Abstract
Monitoring of anti-malarial drug resistance is vital in Northeast India as this region shares its international border with Southeast Asia. Genetic diversity of Plasmodium parasites regulates transmission dynamics, disease severity and vaccine efficacy. P. falciparum chloroquine resistance transporter (Pfcrt), multidrug resistance-1 (Pfmdr-1) and kelch 13 propeller (PfK-13) genes which govern antimalarial drug resistance and three genetic diversity markers, merozoite surface protein 1 and 2 (Pfmsp-1, Pfmsp-2) and glutamate rich protein (Pfglurp) were evaluated from Tripura, Northeast India using molecular tools. In the Pfcrt gene, 87% isolates showed triple mutations at codons M74I, N75E and K76T. 12.5% isolates in Pfmdr-1 gene showed mutation at N86Y. No polymorphism in PfK-13 propeller was found. Polyclonal infections were observed in 53.85% isolates and more commonly in adults (p = 0.0494). In the Pfmsp-1 locus, the K1 allelic family was predominant (71.2%) followed by the 3D7/IC family (69.2%) in the Pfmsp-2 locus. RII region of Pfglurp exhibited nine alleles with expected heterozygosity of 0.85. The multiplicity of infection for Pfmsp-1, Pfmsp-2 and Pfglurp were 1.56, 1.31 and 1.06 respectively. Overall, the study demonstrated a high level of chloroquine resistance and extensive parasite diversity in the region, necessitating regular surveillance in this population group.
Subject terms: Parasite genetics, Malaria
Introduction
Northeast (NE) India, with a population of 45,772,188 (2011 census of India) contributes to approximately 28,341 malaria cases in a year, mostly due to Plasmodium falciparum1. In 2018, almost half (46.15%) of these cases were reported from the state of Tripura1. The NE region is strategically situated and shares a long international border with Southeast Asian countries such as Bhutan, Nepal, China, Myanmar and Bangladesh. This region is highly vulnerable for the importation of anti-malarial drug resistant strains of P. falciparum from surrounding Southeast Asian countries and thereby provides a gateway to the rest of India. The state of Tripura is highly endemic to malaria and a large outbreak was reported in 2014 with around 10,000 malaria cases and more than a hundred deaths2.
Many areas of NE region have stable malaria with a high asymptomatic carrier rate and high levels of transmission which increases the vulnerability of people to polyclonal infections3. Population movement to other parts of India where unstable malaria is prevalent disseminates new parasite strains resulting in malaria outbreaks and spread of drug resistance. Keeping in mind the diversity of the region and its proximity to malaria hot spots of Southeast Asia, drug resistance monitoring is vital. Chloroquine (CQ) was initially the most commonly used drug for presumptive treatment of all fever cases followed by 8-aminoquinoline for radical cure of falciparum malaria and to prevent relapses in vivax malaria4. CQ resistance has been linked to two parasite molecular markers: P. falciparum CQ resistance transporter (Pfcrt) gene located on chromosome 7 and P. falciparum multidrug resistance-1 (Pfmdr-1) gene situated on chromosome 55,6. The Kelch 13-propeller (K-13) domain has been recently linked with artemisinin drug resistance both in vitro and in vivo and has been utilised widely since then for molecular surveillance7.
The information on genetic diversity of Plasmodium parasites is essential in developing an effective malaria vaccine because antigenic variation in these parasites significantly hinders vaccine research due to multiple alleles effectively evading vaccine-induced allele specific immunity. The merozoite surface protein 1 and 2 (msp1 and msp2) and glutamate rich protein (glurp) of P. falciparum are potential candidate antigens for vaccine development8,9. Multiplicity of infection (MOI) is a parameter that is not only related to the transmission intensity of malaria but also predicts disease severity10. Infection by more than one parasite genotype has the potential to naturally select more virulent strains and thereby cause more severe infections10.
The current study examined the genetic diversity of P. falciparum parasites and distribution of anti-malarial drug resistance genes in Tripura state, NE India. The findings will help in understanding the current status of drug resistance genes and genetic structure of P. falciparum parasites which will improve existing and future malaria control strategies in the region.
Results
Study population
A total of 242 patients with fever (133 males, 109 females) were screened for malaria using microscopy and rapid diagnostic tests (RDT). Of these, 84 (males 53, females 31, p = 0.078, OR = 1.67) were found positive for malaria (P. falciparum infection 84.52%, P. vivax 10.71% and mixed infection 4.76%). Children aged ≤15 years had a higher rate of infection as compared to adults (36.11% vs. 28.57%, OR = 1.413), however the difference was not significant statistically (p = 0.2657). All cases positive by RDT and microscopy were also positive by nested Polymerase chain reaction (PCR) for species identification.
Mutation analysis of drug resistance genes
P. falciparum Pfcrt, Pfmdr-1 and PfK-13 genes were amplified using a nested PCR protocol followed by Sanger sequencing.
A total of 64 P. falciparum isolates were sequenced for Pfmdr-1 gene (NCBI Gene Bank Accession: MG641959.1 - MG642022.1). Majority of the isolates were wild type (n = 55, 85.9%); however, eight isolates (12.5%) showed a single nucleotide polymorphism (SNP) at N86Y and one (1.6%) at E130K.
With regard to Pfcrt gene, a total of 46 isolates (NCBI Gene Bank Accession: MH823820.1 - MH823865.1) were successfully sequenced and analysed. Of these, 87% (n = 40) of isolates carried mutant genotypes and only 13% had wild type allele. Among the mutant genotypes, all the isolates (100%, n = 40) showed triple mutations at codons M74I, N75E and K76T. Two haplotypes were identified; CVIET had 40 sequences and CVMNK had 6 sequences. Haplotype diversity was found to be 0.232 (Variance: 0.00544, SD: 0.074).
A total of sixty-two isolates were amplified for PfK-13 by nested PCR and 48 isolates were successfully sequenced and analysed (NCBI Gene Bank accession MG366541.1-MG366588.1). No polymorphism in K-13 propeller gene was found that confer resistance to artemisinin-combination therapy (ACT). At the nucleotide level as well, no mutations were observed.
Linkage disequilibrium (LD) was calculated by estimating r2 values between all possible pairs (15 pairs) of SNPs present in Pfcrt and Pfmdr-1 genes to study the presence of any intergenic or intragenic association. It was observed from the LD analysis that there is a significant intragenic association among all the 4 SNPs of Pfcrt locus whereas no intergenic association was observed between Pfcrt and Pfmdr-1 genes from the analysis (Fig. 1).
Figure 1.
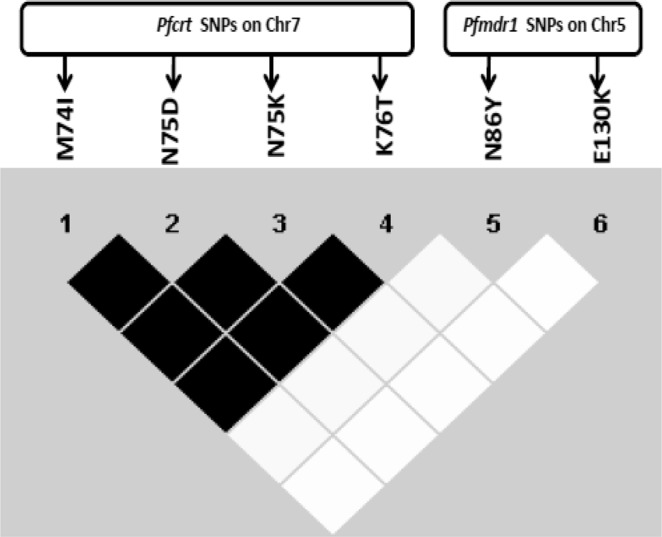
Linkage disequilibrium (LD) plot between the SNPs of Pfcrt and Pfmdr1 gene in P. falciparum isolates collected from Tripura. The strength of LD between the SNPs was determined by the association of statistical significance by calculating the r2 values and represented with the extent of darkness of the boxes (black colour depicts strong LD and white colour depicts weak or no LD).
Genetic diversity of P. falciparum isolates
Allelic frequency, multiplicity of infection (MOI) and expected heterozygosity (HE) were examined in the three genetic diversity genes of P. falciparum, viz: Pfmsp-1, Pfmsp-2 and Pfglurp using PCR.
A total of 52 clinical isolates were successfully amplified for the Pfmsp-1, Pfmsp-2 and Pfglurp genes. Polyclonal infections were observed in 53.85% (n = 28) isolates and were significantly more common in the adult population (71.43% vs. 41.94%, p = 0.0494). The overall MOI for Pfmsp-1, Pfmsp-2 and Pfglurp were 1.56, 1.31 and 1.06, respectively (Table 1).
Table 1.
Multiplicity of infection (MOI), Expected Heterozygosity (HE) and frequency of allelic variants of Pfmsp-1, Pfmsp-2 and Pfglurp genes.
| Genes | Allele frequency (n = 52) | Fragment size range (bp) | Number of genotypes | Monoclonal infection (%) n = 52 |
Polyclonal infection (%) n = 52 |
chi-square p- value (Monoclonal vs. polyclonal infections) |
MOI (n = 52) |
HE |
|---|---|---|---|---|---|---|---|---|
| Pfmsp-1 |
30 (57.7%) |
22 (42.3%) |
0.000086 | 1.56 | 0.89 | |||
| K1 | 37 | 150–290 | 8 | |||||
| MAD20 | 28 | 150–230 | 4 | |||||
| RO33 | 16 | 160 | 1 | |||||
| Pfmsp-2 |
36 (69.2%) |
16 (30.8%) |
1.31 | 0.81 | ||||
| FC27 | 32 | 310–420 | 4 | |||||
| 3D7/IC | 36 | 490–610 | 5 | |||||
| Pfglurp | 55 | 550–1010 | 9 | 49 (94.2%) | 3 (5.8%) | 1.06 | 0.85 | |
The distribution of the allelic variants in children (aged ≤15 years) and adults is summarized in Table 2. MOI for Pfmsp-1and Pfmsp-2 genes was observed to be higher among adults; however, for Pfglurp, it was higher in children (1.1 vs. 1.0).
Table 2.
Difference in Multiplicity of infection (MOI) between adults and children.
| Genes | Children ≤ 15 yrs. | Adults | p-value | |||
|---|---|---|---|---|---|---|
| Allele frequency, n = 31 |
MOI | Allele frequency, n = 21 | MOI | |||
| Pfmsp-1 | K1 | 21 | 1.48 | 16 | 1.67 | 0.5512 |
| MAD20 | 17 | 11 | 1.0 | |||
| RO33 | 8 | 8 | 0.3753 | |||
| Pfmsp-2 | FC27 | 18 | 1.26 | 14 | 1.38 | 0.5746 |
| 3D7/IC | 21 | 15 | 1.0 | |||
| Pfglurp | 34 | 1.1 | 21 | 1.0 | 0.0053 | |
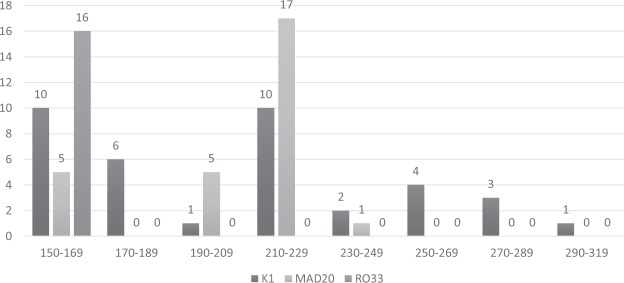
In the Pfmsp-1 locus, frequency of K1 allelic family was observed to be the highest among the isolates (n = 37, 71.2%) with 8 different allele fragments (range: 150–290 bp) of which the 210 bp allele was predominant (Fig. 2). While in the MAD20 and RO33 family, 53.8% (n = 28) and 30.8% (n = 16) isolates showed 4 and 1 allelic variants respectively. In addition, the 220 bp and 160 bp fragments were found to be the most predominant in MAD20 and RO33 family, respectively. The expected heterozygosity (HE) was found to be 0.89.
Figure 2.
Distribution of Pfmsp-1 (K1, MAD20 and RO33) allelic fragments.
Among the Pfmsp-2 allelic family, FC27 was detected in 61.5% (n = 32) isolates with allelic fragments ranging in size from 310–420 bp and the predominant allelic fragment was found to be 310 bp in size (Fig. 3). The 3D7/IC allelic family was detected in 69.2% (n = 36) isolates and the 520 bp fragment was found in 36.1% (n = 13) isolates (Fig. 4). The expected heterozygosity (HE) was found to be 0.81.
Figure 3.
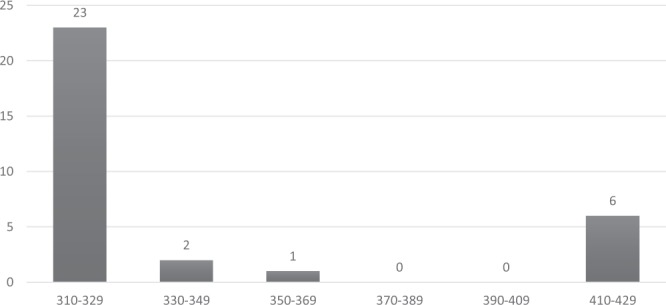
Distribution of Pfmsp-2 (FC) allelic fragments.
Figure 4.
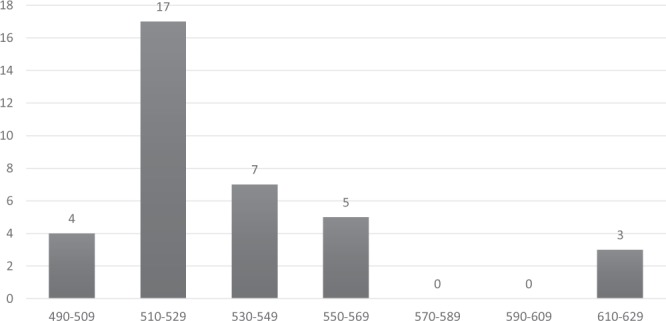
Distribution of Pfmsp-2 (3D7) allelic fragments.
A total of nine different allelic fragments (550–1010 bp) were observed in the RII region of Pfglurp (Fig. 5). The 850 bp fragment was the most commonly detected allele (n = 11, 21.15%). The expected heterozygosity (HE) was found to be 0.85.
Figure 5.
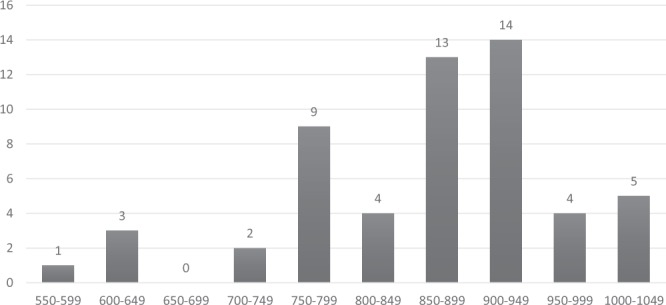
Distribution of Pfglurp allelic fragments.
The frequencies of the allelic variants of the three genetic loci are shown in Table 1. MOI was found to be highest for Pfmsp-1, where the highest number (42.3%) of polyclonal infections was also detected. In the polyclonal infections, the following allelic combinations were observed in Pfmsp-1 gene: K1/MAD20 in 21.2% (n = 11) isolates, K1/RO33 in 17.3% (n = 9) isolates, MAD20/RO33 in 15.4% (n = 8) isolates and K1/MAD20/RO33 (triple allele) in 5.8% (n = 3) clinical isolates (Fig. 6). For Pfmsp-2 gene, the FC27/3D7 alleles in combination were present in 16 isolates. In the Pfglurp locus, 5.8% (n = 3) isolates were found to be polyclonal, all of which occurred in children ≤15 years of age.
Figure 6.
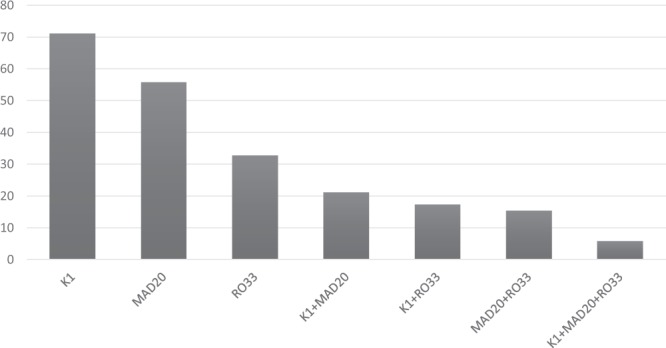
Frequency of Pfmsp-1 allelic fragment combinations (in percentages).
Correlation between Polyclonality of infection and mutation in anti-malarial drug resistance genes
Out of the 71 P. falciparum positive cases, 35 samples with complete data for both the parasite genetic diversity markers (Pfmsp-1, Pfmsp-2 and Pfglurp) and drug resistance gene markers (Pfcrt and Pfmdr-1) were analysed to explore any association between polyclonality and anti-malarial drug resistance. It was observed that 95.5% of the polyclonal infections harboured a Pfcrt mutation whereas for monoclonal infections, the rate was 76.9% (p = 0.26474). Similarly, it was found that 13.6% of the polyclonal infections harboured a mutation in the Pfmdr-1 gene while only 7.7% of the monoclonal infections had the same; the differences were however, not statistically significant (Table 3).
Table 3.
Correlation between Polyclonal infections and mutation in drug resistance genes (Pfcrt and Pfmdr-1).
| Pfcrt Mutation |
p-value (2 Tailed P Test) |
Pfmdr-1 Mutation |
p-value (2 Tailed P Test) |
|||
|---|---|---|---|---|---|---|
| Yes | No | Yes | No | |||
| Polyclonal | 21 | 1 | 0.26474 | 3 | 19 | 1.0 |
| Monoclonal | 10 | 3 | 1 | 12 | ||
Discussion
The current study was carried out to characterize the major molecular markers governing anti-malarial drug resistance and genetic diversity in P. falciparum isolates obtained from a highly malaria endemic area of Tripura in Northeast India. The status of drug resistant parasite strains in a community and the diversity of the parasite population are important determinants in understanding the molecular epidemiology of malaria, especially in high endemic areas.
The Pfcrt protein is localised on the digestive vacuole of the malaria parasite and functions as an anion channel to mediate the efflux of CQ outside the vacuole11. Mutation in the Pfcrt domain, specifically the replacement of lysine 76 with threonine is the most widely described marker for CQ resistance12. This mutation leads to an increase in the lipophilicity and negativity of the Pfcrt protein and specifically favours the efflux of positively charged CQ fractions outside the digestive vacuole. Lysine, having a net positive charge and possessing a bulkier side chain as compared to threonine, inhibits the efflux of CQ fractions from the digestive vacuole11. The Pfmdr-1 protein is also a transporter on the digestive vacuole membrane of the malaria parasite and it normally mediates the transfer of drugs such as CQ from the cytosol into the digestive vacuole. Single nucleotide polymorphisms in the Pfmdr-1 gene such as the N86Y mutation lead to changes in the physicochemical properties of the transporter thereby altering its ability to bind and transfer the target drugs11.
Originating from the Thai-Cambodia border in the 1950s to its appearance in Assam, Northeast India in 1973, CQ resistance has spread extensively in other parts of India12,13. In Assam, during 2006–2007, prevalence of the mutant Pfcrt genotype (K76T) and Pfmdr-1 genotype (N86Y) was 99% and 68%, respectively13. The widespread resistance warranted a change in the anti-malarial drug policy in India with Artemisinin based combination therapy (ACT) replacing CQ for uncomplicated falciparum malaria in 200814. In the present study, prevalence of Pfcrt mutation was observed to be high (87%) as compared to previous studies from India15,16. In addition, majority of our isolates harboured the triple mutant genotype (C72V73I74E75T76) and only 13% isolates carried wild type genotype, which is in agreement with previous Indian studies after CQ withdrawal15,16. In this scenario, the efficacy of implementation of ACT needs to be explored in the region; the possibility that CQ might still be used unofficially for uncomplicated P. falciparum malaria is high and needs to be evaluated. With the current emphasis on the return of CQ sensitive parasites in malaria endemic regions after the withdrawal of drug pressure for sufficient time, the present finding of a high rate of Pfcrt mutations in Tripura represents a challenge for the future control of malaria in the region where resistance to ACT might become a reality. The changing trend of CQ efficacy and Pfcrt mutations after drug withdrawal is interesting. The Comoro islands reported a dramatic reduction in the Pfcrt K76T mutation after CQ withdrawal17. Similar trends were also observed in Ghana, Kenya, Malawi and other African countries18–20. This is encouraging and reinforces the possibility of reintroduction of CQ in the future for malaria control. However, contrasting findings have also been reported from Gabon and Benin where the frequency of Pfcrt K76T mutant was still high after four to seven years of CQ withdrawal21,22. Therefore, a constant monitoring of CQ resistance along with compliance and adherence of anti-malarial drug administration is highly required to assess the dynamics of CQ resistance in an area.
The prevalence of N86Y mutation in the Pfmdr-1 gene was 12.5%, which is lower than other studies from Assam, Chhattisgarh and Puducherry, but comparable to figures reported from Orissa13,15,16. Although the Pfcrt and Pfmdr-1 gene mutations have been linked to CQ resistance, their role is still unclear and many studies have failed to find any association16,23,24.
In this study, LD analysis revealed significant intragenic association between SNPs detected in the Pfcrt locus, while no intergenic association with Pfmdr-1 was noted. This is in agreement with previous studies from other parts of India16,25.
With the current use of ACTs for P. falciparum malaria throughout the world, monitoring anti-malarial drug resistance and treatment efficacy is important for malaria control. In recent years, Kelch 13 propeller domain based molecular surveillance technique has become an important tool and is widely used7. In the current study, no polymorphisms were noted, concurrent with previous reports from other parts of India15,26. This however, needs to be evaluated further as non-synonymous mutations in the K13 propeller domain were reported from this area earlier but were not associated with ACT treatment failure27,28. Recently, Chakrabarti et al., demonstrated reduced artemisinin sensitivity in the north-eastern isolates of P. falciparum by using ring stage survival assay demanding a future investigation on the potential change in the ACT effectiveness in P. falciparum parasites from NE India29.
In the milieu of the prevailing polymorphisms detected in the anti-malarial drug resistance genes, the current study also analysed the diversity patterns in potential vaccine candidate genes of P. falciparum parasites (Pfmsp-1, Pfmsp-2 and Pfglurp) circulating in the region. The genetic polymorphism analysis presents an interesting prospect for the future adoption of a successful vaccine candidate in this part of the country30. In addition, earlier studies have shown that highly complex malaria infections, as demonstrated by high allelic diversities and polyclonal infections have a propensity to select drug resistant parasites and cause more virulent infections10,31,32. In the present study, Pfmsp-1 locus exhibited 13 different genotypes and K1 allelic family was the most frequently observed followed by MAD20 and RO33. This finding is in contrast with previous studies from neighbouring Arunachal Pradesh and Chandigarh, North India where RO33 allelic frequency was highest followed by MAD20 and K133,34. However, MAD20 was found to be more frequent than K1 and RO33 in Central India and Bangladesh35,36.
Hitherto, a wide range of diversity patterns in the block-2 region of Pfmsp-1 and their correlation with disease severity and endemicity patterns have been reported worldwide37,38. Ranjit et al. in Orissa, India demonstrated an association of the 200 bp allelic fragment of MAD20 and 550 bp allele of 3D7 with severe malaria cases39. The block-2 region of Pfmsp-1 contains degenerate tripeptides and repeat sequences which have been shown to participate in recognition of erythrocyte surface and incorporation into the red cell cytoskeleton in malaria pathogenesis40. Specific allelic forms of Pfmsp-1 in isolation or in combination with other markers might favour the expedited entry of the malaria parasite into the red cells thereby favouring rapid multiplication, high parasitaemia and more severe disease39,41. However, this association could not be ascertained in our study as patient follow-up was not a part of the study protocol.
In the present study, the Pfmsp-2 family with 9 different allelic forms was found to be less polymorphic as compared to the Pfmsp-1 allelic family. In addition, within the Pfmsp-2 family, the 3D7 component was more abundant as compared to FC27 with both the allelic variants occurring together in 30.8% isolates, which corroborates with earlier reports from Myanmar and Cameroon42,43. The presence of 13 and 9 different allelic forms for Pfmsp-1 and Pfmsp-2 respectively is concurrent with previous studies from mesoendemic to hypoendemic Asian countries like Thailand and Iran44,45. The R2 region of Pfglurp was observed in all our isolates and showed considerable polymorphism with 9 allelic variants with frequencies ranging from 1.8% to 25.5%; similar findings were obtained from Arunachal Pradesh and Assam, Northeast India33,46. Additionally, the greater number of allelic variants with low allele frequencies of Pfglurp encountered in the present study is suggestive of high endemicity in the area. Regions with low malaria endemicity like Central and South American countries were found to have two to four Pfglurp (R2) alleles47,48. However, most of the studies from highly endemic areas in Asia and Africa have reported eight to twenty Pfglurp (R2) alleles39,44,46,49.
Multiplicity of infection has been shown to be invariably related to transmission intensity and parasite prevalence, but results are still inconclusive50. In the current study, MOI for each of the three selected polymorphic antigenic genes ranged from 1.06 to 1.56, the highest being noted for Pfmsp-1 and lowest for Pfglurp. These results are in agreement with earlier studies from Chhattisgarh, central India and Udalguri district of Assam where malaria is endemic35,46. In recent years, two major changes were recommended in anti-malarial drug policy in Northeast region; the first was the introduction of ACT in 2010 and the second was the replacement of Artemether and sulphadoxine-pyrimethamine combination (A + SP) with Artemether-Lumefantrine (A + L) in 2013. The MOI over this period has remained largely static; the slight reduction might be attributable to the change in drug policy or other vector control measures46. African studies conducted in different transmission settings have reported the lowest MOI rates from areas of low transmission and highest MOI figures from regions where malaria transmission was perennial51. In the present study, we found a higher MOI among adult population with regard to the Pfmsp-1 and Pfmsp-2 antigenic markers, which points to greater exposure to infection with increasing age. To date, studies have reported both similar and conflicting observations and some have found no association of MOI with age52–54.
Heterozygosity values observed in our study for the individual antigenic markers were slightly higher than those reported earlier from Southeast Asia/Pacific and South American countries, but concurrent with those seen in African locations51,54. The HE values of Pfglurp were concurrent with previous reports from Assam46. The high HE values suggest a high transmission rate and a comparatively large parasite population circulating in the region. In this scenario, the possibility of genetic recombination of the parasite strains in mosquito vectors is also expected to be considerable55. The involvement of the indigenous tribes in Jhum, a shifting form of cultivation also results in a high rate of man-mosquito contact in this region thereby facilitating this kind of interaction56.
The current study documented a high rate of polyclonal infections with comparatively higher rates of mutation in Pfcrt and Pfmdr-1 genes as compared to that observed in monoclonal infections. The presence of infections with multiple alleles is indicative of considerable genetic diversity in the parasite population which in turn can lead to the emergence and proliferation of drug resistant clones31,57. Similarly, MOI and polyclonality of P. falciparum can also be considered as an indicator of malaria control efforts in an area, as demonstrated by Hetzel et al., where a higher MOI and polyclonality was found to be associated with an area of no intervention58. A positive association was observed between the rate of polyclonal infections and annual parasite incidence in Indonesia indicating that polyclonality of P. falciparum over an area might provide information on local transmission intensity59. Moreover, in most polyclonal infections, there are invariably large populations of drug sensitive parasites which mask the detection of small populations of drug resistant strains (minority variants) by standard PCR; accurate detection requires more sensitive methods as demonstrated in Malawi60,61.
In the context of high parasite diversity and circulating population of drug resistant strains, whether the genetic structure of the patients themselves residing in this area has a role to play is another interesting aspect. Historically, the indigenous tribes of Tripura belong to the Tibeto – Burman language family which is more or less a homogenous group with high rates of endogamy62,63. Previous studies carried out in African ethnic groups have demonstrated that both immune response and susceptibility to malaria infection might vary depending on the genetic background of the study population. Even drug resistance patterns can vary with genetic polymorphism patterns in host enzymes64. Similar information from malaria endemic areas of India would provide valuable leads for control measures.
Tripura faced an epidemic of malaria in 2014 with an increase of almost seven-fold P. falciparum malaria cases over 201365. It has been observed that malaria in Tripura was in a state of decline till 2014. However, the epidemic has reset the baseline case load of malaria in the state at a higher level than before. A clonal population structure with identical genotype of P. falciparum is expected to circulate during the epidemic period66. However, what drives the changes in the clonal structure and diversity of parasites in the post epidemic period is still not clear.
The limitations of the present study include (i) inability to correlate polymorphism patterns observed in anti-malarial drug resistance genes with treatment response or clinical outcome and (ii) a relatively small sample size. Nevertheless, the strength of the current study lies in the adoption of robust protocols for characterization of the major drug resistance and genetic diversity genes of P. falciparum. Although the number of malaria positive cases included in the characterization of the genetic markers was not very high, there was no obvious sampling bias and sufficient care was taken to cover two of the most highly malaria endemic districts of Tripura. Cases were included following proper ethical guidelines and treatment was also provided to the patients as per national guidelines.
In conclusion, the present study showed a high level of polymorphism in the genes governing CQ resistance in the region even after a significant period of drug withdrawal as compared to other malaria endemic areas of India and Africa. No polymorphisms were found in the PfK13 domain, which, however, needs to be evaluated further in the milieu of extensive genetic diversity observed in the parasite population. The polymorphic regions of Pfmsp-1, Pfmsp-2 and Pfglurp genes exhibited high allelic diversity suggestive of high malaria endemicity and high heterozygosity in the study area. All these factors are conducive to the gradual selection and proliferation of drug resistant parasites and may make malaria more difficult to control in this region in the days to come.
Materials and Methods
Study areas and design
This study was carried out in the malaria endemic areas of North Tripura and Dhalai districts of Tripura state (located between Lat. 23.9408°N and Long. 91.9882°E), Northeast India in May 2015. The population composition in these areas is primarily tribal, with Tripuri, Reang and Chakma tribes predominating. Symptomatic patients were recruited as per the following inclusion criterion: body temperature ≥37.5 °C, age >1 year, history of fever within one week, no recent history of consumption of anti-malarial drugs and absence of severe malnutrition or signs of severe malaria. Symptomatic patients were screened for Plasmodium parasites using rapid diagnostic test (RDT) and microscopic examination of thick and thin blood smears stained with Jaswant Singh and Bhattacharjee (JSB) stain. Two ml of whole blood was collected from malaria positive patients after informed written consent. All malaria positive patients were given treatment with anti-malarial drugs as per the National Vector borne Disease Control Programme (NVBDCP) guidelines for NE India67.
Ethical approval
The current study was approved by the Institutional Ethics Committee (Human) of ICMR - RMRC N. E. Region, Dibrugarh, Assam (Ref: RMRC/Dib./IEC Human/2012/667) and all study protocols and procedures were carried out according to guidelines of Indian Council of Medical Research (ICMR). Written informed consent was obtained from the study participants (parents of participants in case of minors) prior to collection of blood samples.
DNA extraction and nested PCR for malaria parasite species identification
Parasite genomic DNA was extracted from whole blood samples using the QIAamp DNA blood mini kit as per manufacturer’s instructions (Qiagen, CA, USA). The extracted DNA was stored at −20 °C for further analysis. Conventional nested PCR was performed using pan-plasmodium and species-specific primers for molecular identification of the malarial parasites as described previously68.
Genotyping of drug resistance and vaccine candidate genes
All samples which tested positive for P. falciparum were subsequently subjected to further molecular characterisation. The drug resistance markers; PfK13, Pfmdr1 and Pfcrt genes were amplified and sequenced as described previously7,23,69. The PCR products were purified using a column based purification protocol (High Pure PCR Product Purification Kit, Roche) and sequenced using the Sanger’s technique70. The sequences were edited using BioEdit ver 7.2.5 software and aligned using the ClustalW multiple alignment tool built into the software. Edited sequences were submitted to the NCBI GeneBank. The DNA sequences were translated using the ExPASy portal and the amino acid sequences generated were tallied with NCBI database reference sequences using ClustalW71.
The genetic polymorphism in Pfmsp-1, Pfmsp-2 and Pfglurp genes was examined using allele specific nested PCR as previously described without the aid of nucleotide sequencing72. Specifically, the K1, MAD20 and RO33 allelic families of Pfmsp-1 block 2, FC27 and 3D7/IC families of Pfmsp-2 block 3 and Region 2 (RII) of Pfglurp were amplified using primers specific for the individual allelic variants. Amplified PCR products were subjected to agarose gel electrophoresis and stained with EtBr followed by visualisation in a BioRad XR gel documentation system. Laboratory adapted strains, Dd2 and 3D7, were used as controls.
Multiplicity of infection and heterozygosity
The grouping of the different fragments based on amplified DNA product size was done as described previously for determination of multiplicity of infection (MOI); for msp1 and msp2, alleles having a size difference within 20 bp were considered the same, while for Pfglurp; a larger interval of 50 bp was considered73. A monoclonal infection was identified by the presence of a single PCR band for each locus and a polyclonal infection was defined as the presence of multiple PCR fragments for any of the three loci. Multiplicity of infection was defined as the average number of different parasite genotypes (denoted by the maximum number of bands detected for either loci) infecting a single host simultaneously54. Expected heterozygosity (HE) which denotes the possibility of being infected simultaneously by two parasites with different alleles at a given locus was estimated by using the following formula: HE = [n/(n − 1)] [(1 − Σpi2)], where ‘n’ is the total number of samples tested and ‘pi’ is the frequency of the allele (%) at the given locus54.
Statistical analysis
Statistical analyses were performed using EpiInfo ver. 7.2.2.6. Fisher’s exact test and 2 tailed test were used for determining the significance of monoclonal versus polyclonal infections and relationship of the diversity genes with age. Statistical significance was considered at a p value < 0.05. MOI was calculated independently for each gene by dividing the total number of alleles obtained for a given gene (Pfmsp-1, Pfmsp-2, Pfglurp) by the number of isolates positive for that gene by PCR54. To determine the association between the SNPs detected in the drug resistance genes, both intergenic and intragenic linkage disequilibrium (LD) analysis was done using the program Haploview74.
Acknowledgements
This publication was made possible by an NPRP grant (NPRP 5- 098-3-021) from the Qatar National Research Fund (QNRF). The statements made herein are solely the responsibility of the author(s). We thank the study participants, their guardians and relatives for providing their informed consent. The authors also acknowledge the efforts of the health authorities in Tripura for their cooperation in patient recruitment.
Author Contributions
S.J.P., N.P.S., P.K.M., D.B., P.K.B., R.S., J.M. and A.A.S. designed the experiments. S.J.P., N.P.S., N.B., D.K.S., T.N., D.R.B., P.K.M., D.B., P.K.B., R.S., J.M. and A.A.S. performed the experiments. S.J.P., K.S., N.P.S., N.B., D.K.S., T.N., D.B. analyzed the data. S.J.P., K.S., N.P.S., N.B., D.K.S., T.N., D.R.B., P.K.M., D.B., P.K.B., R.S., J.M. and A.A.S. drafted and reviewed the manuscript.
Data Availability
The article text includes all data generated in the course of the study. Submitted sequence IDs have been mentioned in the text and are available in the NCBI database and also with the corresponding author.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Malaria Situation in India from 2015. NVBDCP, https://nvbdcp.gov.in/WriteReadData/l892s/75840419771565787319.pdf (2019).
- 2.Bhattacharjee, U. More Than 100 Die as Tripura Battles Malaria. NDTV, https://www.ndtv.com/india-news/more-than-100-die-as-tripura-battles-malaria-590924 (2014).
- 3.Sharma J, Dutta P, Khan SA. Epidemiological study of malaria cases in North East region of India. Indian J Med Microbiol. 2016;34:261–262. doi: 10.4103/0255-0857.176843. [DOI] [PubMed] [Google Scholar]
- 4.Anvikar AR, et al. Anti-malarial drug policy in India: past, present & future. Indian J Med Res. 2014;139:205–215. [PMC free article] [PubMed] [Google Scholar]
- 5.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng G, et al. Human Immunization With a Polymorphic Malaria Vaccine Candidate Induced Antibodies to Conserved Epitopes That Promote Functional Antibodies to Multiple Parasite Strains. J Infect Dis. 2018;218:35–43. doi: 10.1093/infdis/jiy170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper SJ, et al. Recent advances in recombinant protein-based malaria vaccines. Vaccine. 2015;33:7433–7443. doi: 10.1016/j.vaccine.2015.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacheco MA, et al. Multiplicity of Infection and Disease Severity in Plasmodium vivax. PLoS Negl Trop Dis. 2016;10:e0004355. doi: 10.1371/journal.pntd.0004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibraheem ZO, Abd MR, Noor SM, Sedik HM, Basir R. Role of Different Pfcrt and Pfmdr-1 Mutations in Conferring Resistance to Antimalaria Drugs in Plasmodium falciparum. Malar Res Treat. 2014;2014:950424. doi: 10.1155/2014/950424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in anti-malarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohapatra PK, et al. Molecular evidence of increased resistance to anti-folate drugs in Plasmodium falciparum in North-East India: a signal for potential failure of artemisinin plus sulphadoxine-pyrimethamine combination therapy. PLoS One. 2014;9:e105562. doi: 10.1371/journal.pone.0105562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Country Office for India, World Health Organization (2016). National framework for malaria elimination in India 2016–2030. WHO Country Office for India, https://apps.who.int/iris/handle/10665/246096 (2016).
- 15.Patel P, et al. Prevalence of mutations linked to anti-malarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: A malaria elimination point of view. Sci Rep. 2017;7:16690. doi: 10.1038/s41598-017-16866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antony HA, Das S, Parija SC, Padhi S. Sequence analysis of pfcrt and pfmdr1 genes and its association with chloroquine resistance in Southeast Indian Plasmodium falciparum isolates. Genom Data. 2016;8:85–90. doi: 10.1016/j.gdata.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang B, et al. Prevalence of crt and mdr-1 mutations in Plasmodium falciparum isolates from Grande Comore island after withdrawal of chloroquine. Malar J. 2016;15:414. doi: 10.1186/s12936-016-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duah NO, et al. Increased pfmdr1 gene copy number and the decline in pfcrt and pfmdr1 resistance alleles in Ghanaian Plasmodium falciparum isolates after the change of anti-malarial drug treatment policy. Malar J. 2013;12:377. doi: 10.1186/1475-2875-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mwai L, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 21.Frank M, et al. A thirteen-year analysis of Plasmodium falciparum populations reveals high conservation of the mutant pfcrt haplotype despite the withdrawal of chloroquine from national treatment guidelines in Gabon. Malar J. 2011;10:304. doi: 10.1186/1475-2875-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogouyemi-Hounto A, et al. Prevalence of the molecular marker of Plasmodium falciparum resistance to chloroquine and sulphadoxine/pyrimethamine in Benin seven years after the change of malaria treatment policy. Malar J. 2013;12:147. doi: 10.1186/1475-2875-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djimde A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–891. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan K, Pande V, Das A. DNA sequence polymorphisms of the pfmdr1 gene and association of mutations with the pfcrt gene in Indian Plasmodium falciparum isolates. Infect Genet Evol. 2014;26:213–222. doi: 10.1016/j.meegid.2014.05.033. [DOI] [PubMed] [Google Scholar]
- 25.Sutar SK, Gupta B, Ranjit M, Kar SK, Das A. Sequence analysis of coding DNA fragments of pfcrt and pfmdr-1 genes in Plasmodium falciparum isolates from Odisha, India. Mem Inst Oswaldo Cruz. 2011;106:78–84. doi: 10.1590/S0074-02762011000100013. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee M, et al. No Polymorphism in Plasmodium falciparum K13 Propeller Gene in Clinical Isolates from Kolkata, India. J Pathog. 2015;2015:374354. doi: 10.1155/2015/374354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra N, et al. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob Agents Chemother. 2015;59:2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra N, et al. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar J. 2016;15:583. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti R, et al. Decreased in vitro artemisinin sensitivity of Plasmodium falciparum across India. Antimicrob Agents Chemother. 2019 doi: 10.1128/AAC.00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations: informal consultation organized by the Medicines for Malaria Venture and cosponsored by the World Health Organization, 29–31 May 2007, Amsterdam, The Netherlands, https://apps.who.int/iris/bitstream/handle/10665/43824/9789241596305_eng.pdf?sequence=1 (2008).
- 31.Yakubu B, Longdet IY, Jen TH, Davou DT, Obishakin E. High-Complexity Plasmodium falciparum Infections, North Central Nigeria, 2015-2018. Emerg Infect Dis. 2019;25:1330–1338. doi: 10.3201/eid2507.181614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer CG, May J, Arez AP, Gil JP, Do Rosario V. Genetic diversity of Plasmodium falciparum: asexual stages. Trop Med Int Health. 2002;7:395–408. doi: 10.1046/j.1365-3156.2002.00875.x. [DOI] [PubMed] [Google Scholar]
- 33.Sarmah NP, et al. Molecular characterization of Plasmodium falciparum in Arunachal Pradesh from Northeast India based on merozoite surface protein 1 & glutamate-rich protein. Indian J Med Res. 2017;146:375–380. doi: 10.4103/ijmr.IJMR_291_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur H, et al. Genetic diversity of Plasmodium falciparum merozoite surface protein-1 (block 2), glutamate-rich protein and sexual stage antigen Pfs25 from Chandigarh, North India. Trop Med Int Health. 2017;22:1590–1598. doi: 10.1111/tmi.12990. [DOI] [PubMed] [Google Scholar]
- 35.Patel P, et al. Genetic diversity and antibody responses against Plasmodium falciparum vaccine candidate genes from Chhattisgarh, Central India: Implication for vaccine development. PLoS One. 2017;12:e0182674. doi: 10.1371/journal.pone.0182674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akter J, et al. Genotyping of Plasmodium falciparum using antigenic polymorphic markers and to study anti-malarial drug resistance markers in malaria endemic areas of Bangladesh. Malar J. 2012;11:386. doi: 10.1186/1475-2875-11-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ariey F, et al. Association of severe malaria with a specific Plasmodium falciparum genotype in French Guiana. J Infect Dis. 2001;184:237–241. doi: 10.1086/322012. [DOI] [PubMed] [Google Scholar]
- 38.Robert F, et al. Extensive genetic diversity of Plasmodium falciparum isolates collected from patients with severe malaria in Dakar, Senegal. Trans R Soc Trop Med Hyg. 1996;90:704–711. doi: 10.1016/S0035-9203(96)90446-0. [DOI] [PubMed] [Google Scholar]
- 39.Ranjit MR, et al. Distribution of Plasmodium falciparum genotypes in clinically mild and severe malaria cases in Orissa, India. Trans R Soc Trop Med Hyg. 2005;99:389–395. doi: 10.1016/j.trstmh.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan RC, Farooq U, Dubey ML, Malla N. Genetic polymorphism in Plasmodium falciparum vaccine candidate antigens. Indian J Pathol Microbiol. 2005;48:429–438. [PubMed] [Google Scholar]
- 41.Chotivanich K, et al. Parasite multiplication potential and the severity of Falciparum malaria. J Infect Dis. 2000;181:1206–1209. doi: 10.1086/315353. [DOI] [PubMed] [Google Scholar]
- 42.Kang JM, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131. doi: 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basco L, Tahar R, Escalante A. Molecular epidemiology of malaria in Cameroon. XVIII. Polymorphisms of the Plasmodium falciparum merozoite surface antigen-2 gene in isolates from symptomatic patients. Am J Trop Med Hyg. 2004;70:238–244. doi: 10.4269/ajtmh.2004.70.238. [DOI] [PubMed] [Google Scholar]
- 44.Snounou G, et al. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/S0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 45.Heidari A, Keshavarz H, Rokni MB, Jelinek T. Genetic diversity in merozoite surface protein (MSP)-1 and MSP-2 genes of Plasmodium falciparum in a major endemic region of Iran. Korean J Parasitol. 2007;45:59–63. doi: 10.3347/kjp.2007.45.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar D, et al. Genetic polymorphism and amino acid sequence variation in Plasmodium falciparum GLURP R2 repeat region in Assam, India, at an interval of five years. Malar J. 2014;13:450. doi: 10.1186/1475-2875-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montoya L, et al. Plasmodium falciparum: diversity studies of isolates from two Colombian regions with different endemicity. Exp Parasitol. 2003;104:14–19. doi: 10.1016/S0014-4894(03)00112-7. [DOI] [PubMed] [Google Scholar]
- 48.Haddad D, et al. Limited genetic diversity of Plasmodium falciparum in field isolates from Honduras. Am J Trop Med Hyg. 1999;60:30–34. doi: 10.4269/ajtmh.1999.60.30. [DOI] [PubMed] [Google Scholar]
- 49.A-Elbasit IE, A-Elgadir TM, Elghazali G, Elbashir MI, Giha HA. Genetic fingerprints of parasites causing severe malaria in a setting of low transmission in Sudan. J Mol Microbiol Biotechnol. 2007;13:89–95. doi: 10.1159/000103600. [DOI] [PubMed] [Google Scholar]
- 50.Zhong D, Koepfli C, Cui L, Yan G. Molecular approaches to determine the multiplicity of Plasmodium infections. Malar J. 2018;17:172. doi: 10.1186/s12936-018-2322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nabet C, et al. Genetic diversity of Plasmodium falciparum in human malaria cases in Mali. Malar J. 2016;15:353. doi: 10.1186/s12936-016-1397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takala SL, et al. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayor A, et al. Plasmodium falciparum multiple infections in Mozambique, its relation to other malariological indices and to prospective risk of malaria morbidity. Trop Med Int Health. 2003;8:3–11. doi: 10.1046/j.1365-3156.2003.00968.x. [DOI] [PubMed] [Google Scholar]
- 54.Atroosh WM, et al. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit Vectors. 2011;4:233. doi: 10.1186/1756-3305-4-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Hamidhi S, et al. Genetic diversity of Plasmodium falciparum and distribution of drug resistance haplotypes in Yemen. Malar J. 2013;12:244. doi: 10.1186/1475-2875-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarmah NP, et al. Role of Anopheles baimaii: potential vector of epidemic outbreak in Tripura, North-east India. Journal of Global Health Reports. 2019;3:e2019036. doi: 10.29392/joghr.3.e2019036. [DOI] [Google Scholar]
- 57.Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial Drug Resistance: Literature Review and Activities and Findings of the ICEMR Network. Am J Trop Med Hyg. 2015;93:57–68. doi: 10.4269/ajtmh.15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hetzel MW, et al. Ownership and usage of mosquito nets after four years of large-scale free distribution in Papua New Guinea. Malar J. 2012;11:192. doi: 10.1186/1475-2875-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noviyanti R, et al. Contrasting Transmission Dynamics of Co-endemic Plasmodium vivax and P. falciparum: Implications for Malaria Control and Elimination. PLoS Negl Trop Dis. 2015;9:e0003739. doi: 10.1371/journal.pntd.0003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and CQ resistance, Malawi. Emerg Infect Dis. 2007;13:872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 61.World Health Organization 2005. Susceptibility of Plasmodium falciparum to antimalarial drugs: report on global monitoring 1996–2004, https://apps.who.int/iris/bitstream/handle/10665/43302/9241593466_eng.pdf;jsessionid=4132E55801BB4B79292B041A6C7C2DEF?sequence=1 (2005).
- 62.Indian Genome Variation Consortium Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 63.Gazi NN, et al. Genetic structure of Tibeto-Burman populations of Bangladesh: evaluating the gene flow along the sides of Bay-of-Bengal. PloS one. 2013;8:e75064. doi: 10.1371/journal.pone.0075064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paganotti GM, et al. Human genetic variation is associated with Plasmodium falciparum drug resistance. J Infect Dis. 2011;204:1772–1778. doi: 10.1093/infdis/jir629. [DOI] [PubMed] [Google Scholar]
- 65.National Health Mission. Health and Family Welfare Department. Government of Tripura. Epidemiological Situation Report (w.e.f 2012 up-to July, 2019), http://tripuranrhm.gov.in/NVBDCP.htm (2019).
- 66.Baldeviano GC, et al. Molecular Epidemiology of Plasmodium falciparum Malaria Outbreak, Tumbes, Peru, 2010–2012. Emerg Infect Dis. 2015;21:797–803. doi: 10.3201/eid2105.141427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.National Drug Policy on malaria. Directorate of National Vector Borne Disease Control Programme, Directorate General of Health Services, Ministry of Health and Family Welfare, Government of India; 2013. National Vector Borne Disease Control Programme, https://www.nvbdcp.gov.in/WriteReadData/l892s/National-Drug-Policy-2013.pdf (2013).
- 68.Snounou G, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 69.Bharti PK, et al. Therapeutic efficacy of chloroquine and sequence variation in pfcrt gene among patients with falciparum malaria in central India. Trop Med Int Health. 2010;15:33–40. doi: 10.1111/j.1365-3156.2009.02425.x. [DOI] [PubMed] [Google Scholar]
- 70.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snounou G. Genotyping of Plasmodium spp. Nested PCR. Methods Mol Med. 2002;72:103–116. doi: 10.1385/1-59259-271-6:103. [DOI] [PubMed] [Google Scholar]
- 73.Gupta P, et al. Genetic profiling of the Plasmodium falciparum population using antigenic molecular markers. Scientific World Journal. 2014;2014:140867. doi: 10.1155/2014/140867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The article text includes all data generated in the course of the study. Submitted sequence IDs have been mentioned in the text and are available in the NCBI database and also with the corresponding author.