sizes [43]. For indicators that selectively bind an RNA receptor, CD50 values can be determined in the presence of other nucleic acid competitors to simultaneously assess small molecule selectivity. For example, fluorescently-labeled Tat peptide binds HIV-1 TAR RNA but does not strongly associate with tRNA, allowing for small molecule displacement to be evaluated in the presence of the tRNA competitor [55, 96, 97].
4. Considerations for Implementing an FID Assay
FID assays are a valuable method for identifying and characterizing selective RNA-binding small molecules; however, when setting up an assay, consideration of several parameters must be given to maximize results. Firstly, an indicator whose fluorescence and RNA-binding properties are favorable for a given system should be selected. If small molecules in the screening library are suspected to be intrinsically fluorescent, then the use of red-shifted probes would be favorable to limit small molecule fluorescence interference and avoid removal of compounds from evaluation [61]. Obtaining excitation and emission spectra of small molecules at the highest anticipated concentration for screening could help to inform the selection of an indicator whose wavelengths do not overlap. RNA-binding properties to consider for selecting an indicator include the stoichiometry and binding affinity to the receptor. If an indicator binds an RNA at multiple sites, then displacement by a small molecule may not result in a significant change in fluorescence unless the ligand also binds at multiple sites or induces a large RNA conformational change, leading to the displacement of several dye molecules. For the evaluation of monovalent small molecules, an indicator that binds stoichiometrically and near the anticipated binding site of small molecules would be favorable towards the development of a high-quality assay. In addition, the binding affinity of the indicator to the RNA target should be considered as FID assays rely on the competition between a small molecule and indicator for signal modulation. Therefore, the binding affinity between the indicator and RNA should be comparable to that between the small molecule and RNA [1, 103]. High affinity indicators allow for the use of lower amounts of RNA but may restrict the detection of moderate or weak binding small molecules [54].
Secondly, the concentrations of both the indicator and RNA target should be considered as the relative ratio will influence the maximum signal and quality of the assay. Commonly, the quality of an assay is defined by its Z’-factor that describes the signal variation for and separation between the free and bound states of an indicator [105]. Z’-factors can range from 0 to 1, where values equal to or greater than 0.5 describe reliable and high-quality assays. It is important to mention that a selected indicator with a given RNA may not always result in an acceptable Z’-factor. For example, the Z’-factor for an FID assay may suffer from high signal background interference when using a turn-OFF indicator in comparison to a turn-ON indicator [54]. Assay signal prior to the addition of test compound will be highly dependent on the initial fraction of indicator bound to an RNA (Fb0), calculated using total RNA/indicator concentrations and the experimentally determined indicator binding constant for an RNA [106]. The impact of Fb0 on FID assays has been demonstrated by del Villar-Guerra, Chaires, and coworkers, where simulations of the expected indicator displacement as a function of test compound binding affinity revealed a minimum affinity required to onset indicator displacement for their system [4]. Furthermore, these simulations can assist assay development to inform the selection of total RNA and indicator concentrations, resulting in an Fb0 that provides suitable stringency for small molecule screening.
Lastly, upon rational selection of assay parameters, the concentration(s) of small molecules for screening should be considered. Commonly, HTS FID assays have utilized 1, 10, 50, and/or 100 µM and generally ≥ 25% displacement has designated a small molecule as a hit [7, 18, 19, 31, 32, 34, 35, 54, 56]. It is important to note that the observance of little to no displacement does not necessarily confirm that a small molecule does not bind to an RNA as the small molecule may bind to a non-competitive site resulting in the formation of a ternary complex (RNA:indicator:small molecule) (Fig. 5) [4, 6]. Comparably, an indicator may bind an RNA at a second or third site albeit with weaker affinity when displaced from its primary binding site by a small molecule. Using ESI-MS, the displacement of indicator from the RNA receptor and/or formation of a ternary complex can be monitored as small molecule analyte is added, providing valuable insights on experimental results [6]. Alternatively, the small molecule may interact with the indicator [6], leading to either a false positive or negative result (Fig. 5). Therefore, controls that measure for potential interactions between a small molecule and indicator should be implemented. Additionally, small molecules should be evaluated by a secondary method such as ESI-MS, ITC, or SPR to validate binding interactions and improve interpretation of results. If possible, similar assay and buffer conditions should be used to allow for appropriate and meaningful comparisons. Altogether, careful consideration of each guideline described in this section will help to facilitate the selection of an indicator, optimization of assay conditions, and interpretation of FID assay results.
Fig. 5.
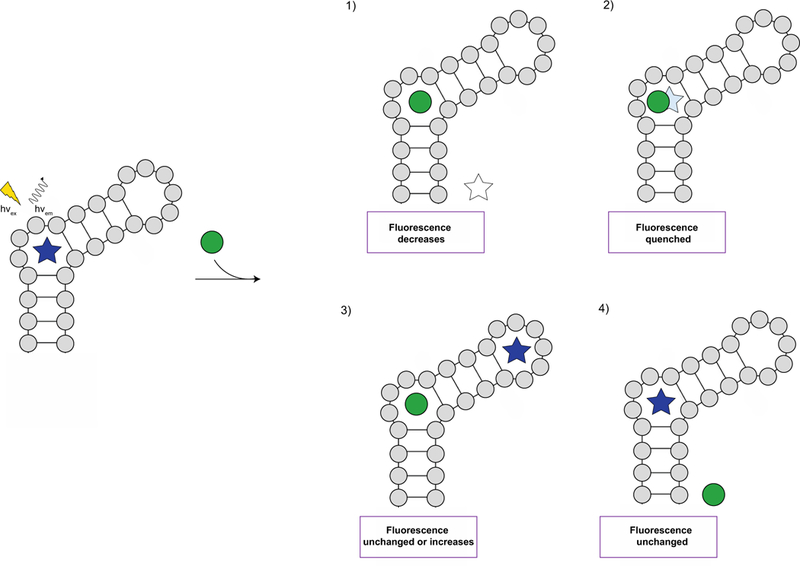
Potential outcomes of an FID assay [6]. Initially, fluorescence is observed as the indicator (star) is bound to the RNA receptor. Upon addition of the small molecule analyte (green circle), one of four scenarios can occur. Scenario 1): the small molecule analyte displaces the indicator, leading to a decrease in fluorescence. Scenario 2): the binding site of the small molecule analyte overlaps with the indicator, resulting in quenched fluorescence. Scenario 3): the binding site of the small molecule analyte is different than that of the indicator, leading to unchanged fluorescence or an increase in fluorescence if binding of the small molecule analyte to the RNA receptor enhances the binding of the indicator. Scenario 4): the small molecule analyte does not bind the RNA receptor or cannot outcompete the indicator, leading to unchanged fluorescence.
5. Concluding Remarks
Small molecule targeting of RNA has been largely recognized by the scientific community as a strategic approach to probe the unknown RNA functions in biological processes and disease [16, 107, 108]. Interest towards the rational development of selective RNA-binding ligands has been heightened in both academic and industrial settings as examples of small molecule probes used to attenuate RNA-specific disease-state interactions emerge [17, 109–112]. Additionally, great efforts have been made to fundamentally understand small molecule recognition of RNA and identify key properties that lead to selective interactions that could in turn provide guiding principles for targeting RNA [107, 113]. As a result, methods that can accelerate the identification and characterization of unique RNA:small molecule interactions are critical for the advancement of next generation chemical probes. In this review, we have surveyed the successful application of FID assays to study diverse RNA:small molecule interactions, provided a comprehensive summary of well-characterized RNA-binding indicators, and exemplified the valuable insights that RNA-based FID assays have provided towards small molecule recognition. Future design of novel indicators that are specific for an RNA structure and/or sequence may be advantageous for evaluating small molecule selectivity in the presence of multiple RNAs as well as potentially monitoring displacement of highly specific indicators in cells through standard fluorescence or FRET. Furthermore, we hope that the collection of promising results described here along with guidelines for experimental design will facilitate the use of FID assays to rapidly screen small molecules against underexplored RNAs and aid the identification of first-in-class chemical probes.
Highlights.
Reviews fluorescent indicator displacement (FID) assays with RNA and small molecules.
Surveys the fluorescence and RNA-binding properties of known indicators.
Provides examples where use of FID assays aided high-throughput screening efforts.
Highlights use of FID assays to quantify small molecule affinity/selectivity for RNA.
Describes practical considerations for the implementation of RNA-based FID assays.
Acknowledgements
We thank members of the Hargrove lab for providing constructive feedback during the preparation of the manuscript. A. E. H. wishes to acknowledge Duke University and the National Institute of Health (NIH) Maximizing Investigator’s Research Award (MIRA), Grant/Award Number: R35GM124785 for financial support. S. L. W. was supported in part through the U.S. Department of Education GAANN Fellowship (P200A150114). Apologies are given to the many scientists whose great work could not be included within this manuscript due to space restriction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nguyen BT, Anslyn EV, Indicator-displacement assays, Coord. Chem. Rev 250 (2006) 3118–3127. 10.1016/j.ccr.2006.04.009. [DOI] [Google Scholar]
- [2].Inouye M, Hashimoto K, Isagawa K, Nondestructive Detection of Acetylcholine in Protic Media: Artificial-Signaling Acetylcholine Receptors, J. Am. Chem. Soc 116 (1994) 5517–5518. 10.1021/ja00091a085. [DOI] [Google Scholar]
- [3].Wiskur SL, Ait-Haddou H, Lavigne JJ, Anslyn EV, Teaching old indicators new tricks, Acc. Chem. Res 34 (2001) 963–972. 10.1021/ar9600796. [DOI] [PubMed] [Google Scholar]
- [4].del Villar-Guerra R, Gray RD, Trent JO, Chaires JB, 2018. A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand-nucleic acid structural selectivity. Nucleic Acids Res 46, e41 10.1093/nar/gky019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP, A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity, J. Am. Chem. Soc 123 (2001) 5878–5891. 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- [6].Asare-Okai PN, Chow CS, A modified fluorescent intercalator displacement assay for RNA ligand discovery, Anal. Biochem 408 (2011) 269–276. 10.1016/j.ab.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang JH, Umemoto S, Nakatani K, Fluorescent Indicator Displacement Assay for Ligand-RNA Interactions, J. Am. Chem. Soc 132 (2010) 3660–3661. 10.1021/ja100089u. [DOI] [PubMed] [Google Scholar]
- [8].Watkins D, Norris FA, Kumar S, Arya DP, A fluorescence-based screen for ribosome binding antibiotics, Anal. Biochem 434 (2013) 300–307. 10.1016/j.ab.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Metzger A, Lynch VM, Anslyn EV, A synthetic receptor selective for citrate, Angew. Chem. Intl. Ed 36 (1997) 862–865. 10.1002/anie.199708621. [DOI] [Google Scholar]
- [10].Bourson J, Pouget J, Valeur B, Ion-Responsive Fluorescent Compounds. 4. Effect of Cation Binding on the Photophysical Properties of a Coumarin Linked to Monoaza-and Diaza-Crown Ethers, J. Phys. Chem 97 (1993) 4552–4557. 10.1021/j100119a050. [DOI] [Google Scholar]
- [11].Chen CT, Huang WP, A highly selective fluorescent chemosensor for lead ions, J. Am. Chem. Soc 124 (2002) 6246–6247. 10.1021/ja025710e. [DOI] [PubMed] [Google Scholar]
- [12].Hickey SF, Hammond MC, Structure-Guided Design of Fluorescent S-Adenosylmethionine Analogs for a High-Throughput Screen to Target SAM-I Riboswitch RNAs, Chem. Biol 21 (2014) 345–356. 10.1016/j.chembiol.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hortala MA, Fabbrizzi L, Marcotte N, Stomeo F, Taglietti A, Designing the selectivity of the fluorescent detection of amino acids: A chemosensing ensemble for histidine, J. Am. Chem. Soc 125 (2003) 20–21. 10.1021/ja0271101. [DOI] [PubMed] [Google Scholar]
- [14].Breaker RR, Joyce GF, The Expanding View of RNA and DNA Function, Chem. Biol 21 (2014) 1059–1065. 10.1016/j.chembiol.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Esteller M, Non-coding RNAs in human disease, Nat. Rev. Genet 12 (2011) 861–874. 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- [16].Connelly CM, Moon MH, Schneekloth JS, The Emerging Role of RNA as a Therapeutic Target for Small Molecules, Cell Chem. Biol 23 (2016) 1077–1090. 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, Murgolo N, Barbieri CM, Mann PA, Carr D, Xia E, Zuck P, Riley D, Painter RE, Walker SS, Sherborne B, de Jesus R, Pan WD, Plotkin MA, Wu J, Rindgen D, Cummings J, Garlisi CG, Zhang RM, Sheth PR, Gill CJ, Tang HF, Roemer T, Selective small-molecule inhibition of an RNA structural element, Nature 526 (2015) 672–677. 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- [18].Su ZM, Zhang YJ, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh SY, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, Morelli C, Ratti A, Silani V, Rademakers R, Brown RH, Rothstein JD, Boylan KB, Petrucelli L, Disney MD, Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS, Neuron 83 (2014) 1043–1050. 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Garcia-Lopez A, Tessaro F, Jonker HRA, Wacker A, Richter C, Comte A, Berntenis N, Schmucki R, Hatje K, Petermann O, Chiriano G, Perozzo R, Sciarra D, Konieczny P, Faustino I, Fournet G, Orozco M, Artero R, Metzger F, Ebeling M, Goekjian P, Joseph B, Schwalbe H, Scapozza L, 2018. Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes. Nat. Commun 9, 2032 10.1038/s41467-018-04110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morgan BS, Forte JE, Hargrove AE, Insights into the development of chemical probes for RNA, Nucleic Acids Res 46 (2018) 8025–8037. 10.1093/nar/gky718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Blakeley BD, DePorter SM, Mohan U, Burai R, Tolbert BS, McNaughton BR, Methods for identifying and characterizing interactions involving RNA, Tetrahedron 68 (2012) 8837–8855. 10.1016/j.tet.2012.07.001. [DOI] [Google Scholar]
- [22].Thomas JR, Hergenrother PJ, Targeting RNA with small molecules, Chem. Rev 108 (2008) 1171–1224. 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- [23].Eubanks CS, Hargrove AE, RNA Structural Differentiation: Opportunities with Pattern Recognition, Biochemistry 58 (2019) 199–213. 10.1021/acs.biochem.8b01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Murata A, Fukuzumi T, Umemoto S, Nakatani K, Xanthone derivatives as potential inhibitors of miRNA processing by human Dicer: Targeting secondary structures of pre-miRNA by small molecules, Bioorg. Med. Chem. Lett 23 (2013) 252–255. 10.1016/j.bmcl.2012.10.108. [DOI] [PubMed] [Google Scholar]
- [25].Lipfert J, Doniach S, Das R, Herschlag D, Understanding Nucleic Acid-Ion Interactions, Annu. Rev. Biochem 83 (2014) 813–841. 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Krishnamurthy M, Schirle NT, Beal PA, Screening helix-threading peptides for RNA binding using a thiazole orange displacement assay, Bioorg. Med. Chem 16 (2008) 8914–8921. 10.1016/j.bmc.2008.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pei R, Stojanovic MN, Study of thiazole orange in aptamer-based dye-displacement assays, Anal. Bioanal. Chem 390 (2008) 1093–1099. 10.1007/s00216-007-1773-2. [DOI] [PubMed] [Google Scholar]
- [28].Sarpong K, Datta B, 2012. Nucleic-Acid-binding chromophores as efficient indicators of aptamer-target interactions. J. Nucleic Acids 2012, 247280 10.1155/2012/247280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pei R, Rothman J, Xie YL, Stojanovic MN, 2009. Light-up properties of complexes between thiazole orange-small molecule conjugates and aptamers. Nucleic Acids Res 37, e59 10.1093/nar/gkp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang ZF, Ursu A, Childs-Disney JL, Guertler R, Yang WY, Bernat V, Rzuczek SG, Fuerst R, Zhang YJ, Gendron TF, Yildirim I, Dwyer BG, Rice JE, Petrucelli L, Disney MD, The Hairpin Form of r(G4C2)(exp) in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules, Cell Chem. Biol (2018) 179–190. 10.1016/j.chembiol.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tran T, Disney MD, 2012. Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun 3, 1125 10.1038/ncomms2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Haniff HS, Graves A, Disney MD, Selective Small Molecule Recognition of RNA Base Pairs, ACS Comb. Sci 20 (2018) 482–491. 10.1021/acscombsci.8b00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Umemoto S, Im S, Zhang JH, Hagihara M, Murata A, Harada Y, Fukuzumi T, Wazaki T, Sasaoka S, Nakatani K, Structure-Activity Studies on the Fluorescent Indicator in a Displacement Assay for the Screening of Small Molecules Binding to RNA, Chem. Eur. J 18 (2012) 9999–10008. 10.1002/chem.201103932. [DOI] [PubMed] [Google Scholar]
- [34].Murata A, Harada Y, Fukuzumi T, Nakatani K, Fluorescent indicator displacement assay of ligands targeting 10 microRNA precursors, Bioorg. Med. Chem 21 (2013) 7101–7106. 10.1016/j.bmc.2013.09.007. [DOI] [PubMed] [Google Scholar]
- [35].Fukuzumi T, Murata A, Aikawa H, Harada Y, Nakatani K, Exploratory Study on the RNA-Binding Structural Motifs by Library Screening Targeting pre-miRNA-29a, Chem. Eur. J 21 (2015) 16859–16867. 10.1002/chem.201502913. [DOI] [PubMed] [Google Scholar]
- [36].Wang Y, Hamasaki K, Rando RR, Specificity of aminoglycoside binding to RNA constructs derived from the 16S rRNA decoding region and the HIV-RRE activator region, Biochemistry 36 (1997) 768–779. 10.1021/bi962095g. [DOI] [PubMed] [Google Scholar]
- [37].Wang Y, Rando RR, Specific Binding of Aminoglycoside Antibiotics to RNA, Chem. Biol 2 (1995) 281–290. 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- [38].Ramos LMG, Degtyareva NN, Kovacs NA, Holguin SY, Jiang LW, Petrov AS, Biesiada M, Hu MY, Purzycka KJ, Arya DP, Williams LD, Eukaryotic Ribosomal Expansion Segments as Antimicrobial Targets, Biochemistry 56 (2017) 5288–5299. 10.1021/acs.biochem.7b00703. [DOI] [PubMed] [Google Scholar]
- [39].Watkins D, Jiang LW, Nahar S, Maiti S, Arya DP, 2015. A pH Sensitive High-Throughput Assay for miRNA Binding of a Peptide-Aminoglycoside (PA) Library. PLoS One 10, e0144251 10.1371/journal.pone.0144251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Y, Killian J, Hamasaki K, Rando RR, RNA molecules that specifically and stoichiometrically bind aminoglycoside antibiotics with high affinities, Biochemistry 35 (1996) 12338–12346. 10.1021/bi960878w. [DOI] [PubMed] [Google Scholar]
- [41].Lemay JF, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine DA, Folding of the adenine riboswitch, Chem. Biol 13 (2006) 857–868. 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- [42].Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R, Novel Ligands for a Purine Riboswitch Discovered by RNA-Ligand Docking, Chem. Biol 18 (2011) 324–335. 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Meyer ST, Hergenrother PJ, Small Molecule Ligands for Bulged RNA Secondary Structures, Org. Lett 11 (2009) 4052–4055. 10.1021/ol901478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nelson JW, Tinoco I, Intercalation of Ethidium Ion into DNA and RNA Oligonucleotides, Biopolymers 23 (1984) 213–233. 10.1002/bip.360230205. [DOI] [PubMed] [Google Scholar]
- [45].Luedtke NW, Tor Y, Fluorescence-based methods for evaluating the RNA affinity and specificity of HIV-1 Rev-RRE inhibitors, Biopolymers 70 (2003) 103–119. 10.1002/bip.10428. [DOI] [PubMed] [Google Scholar]
- [46].Kumar S, Kellish P, Robinson EW, Wang D, Appella D, Arya DP, Click Dimers To Target HIV TAR RNA Conformation, Biochemistry 51 (2012) 2331–2347. 10.1021/bi201657k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bai LP, Hagihara M, Nakatani K, Jiang ZH, 2014. Recognition of Chelerythrine to Human Telomeric DNA and RNA G-quadruplexes. Sci. Rep 4, 6767 10.1038/srep06767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sato Y, Rokugawa M, Ito S, Yajima S, Sugawara H, Teramae N, Nishizawa S, Fluorescent Trimethylated Naphthyridine Derivative with an Aminoalkyl Side Chain as the Tightest Non-aminoglycoside Ligand for the Bacterial A-site RNA, Chem. Eur. J 24 (2018) 13862–13870. 10.1002/chem.201802320. [DOI] [PubMed] [Google Scholar]
- [49].DeJong ES, Chang CE, Gilson MK, Marino JP, Proflavine acts as a Rev inhibitor by targeting the high-affinity Rev binding site of the Rev responsive element of HIV-1, Biochemistry 42 (2003) 8035–8046. 10.1021/bi0345252z. [DOI] [PubMed] [Google Scholar]
- [50].Qi L, Fan YY, Wei H, Zhang D, Zhang ZQ, Graphene oxide-enhanced and proflavine-probed fluorescence polarization biosensor for ligand-RNA interaction assay, Sens. Actuators B Chem 257 (2018) 666–671. 10.1016/j.snb.2017.11.036. [DOI] [Google Scholar]
- [51].Qi L, Wei JR, Lv XJ, Huo Y, Zhang ZQ, A ratiometric fluorescence RRE RNA-targeted assay for a new fluorescence ligand, Biosens. Bioelectron 86 (2016) 287–292. 10.1016/j.bios.2016.06.051. [DOI] [PubMed] [Google Scholar]
- [52].Li X, Chen B, Lan L, Wang R, Luo D, Liu L, Cheng L, Selective recognition of HIV RNA by dinuclear metallic ligands, Chin. Chem. Lett 29 (2018) 1637–1640. 10.1016/j.cclet.2018.06.003. [DOI] [Google Scholar]
- [53].Matsumoto C, Hamasaki K, Mihara H, Ueno A, A high-throughput screening utilizing intramolecular fluorescence resonance energy transfer for the discovery of the molecules that bind HIV-1 TAR RNA specifically, Bioorg. Med. Chem. Lett 10 (2000) 1857–1861. 10.1016/S0960-894x(00)00359-0. [DOI] [PubMed] [Google Scholar]
- [54].Patwardhan NN, Cai Z, Newson CN, Hargrove AE, Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA, Org. Biomol. Chem 17 (2019) 1778–1786. 10.1039/c8ob02467g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Patwardhan NN, Ganser LR, Kapral GJ, Eubanks CS, Lee J, Sathyamoorthy B, Al-Hashimi HM, Hargrove AE, Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR, MedChemComm 8 (2017) 1022–1036. 10.1039/c6md00729e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Prado S, Beltran M, Moreno A, Bedoya LM, Alcami J, Gallego J, A small-molecule inhibitor of HIV-1 Rev function detected by a diversity screen based on RRE-Rev interference, Biochem. Pharmacol 156 (2018) 68–77. 10.1016/j.bcp.2018.07.040. [DOI] [PubMed] [Google Scholar]
- [57].Kirk SR, Luedtke NW, Tor Y, Neomycin-acridine conjugate: A potent inhibitor of Rev-RRE binding, J. Am. Chem. Soc 122 (2000) 980–981. 10.1021/ja993387i. [DOI] [Google Scholar]
- [58].Shindy HA, Fundamentals in the chemistry of cyanine dyes: A review, Dye. Pigment 145 (2017) 505–513. 10.1016/j.dyepig.2017.06.029. [DOI] [Google Scholar]
- [59].Armitage BA, Cyanine Dye-Nucleic Acid Interactions, Top. Heterocycl. Chem 14 (2008) 11–29. 10.1007/7081_2007_109. [DOI] [Google Scholar]
- [60].Lee LG, Chen CH, Chiu LA, Thiazole Orange - a New Dye for Reticulocyte Analysis, Cytometry 7 (1986) 508–517. 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- [61].Simeonov A, Jadhav A, Thomas CJ, Wang YH, Huang RL, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J, Fluorescence spectroscopic profiling of compound libraries, J. Med. Chem 51 (2008) 2363–2371. 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- [62].Petty JT, Bordelon JA, Robertson ME, Thermodynamic characterization of the association of cyanine dyes with DNA, J. Phys. Chem. B 104 (2000) 7221–7227. 10.1021/jp000916s. [DOI] [Google Scholar]
- [63].Shagufta I Ahmad, Recent insight into the biological activities of synthetic xanthone derivatives, Eur. J. Med. Chem 116 (2016) 267–280. 10.1016/j.ejmech.2016.03.058. [DOI] [PubMed] [Google Scholar]
- [64].Pace TCS, Monahan SL, MacRae AI, Kaila M, Bohne C, Photophysics of aminoxanthone derivatives and their application as binding probes for DNA, Photochem. Photobiol 82 (2006) 78–87. 10.1562/2005-05-16-Ra-529. [DOI] [PubMed] [Google Scholar]
- [65].Malim MH, Tiley LS, Mccarn DF, Rusche JR, Hauber J, Cullen BR, HIV-1 Structural Gene Expression Requires Binding of the Rev Trans-Activator to Its RNA Target Sequence, Cell 60 (1990) 675–683. 10.1016/0092-8674(90)90670-A. [DOI] [PubMed] [Google Scholar]
- [66].Battiste JL, Mao HY, Rao NS, Tan RY, Muhandiram DR, Kay LE, Frankel AD, Williamson JR, alpha helix-RNA major groove recognition in an HIV-1 Rev peptide RRE RNA complex, Science 273 (1996) 1547–1551. 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- [67].Lacourciere KA, Stivers JT, Marino JP, Mechanism of neomycin and Rev peptide binding to the Rev responsive element of HIV-1 as determined by fluorescence and NMR spectroscopy, Biochemistry 39 (2000) 5630–5641. 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- [68].Dambach MD, Winkler WC, Expanding roles for metabolite-sensing regulatory RNAs, Curr. Opin. Microbiol 12 (2009) 161–169. 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Breaker RR, 2012. Riboswitches and the RNA World Cold Spring Harb: Perspect. Biol; 4, a003566 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Barrick JE, Breaker RR, 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8, R239 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Blount KF, Breaker RR, Riboswitches as antibacterial drug targets, Nat. Biotechnol 24 (2006) 1558–1564. 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- [72].Deigan KE, Ferre-D’Amare AR, Riboswitches: Discovery of Drugs That Target Bacterial Gene-Regulatory RNAs, Acc. Chem. Res 44 (2011) 1329–1338. 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Winkler WC, Cohen-Chalamish S, Breaker RR, An mRNA structure that controls gene expression by binding FMN, Proc. Natl. Acad. Sci. USA 99 (2002) 15908–15913. 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Serganov A, Patel DJ, Metabolite Recognition Principles and Molecular Mechanisms Underlying Riboswitch Function, Annu. Rev. Biophys 41 (2012) 343–370. 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gilbert SD, Stoddard CD, Wise SJ, Batey RT, Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain, J. Mol. Biol 363 (2006) 754–768. 10.1016/j.jmb.2006.04.075. [DOI] [PubMed] [Google Scholar]
- [76].Ward DC, Reich E, Fluorescence Studies of Nucleotides and Polynucleotides I. Formycin, 2-aminopurine, 2,6-diaminopurine riboside, and their derivatives, J. Biol. Chem 244 (1969) 1228–1237. [PubMed] [Google Scholar]
- [77].Ottink OM, Nelissen FHT, Derks Y, Wijmenga SS, Heus HA, Enzymatic stereospecific preparation of fluorescent S-adenosyl-L-methionine analogs, Anal. Biochem 396 (2010) 280–283. 10.1016/j.ab.2009.09.013. [DOI] [PubMed] [Google Scholar]
- [78].Luedtke NW, Liu Q, Tor Y, RNA-ligand interactions: Affinity and specificity of aminoglycoside dimers and acridine conjugates to the HIV-1 rev response element, Biochemistry 42 (2003) 11391–11403. 10.1021/bi034766y. [DOI] [PubMed] [Google Scholar]
- [79].Lombardo CM, Welsh SJ, Strauss SJ, Dale AG, Todd AK, Nanjunda R, Wilson WD, Neidle R, A novel series of G-quadruplex ligands with selectivity for HIF-expressing osteosarcoma and renal cancer cell lines, Bioorg. Med. Chem. Lett 22 (2012) 5984–5988. 10.1016/j.bmcl.2012.07.009. [DOI] [PubMed] [Google Scholar]
- [80].Hamasaki K, Rando RR, A high-throughput fluorescence screen to monitor the specific binding of antagonists to RNA targets, Anal. Biochem 261 (1998) 183–190. 10.1006/abio.1998.2740. [DOI] [PubMed] [Google Scholar]
- [81].Velagapudi SP, Seedhouse SJ, French J, Disney MD, Defining the RNA Internal Loops Preferred by Benzimidazole Derivatives via 2D Combinatorial Screening and Computational Analysis, J. Am. Chem. Soc 133 (2011) 10111–10118. 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Velagapudi SP, Pushechnikov A, Labuda LP, French JM, Disney MD, Probing a 2-Aminobenzimidazole Library for Binding to RNA Internal Loops via Two-Dimensional Combinatorial Screening, ACS Chem. Biol 7 (2012) 1902–1909. 10.1021/cb300213g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chittapragada M, Roberts S, Wan Ham Y, Aminoglycosides: Molecular Insights on the Recognition of RNA and Aminoglycoside Mimics, Perspect. Med. Chem 3 (2009) 21–37. 10.4137/PMC.S2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lynch SR, Puglisi JD, Structural origins of aminoglycoside specificity for prokaryotic ribosomes, J. Mol. Biol 306 (2001) 1037–1058. 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- [85].Schatz A, Bugie E, Waksman SA, Streptomycin, a substance exhibiting antibiotic activity against gram positive and gram-negative bacteria, Proc. Soc. Exp. Biol. Med 55 (1944) 66–69. 10.3181/00379727-55-14461. [DOI] [PubMed] [Google Scholar]
- [86].Arya DP, Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery, first ed., Wiley-Intersciecne, Hoboken, NJ, 2007. [Google Scholar]
- [87].Wang H, Tor Y, Electrostatic interactions in RNA aminoglycosides binding, J. Am. Chem. Soc 119 (1997) 8734–8735. 10.1021/ja9715105. [DOI] [Google Scholar]
- [88].Jiang LC, Patel DJ, Solution structure of the tobramycin-RNA aptamer complex, Nat. Struct. Biol 5 (1998) 769–774. 10.1038/1804. [DOI] [PubMed] [Google Scholar]
- [89].Sannes-Lowery KA, Mei HY, Loo JA, Studying aminoglycoside antibiotic binding to HIV-1 TAR RNA by electrospray ionization mass spectrometry, Int. J. Mass. Spectrom 193 (1999) 115–122. 10.1016/S1387-3806(99)00111-6. [DOI] [Google Scholar]
- [90].Vonahsen U, Davies J, Schroeder R, Noncompetitive Inhibition of Group-I Intron Rna Self-Splicing by Aminoglycoside Antibiotics, J. Mol. Biol 226 (1992) 935–941. 10.1016/0022-2836(92)91043-O. [DOI] [PubMed] [Google Scholar]
- [91].Zhang J, Tamilarasu N, Hwang SW, Garber ME, Huq I, Jones KA, Rana TM, HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1, J. Biol. Chem 275 (2000) 34314–34319. 10.1074/jbc.M006804200. [DOI] [PubMed] [Google Scholar]
- [92].Aboulela F, Karn J, Varani G, The Structure of the Human Immunodeficiency Virus Type-1 TAR RNA Reveals Principles of RNA Recognition by Tat Protein, J. Mol. Biol 253 (1995) 313–332. 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- [93].Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM, Fragments of the HIV-1 Tat Protein Specifically Bind Tar RNA, Science 249 (1990) 1281–1285. 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- [94].Weeks KM, Crothers DM, RNA Recognition by Tat-Derived Peptides: Interaction in the Major Groove?, Cell 66 (1991) 577–588. 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- [95].Ludwig V, Krebs A, Stoll M, Dietrich U, Ferner J, Schwalbe H, Scheffer U, Durner G, Gobel MW, Tripeptides from synthetic amino acids block the Tat-TAR association and slow down HIV spread in cell cultures, ChemBioChem 8 (2007) 1850–1856. 10.1002/cbic.200700232. [DOI] [PubMed] [Google Scholar]
- [96].Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM, Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble, Nat. Chem. Biol 7 (2011) 553–559. 10.1038/Nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ganser LR, Lee J, Rangadurai A, Merriman DK, Kelly ML, Kansal AD, Sathyamoorthy B, Al-Hashimi HM, High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble, Nat. Struct. Mol. Biol 25 (2018) 425–434. 10.1038/s41594-018-0062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR, The HIV-1 Rev Trans-Activator Acts through a Structured Target Sequence to Activate Nuclear Export of Unspliced Viral mRNA, Nature 338 (1989) 254–257. 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- [99].Gonzalez-Bulnes L, Ibanez I, Bedoya LM, Beltran M, Catalan S, Alcami J, Fustero S, Gallego J, Structure-Based Design of an RNA-Binding p-Terphenylene Scaffold that Inhibits HIV-1 Rev Protein Function, Angew. Chem. Int. Ed 52 (2013) 13405–13409. 10.1002/anie.201306665. [DOI] [PubMed] [Google Scholar]
- [100].Prado S, Beltran M, Coiras M, Bedoya LM, Alcami J, Gallego J, Bioavailable inhibitors of HIV-1 RNA biogenesis identified through a Rev-based screen, Biochem. Pharmacol 107 (2016) 14–28. 10.1016/j.bcp.2016.02.007. [DOI] [PubMed] [Google Scholar]
- [101].Kjems J, Calnan BJ, Frankel AD, Sharp PA, Specific Binding of a Basic Peptide from Hiv-1 Rev, EMBO J 11 (1992) 1119–1129. 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Luedtke NW, Tor Y, A novel solid-phase assembly for identifying potent and selective RNA ligands, Angew. Chem. Int. Ed 39 (2000) 1788–1790. . [DOI] [PubMed] [Google Scholar]
- [103].Hargrove AE, Zhong ZL, Sessler JL, Anslyn EV, Algorithms for the determination of binding constants and enantiomeric excess in complex host : guest equilibria using optical measurements, New J. Chem 34 (2010) 348–354. 10.1039/b9nj00498j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jiang LW, Watkins D, Jin Y, Gong C, King A, Washington AZ, Green KD, Garneau-Tsodikova S, Oyelere AK, Arya DP, Rapid Synthesis, RNA Binding, and Antibacterial Screening of a Peptidic-Aminosugar (PA) Library, ACS Chem. Biol 10 (2015) 1278–1289. 10.1021/cb5010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang JH, Chung TDY, Oldenburg KR, A simple statistical parameter for use in evaluation and validation of high throughput screening assays, J. Biomol. Screen 4 (1999) 67–73. 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [106].Roehrl MHA, Wang JY, Wagner G, A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization, Biochemistry 43 (2004) 16056–16066. 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- [107].Warner KD, Hajdin CE, Weeks KM, Principles for targeting RNA with drug-like small molecules, Nat. Rev. Drug Discov 17 (2018) 547–558. 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Donlic A, Hargrove AE, 2018. Targeting RNA in mammalian systems with small molecules. WIREs RNA 9, e1477 10.1002/wrna.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang XL, Van Hoosear M, Shin Y, Chin DN, Keller G, Beibel M, Renaud NA, Smith TM, Salcius M, Shi XY, Hild M, Servais R, Jain M, Deng L, Bullock C, McLellan M, Schuierer S, Murphy L, Blommers MJJ, Blaustein C, Berenshteyn F, Lacoste A, Thomas JR, Roma G, Michaud GA, Tseng BS, Porter JA, Myer VE, Tallarico JA, Hamann LG, Curtis D, Fishman MC, Dietrich WF, Dales NA, Sivasankaran R, SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice, Nat. Chem. Biol 11 (2015) 511–517. https://doi.org/DOI 10.1038/nchembio0915-741a. [DOI] [PubMed] [Google Scholar]
- [110].Sivaramakrishnan M, McCarthy KD, Campagne S, Huber S, Meier S, Augustin A, Heckel T, Meistermann H, Hug MN, Birrer P, Moursy A, Khawaja S, Schmucki R, Berntenis N, Giroud N, Golling S, Tzouros M, Banfai B, Duran-Pacheco G, Lamerz J, Liu YH, Luebbers T, Ratni H, Ebeling M, Clery A, Paushkin S, Krainer AR, Allain FHT, Metzger F, 2017. Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers. Nat. Commun 8, 1476 10.1038/s41467-017-01559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, Disney MD, Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit, J. Am. Chem. Soc 139 (2017) 3446–3455. 10.1021/jacs.61311273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Fedorova O, Jagdmann GE, Adams RL, Yuan L, Van Zandt MC, Pyle AM, Small molecules that target group II introns are potent antifungal agents, Nat. Chem. Biol 14 (2018) 1073–1078. 10.1038/s41589-018-0142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Morgan BS, Forte JE, Culver RN, Zhang YQ, Hargrove AE, Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-Targeted Bioactive Ligands, Angew. Chem. Int. Ed 56 (2017) 13498–13502. 10.1002/anie.201707641. [DOI] [PMC free article] [PubMed] [Google Scholar]


