Abstract
Unlike any protein studied so far, the active site of bilirubin oxidase from Myrothecium verrucaria contains a unique type of covalent link between tryptophan and histidine side chains. The role of this post-translational modification in substrate binding and oxidation is not sufficiently understood. Our structural and mutational studies provide evidence that this Trp396–His398 adduct modifies T1 copper coordination and is an important part of the substrate binding and oxidation site. The presence of the adduct is crucial for oxidation of substituted phenols and it substantially influences the rate of oxidation of bilirubin. Additionally, we bring the first structure of bilirubin oxidase in complex with one of its products, ferricyanide ion, interacting with the modified tryptophan side chain, Arg356 and the active site-forming loop 393-398. The results imply that structurally and chemically distinct types of substrates, including bilirubin, utilize the Trp–His adduct mainly for binding and to a smaller extent for electron transfer.
Subject terms: Enzyme mechanisms, X-ray crystallography
Introduction
Bilirubin oxidase (MvBOx; EC 1.3.3.5) from the ascomycete plant pathogen Myrothecium verrucaria (Albifimbria verrucaria) is a member of the blue multicopper oxidase family (MCO). MCOs are capable of oxidizing various organic and/or inorganic substrates and reducing oxygen to water without release of reactive oxygen species1–6. MvBOx, composed of 534 amino acid residues, consists of three cupredoxin-like domains with four copper ions forming two active sites7,8. These copper ions can be divided into three classes according to their spectroscopic properties9,10. One copper ion is of type I (T1Cu) and is present at the so called T1Cu site near the protein surface. Coordination of this copper ion is responsible for the distinctive blue color of MvBOx and all MCOs (absorption at 600 nm) and for oxidation of substrates with the Cu2+ ion being an electron acceptor9–11. T1Cu is connected with the trinuclear cluster (TNC), composed of one type II (T2Cu) and two type III (T3Cu) copper ions, via a conserved His–Cys–His motif (serving as electron transfer path). The binuclear T3Cu site with a bridging hydroxyl or dioxygen is responsible for a characteristic shoulder at approximately 330 nm in UV–VIS absorption spectrum. At the TNC one molecule of O2 is reduced to two molecules of water using four electrons supplied by the T1Cu site9–18.
MvBOx can oxidize a variety of substrates (Fig. S1) including bilirubin, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), substituted phenols, or ferrocyanide ([Fe(CN)6]4−), with different pH optima for different classes of substrates6,19. MvBOx can be utilized for a range of purposes. In medicine, it is used for diagnostics of bilirubin in serum19–21, in biotechnology for decolorization of synthetic dyes or detoxification of the environment19,22,23. It also shows a potential for use in biosensors and biofuel technology24–33.
The mechanism of dioxygen reduction inside the TNC as well as the electron transfer path between the T1Cu site and the TNC are very similar within the MCO family and are well understood. They were intensively studied using biochemical, structural, and computational methods12,16,34–36. However, the mechanism of substrate binding and oxidation at the T1Cu site varies among MCOs and is known only for several representatives (e.g. laccases37–39). In most laccases and many other MCOs the T1Cu site can be directly accessed by substrate, usually with a direct contact (or with a very short distance) of the oxidized moiety to one of the T1Cu-coordinating histidine side chains. In MvBOx the T1Cu site is separated from bulk solvent by additional amino acid residues, including Trp396. This raises questions regarding the separation of the substrate binding site and the T1Cu site in relation to the enzyme function.
Five structures of MvBOx have been published so far7,8,40, however, they do not show binding of any ligands besides water molecules in the proximity of the T1Cu site. Here we present the first complex of MvBOx with a product, ferricyanide ion ([Fe(CN)6]3−), binding close to the T1Cu site, together with the structure-function analysis of the protein layer between the T1Cu site and the identified substrate binding site containing a natural post-transitional modification, the covalent crosslink between T1Cu-bound His398 and Trp39640. We examined its role in the reaction mechanism by mutagenesis connected with structure-function analysis.
Results
Structure of MvBOx wild type from acidic pH
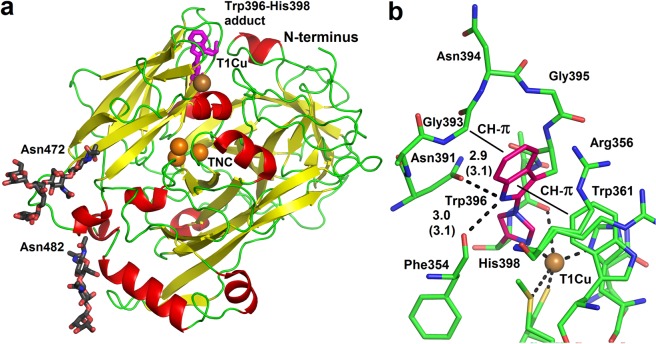
The crystal structure of MvBOx wild type (MvBOxWT) in complex with ferricyanide (PDB ID 6I3J; WT:FECN) was obtained from a strongly acidic crystallization condition (pH 3.1). MvBOx crystallized in the space group F222 with two monomers in the asymmetric unit (ASU). Nevertheless, the protein fold is very similar to the previously reported structures of MvBOx (Fig. 1a), one from a basic condition with a positional r.m.s.d. of 533 Cα atoms of 0.30 Å (PDB ID 2XLL, crystallization at pH 8.7, space group P1, four monomers in ASU8), and the second one from an acidic condition (PDB ID 6IQZ, pH 5, space group C2, one monomer in ASU40) with an r.m.s.d. of 0.25 Å. The structures were superimposed using the Secondary structure matching algorithm (SSM) in Coot41. Glycosylation of MvBOx in the strongly acidic condition (at Asn472 and Asn482) is preserved and was modelled (Fig. 1a). Conformations of all residues (including the side chains) around the T1Cu site are basically the same in all three structures (Fig. S2).
Figure 1.
(a) Structure of MvBOxWT from strongly acidic condition. The structure (6I3J) is shown in secondary structure representation (helices are colored red, β–strands yellow, loops green). Copper ions are shown as orange spheres. The Trp396–His398 adduct (shown as magenta sticks), oligosaccharides modifying asparagine side chains (black sticks), the N-terminus, T1 copper and the trinuclear copper cluster are labeled. (b) Chemical environment of the Trp–His adduct in MvBOx. Hydrogen bond distances are given in Ångströms. Values in parentheses are for chain B of the structure 6I3J. The CH – π interactions of Trp396 are marked. The indole–imidazole moiety of the adduct is shown with carbon colored magenta. T1Cu is shown as orange sphere. Molecular graphics were created using PyMOL (Schrödinger, LLC).
Trp396–His398 crosslink
The covalent crosslink between the Trp396 and His398 side chains was initially identified due to the observed close contact between Trp396-Cδ1 and His398-Nε2 supported by the electron density (Fig. 1). The existence of the Trp396–His398 crosslink was confirmed afterwards by mass spectrometry observing an ion at m/z 649.8327 (4+) corresponding to the modified WELINAGNGWTHPIHIHLVDFK peptide (Fig. S3). The correct assignment of the peptide ion was confirmed by collision induced dissociation.
Refinement of this type of covalent link required a proper definition of its geometry. There were several examples of X-ray structures of small molecules in the Cambridge Structural Database (CSD, see Materials and Methods for details) containing this type of bond and suitable for extraction of geometrical restraints. The initial refinement of the structure imposing only a covalent link restraint with the target value of the distance between Trp396-Cδ1 and His398-Nε2 of 1.41 Å led to small distortions of the planar side chains of both residues and a slight deviation of the new bond from the planes of both side chains (with opposite signs in chains A and B). Therefore, co-planarity of the bond with each side chain (independently) was further restrained, with target values of bond distances and angles in the nearest vicinity derived from the CSD entries. In this way the distance was refined to 1.42 Å and 1.40 Å (in chains A and B, respectively), following closely the set target values for the bond distance (1.41 Å) and the nearby environment. This approach led to a successful refinement of this moiety and to a good agreement with electron density without any difference peaks at this site.
The Trp396 side chain of the Trp–His adduct in MvBOx is involved in several interactions (Fig. 1b). Trp396-Nε1 forms hydrogen bonds to the main chain oxygen of Phe354 and the side chain oxygen of Asn391. It is also involved in CH–π interactions with Gly393-Cα and the Trp361 side chain. The closest surroundings of Trp–His in MvBOx is further formed by the side chain of Arg356 and the main chain atoms of the 393–395 loop (Gly393, Asn394, Gly395).
Functional mutants of MvBOx Trp396
To analyze the role of the Trp396–His398 adduct, three functional variants of MvBOx were designed and prepared. Trp396 was mutated to alanine (MvBOxW396A) in order to enable direct solvent access to His398, to phenylalanine (MvBOxW396F) in order to introduce an aromatic residue not capable of crosslink formation, and to aspartic acid (MvBOxW396D) in order to disrupt this site by introducing negative charge. All three MvBOx variants have the same composition of the secondary structure elements (overall structure) as the wild type (See Fig. S4 for CD spectrometry results) and all have the T1Cu site preserved, which is clear from the blue color of their solutions (See Fig. S5 for UV-VIS spectra).
Structures of MvBOx mutants Trp396Ala and Trp396Phe from acidic pH
Similarly to the structure of WT:FECN, also the crystal structure of MvBOxW396A in complex with ferricyanide (PDB ID 6I3K; W396A:FECN) and the structure of MvBOxW396F without any ligand near the T1Cu site (PDB ID 6I3L; W396F) were obtained from the same acidic crystallization condition. Both proteins crystallized in the space group F222. In neither mutant the protein fold was affected by the mutation. The positional r.m.s.d. of 533 Cα atoms between the structures WT:FECN (PDB ID 6I3J, chain A) and W396A:FECN (PDB ID 6I3K, chain A) or W396F (PDB ID 6I3L, chain A) was 0.23 Å and 0.21 Å, respectively. The surroundings of the T1Cu site were also unaffected (Figs S6 and S7).
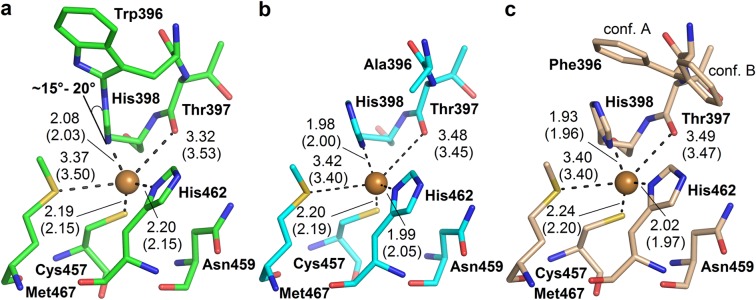
Coordination of T1Cu
The coordination of T1Cu in WT:FECN differs in two features when compared to those of MCOs without the Trp–His adduct (e.g. CotA42). The first difference lies in the fact that T1Cu is no longer in the imidazole plane of coordinating His398, but the histidine side chain is tilted by about 15–20°. The second difference is a longer coordination distance (~2.2 Å) between T1Cu and His462-Nδ1 (Fig. 2). These features are present only in the structure WT:FECN (PDB ID 6I3J). In the structures of both mutants (PDB ID 6I3K and 6I3L) T1Cu is coordinated in the plane of the His398 side chain and the distance between T1Cu and His462-Nδ1 is shorter (~2.0 Å) and similar with that in CotA or other MCOs.
Figure 2.
Coordination of T1Cu: (a) in the structure of WT:FECN (PDB ID 6I3J, carbon green), (b) in W396A:FECN (PDB ID 6I3K, carbon light blue), and (c) in W396F (PDB ID 6I3L, carbon pale orange). Distances are given in Ångströms with 0.01 Å precision to support the discussion of the T1Cu environment changes. Values in parentheses are for chain B of the corresponding structure. T1 copper is shown as orange sphere. The Phe396 side chain in the structure W396F adopts two conformations marked conf. A and conf. B. Molecular graphics were created using PyMOL (Schrödinger, LLC).
Binding of ferricyanide in the active site of MvBOxWT and MvBOxW396A
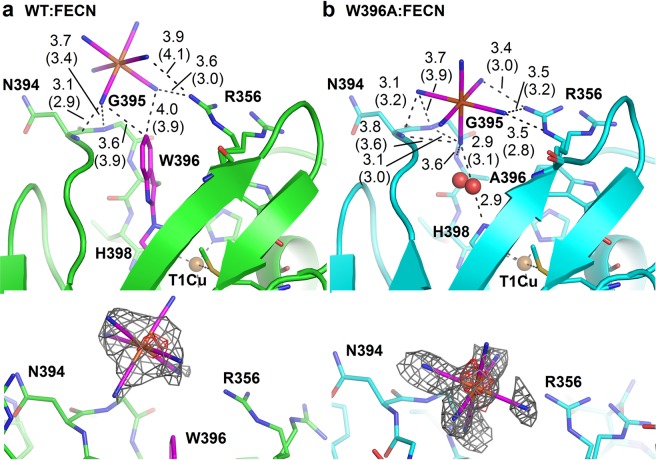
A ferricyanide ion was identified in the active site of both WT:FECN and W396A:FECN (PDB ID 6I3J and 6I3K, respectively) using difference electron density (mFo-DFc) after the phase problem solution. Its presence was confirmed by a peak in anomalous difference Fourier at the iron atom and composite omit map (Fig. 3). Complexes were prepared by soaking crystals in the solution containing ferrocyanide (substrate, see Materials and Methods section). During this process crystals of both MvBOxWT and MvBOxW396A gradually changed their blue color to transparent which proved the reduction of the T1Cu site in parallel with ongoing oxidation of ferrocyanide in the crystals (Supplementary video sequences 1 and 2).
Figure 3.
Binding of ferricyanide in the active site of MvBOx wild type and its W396A mutant. (a) Binding of ferricyanide in OS1 of the WT:FECN structure (6I3J, carbon green). The ferricyanide ion and Trp–His adduct are shown with carbon colored magenta. (b) Binding of ferricyanide (magenta) in OS1 of the W396A:FECN structure (6I3K, carbon light blue). One water molecule (shown as red sphere, in two alternative positions) connects ferricyanide and His398. Interacting residues are marked. Distances are given in Ångströms. If values differ in chain A and B, they are given in parentheses for chain B of the corresponding structure. T1Cu is shown as orange sphere. The composite omit electron density map (2mFo-DFc) is shown as grey mesh and contoured at 1.0 σ level around the ferricyanide ion at the bottom of each panel. The map was calculated using Phenix67. Anomalous difference Fourier is shown as red mesh and contoured at 2.5 σ level around iron. Molecular graphics were created using PyMOL (Schrödinger, LLC).
In WT:FECN, the ferricyanide ion was bound in close proximity of the Trp396–His398 adduct, interacting with the main chain nitrogen atoms of Asn394 and Gly395 through one of its cyanide moieties and with the Arg356 side chain through two adjacent cyanide moieties (Fig. 3a). The shortest distance observed between ferricyanide and T1Cu is 11.4 Å (Fig. S8a). This is well within the 14 Å limit identified as the boundary for efficient electron transfer in proteins43. Moreover, ferricyanide in this position is not involved in any symmetry (crystal-induced) contacts, which supports the suggestion that this is a genuine substrate/product binding site. Therefore, we refer to this site as oxidation site 1 (OS1). For visualization of the MvBOx surface belonging to OS1 refer to Fig. S9.
In the structure W396A:FECN, a ferricyanide ion is bound in the same site but shifted towards the T1Cu site, with the closest observed distance to T1Cu now being 9.2 Å (Fig. S8b), and interacting through a water molecule with His398 which coordinates T1Cu. It still interacts with the main chain nitrogen atoms of Asn394 and Gly395 but now through two of its cyanide moieties. It also interacts with the main chain nitrogen atom of Ala396 and with Arg356 via two adjacent cyanide moieties (Fig. 3b).
Electron density for several other ferricyanide ions was identified and modeled in both WT:FECN and W396A:FECN (not shown) but none of them in close proximity of the T1Cu site. Some of these additional ferricyanide ions are involved in the formation of crystal contacts. A similar behavior of ferricyanide was observed in crystals of another MCO, two-domain laccase from Streptomyces coelicolor44.
Analysis of enzymatic activity of WT and mutant MvBOx
Enzymatic activities of MvBOxWT and of the variants W396A, W396F, and W396D were analyzed using four different substrates: inorganic ([Fe(CN)6]4− (ferrocyanide); substituted phenol 2,6-dimethoxyphenol (DMP); 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonate (ABTS) as a standard substrate for MCOs analysis; and bilirubin as the canonical substrate for bilirubin oxidase (Fig. S1). Kinetic profiles are presented in Fig. 4 and kinetic parameters are summarized in Table 1. The kinetic profile for oxidation of bilirubin always showed sigmoidal dependence and the parameters were calculated using Eq. 1 (Materials and Methods). Kinetic parameters for oxidation of ferrocyanide, ABTS, and DMP were calculated using the Michaelis-Menten equation, providing the data converge. Otherwise the kinetic parameters remained uncalculated and a simple connecting line was used in the graphs.
Figure 4.
Oxidation of (a) bilirubin; (b) K4Fe(CN)6; (c) ABTS; and (d) DMP by MvBOxWT (solid line, ●), MvBOxW396A (dotted line, ▲), MvBOxW396F (dot-dash line, ▢), and MvBOxW396D (dashed line, ◆). Plots show the reaction velocity v as a function of substrate concentration. The curves were fitted using the program GraphPad Prism 7.02 (GraphPad Software). Standard deviations are marked as error bars. In the case of the mutant MvBOxW396D (all substrates) and all mutants (only DMP), a simple connecting line was used to link the mean values.
Table 1.
Kinetic parameters for oxidation of bilirubin, K4Fe(CN)6, ABTS, and DMP calculated for the measurements shown in Fig. 4. The parameters were calculated with use of the Michaelis-Menten equation (KM, Vmax) for K4Fe(CN)6, ABTS, and DMP. The allosteric sigmoidal equation (Equation 1, K1/2, Vmax, h) was used for bilirubin oxidation. The parameters for DMP as substrate and mutant enzymes could not be calculated due to almost zero activity.
| Substrate Enzyme variant | KM (mM) | Vmax (nmol·min−1 μg−1) | K1/2 (mM) | Vmax (nmol·min−1·μg−1) | h |
|---|---|---|---|---|---|
| Bilirubin | |||||
| MvBOxWT | 0.060 ± 0.002 | 15.8 ± 0.4 | 1.9 ± 0.1 | ||
| W396A | 0.160 ± 0.050 | 17.0 ± 6.0 | 2.2 ± 0.3 | ||
| W396F | 0.079 ± 0.004 | 8.8 ± 0.5 | (4.1 ± 0.7)# | ||
| K 4 Fe(CN) 6 | |||||
| MvBOxWT | 1.2 ± 0.2 | 460 ± 20 | |||
| W396A | 1.2 ± 0.2 | 490 ± 20 | |||
| W396F | 1.5 ± 0.2 | 590 ± 20 | |||
| ABTS | |||||
| MvBOxWT | 0.30 ± 0.02 | 30.3 ± 0.3 | |||
| W396A | 3.1 ± 0.2 | 37.5 ± 0.9 | |||
| W396F | 6.8 ± 0.4 | 33.8 ± 0.9 | |||
| DMP | |||||
| MvBOxWT | 20.1 ± 0.9 | 3.78 ± 0.08 | |||
#The value of h lies within a range of 2–4. Exact value cannot be determined due to the high error present in some of the points obtained for the measurement of bilirubin oxidation by mutant W396F.
Oxidation rate and affinity of bilirubin are affected by mutations of Trp396
Oxidation of bilirubin was measured by detection of decrease of bilirubin concentration over time. For calculations of kinetic parameters, the values of absorbance decrease with an inverted sign were used. Since the F test and the AICc (corrected Akaike’s Information Criterion) calculations test confirmed that the measured data should be interpreted by a sigmoidal dependence on bilirubin concentration, the allosteric sigmoidal equation (Eq. 1 in Materials and Methods) was used to fit the data (Fig. 4a). The K1/2 values for the wild type and the W396F variant are comparable (0.060 ± 0.002 mM and 0.079 ± 0.004 mM, respectively) but Vmax of W396F (8.8 ± 0.5 nmol·min−1 ·μg−1) is roughly a half of the wild type value (15.8 ± 0.4 nmol·min−1 ·μg−1). Kinetic parameters for the oxidation of bilirubin using the W396A variant are determined with considerably greater errors in comparison with the other MvBOx variants. In this case, kinetic measurements at saturating bilirubin concentrations (>160 µM) were attempted but yielded high errors and measurements were not interpretable by mean values. The W396D variant showed practically zero activity ([−0.4 ± 2.8] % of MvBOxWT reaction velocity with 120 μM bilirubin for 2 min). The results are summarized in Fig. 4a and Table 1.
Oxidation of ferrocyanide is significantly affected only for mutant Trp396Asp
MvBOxWT, MvBOxW396A, and MvBOxW396F showed similar kinetic parameters for oxidation of ferrocyanide (Fig. 4b, Table 1). The mutation W396D led to a considerable decrease in the enzymatic activity ([1.27 ± 0.06] % of reaction velocity of MvBOxWT with 10 mM K4Fe(CN)6 as substrate for 3 minutes).
Oxidation of ABTS is affected by mutations of Trp396
The affinity to the substrate ABTS significantly decreased for variants W396A and W396F (3.1 ± 0.2 mM and 6.8 ± 0.4 mM, respectively) compared to MvBOxWT (0.30 ± 0.02 mM). The Vmax value (30.3 ± 0.3 nmol·min−1·μg−1 for MvBOxWT) slightly increased for both W396A and W396F (37.5 ± 0.9 nmol·min−1·μg−1 and 33.8 ± 0.9 nmol·min−1·μg−1, respectively). The mutation W396D led to null activity ([−0.8 ± 0.3] % of MvBOxWT reaction velocity for 20 mM ABTS as substrate for 3 min). The results are summarized in Fig. 4c and Table 1.
All mutations of Trp396 diminish oxidation of DMP
All mutants show negligible activity compared to MvBOxWT. Therefore, a simple connecting line was used to connect the mean values in the kinetic profile (Fig. 4d). The variants W396A, W396F, and W396D reached 5.0 ± 2.0%, 10.0 ± 3.0%, and 0.6 ± 0.3% of the MvBOxWT reaction velocity for 30 mM DMP as substrate in 4 min, respectively. The results are summarized in Fig. 4d and Table 1.
Discussion
Structure of MvBOx is maintained under extreme pH
The comparison of the MvBOx structures from acidic (pH 3.1, PDB ID 6I3J and pH 5.0, PDB ID 6IQZ) and basic (pH 8.7, PDB ID 2XLL) crystallization conditions proves that the MvBOx structure did not change with the change of pH (Fig. S2). It should be noted that the same glycosylation pattern (at Asn472 and Asn482) is present under all the studied pH values and that the position of glycosylation differs from fungal laccases45. This lack of pH-dependent structural differences suggests that different pH optima for the individual types of substrates cannot be connected with structural changes (e.g. pH 4 for oxidation of ABTS;6 pH around 8.4 for oxidation of bilirubin18). This observation is in agreement with the findings of Otsuka et al., according to which the pH optimum of MvBOx is predominantly determined by the difference between the redox potentials of MvBOx and of the particular substrate6.
Trp396–His398 covalent crosslink is natively present in MvBOx and its existence is not pH dependent
As the existence of the Trp396–His398 covalent crosslink in MvBOx was confirmed using LC–MS/MS (Fig. S3) we can conclude that it is present in the enzyme in solution. Moreover, it is present not only in the structure from the strongly acidic crystallization condition reported here (PDB ID 6I3J), but also in the previously reported structures of MvBOx from acidic (6IQZ) and basic condition (2XLL), as discussed by Akter et al.40. Based on these results it is safe to conclude that the Trp–His adduct is present in native MvBOx in solution over a broad range of functionally relevant pH values. This is important for relevance of the mutagenesis–activity studies presented in this work.
Trp396 and His398, along with several residues in their proximity, are conserved in many homologues of MvBOx from fungi and bacteria (Fig. S10). Conservation of these residues indicates the possible existence of the same Trp–His crosslink in bilirubin oxidases or closely related enzymes from other organisms.
Trp396–His398 adduct modifies coordination of T1Cu
The presence of the Trp396–His398 adduct in MvBOx has measurable effects on the coordination of T1 copper as described in the Results section. The significant tilt of the His398 imidazole moiety coordinating T1Cu is also observable in electron density in the structure of MvBOx wild type 2XLL8 and present in the structure of MvBOx wild type determined by Akter et al. (PDB ID 6IQZ40). However, this tilt is not in the structure of MvBOx M467Q which, due to the mutation, does not contain the Trp396–His398 crosslink (PDB ID 6IQY40). As the T1Cu site in the structures W396A:FECN (PDB ID 6I3K) and W396F (6I3L) did not show this tilt either, we conclude that it is caused by the coordination of T1Cu by the Trp396–His398 adduct (Fig. 2).
The longer coordination distance of T1Cu to His462-Nδ1 observed in the case of WT:FECN (Fig. 2) is also present in the structure 2XLL (refined to approximately 2.2 Å8), but not in the structure of MvBOx wild type determined by Akter et al.40. And so this change of T1Cu coordination distance in connection with the adduct presence requires further investigation.
Trp396–His398 crosslink formation
To the best of our knowledge, the Trp–His crosslink has been observed only in MvBOx, but covalent crosslinks between side chains of residues Tyr and Cys, Tyr and Met, Tyr and Trp, Tyr and His, and Cys and His were identified in several enzymes, with three types of copper-containing oxidases among them46. Tryptophan side chain, especially the indole group, is reactive and susceptible to chemical modifications47. It can undergo electrochemical oxidation on carbon Cδ1 48 with an oxidation peak potential of 0.64 V at pH 749. The redox potential of the T1Cu site was identified as the main reason for the Trp396–His398 adduct formation in MvBOx also by Akter et al. in their recent work40.
According to our analysis, the observed rotamer of the Trp396 side chain is the only standard tryptophan rotamer acceptable for this site without any clashes with the surrounding residues. This is true for both, the Trp–His adduct (PDB ID 6I3J, this work) and also for the structure without the crosslink between Trp396 and His398 (6IQY40). Therefore, we can conclude that the particular fold of the enzyme in this region is actually in favor of the adduct formation by placing the indole group in a close contact with the His398 side chain.
Ferricyanide binds to positively charged site near Trp396–His398 adduct
The surface of MvBOx near the T1Cu site is positively charged (Fig. S11a), as observed previously in investigations of the orientation of MvBOx molecules on negatively charged electrodes for direct electron transfer50–52. OS1, the ferricyanide binding site of MvBOx (Fig. 3a), is a part of this positively charged surface near the T1Cu site. Thus, this crystallographically identified site OS1 is the central site for substrate oxidation of MvBOx substrates and its properties must influence substrate binding and catalytic efficiency of the enzyme.
Trp396–His398 adduct has no significant structural role in MvBOx
The structure of MvBOx was not affected by the mutation of Trp396 to Ala and Phe (Figs S6 and S7). An X-ray structure of MvBOxW396D could not be determined, because this variant did not crystallize. Nevertheless, based on the CD (Fig. S4), and UV-VIS (Fig. S5) spectra it can be concluded that also this mutation did not change the secondary structure composition of MvBOx and the existence of the T1Cu site. So neither the elimination of the Trp396–His398 adduct, nor functional changes of the T1Cu site environment (direct solvent access to His398, aromatic residue without the crosslink or introduction of a negatively charged residue) have significant effects on the MvBOx structure. The presence of the Trp396–His398 adduct in MvBOx plays a minor role in its thermal stability (Fig. S12) as follows from the slight decrease of the melting temperature (Tm) of the W396A and W396F mutants (by about 5 °C and 7 °C, respectively). The W396D mutation caused a significant decrease of Tm (by about 20 °C). This can be explained by exchanging a hydrophobic residue (Trp) for a hydrophilic one (Asp) and also possibly by introducing negative charge to the otherwise positively charged site (Fig. S11b).
Trp396–His398 adduct participates in substrate binding and oxidation, depending on substrate type
The kinetic data measured for MvBOxWT and the Trp396 mutants (Table 1, Fig. 4) clearly show that different substrates utilize the Trp396–His398 adduct in different ways and that the adduct (or possibly Trp396) is crucial only in the case of DMP as substrate. Simultaneously, mutation of Trp396 to Asp disabled oxidation of all substrates as this mutation changed the electrostatic potential distribution in OS1 (Fig. S11b).
Trp396–His398 adduct is important in bilirubin oxidation
Our kinetic data for bilirubin show that the Trp396–His398 adduct is most probably involved in both, the substrate binding and its oxidation. All the investigated mutations always led to a significant decrease of the catalytic efficiency (Vmax/K1/2) for bilirubin. The W396A mutant binds bilirubin with a lower affinity when compared to the wild type. The W396F mutant retained affinity comparable with that of the wild type, but with a lower maximal reaction velocity. In addition, both mutants showed much more distinctive allosteric effect (Table 1, Fig. 4a). On the structural level, the mutation of Trp396 to Phe allows for conservation of the aromatic moiety in the proximity of the T1Cu site (Fig. 2c), whereas the mutation to Ala disposes of the aromatic moiety and instead allows for solvent access to His398 coordinating T1Cu (Fig. 3b). Considering these differences (both structural and in the enzymatic activity) it is clear that the Trp396–His398 adduct is involved in binding of bilirubin, although bilirubin can still bind to MvBOx mutants not containing tryptophan at the position 396. One of the possible explanations for the substantial decrease in bilirubin oxidation observed in the M467Q mutant5, which contains Trp396 but without the crosslink to His39840, is the usage of low substrate concentration (27 μM). At this concentration, the activity may be influenced by the allosteric effect observed in the mutants W396A and W396F (Fig. 4a).
Trp396–His398 adduct and ferrocyanide oxidation
In the case of ferrocyanide, the kinetic parameters of MvBOxWT and both mutants W396A and W396F are similar. Ferricyanide (product) binds in the same site in both MvBOxWT and MvBOxW396A, although in two different poses. And even if Trp396 forms a part of the ferricyanide binding site in the wild type, it is not necessary for its binding. The main structural features, which ferricyanide utilizes, are the main chain nitrogen atoms of Asn394 and Gly395 together with the side chain of Arg356 (Fig. 3). The main difference in binding of ferricyanide between MvBOxWT and MvBOxW396A lies in the fact that the replacement of Trp by Ala allows ferricyanide to bind closer to T1Cu. Ferricyanide in the W396A mutant interacts with T1Cu-coordinating His398 through a water molecule, possibly mediating electron transfer. The W396D mutant is almost inactive (~1% of the wild type activity). Possibly, the negatively charged aspartic acid side chain either compensates the partial positive charges of the Asn394/Gly395 main chain nitrogen atoms or directly repels the negatively charged ferrocyanide ion and so interferes with its binding. Preservation of ferrocyanide activity was also reported for the M467Q mutant5. From these results, it can be concluded that the Trp396–His398 adduct itself is not important for binding of ferrocyanide. The geometry and positive electrostatic potential of OS1 are sufficient for its binding close enough in the proximity of T1Cu for efficient electron transfer (within 14 Å limit43; see Fig. S8). Although the adduct is probably involved in electron transfer in MvBOxWT, its presence is not crucial in the case of ferrocyanide oxidation as electron transfer can be realized using another path, possibly via the protein main chain of the loop forming OS1 (residues 394–398) or via water molecule in the case of W396A:FECN.
Trp396–His398 adduct and ABTS oxidation
The mutations of Trp396 to Ala or Phe have similar effects. ABTS binds to both mutants with a significantly lower affinity, but the maximal velocity is not affected. So, in the case of ABTS, the Trp396–His398 adduct plays a role in the substrate binding. The electron transfer in this case can be realized via the adduct or via another path (possibly main chain of the loop forming OS1).
Trp396–His398 adduct and DMP oxidation
As all the mutants studied here had a significantly lower activity toward DMP when compared to the wild type of MvBOx, the kinetic parameters could not be calculated. Attempts to obtain structural information for a complex of MvBOx and DMP failed. It remains to be deciphered if the observed lack of activity towards DMP in the mutants is caused by a lower substrate affinity, disturbance of the electron transfer path upon elimination of the Trp396–His398 adduct or simply by the unfavorable difference in the redox potential between DMP and the individual enzyme variants.
OS1, Trp–His adduct, and electron transfer
Based on the presented results, we propose that all the substrates studied here utilize the oxidation site 1 (OS1), although the different substrates very likely bind at or near OS1 in a different way. OS1 comprises of residues which contribute only toward substrate binding (Asn197, Arg356, and Asn394) and residues which also participate in electron transfer from substrate to T1Cu (Trp396 of the Trp–His adduct, Gly395, and possibly also Asn394). In the case of ferrocyanide oxidation, the structures of the complexes enable a deeper analysis of the mechanism of substrate binding and oxidation, including the details of ferro/ferricyanide interactions with MvBOx.
For all substrate types, it seems that oxidation always relies, besides the difference between the redox potentials of substrate and MvBOx, on the interplay between the Trp396–His398 adduct and the main chain nitrogen atoms of the 393–395 loop forming the positively charged binding site. Other residues in the vicinity of the Trp396–His398 adduct may also play roles in substrate binding and/or oxidation (most likely including Arg356, Trp361, and Asn197). The Trp–His adduct is not the only possible electron transfer route in MvBOx. The whole loop 393–396 is important in substrate binding, adduct formation, and electron transfer. For some substrates, it likely participates in electron transfer via its main chain atoms.
Conclusion
Formation of the covalent link between the side chains of Trp396 and T1 copper-coordinating His398, confirmed in M. verrucaria bilirubin oxidase, is facilitated by the enzyme fold and local organization of the protein chain. The Trp396 indole ring effectively mimics the position of substrates in other multicopper oxidases and is activated by the T1Cu site redox potential. The adduct participates in formation of the oxidation site 1 involved in substrate binding and oxidation of all substrates including larger and/or aromatic compounds and bilirubin. Mutations of Trp396 influence the enzyme activity but not the enzyme structure (except the replaced residue). Based on the mutagenesis and kinetics results, different substrate types must bind in the proximity of the Trp–His adduct, while at the same time utilize this unique substrate oxidation site differently. As most of the studied substrates, including bilirubin, are oxidized even in the absence of this adduct, its role in electron transfer is not crucial. In the case of ferricyanide binding, except the Trp–His adduct, also Arg356 and the loop 393–398 are important for substrate/product-enzyme interactions.
Materials and Methods
Cloning of bilirubin oxidase wild type
Gene for bilirubin oxidase was amplified from M. verrucaria (A. verrucaria) strain ATCC24571 by primers 5′-AGAGCGAUACCATGTTCAAACACACAC and 5′-AACGTCACGUCTACTCGTCAGCTGCGGC having overhangs that incorporated a single deoxyuracil residue (dU) flanking the 3′ end of the homology region. The amplified DNA (band of 2059 base pairs) was used for USER® cloning into an expression vector.
Construction of mutated variants of bilirubin oxidase
Genes of all variants were generated by spliced overlap extension (SOE) polymerase chain reaction (PCR) with flanking primers 5′-AGAGCGAUACCATGTTCAAACACACAC (forward) and 5′-AACGTCACGUCTACTCGTCAGCTGCGGC (reverse) and hybrid primers containing the desired codon change. The resulting oxidase variant genes were cloned into an expression vector by USER®.
Expression and purification
All samples were expressed and purified similarly as described in Kovaľ et al.53. In detail, constructs were verified by DNA sequencing and transformed into protoplasts of Aspergillus oryzae for expression driven by the TAKA amylase promoter. The transformed strain of A. oryzae was grown for 3 days at 30 °C and 200 rpm in shake flasks containing MDU-2BP (45 g of maltose, 1 g of MgSO4.7H2O, 1 g of NaCl, 2 g of K2SO4, 12 g of KH2PO4, 7 g of yeast extract, 0.5 ml of trace elements, and 1% (w/v) urea per l). Additional CuSO4 was added to the shake flasks to a final concentration of 0.5 mM. The fermentation broth was sterile filtered to remove fungal hyphae. Salts and other low molecular weight solutes were removed by ultrafiltration. 1 M Tris/HCl, pH 7.5 was added to the resulting retentate to a final concentration of 25 mM. pH and ionic strength were determined to be within the acceptable range for anion exchange chromatography. The chromatography was then conducted with an ÄKTA Prime instrument (Amersham Biosciences). Briefly, the protein was bound to a column with 20 ml Q Sepharose High Performance pre-equilibrated with 25 mM Tris/HCl, pH 7.5. After a thorough wash with the equilibration buffer, the bound protein was eluted from the column with a linear NaCl gradient (0–0.5 M) in the equilibration buffer over ten column volumes. MvBOx eluted at approximately 250 mM NaCl. Collected fractions containing pure MvBOx, as estimated by SDS-PAGE, were pooled. All purification steps were carried out at room temperature.
Mass spectrometry
MvBOxWT was digested by trypsin. Peptides were further analyzed by LC-MS/MS using a 15 T solariX FT-ICR mass spectrometer (Bruker Daltonics) operating in positive mode.
MvBOx activity assay
The steady-state kinetic parameters for all MvBOx variants were determined using four distinct substrates: potassium ferrocyanide (further referred to as ferrocyanide because only [Fe(CN)6]4− anion undergoes oxidation), 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), bilirubin, and 2,6-dimethoxyphenol (DMP). All reactions were monitored spectrophotometrically using a CLARIOstar Monochromator Microplate Reader (BMG Labtech, Ortenberg, GE). Reactions were monitored for substrates [Fe(CN)6]4− and ABTS at 420 nm (ε420 = 1.04 mM−1 cm−1 54 and ε420 = 36 mM−1 cm−1 55, respectively), for DMP at 468 nm (ε468 = 14.8 mM−1 cm−1)56, and for bilirubin at 440 nm (ε440 = 56.3 mM−1 cm−1)57. Single reactions (100 μl total volume) were prepared in triplicates and run in black 96-well plates (BRAND, Wertheim, GE). All reactions were carried out at 27 °C.
Oxidation of K4Fe(CN)6 (0.1–10 mM) was done using 50 mM Bis-Tris, pH 6 and 25 mM NaCl with 0.15 μg of enzyme for 3 min (total volume 100 μl). Oxidation of ABTS (0.2–20 mM) was done using 100 mM sodium acetate, pH 4 with 0.06 μg of enzyme for 3 min. Oxidation of bilirubin (1–180 μM) was done using 200 mM Tris-HCl, pH 8.7 with 0.02 μg of enzyme for 3 min. Oxidation of DMP (0.5–60 mM) was done using 50 mM Bis-Tris, pH 6.8 and 25 mM NaCl with 0.4 μg of enzyme for 4 min.
Steady-state kinetic parameters (maximal velocity Vmax and Michaelis-Menten constant KM) were calculated using the Michaelis-Menten non-linear regression equation with GraphPad Prism version 7.02 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). In the case of bilirubin, the non-hyperbolic data with “S-shaped” sigmoidal behavior were taken into consideration by application of the Hill equation (Eq. 1, according to GraphPad Prism 7.02 Software):
| 1 |
K1/2 is the concentration of substrate at the half of the maximal velocity Vmax, h is a Hill slope. When h = 1, K1/2 is the KM value. To evaluate the best fit, the F test and the corrected Akaike’s Information Criterion (AICc) calculations were performed using the GraphPad QuickCalcs Web (http://www.graphpad.com/quickcalcs/ConfInterval1.cfm, accessed May 2017). The equation best fitting the particular data was used for the final calculation of the kinetic parameters.
Crystallization, data collection, structure solution, and refinement
For crystallization, MvBOxWT, MvBOxW396A, and MvBOxW396F were concentrated in the storage buffer (25 mM Tris/HCl, pH 7.5 with 250 mM NaCl) to 25 mg ml−1 using a 10 kDa cut–off Nanosep centrifugal device (Pall Corporation). Initial screening for crystallization conditions was done only for MvBOxWT using the hanging drop vapor diffusion setup with a protein to reservoir drop volume ratio of 1:1 (0.5 μl + 0.5 μl). Screening using several commercially available crystallization screens did not yield hits. As the theoretical pI of mature MvBOx is 4.97, a limited crystallization screen containing acidic conditions was used58. Initial hits were observed after 1–3 days using 25% (w/v) PEG 3350, 0.1 M citric acid, pH 3 as a reservoir solution and this condition was further optimized. Crystal of MvBOxWT used for X-ray analysis was obtained using the hanging drop vapor diffusion method. The drop was composed of 1 μl of protein mixed with 1 μl of reservoir solution (0.1 M succinic acid, 14% (w/v) polyethylene glycol 3350). Succinic acid was not titrated. Solution obtained by mixing the reservoir solution with the storage buffer in ratio 1:1 had pH 3.1. Crystals of MvBOxW396A and MvBOxW396F were obtained using the same reservoir solution and sitting drop setup with 0.5 μl + 0.5 μl drop volume. All crystallization trials were done at 18 °C.
Prior to vitrification using liquid nitrogen, crystals of MvBOxWT and MvBOxW396A were soaked in reservoir solution containing 10 mM K4Fe(CN)6 and cryoprotectant for 90 seconds (for MvBOxWT, a combination of 15% (w/v) polyethylene glycol 200, 1% (v/v) glycerol, 1% (v/v) ethylene glycol, and 1% (v/v) propylene glycol; for MvBOxW396A 25% (v/v) glycerol). Both crystals changed appearance from blue to transparent indicating reduction of T1Cu (Supplementary video sequences 1 and 2). Soaking solution changed color from pale yellow to darker yellow indicating oxidation of ferrocyanide ([Fe(CN)6]4−) to ferricyanide ([Fe(CN)6]3−). An MvBOxW396F crystal was soaked in solution containing 25% (v/v) glycerol and 10 mM pyrogallol in reservoir solution for 60 seconds. An MvBOxWT crystal was mounted in nylon CryoLoop (Hampton Research), MvBOxW396A and MvBOxW396F crystals in round LithoLoop (Molecular Dimensions). For WT and W396A crystals, diffraction data were collected with 0.91841 Å wavelength at beamline BL 14.1 of the BESSY II synchrotron radiation source (Helmholtz Zentrum Berlin, DE) at 100 K. Data for WT:FECN were collected using a MAR Mosaic CCD 225 detector and a mini kappa goniometer, data for W396A:FECN were collected using a Dectris Pilatus 6 M detector and a mini kappa goniometer59. Data for W396F were collected at 100 K and with 1.3418 Å wavelength (gallium Kα) using a D8 VENTURE diffractometer, a Photon II detector (Bruker) and a METALJET X-ray source (Excillum). Data were processed and scaled using XDS or XDSGui60 and merged using Aimless61. For all structures, the phase problem was solved by molecular replacement using Molrep62 and the structure of Myrothecium verrucaria bilirubin oxidase (PDB code 2XLL)8 as a template.
All three structures were manually edited using Coot63 and refined using restrained refinement in REFMAC564 with Rfree as a cross validation method. The last refinement cycle was done using all reflections. Structures were validated using the tools implemented in Coot, the structure-validation web service Molprobity65 and the wwPDB Validation service66. Data collection and processing statistics are reported in Table 2.
Table 2.
Statistics of data collection and processing and structure refinement parameters for MvBOx and its variants. Values in parentheses are for the highest resolution shell.
| Structure | Wild type + ferricyanide | W396A mutant + ferricyanide | W396F mutant |
|---|---|---|---|
| PDB ID | 6I3J | 6I3K | 6I3L |
| Data collection | |||
| X-ray source | BESSY II, BL14.1 | BESSY II, BL14.1 | MetalJet D2 |
| Wavelength (Å) | 0.91841 | 0.91841 | 1.3418 |
| Detector | MAR mosaic CCD | Pilatus 6 M | Photon II |
| Detector distance (mm) | 313.5 | 266.7 | 75 |
| No. of oscillation images | 217 | 1000 | 400 |
| Exposure time per image (s) | 2 | 0.2 | 90 |
| Oscillation width (°) | 0.5 | 0.1 | 0.3 |
| Space group | F222 | F222 | F222 |
| Unit-cell parameters a, b, c (Å) | 134.4, 203.9, 226.7 | 136.9, 201.8, 217.9 | 136.3, 200.7, 217.1 |
| Resolution range (Å) | 47.30–2.59 (2.67–2.59) | 47.93–1.60 (1.63–1.60) | 45.54–2.10 (2.14–2.10) |
| No. of observations | 313528 (21715) | 739043 (37457) | 400760 (13579) |
| No. of unique reflections | 48305 (4411) | 195423 (9686) | 83782 (3866) |
| Data completeness (%) | 100.0 (99.9) | 99.6 (99.8) | 97.3 (82.4) |
| Redundancy | 6.5 (4.9) | 3.8 (3.9) | 4.8 (3.5) |
| Mosaicity (°) | 0.22 | 0.07 | 0.13 |
| Average I/σ(I) | 13.5 (2.0) | 9.2 (1.5) | 7.3 (2.1) |
| Solvent content (%) | 62.2 | 61.0 | 60.4 |
| R merge | 0.144 (0.858) | 0.074 (0.635) | 0.188 (0.649) |
| R meas | 0.157 (0.963) | 0.086 (0.738) | 0.212 (0.759) |
| CC(1/2) | 0.994 (0.608) | 0.998 (0.700) | 0.990 (0.697) |
| Wilson B factor (Å2) | 30.7 | 10.9 | 16.7 |
| Refinement | |||
| R work | 0.160 | 0.131 | 0.159 |
| R free | 0.226 | 0.154 | 0.196 |
| Average B factor (Å2) | 36.1 | 15.5 | 19.8 |
| R.m.s.d. bonds from ideal (Å) | 0.009 | 0.011 | 0.009 |
| R.m.s.d. angles from ideal (°) | 1.350 | 1.670 | 1.539 |
| Ramachandran favoured (%) | 94.45 | 95.77 | 95.30 |
| Ramachandran outliers (%) | 0 | 0 | 0 |
The complexes of MvBOxWT and MvBOxW396A with ferricyanide ([Fe(CN)6]3−, product) reported here (PDB ID 6I3J and 6I3K, respectively) were obtained by soaking of the crystals in excess of ferrocyanide ([Fe(CN)6]4−, substrate). The oxidation of ferrocyanide and reduction of MvBOx during the soaking of the crystal was clearly observable (see above). However, due to the fact that the trinuclear copper cluster in WT:FECN is fully reduced (Fig. S13), the observed ligand could still be ferrocyanide. Unfortunately, ferrocyanide and ferricyanide are not distinguishable at the obtained resolutions. We chose to model product (ferricyanide, [Fe(CN)6]3−) in both structures. The geometrical restraint library for the link between Trp396 and His398 was built based on data found in the crystallographic database of organic compounds (Cambridge Structural Database, The Cambridge Crystallographic Data Centre). The particular geometry of the link was identified in 3 records of CSD with ID codes CIMGUC (occurrence 4×), CIMHAJ (1×) and SEPXOC (1×). Records CIMGUC and CIMHAJ were chosen for extraction of averaged geometrical parameters, which were directly used for construction of the restraints. Orientation of the new C-N bond was restrained independently in the planes of the two corresponding planar side chains.
Supplementary information
Acknowledgements
This work was supported by the institutional support of IBT CAS, v. v. i. (RVO: 86652036), by the European Regional Development Fund (CZ.02.1.01/0.0/0.0/15_003/0000447, CZ.02.1.01/0.0/0.0/16_013/0001776, and CZ.1.05/1.1.00/02.0109), and by the Ministry of Education, Youth and Sports of the Czech Republic (LM2015043, support of Biocev-CMS). We would like to thank Drs Jiri Pavlicek, Petr Pompach and Tatsiana Charnavets from the Centre of Molecular Structure, Biocev for assistance with data acquisition.
Author Contributions
T.K. and J.Do. designed research; L.H.O., T.K. and L.S. prepared samples; T.K. performed crystallization and biophysical characterisation; T.K., L.S., T.S., P.K., J.S. and J.Do. performed data collection and processing; T.K. and L.S. performed structure solution and refinement, T.K., L.S., P.K. and J.Do. performed structure validation; L.S. performed kinetic measurements; T.K., L.S., T.S., P.K., J.Du., J.H., M.T., K.F. and J.Do. analyzed and interpreted the data; J.Du. secured laboratory background; T.K., L.S., and J.Do. wrote the manuscript with contributions from all authors.
Data Availability
The crystal structures and corresponding data were deposited in the Protein Data Bank under the codes 6I3J (wild type in complex with ferricyanide) 6I3K (mutant W396A with ferricyanide) and 6I3L (mutant W396F). All other source data are available upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tomáš Kovaľ and Leona Švecová contributed equally.
Contributor Information
Tomáš Kovaľ, Email: tomas.koval@ibt.cas.cz.
Jan Dohnálek, Email: jan.dohnalek@ibt.cas.cz.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50105-3.
References
- 1.Murao S, Tanaka N. A new enzyme bilirubin oxidase produced by Myrothecium verrucaria MT-1. Agric. Biol. Chem. 1981;45:2383–2384. [Google Scholar]
- 2.Tanaka N, Murao S. Difference between various copper-containing enzymes (Polyporus laccase, mushroom tyrosinase and cucumber ascorbate oxidase) and bilirubin oxidase. Agric. Biol. Chem. 1983;47:1627–1628. [Google Scholar]
- 3.Guo J, Liang XX, Mo PS, Li GX. Purification and properties of bilirubin oxidase from Myrothecium verrucaria. Appl. Biochem. Biotechnol. 1991;31:135–143. doi: 10.1007/BF02921784. [DOI] [PubMed] [Google Scholar]
- 4.Xu F, et al. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta. 1996;1292:303–311. doi: 10.1016/0167-4838(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 5.Kataoka K, et al. Point mutations at the type I Cu ligands, Cys457 and Met467, and at the putative proton donor, Asp105, in Myrothecium verrucaria bilirubin oxidase and reactions with dioxygen. Biochemistry. 2005;44:7004–7012. doi: 10.1021/bi0476836. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka K, Sugihara T, Tsujino Y, Osakai T, Tamiya E. Electrochemical consideration on the optimum pH of bilirubin oxidase. Anal Biochem. 2007;370:98–106. doi: 10.1016/j.ab.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Mizutami K, et al. X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 Å resolution using a twinned crystal. Acta Cryst. 2010;F66:765–770. doi: 10.1107/S1744309110018828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cracknell JA, McNamara TP, Lowe ED, Blanford CF. Bilirubin oxidase from Myrothecium verrucaria: X-ray determination of the complete crystal structure and a rational surface modification for enhanced electrocatalytic O2 reduction. Dalton Trans. 2011;40:6668–6675. doi: 10.1039/c0dt01403f. [DOI] [PubMed] [Google Scholar]
- 9.Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem. Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem. Record. 2007;7:220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- 11.Quintanar L, et al. Shall We Dance? How A Multicopper Oxidase Chooses Its Electron Transfer Partner. Acc. Chem. Res. 2007;40:445–452. doi: 10.1021/ar600051a. [DOI] [PubMed] [Google Scholar]
- 12.Bento I, Martins LO, Gato Lopes G, Carrondo MA, Lindley PF. Dioxygen reduction by multi-copper oxidases; a structural perspective. Dalton Trans. 2005;7:3507–3513. doi: 10.1039/b504806k. [DOI] [PubMed] [Google Scholar]
- 13.Yoon J, Solomon EI. Electronic Structure of the Peroxy Intermediate and Its Correlation to the Native Intermediate in the Multicopper Oxidases: Insights into the Reductive Cleavage of the O-O bond. J. Am. Chem. Soc. 2007;129:13127–13136. doi: 10.1021/ja073947a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataoka K, et al. Four-electron Reduction of Dioxygen by a Multicopper Oxidase, CueO, and Roles of Asp112 and Glu506 Located Adjacent to the Trinuclear Copper Centre. The journal of biological chemistry. 2009;284:14405–14413. doi: 10.1074/jbc.M808468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.dos Santos L, Climent V, Blanford CF, Armstrong FA. Mechanistic studies of the ‘blue’ Cu enzyme, bilirubin oxidase, as a highly efficient electrocatalyst for the oxygen reduction reaction. Phys. Chem. Chem. Phys. 2010;12:13962–13974. doi: 10.1039/c0cp00018c. [DOI] [PubMed] [Google Scholar]
- 16.Bento I, et al. Mechanisms underlying dioxygen reduction in laccases. Structural and modelling studies focusing on proton transfer. BMC Structural Biology. 2010;10:28. doi: 10.1186/1472-6807-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwaki M, et al. ATR-FTIR study of the protonation states of the Glu residue in the multicopper oxidases, CueO and bilirubin oxidase. FEBS Lett. 2010;584:4027–4031. doi: 10.1016/j.febslet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu A, et al. Myrothecium verrucaria bilirubin oxidase and its mutants for potential copper ligands. Biochemistry. 1999;38:3034–3042. doi: 10.1021/bi9819531. [DOI] [PubMed] [Google Scholar]
- 19.Mano N. Features and applications of bilirubin oxidases. Appl Microbiol Biotechnol. 2012;96:301–307. doi: 10.1007/s00253-012-4312-9. [DOI] [PubMed] [Google Scholar]
- 20.Doumas BT, Yein F, Perry B, Jendrzejczak B, Kessner A. Determination of the sum of bilirubin sugar conjugates in plasma by bilirubin oxidase. Clin. Chem. 1999;45:1255–1260. [PubMed] [Google Scholar]
- 21.Kimura S, Iyama S, Yamaguchi Y, Hayashi S, Yanagihara T. Enzymatic assay for conjugated bilirubin (Bc) in serum using bilirubin oxidase (BOD) J Clin Lab Anal. 1999;13:219–223. doi: 10.1002/(SICI)1098-2825(1999)13:5<219::AID-JCLA5>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Huang J, Zhang X. Decolorization and biodegradation of remazol brilliant blue R by bilirubin oxidase. J Biosci Bioeng. 2009;108:496–500. doi: 10.1016/j.jbiosc.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Han X, Zhao M, Lu L, Liu Y. Purification, characterization and decolorization of bilirubin oxidase from Myrothecium verrucaria 3.2190. Fungal. Biol. 2012;116:863–871. doi: 10.1016/j.funbio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez P, et al. Direct electron transfer from graphite and functionalized gold electrodes to T1 and T2/T3 copper centers of bilirubin oxidase. Biochim. Biophys. Acta, Bioenerg. 2008;1777:1364–1369. doi: 10.1016/j.bbabio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Brocato S, Lau C, Atanassov P. Mechanistic study of direct electron transfer in bilirubin oxidase. Electrochim. Acta. 2012;61:44–49. doi: 10.1016/j.electacta.2011.11.074. [DOI] [Google Scholar]
- 26.Leech D, Kavanagh P, Schuhmann W. Enzymatic fuel cells: Recent progress. Electrochim. Acta. 2012;84:223–234. doi: 10.1016/j.electacta.2012.02.087. [DOI] [Google Scholar]
- 27.Filip J, Sefcovicova J, Gemeiner P, Tkac J. Electrochemistry of bilirubin oxidase and its use in preparation of a low cost enzymatic biofuel cell based on a renewable composite binder chitosan. Electrochim. Acta. 2013;87:366–374. doi: 10.1016/j.electacta.2012.09.054. [DOI] [Google Scholar]
- 28.Mano N, Edembe L. Bilirubin oxidases in bioelectrochemistry: Features and recent findings. Biosensors and Bioelectronics. 2013;50:478–485. doi: 10.1016/j.bios.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Santoro C, Babanova S, Erable B, Schuler A, Atanassov P. Bilirubin oxidase based enzymatic air-breathing cathode: Operation under pristine and contaminated conditions. Bioelectrochemistry. 2016;108:1–7. doi: 10.1016/j.bioelechem.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Mazurenko I, et al. How the Intricate Interactions between Carbon Nanotubes and Two Bilirubin Oxidases Control Direct and Mediated O2 Reduction. ACS Appl. Mater. Interfaces. 2016;8:23074–23085. doi: 10.1021/acsami.6b07355. [DOI] [PubMed] [Google Scholar]
- 31.Filip J, Andicsova-Eckstein A, Vikartovska A, Tkac J. Immobilization of bilirubin oxidase on graphene oxide flakes with different negative charge density for oxygen reduction. The effect of GO charge density on enzyme coverage, electron transfer rate and current density. Biosens Bioelectron. 2017;89:384–389. doi: 10.1016/j.bios.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Mano N, de Poulpiquet A. O2 Reduction in Enzymatic Biofuel Cells. Chem. Rev. 2018;118:2392–2468. doi: 10.1021/acs.chemrev.7b00220. [DOI] [PubMed] [Google Scholar]
- 33.Lopez F, Zerria S, Ruff A, Schuhmann W. An O2 Tolerant Polymer/Glucose Oxidase Based Bioanode as Basis for a Self-powered Glucose Sensor. Electroanalysis. 2018;30:1311–1318. doi: 10.1002/elan.201700785. [DOI] [Google Scholar]
- 34.Cracknell JA, Blanford CF. Developing the mechanism of dioxygen reduction catalyzed by multicopper oxidases using protein film electrochemistry. Chemical Science. 2012;3:1567–1581. doi: 10.1039/c2sc00632d. [DOI] [Google Scholar]
- 35.Agbo P, Heath JR, Gray HB. Modeling Dioxygen Reduction at Multicopper Oxidase Cathodes. J. Am. Chem. Soc. 2014;136:13882–13887. doi: 10.1021/ja5077519. [DOI] [PubMed] [Google Scholar]
- 36.Komori H, Higuchi Y. Structural insights into the O2 reduction mechanism of multicopper oxidase. The Journal of Biochemistry. 2015;158:293–298. doi: 10.1093/jb/mvv079. [DOI] [PubMed] [Google Scholar]
- 37.Bertrand T, et al. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41:7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- 38.Enguita FJ, et al. Substrate and Dioxygen Binding to the Endospore Coat Laccase from Bacillus Subtilis. J. Biol. Chem. 2004;279:23472–23476. doi: 10.1074/jbc.M314000200. [DOI] [PubMed] [Google Scholar]
- 39.Kallio JP, et al. Structure Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds. J. Mol. Biol. 2009;392:895–909. doi: 10.1016/j.jmb.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 40.Akter M, et al. Redox potential-dependent formation of an unusual His-Trp bond in bilirubin oxidase. Chemistry. 2018;24:18052–18058. doi: 10.1002/chem.201803798. [DOI] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Enguita FJ, Martins LO, Henriques AO, Carrondo MA. Crystal Structure of a Bacterial Endospore Coat Component. J. Biol. Chem. 2003;278:19416–19425. doi: 10.1074/jbc.M301251200. [DOI] [PubMed] [Google Scholar]
- 43.Moser CC, Anderson JL, Dutton PL. Guidelines for tunneling in enzymes. Biochim Biophys Acta. 2010;1797:1573–1586. doi: 10.1016/j.bbabio.2010.04.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skalova T, et al. Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 Å. Acta Crystallogr F Struct. Biol. Cryst. Commun. 2011;67:27–32. doi: 10.1107/S1744309110046099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlikowska M, et al. Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus. Int. J. Biol. Macromol. 2018;107:1629–1640. doi: 10.1016/j.ijbiomac.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Ravikiran B, Mahalakshmi R. Unusual post-translational protein modification: the benefits of sophistication. RSC Adv. 2014;4:33958–33974. doi: 10.1039/C4RA04694C. [DOI] [Google Scholar]
- 47.Ehrenshaft M, Deterding LJ, Mason RP. Tripping Up Trp: Modification of Protein Tryptophan Residues by Reactive Oxygen Species, Modes of Detection, and Biological Consequences. Free Radic Biol Med. 2015;89:220–228. doi: 10.1016/j.freeradbiomed.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen NT, Wrona MZ, Dryhurst G. Electrochemical oxidation of tryptophan. J. Electroanal. Chem. 1986;199:101–126. doi: 10.1016/0022-0728(86)87045-0. [DOI] [Google Scholar]
- 49.Enache TA, Oliveira-Brett AM. Pathways of Electrochemical Oxidation of Indol Compounds. Electroanalysis. 2011;23:1337–1344. doi: 10.1002/elan.201000671. [DOI] [Google Scholar]
- 50.Yang S, Liu J, Quan X, Zhou J. Bilirubin Oxidase Adsorption onto Charged Self-Assembled Monolayers: Insights from Multiscale Simulations. Langmuir. 2018;34:9818–9828. doi: 10.1021/acs.langmuir.8b01974. [DOI] [PubMed] [Google Scholar]
- 51.Xia H, Kitazumi Y, Shirai O, Kano K. Enhanced direct electron transfer-type bioelectrocatalysis of bilirubin oxidase on negatively charged aromatic compound-modified carbon electrode. J. Electroanal. Chem. 2016;763:104–109. doi: 10.1016/j.jelechem.2015.12.043. [DOI] [Google Scholar]
- 52.Sakai K, Xia H, Kitazumi Y, Shirai O, Kano K. Assembly of direct-electron-transfer-type bioelectrodes with high performance. Electrochimica Acta. 2018;271:305–311. doi: 10.1016/j.electacta.2018.03.163. [DOI] [Google Scholar]
- 53.Kovaľ T, et al. Structural and Catalytic Properties of S1 Nuclease from aspergillus oryzae responsible for substrate recognition, cleavage, non–specificity, and inhibition. PLoS ONE. 2016;11:e0168832. doi: 10.1371/journal.pone.0168832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, et al. A high effective NADH-ferricyanide dehydrogenase coupled with laccase for NAD+ regeneration. Biotechnol. Lett. 2016;38:1315–1320. doi: 10.1007/s10529-016-2106-3. [DOI] [PubMed] [Google Scholar]
- 55.Reiss R, Ihssen J, Thöny-Meyer L. Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnology. 2011;11:9. doi: 10.1186/1472-6750-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand F, et al. Bilirubin oxidase from Bacillus pumillus: A promising enzyme for elaboration of efficient cathodes in Biofuel cells. Biosens Bioelectron. 2012;35:140–146. doi: 10.1016/j.bios.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sigma-Aldrich bilirubin oxidase assay protocol, www.sigmaaldrich.com.
- 58.Fejfarová K, et al. Crystallization of nepenthesin I using a low-pH crystallization screen. Acta Crystallogr F Struct. Biol. Cryst. Commun. 2016;72:24–28. doi: 10.1107/S2053230X15022323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz–Zentrum Berlin. J Synchrotron Radiat. 2012;19:442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Murshudov GN, Vagin A, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 65.Chen VB, Arendall WB, Headd JJ, Keedy DA. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nature Structural Biology. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 67.Afonine PV, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structures and corresponding data were deposited in the Protein Data Bank under the codes 6I3J (wild type in complex with ferricyanide) 6I3K (mutant W396A with ferricyanide) and 6I3L (mutant W396F). All other source data are available upon reasonable request.