Abstract
This review highlights three recent trends in the field of kinetochore biology: the proliferation of structural data for kinetochore protein complexes (including CBF3, Dam1c, Mis12cMIND, and CENP-NLChl4/Iml3); the growing consensus that the kinetochore is a dynamic structure whose composition changes as the cell cycle progresses; and the mounting evidence of multiple pathways whereby the microtubule binding elements of the outer kinetochore may be recruited by inner kinetochore proteins. Our focus is on the two best-studied systems in the field: human and budding yeast kinetochores. This review will demonstrate the remarkable similarity of these two systems, as well as their intriguing differences.
Introduction
How are duplicated chromosomes partitioned between daughter cells during mitosis and meiosis? How is this process regulated? How does it go awry in cancer? The kinetochore is central to all of these questions.
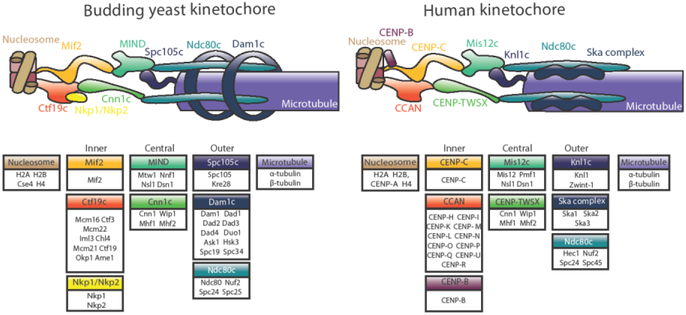
A macromolecular complex of about forty core proteins (not including regulatory proteins and motors) assembled on the centromere[1], the kinetochore mediates the process of chromosome segregation by coupling microtubule dynamics to chromosome movement (Figure 1). It also serves as a tension-sensitive regulatory hub for destabilization of improper microtubule-kinetochore attachments [2,3]. The kinetochore may be conceptually divided into the microtubule-proximal outer kinetochore and the centromere-proximal inner kinetochore.
Figure 1.
Model for the budding yeast (S.cerevisiae) and human (H. sapiens) kinetochores. The individual proteins of each subcomplex are shown in boxes. Inner kinetochore proteins, called the constitutively centromere-associated network (CCAN), contact the centromeric nucleosome and recruit central kinetochore proteins. These contact the microtubule-binding elements that make up the outer kinetochore. Homologous subcomplexes are colored identically, with the exception of the Dam1 and Ska complexes, which are non-homologous functional counterparts.
Poised at the intersection of genetics, protein biochemistry, and molecular biophysics, pertinent to cancer biology and human genetic disease, the field of kinetochore research has a rich history, which is beyond the scope of this review. (Interested readers are directed to Musacchio and Desai 2017 [4].) Rather, this review will highlight recent trends in the field, specifically: a proliferation of structural data for kinetochore proteins; a growing consensus that the composition of the kinetochore changes as the cell cycle progresses; and the discovery of multiple means by which microtubule-binding elements are recruited by the inner kinetochore— paths whose primacy may vary between organisms and between mitotic stages.
Structures, structures, everywhere
Direct electron detector technology for cryo-electron microscopy (cryo-EM) has revolutionized the field of structural biology [5,6], bringing a wave of high-resolution macromolecular structures — including some protein subcomplexes of the kinetochore. In the past several years, these structures have provided unprecedented insight into the physical makeup of that molecular machine.
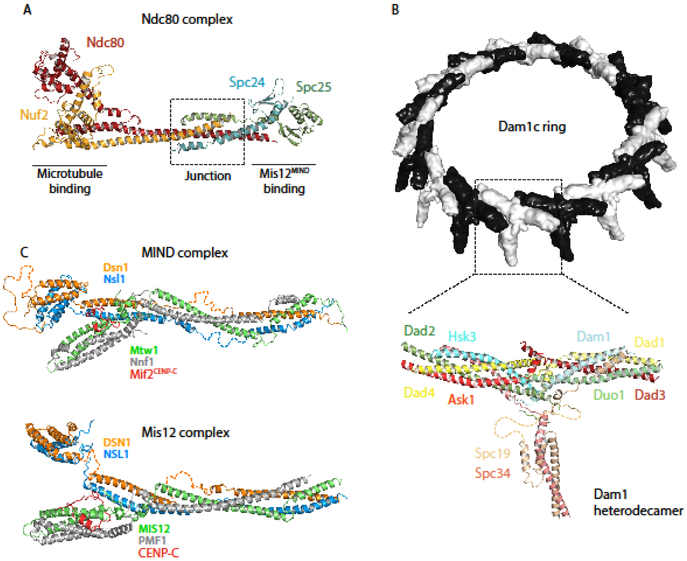
For years, kinetochore proteins have eluded structural analysis due to their flexible architectures. Severe truncations and simplifications were necessary to obtain constructs that would crystallize. An example of this approach is the creation and crystallization of the “bonsai” Ndc80c complex [7]. The N-terminus of Ndc80 was fused to the C-terminus of Spc25, and the N-terminus of Nuf2 was fused to the C-terminus of Spc24, abrogating the tetramerization domain and most of the coiled coils in order to allow crystallization of the globular microtubule-binding and kinetochore-binding elements of Ndc80c [7]. It was rare and hard-won triumph for kinetochore protein crystallographers and was instrumental to our understanding of how Ndc80c binds microtubules.
It was eight years before the intact tetramerization domain was crystalized, revealing a junction between the Ndc80-Nuf2 and Spc24-Spc25 coiled coils [8]. This junction may be subdivided into a region of three-chain overlap and one of four-chain overlap, which buries an aromatic side-chain stack to which all four proteins contribute [8]. A network of conserved polar contacts further stabilizes the junction. This structure of Ndc80c’s tetramerization domain, taken together with the globular head structures originally elucidated by “bonsai” Ndc80c, allow visualization of the intra-complex interactions that hold Ndc80c together, as well as the surfaces that interact with its binding partners— a significant step toward understanding the structures responsible for microtubule binding.
Just last year, cryo-EM cracked the structure of another key microtubule-binding complex (at least in fungi). A ~4.5 Å structure of the Dam1 complex (Dam1c) of Chaetomium thermophilum reveals how individual T-shaped heterodecamers come together at conserved polar and non-polar contact points to form a microtubule-encircling ring [9,10](Figure 2B). Orthogonal to both arms of the T, the protrusion domain is comprised of the C-terminal domains of Spc19 and Spc34; it is posited to contact the Ndc80-Nuf2 coiled-coil. In agreement with the conclusions of previously published cross-linking experiments [11,12], the C-termini of Dam1 and Duo1 are oriented towards the microtubule lattice, which they are predicted to bind. While the residues that actually contact Ndc80c (based on cross-linking data [13]) were not present in the reconstruction, Jenni and coworkers conclude that all three contacts with Ndc80c could be made by a single Dam1c ring— in contrast to Kim et al.’s conclusion that Ndc80c bridges two Dam1c rings at the kinetochore [9,13]. Using electron cryotomography to examine the outer kinetochore in vivo, Ng et al. found that while a few kinetochore microtubules do have two rings, most have only one; and though some complete 17-membered rings were observed, the majority of Dam1c oligomers formed only partial rings [14].
Figure 2.
Recently published structures of outer kinetochore protein complexes. (A) Ndc80cDwarf is an elongated heterotetramer comprised of a microtubule-binding Hec1Ndc80-Nuf2 dimer and Mis12MIND-binding Spc24-Spc25 dimer. The structure shows the complete junction region of the heterotetramer (PDB ID: 5TCS) [7,8]. (B) (top) Chaetomium thermophilum Dam1c heterodecamers oligomerize into 17-membered rings with an outer diameter of about 560 Å [9]. (bottom) Ribbon diagram of a heterodecamer subunit, containing amino acids 13-78 of Ask1, 18-76 of Dad1, 25-95 and 109-116 of Dad2, 18-82 of Dad3, 3-70 of Dad4, 53-107 of Dam1, 49-121 of Duo1, 22-77 of Hsk3, 7-112 of Spc19, 3-48 and 112-199 of Spc34 (PDB ID: 6CFZ) [9]. (C) (top) K.lactis MIND is a Y-shaped heterotetramer, the stem of which binds Spc24-Spc25 of Ndc80c. One globular head binds inner kinetochore proteins CENP-CMif2 and CENP-UAme1, while a N-terminal extension of Dsn1 from the other head inhibits these interactions except when phosphorylated by Aurora B kinase (PDB ID: 5T58 and 5T51) [19]. (bottom) H.sapiens Mis12c with CENP-C shares a conserved structure with its budding yeast homolog; it also forms Y-shaped heterotetramer about 200 Å long, with conserved sites of interaction with CENP-CMif2 and Ndc80(PDB ID:5LSK) [20].
While no vertebrate homolog of Dam1c has been identified, it was proposed a decade ago that the Ska complex might be Dam1c’s “metazoan functional counterpart [15].” And despite the fact that the W-shaped, dimeric Ska complex looks nothing like the Dam1c ring [16], this was confirmed by recent reports that the Ska complex, like Dam1c, can bind Ndc80c [17], bear load at microtubule tips [18], and strengthen Ndc80c-mediated microtubule binding [18].
Moving away from the microtubule interface, we come to the Mis12cMIND complex, which bridges the inner and outer kinetochore. In 2016, both the budding yeast Kluyveromycces lactis (K. lactis) [19] and human [20] Mis12cMIND structures were published, revealing a conserved Y-shaped tetramer in which the N-termini of all four substituents (Dsn1, Ns11, Pmf1Nnf1, & Mis12Mtw1) are oriented towards the inner kinetochore (Figure 2C). The N-termini of Pmf1Nnf1 and Mis12Mtw11 comprise “head I” of the Y; the N-termini of Dsn1 and Ns11 comprise “head II.” Inner kinetochore proteins CENP-UAmc1 and CENP-CMif2 both bind to head I, while head II serves a regulatory function auto-inhibiting this interaction unless phosphorylated by Aurora B kinase [19]. The same mechanism regulates the Mis12cMIND-CENP-CMif2 interaction in humans [20], indicating that the interface between Mis12MIND and the inner kinetochore is organized and regulated by a conserved mechanism.
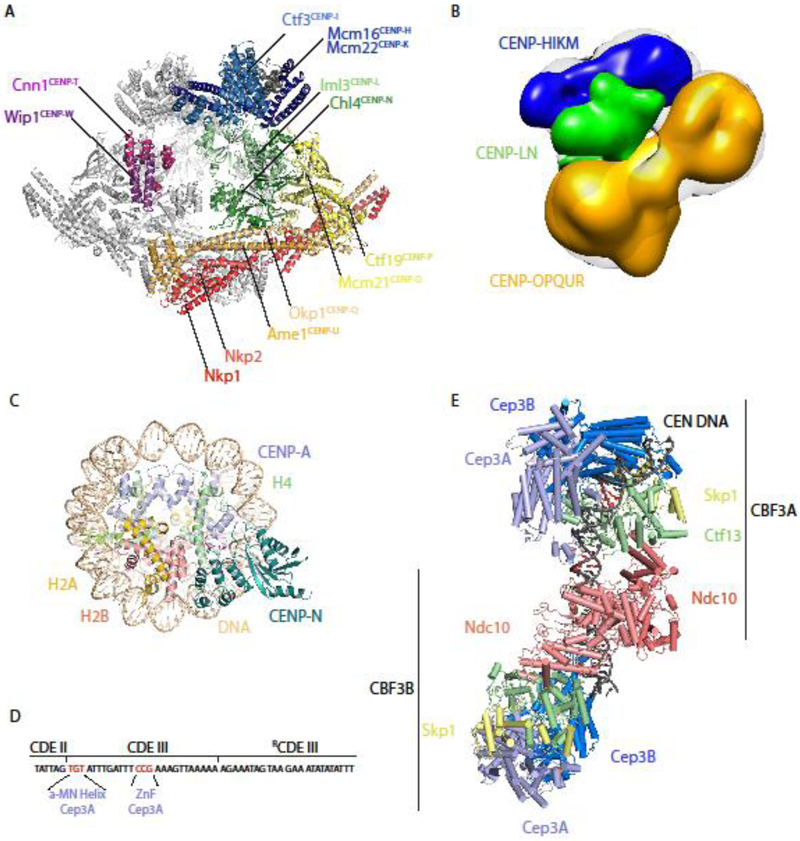
Considerable progress has also been made towards visualizing the inner kinetochore. Building upon their previously published structures of the RWD domains of K. lactis CENP-Pctf19 and CENP-OMcm21 [21], Schmitzberger and colleagues used crystallography, hydrogen-deuterium exchange, and mass spectroscopy to demonstrate that CENP-QOkp1 binds CENP-UAme1, Nkp1/Nkp2, & CENP-POctf19/Mcm21 through three distinct interfaces, which are separated by flexible elements [22].
CENP-NLch14/Im13 is an inner kinetochore protein complex of particular interest because like CENP-CMif2 it binds directly and specifically to centromeric, CENP-ACse4-containing nucleosomes [23-25]. Since 2013, we have had a partial structure of the budding yeast Saccharomyces cerevisiae (S. cerevisiae) CENP-NLch14/Im13, including the heterodimer interface [26], but it was several years before the basis of centromeric nucleosome recognition by this complex was revealed by two independently obtained cryo-EM structures of a human CENP-ACse4 nucleosome in complex with an N-terminal domain of CENP-Nch14 [27,28]. These structures revealed that CENP-Nch14 recognizes centromeric nucleosomes through specific interactions with charged residues in the L1 loop of CENP-ACse4 that are absent from histone H3; this interaction is strengthened through electrostatic interactions between basic amino acids on CENP-Nch14 and the phosphate backbone of DNA [27,28](Figure 3C).
Figure 3. Recently published structures of inner kinetochore protein complexes.
(A) Dimers of the 13-protein S. cerevisiae Ctf19c flank a central cavity. CENP-NLCh14/Im13 occupies the middle of each protomer, and its CENP-ACse4-binding domain extends into the central cavity (PDB ID: 6NUW) [29]. (B) An ~22 Å negative stain 3D reconstruction of a reconstituted 11-protein human CCAN, in which CENP-HIKM (density of subcomplex in blue) and CENP-OPQUR (density of sub-complex in yellow) sandwich CENP-NL (density of subcomplex in green) [32]. (C) The N-terminus of human CENP-N specifically recognizes centromeric, CENP-A-containing nucleosomes by binding to the unique L1 loop (indicated in red) of CENP-A [27, 28]. (Model shown from PDB ID: 6C0W, related structures: 6BUZ and 6EQT) (D) Schematic of the 56 bp nuclease-resistant sequence bound to CBF3. Conserved TGT and CCG motifs in Centromere Determining Element III (CDEIII) are specifically bound by the α-MN helix and zinc finger (ZnF) motifs, respectively, of the same Cep3A subunit [32]. (E) Budding yeast CBF3 binds centromeric DNA as a dimer, recognizing specific DNA sequences (colored red) prior to Cse4 deposition [32]. Orange spheres indicate zinc atoms. (Model shown from PDB ID: 6GYS, similar structures: 6GSA and 6F07).
These structures facilitated residue assignment in a recent high-resolution (~4 Å) reconstruction of the 13-subunit budding yeast Ctf19 complex (Ctf19c) [29](Figure 3A). Two protomers containing one copy each of nearly every inner kinetochore protein (save CENP-CMif2) flank a central cavity. The CENP-ACse4-binding β3- β4 loop of CENP-Nch14 extends into this cavity, which is, however, too small to accommodate a CENP-ACse4 nucleosome without Ctf19c dimer dissociation, significant conformational rearrangement, or partial nucleosome unwrapping [29]. The overall organization of the Ctf19c reflects published recruitment hierarchies, in which CENP-QUOA is foundational, upstream of all other inner kinetochore components, while CENP-TWCnn1/Wip1 requires nearly all other Ctf19c proteins for proper kinetochore localization [29-31]. The organization may also be conserved; Pesenti and coworkers published a low-resolution (~22 Å) negative stain 3D reconstruction of a reconstituted 11-protein human CCAN and concluded that CENP-HIKM and CENP-OPQUR sandwich CENP-NL, just as the yeast homologs of CENP-HIK and CENP-OPQU sandwich CENP-NLch14/Im13 in the Ctf19c (Figure 3B) [32].
There have also been not one but three recent structures of the CBF3 complex, an essential complex that binds the genetically specified centromere of budding yeast in the first step of CENP-ACse4 deposition and kinetochore assembly. Leber, Nans, and Singleton published a 3.6 Å cryo-EM reconstruction of a Cep3 dimer in complex with Ctf13 and Skp1[33]. The entire complex is U-shaped, with a charged central channel in which DNA was postulated to bind. Ctf13 was characterized as an F-box-leucine-rich-repeat protein for the first time, a finding corroborated in 2018 by the publication of a similar “core CBF3” structure, this time including the essential DNA-binding protein Ndc10 [34]. Intriguingly, the central channel formed by Cep3 dimers and the DNA-binding domain of Ndc10 are perpendicular, leading to the proposal that CBF3 bends centromeric DNA into a loop, perhaps to facilitate loading of the centromeric nucleosome [34]. Yan and colleagues then published a structure of the complete CBF3 in complex with a 147-bp yeast centromere, revealing how Cep3, Ctf13, and Ndc10 interact with DNA [35] (Figures 4D and 4E). Of particular interest are the two sequence-specific Centromere Determining Element III (CDEIII)-binding sites, both of which occur in the same copy of Cep3 in one protomer of a CBF3 dimer; a zinc finger of this Cep3 interacts with a conserved CCG motif, while the α-MN helix contacts a conserved TGT motif [35].
Together these structures may be assembled into a near-atomic view of much of the kinetochore, although we cannot yet visualize what an entire yeast or human kinetochore might look like. It is unclear how many copies of each complex to place in such a reconstruction; absolute protein copy numbers remain controversial [36-40]. We also lack structural information on CENP-CMif2, the essential DNA-binding protein conserved even in highly simplified kinetochores [41]. CENP-CMif2 has been described as the “blueprint” of inner kinetochore assembly [42], but comparatively little is known about what that blueprint might look like. Possibly this is because CENP-CMif2 (except for its C-terminal dimerization domain) remains largely disordered and flexible when not in contact with its binding partners [43]. This has been suggested of other kinetochore proteins, including Ame1[22], Okp1 [22], Knl1 [44], Dad1 [45], and BubR1[46]. Thus the reconstitution of increasing complete kinetochore particles may be the necessary groundwork for another leap in our structural understanding of the kinetochore [47,48].
Cell-cycle dependent kinetochore composition
As a field, we have been unable to arrive at a single answer to one of the most basic questions about the kinetochore: how many copies of each protein complex are in it? Published estimates of the number of Ndc80c at a budding yeast kinetochore range from 6 to 20 [36,37,40]. A growing trend in the field is to view these divergent answers not merely as an indication of the inherent difficulty of counting individual proteins in a living cell, but as a reflection of a dynamic kinetochore whose composition and architecture change as the cell cycle progresses.
Controversy exists at the very foundation of the kinetochore: the CENP-ACse4-containing nucleosome. While there is consensus that a single Cse4-containing nucleosome is present at each budding yeast centromere, the exact composition of that centromere is a point of controversy [39,49-53]. A widely accepted model is the most conventional one: an octameric nucleosome containing two copies each of Cse4, H2A, H2B, and H4 wrapped with DNA in a left-handed manner [53-55]. But evidence has also been published supporting “hemisome [52],” “tetrasome [56],” “hexasome [57],” and “trisome [58]” models; in 2011, Black and Cleveland proposed to reconcile this confusion with a model in which centromeric nucleosomes mature throughout the cell cycle [59]. In this model, CENP-ACse4 occupies a pre-nucleosomal trisome or hexasome during HJURPScm3-mediated deposition, a tetrasomal intermediate prior to H2A:H2B addition, and a standard octameric nucleosome for the rest of the cell cycle [59]. Within a year, biophysical evidence of centromeric nucleosomes undergoing cell cycle-coupled structural transitions in both yeast and humans was published [38,60]. However, in 2017 the Cleveland Lab — early proponents of centromeric nucleosome maturation — mapped all of the sequences bound by CENP-A onto individual α-satellite arrays in centromere reference models and concluded that CENP-A centromeric chromatin is made up of conventional octameric nucleosomes throughout the cell cycle, a conclusion supported by their biochemical, hydrodynamic, and solid-state nanopore analyses [61].
But the centromeric nucleosome maturation hypothesis still profoundly influences kinetochore research. And because CENP-ACse4 fluorescence has been used as the calibration standard for estimates of kinetochore protein copy number in methods using genetically encoded fluorescent proteins [40,62], reevaluating the number of CENP-ACse4 present at the kinetochore means reevaluating all estimates of kinetochore protein copy numbers. Indeed, building on their finding that a second CENP-ACse4 is not deposited at the centromeric nucleosome until anaphase, the Gerton Lab has recently published that nearly all kinetochore components (save Dam1c) double in abundance during anaphase [36].
While this finding has yet to be corroborated, there has long been consensus that levels of Ndc80c receptor CENP-TCnn1 increase at kinetochore during anaphase, at the same time that an Ndc80c- CENP-TCnn1 becomes detectable [31,63,64]. This phenomenon was recently explained by the Musacchio Lab’s demonstration that CENP-TCnn1 binds two Ndc80c and one Mis12cMIND at three distinct sites upon phosphorylation by the CDK1:Cyclin B complex [65]. That a cyclin-dependent kinase should regulate the association of CENP-TCnn1 with outer kinetochore components explains neatly how levels on Ndc80c at the kinetochore increase as mitosis progresses (although it is puzzling that Ndc80c-CENP-TCnn1 levels increase in anaphase, when Cyclin B is being degraded [66]). As to the why, the fact that CENP-TCnn1 recruits additional Ndc80c to the kinetochore only after the spindle assembly checkpoint (SAC) is satisfied and cells have irreversibly committed to chromosome segregation suggests that the relative importance of different Ndc80c recruitment paths may change as the cell cycle progresses.
Thus the kinetochore may change its organization and composition throughout the cell cycle in order to perform in turns its mitotic functions as tension sensor, regulatory hub, and molecular tether.
Parallel paths of Ndc80c recruitment
Over the past 15 years, the Davis and Asbury Labs have demonstrated using optical tweezers that Ndc80c and Dam1c in cooperation are the load-bearing, microtubule-binding elements of the budding yeast kinetochore and that their binding to the microtubule tip and one another is multivalent and phospho-regulated [14,31,65,67-71]. This year, Volkov and coworkers recapitulated the finding that Ndc80c multivalency is required for efficient microtubule coupling using a novel Ndc80c oligomerization platform [72]. This highlights another persistent puzzle in our field: how does a single centromeric nucleosome (in the case of budding yeast) recruit many copies of Ndc80c?
The final trend we wish to highlight in this review is the growing consensus that there are multiple, parallel pathways of Ndc80c recruitment in both yeast and human kinetochores.
In the previous section, we highlighted recent work demonstrating that each CENP-TCnn1 recruits up to three Ndc80c [65]. Using a novel kinetochore assembly assay to examine recruitment dependencies, Lang, Barber, and Biggins further elucidated this pathway in budding yeast; they determined that CENP-TCnn1 is downstream of most inner kinetochore proteins save CENP-CMif2 [31]. As both of these inner kinetochore proteins can recruit the outer kinetochore (either directly or through Mis12cMIND), Lang, Barber, and Biggins conclude that they represent two distinct Ndc80c recruitment pathways [31].
Recent work complicates this picture further. Building on the demonstration that K. lactis CENP-CMif2 and CENP-UAmc1 both bind to Mis12cMIND [19], Fischboeck and colleagues demonstrated that S. cerevisiae CENP-QUOkp1/Ame1 (like CENP-CMif2) also binds specifically to Cse4-containing nucleosomes [73]. The Ehrenhofer-Murray Lab corroborated this finding, further demonstrating that two post-translational modifications to the N-terminus of Cse4 (R37Me and K49Ac) inhibit its interaction with CENP-QUOkp1/Ame1 [74]. In light of the Musacchio Lab’s report that H. sapiens CENP-QUOkp1/Ame1 binds microtubules but not centromeric nucleosomes [47], it appears that CENP-QUOkp1/Ame1’s function may vary between organisms.
Taken together, there is now evidence for two conserved pathways of Ndc80c recruitment and hints of a possible third pathway in fungi. Multiple microtubule binding elements have been key to our conception of the kinetochore since the publication of Hill’s seminal “sleeve” model over 30 years ago [75]; only now are we beginning to understand how they are recruited.
Conclusion
Recent work has reshaped the field of kinetochore biology. For the first time we are able to visualize the protein complexes that make up this marvelous molecular machine. There is a growing sense that the organization of that machine may change throughout the cell cycle. And our understanding of the kinetochore as a multivalent microtubule coupler has been complicated by mounting evidence that there are distinct pathways whereby the microtubule-binding elements of the kinetochore are recruited.
Acknowledgements
T.D. and G.H. thank National Institutes of Health grants R01 GM040506, R35 GM130293 and T32 GM008268.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors declare no conflicts of interest.
References
- 1.Cheeseman IM, Desai A: Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008, 9:33–46. [DOI] [PubMed] [Google Scholar]
- 2.Nicklas RB, Ward SC: Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol 1994, 126:1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S: Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A, Desai A: A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai XC, McMullan G, Scheres SHW: How cryo-EM is revolutionizing structural biology. Trends in Biochemical Sciences 2015, 40:49–57. [DOI] [PubMed] [Google Scholar]
- 6.Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, et al. : Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016, 165:1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. : Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 2008, 133:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valverde R, Ingram J, Harrison SC: Conserved Tetramer Junction in the Kinetochore Ndc80 Complex. Cell Rep 2016, 17:1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Jenni S, Harrison SC: Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science 2018, 360:552–558.The authors present a cryo-EM structure of the 10-component Dam1c (from C. thermophilum) assembled into rings. In this structure, each T-shaped heterodecamer interacts with neighboring heterodecamers through two conserved interfaces and extends a "protrusion domain" towards the plus end of the microtubule and the C-termini of the Ndc80/Nuf2 dimer. The author credit flexible extensions not present in the structure with binding to Ndc80c and the microtubule lattice.
- 10.Miranda JL, De Wulf P, Sorger PK, Harrison SC: The yeast DASH complex forms closed rings on microtubules. Nature Structural & Molecular Biology 2005, 12:138. [DOI] [PubMed] [Google Scholar]
- 11.Legal T, Zou J, Sochaj A, Rappsilber J, Welburn JP: Molecular architecture of the Dam1 complex-microtubule interaction. Open Biol 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelter A, Bonomi M, Kim J, Umbreit N, Hoopmann M, Johnson R, Riffle M, Jaschob D, Maccoss M, Moritz R, et al. : The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nature Communications 2015, 6:8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Kim JO, Zelter A, Umbreit NT, Bollozos A, Riffle M, Johnson R, MacCoss MJ, Asbury CL, Davis TN: The Ndc80 complex bridges two Dam1 complex rings. Elife 2017, 6.Working with recombinantly expressed and purified yeast proteins, Kim et al. use a combination of TIRF microscopy, optical tweezers, and electron microscopy to demonstrate that in vitro the Ndc80c complex can bridge two Dam1c rings through three interfaces, each of which is regulated by Aurora B kinase.
- **14.Ng CT, Deng L, Chen C, Lim HH, Shi J, Surana U, Gan L: Electron cryotomography analysis of Dam1C/DASH at the kinetochore-spindle interface in situ. The Journal of Cell Biology 2018:jcb.201809088.Using electron cryotomography, Ng et al. find that there are both partial and complete Dam1c rings in vivo, both of which bind microtubules. They observed that complete rings have 17-fold symmetry (as seen in the recently published cryo-EM structure from the Harrison Lab) and found that a while a small fraction of kinetochore microtubules have Dam1c rings, most have only one.
- 15.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM: The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell 2009, 16:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E: Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell 2012, 46:274–286. [DOI] [PubMed] [Google Scholar]
- 17.Janczyk P, Skorupka KA, Tooley JG, Matson DR, Kestner CA, West T, Pornillos O, Stukenberg PT: Mechanism of Ska Recruitment by Ndc80 Complexes to Kinetochores. Dev Cell 2017, 41:438–449.e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **18.Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN: Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc Natl Acad Sci U S A 2018, 115:2740–2745.Using optical tweezers, Helgeson et al. demonstrate that the Ska complex can bear load at the microtubule plus end, bind Ndc80c through the C-terminal of Ska3, strengthen Ndc80c-based microtubule attachments, and affect microtubule dynamics.
- 19.Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC: Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell 2016, 167:1014–1027.el 012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovic A, Keller J, Liu Y, Overlack K, John J, Dimitrova YN, Jenni S, van Gerwen S, Stege P, Wohlgemuth S, et al. : Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell 2016, 167:1028–1040.el015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitzberger F, Harrison SC: RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep 2012, 13:216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitzberger F, Richter MM, Gordiyenko Y, Robinson CV, Dadlez M, Westermann S: Molecular basis for inner kinetochore configuration through RWD domain-peptide interactions. Embo j 2017, 36:3458–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll CW, Milks KJ, Straight AF: Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 2010, 189:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Q, Tao Y, Liu H, Teng M, Li X: Structural insights into the role of the Ch14-Im13 complex in kinetochore assembly. Acta Crystallogr D Biol Crystallogr 2013, 69:2412–2419. [DOI] [PubMed] [Google Scholar]
- 25.McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM: The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol Cell 2015, 60:886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinshaw SM, Harrison SC: An Im13-Ch14 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep 2013, 5:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, Subramaniam S: Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science (New York, N.Y.) 2017, 359.The authors present a ~3.9 Å cryo-EM reconstruction of the N-terminus of human CENP-N in complex with a CENP-A nucleosome core particle. They identify specific residues that mediate recognition of the L1 loop of CENP-A by CENP-N and confirm the importance of these residues by mutational analysis of the homologous Xenopus proteins and residue swapping experiments using the L1 loop.
- *28.Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, Luger K: Decoding the centromeric nucleosome through CENP-N. Elife 2017, 6.The authors present a crystal structure of the CENP-A-binding N-terminus of human CENP-N, as well as a ~4 Å cryo-EM reconstruction of that domain in complex with a CENP-A nucleosome core particle. The authors identify the interface between N-terminus of CENP-N and the L1 loop of CEP-A (which is absent in histone H3) as the basis for CENP-LN’s ability to recognize and bind centromeric nucleosomes.
- **29.Hinshaw SM, Harrison SC: The structure of the Ctf19c/CCAN from budding yeast. Elife 2019, 8.Thirteen inner kinetochore components of budding yeast were reconstituted and assembled into a Ctf13c complex, the structure of which was determined using cryo-EM microscopy to an overall resolution of 4.2 Å. Two protomers, each containing one copy of every protein, interface around a central cavity that in theory accommodate a partially unwrapped centromeric nulceosome. Although the Mis12cMIND-binding domains of these proteins are not present in the reconstruction, this Ctf19c structure elucidates both the molecular basis of previously reported recruitment hierarchies among inner kinetochore proteins and how the nucleosome-binding and outer kinetochore-recruiting activities of the inner kinetochore are spatially organized.
- 30.Pekgöz Altunkaya G, Malvezzi F, Demianova Z, Zimniak T, Litos G, Weissmann F, Mechtler K, Herzog F, Westermann S: CCAN Assembly Configures Composite Binding Interfaces to Promote Cross-Linking of Ndc80 Complexes at the Kinetochore. Curr Biol 2016, 26:2370–2378. [DOI] [PubMed] [Google Scholar]
- **31.Lang J, Barber A, Biggins S: An assay for de novo kinetochore assembly reveals a key role for the CENP-T pathway in budding yeast. Elife 2018, 7.The authors assemble kinetochore particles on centromeric DNA templates incubated with yeast whole-cell extracts. These kinetochore particles assemble through deposition of a centromeric nucleosome and recruitment of kinetochore proteins in a cell cycle-dependent manner. They are capable of binding microtubules. Using this platform, the authors elucidate recruitment hierarchies in the budding yeast kinetochore, ultimately concluding that there are two parallel paths of Ndc80c recruitment: the Mis12c pathway and the CENP-TCnn1 pathway.
- 32.Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, et al. : Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80. Mol Cell 2018, 71:923–939.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leber V, Nans A, Singleton MR: Structural basis for assembly of the CBF3 kinetochore complex. Embo j 2018, 37:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Lukoyanova N, Miah S, Lucas J, Vaughan CK: Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10. Cell Rep 2018, 24:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Yan K, Zhang Z, Yang J, McLaughlin SH, Barford D: Architecture of the CBF3-centromere complex of the budding yeast kinetochore. Nat Struct Mol Biol 2018, 25:1103–1110.Yan et al. present the most complete of three recently published CBF3 structures: a cryo-EM reconstruction of a CBF3-CEN3 complex, revealing the structural basis of two sequence-specific CBF3-CDEIII interactions.
- *36.Dhatchinamoorthy K, Shivaraju M, Lange JJ, Rubinstein B, Unruh JR, Slaughter BD, Gerton JL: Structural plasticity of the living kinetochore. The Journal of cell biology 2017, 216:3551.Using FRAP, calibrated imaging, and photoconversion to calculate absolute numbers of budding yeast kinetochore protein in vivo, the authors reach the unprecedented conclusion that nearly all subcomplexes (save Dam1c) add additional copies to the kinetochore during anaphase.
- 37.Lawrimore J, Bloom KS, Salmon ED: Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol 2011, 195:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL: Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 2012, 150:304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aravamudhan P, Felzer-Kim I, Joglekar AP: The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr Biol 2013, 23:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED: Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol 2006, 8:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drinnenberg IA, Henikoff S, Malik HS: Evolutionary Turnover of Kinetochore Proteins: A Ship of Theseus? Trends Cell Biol 2016, 26:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A: CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol 2015, 210:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT: Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Molecular biology of the cell 2008, 19:4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghongane P, Kapanidou M, Asghar A, Elowe S, Bolanos-Garcia VM: The dynamic protein Knl1 - a kinetochore rendezvous. J Cell Sci 2014, 127:3415–3423. [DOI] [PubMed] [Google Scholar]
- 45.Waldo JT, Greagor SA, Iqbal AJ, Gittens AS, Grant KK: The Dad1 subunit of the yeast kinetochore Dam1 complex is an intrinsically disordered protein. Biochem Biophys Res Commun 2010, 400:313–317. [DOI] [PubMed] [Google Scholar]
- 46.D'Arcy S, Davies OR, Blundell TL, Bolanos-Garcia VM: Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J Biol Chem 2010, 285:14764–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, et al. : Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80. Molecular Cell 2018, 71:923–939.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischböck J, Pentakota S, Keller J, Pesenti ME, Pan D, et al. : Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 2016, 537:249–253. [DOI] [PubMed] [Google Scholar]
- 49.Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M: Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol 1999, 19:6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coffman VC, Wu P, Parthun MR, Wu JQ: CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol 2011, 195:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henikoff S, Henikoff JG: "Point" centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics 2012, 190:1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henikoff S, Ramachandran S, Krassovsky K, Bryson TD, Codomo CA, Brogaard K, Widom J, Wang J-P, Henikoff JG, Struhl K: The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL: Cse4 Is Part of an Octameric Nucleosome in Budding Yeast. Molecular Cell 2009, 35:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekulic N, Bassett EA, Rogers DJ, Black BE: The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 2010, 467:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park S-Y, et al. : Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 2011, 476:232. [DOI] [PubMed] [Google Scholar]
- 56.Williams JS, Hayashi T, Yanagida M, Russell P: Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 2009, 33:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C: Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 2007, 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- 58.Furuyama T, Henikoff S: Centromeric nucleosomes induce positive DNA supercoils. Cell 2009, 138:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Black BE, Cleveland DW: Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 2011, 144:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bui M, Dimitriadis Emilios k, Hoischen C, An E, Quénet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y: Cell-Cycle-Dependent Structural Transitions in the Human CENP-A Nucleosome In Vivo. Cell 2012, 150:317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AY, Nguyen K, Dekker C, et al. : Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. The Journal of Cell Biology 2017, 216:607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joglekar AP, Bloom K, Salmon ED: In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol 2009, 19:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, Alfieri C, Golfieri C, Oldani A, Maschio MD, et al. : Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nature Cell Biology 2012, 14:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S: CENP- T proteins are conserved centromere receptors of the Ndc80 complex. Nature Cell Biology 2012, 14:604. [DOI] [PubMed] [Google Scholar]
- 65.Huis In 't Veld PJ, Jeganathan S, Petrovic A, Singh P, John J, Krenn V, Weissmann F, Bange T, Musacchio A: Molecular basis of outer kinetochore assembly on CENP-T. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minshull J, Pines J, Golsteyn R, Standart N, Mackie S, Colman A, Blow J, Ruderman J, Wu M, Hunt T: The role of cyclin synthesis, modification and destruction in the control of cell division. Journal of Cell Science 1989, 1989:77–97. [DOI] [PubMed] [Google Scholar]
- 67.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN: The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A 2006, 103:9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Charles LA, Trisha ND: Phosphoregulation and depolymerization- driven movement of the Dam1 complex do not require ring formation. Nature Cell Biology 2008, 10:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL: The Ndc80 Kinetochore Complex Forms Load-Bearing Attachments to Dynamic Microtubule Tips via Biased Diffusion. Cell 2009, 136:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN: Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol 2010, 189:713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Umbreit NT, Miller MP, Tien JF, Ortola JC, Gui L, Lee KK, Biggins S, Asbury CL, Davis TN: Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat Commun 2014, 5:4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *72.Volkov VA, Huis In 't Veld PJ, Dogterom M, Musacchio A: Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. Elife 2018, 7.Using TIRF microscopy and a multimerization platform for human Ndc80c, the authors demonstrate that multivalency is essential for the microtubule-coupling ability of this complex.
- 73.Fischboeck J, Singh S, Potocnjak M, Hagemann G, Solis V, Woike S, Ghodgaonkar M, Andreani J, Herzog F: The COMA complex is required for positioning Ipl1 activity proximal to Cse4 nucleosomes in budding yeast. bioRxiv 2018:444570. [Google Scholar]
- 74.Anedchenko EA, Samel-Pommerencke A, Tran Nguyen TM, Shahnejat-Bushehri S, Pöpsel J, Lauster D, Herrmann A, Rappsilber J, Cuomo A, Bonaldi T, et al. : The kinetochore module Okp1 CENP-Q /Ame1 CENP-U is a reader for N-terminal modifications on the centromeric histone Cse4 CENP-A. EMBO Journal 2019, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hill TL: Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A 1985, 82:4404–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]