Abstract
Background
To investigate the secretion of interleukin‐1β (IL‐1β), IL‐6, IL‐10, IL‐8, and soluble intercellular adhesin molecule 1 (sICAM‐1) from THP‐1 monocytes stimulated by different Streptococcus pneumoniae (S pneumoniae) strains.
Methods
Fifty‐eight strains of S pneumoniae were collected: ATCC49619, 23 from sputum (sd‐SP), 23 from blood (bd‐SP), and 11 from cerebrospinal fluid (CSF; cd‐SP). Such strains were cultured and suspended at 0.5 McFarland. THP‐1 monocytes were cultured and resuspended at 5.0 × 108/L, which were stimulated by S pneumoniae for 4, 8, and 12 hours, respectively. The suspensions were analyzed for IL‐1β, IL‐6, IL‐10, IL‐8, and sICAM‐1 using an ELISA method. The data were assayed with SPSS 19.0.
Results
Contrary to IL‐10, the concentrations of IL‐1β, IL‐6, IL‐8, and sICAM‐1 all increased first and then decreased. IL‐1β and sICAM‐1 levels in the ATCC49619 group were both higher than all the clinical S pneumoniae groups (sd‐SP, bd‐SP, and cd‐SP), IL‐6 and IL‐8 versa, and IL‐10 equal. The difference among clinical S pneumoniae groups lay only in sICAM‐1. cd‐SP group showed lower sICAM‐1 concentrations than sd‐SP and bd‐SP groups at both 4 and 8 hours. However, they became equal at 12 hours.
Conclusions
The secretion summit is 8 hours for IL‐1β, IL‐6, IL‐8, and sICAM‐1, bottom for IL‐10. Different clinical S pneumoniae strains show similar ability to induce THP‐1 cells secreting interleukins. However, cd‐SP induces THP‐1 cells secreting lower sICAM‐1 than sd‐SP and bd‐SP, which may in turn facilitate its invasion into CSF.
Keywords: adhesin, ELISA, interleukin, Streptococcus pneumoniae
1. INTRODUCTION
Streptococcus pneumoniae (S pneumoniae) is a dominant agent worldwide for pneumonia, bacteremia, and meningitis, which accounts for about one million deaths in children under 5 years1 each year. Bacteremia and meningitis are more fatal than pneumonia, which lead to children's deaths mainly due to nervous system complications and the old people's deaths primarily due to systematic complications.2, 3 S pneumoniae is frequently localized in human's respiratory tract before its pathogenic transformation.4 Due to the poor immune response against polysaccharide of S pneumoniae, infants and the aged are more susceptible to various S pneumoniae infections.5, 6 Innate immune is vital for host's recognizing pathogenic organisms and consequent initializing protection response,7 deficit of which reduces various infections. In addition, the amount and pathogenicity of S pneumoniae constitute the other key factors. We previously verified that certain S pneumoniae virulence factors such as pneumolysin (Ply) may facilitate S pneumoniae's invading blood system.8, 9 However, other differences remain still uncertain among different S pneumoniae strains responsible for the former three types of infections. Dendritic cells, monocytes, and neutrophils are all primary guards for host's fighting S pneumoniae, the former two of which were used for research more often.10, 11 The first immune cell type to combat S pneumoniae during early infection is the alveolar macrophage,12 which derives from monocytes and is responsible for the initial detection of bacteria and subsequent modulation of the inflammatory response.13 In this study, we used THP‐1 monocytes to be cultured with S pneumoniae strains from sputum, blood, and cerebrospinal fluid (CSF), and analyzed the consequent interleukin‐1β (IL‐1β), IL‐6, IL‐10, IL‐8, and soluble intercellular adhesion molecule (sICAM‐1) levels so as to unveil the differences among different S pneumoniae strains.
2. MATERIALS AND METHODS
2.1. Strains and culture
A total of 58 S pneumoniae strains were collected, of which 23 were isolated from sputum samples from patients with pneumonia at Taizhou Municipal Hospital during 2017, and 23 and 11 from blood and CSF samples, respectively, at Tangshan Maternal and Child Health Hospital in 2016 and 2017. Pneumonia was diagnosed based on the results of physical examinations, X‐ray, and laboratory medicine. Another strain was ATCC49619, provided by the Chinese National Center for Medical Culture Collections. All the strains were non‐repeated, that is, only one strain from each patient. They were all identified using VITEK‐2 automatic microorganism analyzer (bioMérieux Co.), followed by a specific PCR method according to du Plessis.14
All the 58 strains were cultured on blood agar plates at 37°C in a 5% CO2 incubator (Thermo Electron Co.) overnight and then adjusted to 0.5 McFarland in normal saline, which were used for stimulation experiments.
2.2. pbp2B detection
Ten to fifteen pure colonies of each S pneumoniae strain were collected from a blood agar plate and suspended in a 0.5‐mL centrifuge tube containing 200 µL ddH2O. The tube was then placed in a boiling water bath for 30 minutes and transferred into a −20°C freezer for 10 minutes, followed by thawing and the centrifugation at 16 099 g for 30 seconds. The subsequent supernatant was transferred into a new 0.5‐mL centrifuge tube for regular PCR of pbp2B with primers as following: forward: 5′‐CTGACCATTGATTTGGCTTTCCAA‐3′ and reverse: 5′‐TTTGCAATAGTTGCTACATACTG‐3′.14 The PCR product was 682 bp. The PCR conditions were as following: 94°C denaturation for 5 minutes, 94°C for 30 seconds, 55°C for 1 minute and 72°C for 1 minute for 30 cycles, and 72°C for 10 minutes. Electrophoresis was conducted with 5 µL PCR products and 1 µL 6× loading buffer. The mixture was then loaded into 2.0% agarose gel and electrophoresed at 150 V and 75 mA for 25 minutes. The electrophoresis results were acquired by a gel camera system (Bio‐Rad).
2.3. Cell line and culture
Human THP‐1 monocytes were bought from the Cell Bank of Chinese Academy of Sciences and cultured with RPMI 1640 medium (GIBCO) and 10% fetal bovine serum (Zhejiang Tianhang Biological Technology Co.) plus 1% strep‐penicillin (Beijing Solarbio Science &Technology Co.) at 37°C in a humidified incubator (Thermo Electron Co.) with 95% air and 5% CO2. THP‐1 monocytes under seven passages were then collected and resuspended at 5.0 × 108/L using RPMI 1640 medium and 10% fetal bovine serum inactivated at 60°C for 30 minutes. Viability of THP‐1 monocytes was over 95% as determined by trypan blue exclusion. This study was conducted between April and June 2018 at Zhejiang Provincial Demonstration Center of Laboratory Medicine Experimental Teaching, Wenzhou Medical University.
2.4. Infection of THP‐1 cell line with S pneumoniae cultures
One hundred and seventy‐seven milliliters of THP‐1 cells were inoculated in 177 wells of 12‐well culture plates, which were 5.0 × 108/L, one milliliter per well. One hundred microliters of the aforementioned S pneumoniae suspensions at 0.5 McFarland was then seeded into each one of 174 wells. Normal saline was used as control, 100 µL per well for the other three wells. All the wells were cultured at 37°C in 5% CO2 for 4 hours (59 wells), 8 hours (another 59 wells), and 12 hours (another 59 wells). At the end of the experiments, all of the cell cultures plus controls were collected in 1.5‐mL centrifuge tubes and centrifuged for 10 minutes at 4500 g. The supernatants were for the analysis of interleukins and adhesin. The experiments were repeated thrice.
2.5. Analysis of interleukins and adhesin
IL‐1β, IL‐6, IL‐10, IL‐8, and sICAM‐1 concentrations were analyzed using an ELISA method, according to the manufacturer's protocols (Shanghai Xitang Biotechnology Co.).
2.6. Statistical analysis
All the data analyses were made using SPSS 19.0 statistical software (SPSS). The normality analysis was performed by Kolmogorov‐Smirnov test and homogeneity of variance analysis between two groups was by Levene's test. The mean difference analysis was conducted by the single‐sample t test or analysis of variance for factorial designs. P > 0.10 in the normality test and P < 0.05 in mean comparison were considered statistically significant.
3. RESULTS
As shown in Figure 1, the concentrations of IL‐1β in clinical S pneumoniae groups (sd‐SP, bd‐SP, and cd‐SP) were all higher than ATCC49619 by the single‐sample t test. Variance for factorial designs was used to determine the differences among the three clinical S pneumoniae groups. No significant difference was found among three clinical groups (F = 2.791, P = 0.064), which was found among three stimulation time points (F = 70.559, P = 0.000). Interaction effect was not found in three clinical groups and tree stimulation time points (F = 0.457, P = 0.767), suggesting no interaction effect between S pneumoniae groups and stimulation time points.
Figure 1.
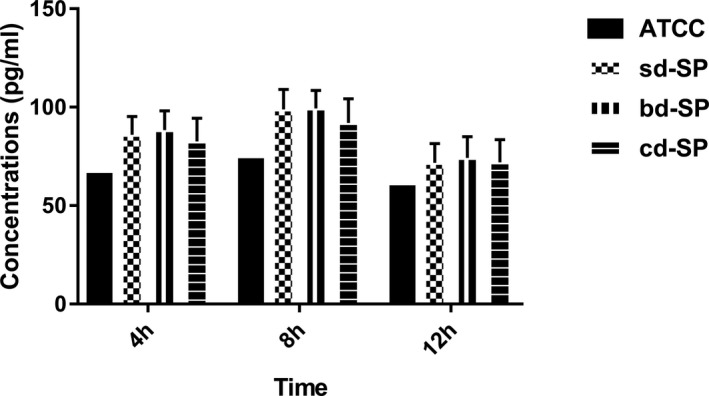
Concentrations of IL‐1β secreted by THP‐1 cells stimulated by S pneumoniae groups. (ATCC, sd‐SP, bd‐SP, and cd‐SP). ATCC, ATCC49619; bd‐SP, blood‐derived SP; cd‐SP, cerebrospinal fluid‐derived SP; IL‐1β, interleukin‐1β; sd‐SP, sputum‐derived SP
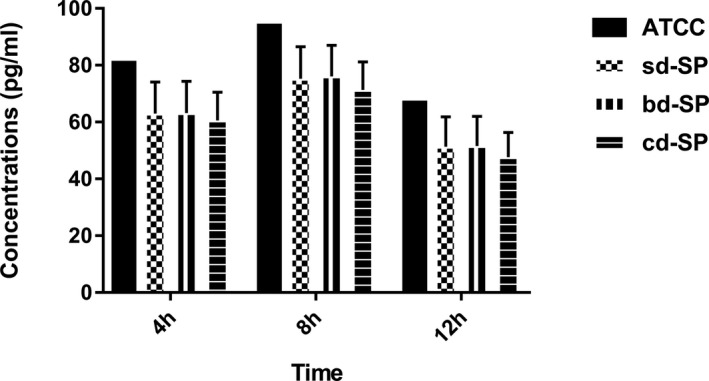
As shown in Figure 2, the concentrations of IL‐6 in clinical S pneumoniae groups were lower than ATCC49619 except at 8 hours for sd‐SP group by the single‐sample t test. Variance for factorial designs was used to determine the differences among the three clinical S pneumoniae groups. No significant difference was found among three clinical groups (F = 2.841, P = 0.061), which was found among three stimulation time points (F = 67.312, P = 0.000). Interaction effect was not found in three clinical groups and tree stimulation time points (F = 0.334, P = 0.855), showing no interaction effect between S pneumoniae groups and stimulation time points.
Figure 2.
Concentrations of IL‐6 secreted by THP‐1 cells stimulated by S pneumoniae groups
As shown in Figure 3, no significant difference was found between the concentrations of IL‐10 in clinical S pneumoniae groups and ATCC49619 group by the single‐sample t test. Variance for factorial designs was used to determine the differences among the three clinical S pneumoniae groups. No significant difference was found among three clinical groups (F = 0.968, P = 0.382), which was found among three stimulation time points (F = 170.524, P = 0.000). Interaction effect was not found in three clinical groups and tree stimulation time points (F = 0.019, P = 0.999), suggesting no interaction effect between S pneumoniae groups and stimulation time points.
Figure 3.

Concentrations of IL‐10 secreted by THP‐1 cells stimulated by S pneumoniae groups
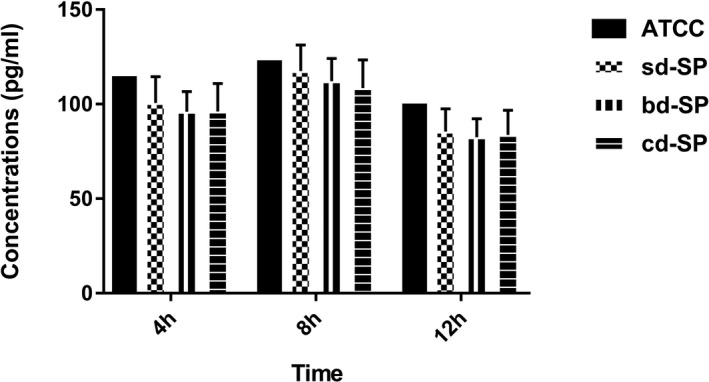
As shown in Figure 4, the concentrations of IL‐8 in clinical S pneumoniae groups were lower than ATCC49619 except at 8 hours point for bd‐SP group by the single‐sample t test. Variance for factorial designs was used to determine the differences among the three clinical S pneumoniae groups. No significant difference was found among three clinical groups (F = 1.485, P = 0.230), which was found among three stimulation time points (F = 64.852, P = 0.000). Interaction effect was not found in three clinical groups and tree time points (F = 0.040, P = 0.997), showing no interaction effect between S pneumoniae groups and stimulation time points.
Figure 4.
Concentrations of IL‐8 secreted by THP‐1 cells stimulated by S pneumoniae groups
As shown in Figure 5, the concentrations of sICAM‐1 in clinical S pneumoniae groups were all higher than ATCC49619 by the single‐sample t test. Variance for factorial designs was used to determine the differences among the three clinical S pneumoniae groups. Significant difference was found among three clinical groups (F = 7.669, P = 0.001), which was also found among three stimulation time points (F = 60.898, P = 0.000). Interaction effect was not found in three clinical groups and tree time points (F = 0.751, P = 0.559), suggesting no interaction effect between S pneumoniae groups and stimulation time points. However, Figure 5 shows lower sICAM‐1 concentrations in cd‐SP group than sd‐SP and bd‐SP groups at 4 and 8 hours points, equal at 12 hours.
Figure 5.
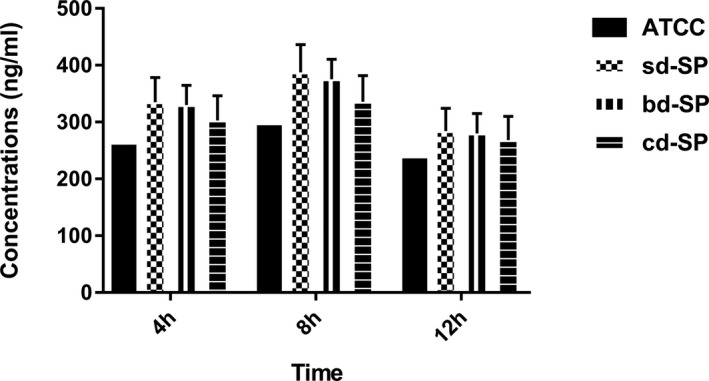
Concentrations of sICAM‐1 secreted by THP‐1 cells stimulated by S pneumoniae groups
As shown in Figure 6, rapid accumulation of THP‐1 cells was captured after S pneumoniae was added. With the stimulation time progressing, THP‐1 cells tended to show similar morphisms while S pneumoniae kept decreasing. No significant difference was found among the three clinical S pneumoniae groups for morphological changes.
Figure 6.
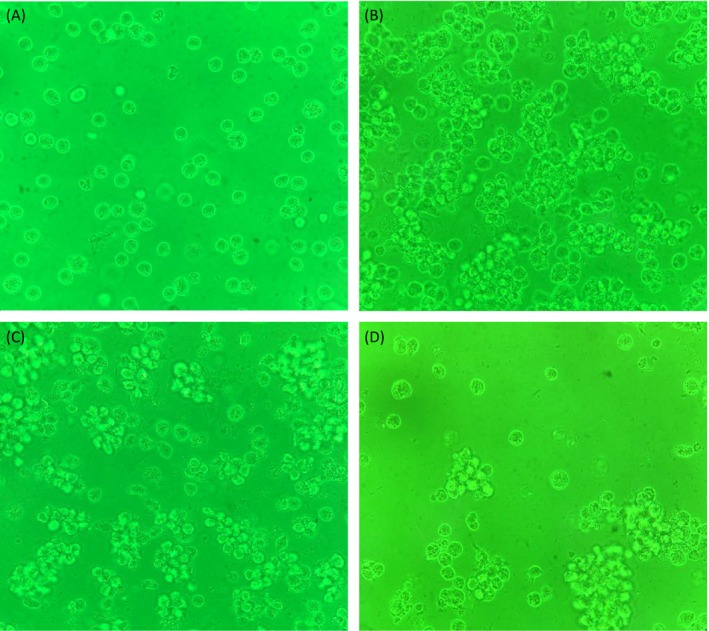
A, THP‐1 cells before stimulation (400×). B, THP‐1 cells after 4‐h stimulation (400×). C, THP‐1 cells after 8‐h stimulation (400×). D, THP‐1 cells after 12‐h stimulation (400×)
4. DISCUSSION
Our data showed the different impact on THP‐1 cells by different S pneumoniae strains. ATCC49619 was remarkably different from clinical S pneumoniae strains, while the latter themselves showed similar traits except the reduction of sICAM‐1.
The forebody of IL‐1β is 34 KD, which is spliced as 17KD. IL‐1β plays an important role in the process of various infectious and chronic inflammatory diseases.15, 16, 17 IL‐1β is primarily secreted by neutrophils, monocytes/macrophages, and dendritic cells, which is also induced by Ply.18 IL‐6 is mainly secreted by neutrophils and monocytes/macrophages, the lack of which could result in the deficit of innate CNS immune against S pneumoniae and consequent rising mortality.19 IL‐10 is a kind of anti‐inflammation cytokine that is predominantly secreted by neutrophils, monocytes/macrophages, dendritic cells, and lymphocytes and is vital in acute and persistent bacterial infections.20, 21, 22 IL‐10 could prevent pulmonary inflammation from aggravation induced by S pneumoniae, the lack of which may cause the rising mortality.23 IL‐8 is also called chemokine 8, which is secreted by monocytes/macrophages and dendritic cells. IL‐8 could promote inflammatory cells’ accumulation and activation, the release of inflammatory mediators, and chemotaxis of neutrophils to inflammatory sites so as to regulate host's inflammatory response.24, 25 Cytokines are classified as proinflammatory cytokines (IL‐1β, IL‐6, and IL‐8, etc) and anti‐inflammatory ones (IL‐4 and IL‐10, etc).26 ICAM‐1 could bind the lymphocyte function–associated antigen 1 (LFA‐1)27 widely distributed in lymph node and tonsil vascular endothelial cells, thymic dendritic cells, tonsil and glomerular epithelial cells, leukocytes, macrophages, and fibroblasts. ICAM‐1 could also promote leukocytes’ accumulation toward S pneumoniae‐infected sites, especially extravascular sites.28, 29
As opposed to IL‐10, as shown in Figures 1, 2, 3, 4, 5, IL‐1β, IL‐6, IL‐8, and sICAM‐1 levels all increased first and then decreased, which reflects the expanding to diminishing of inflammatory response. IL‐10 promoted such response transformation to prevent host from severe lesions.23, 30, 31, 32 The concentrations of IL‐1β and sICAM‐1 in the group ATCC49619 were both higher than all the clinical S pneumoniae groups, IL‐10 equal, IL‐6 and IL‐8 versa. ATCC49619 is a standard S pneumoniae strain, which is less virulent than clinical ones as shown in our previous study.8, 9, 33 The intriguing difference among clinical S pneumoniae groups lies in sICAM‐1 except IL‐1β, IL‐6, IL‐10, and IL‐8. cd‐SP group showed lower sICAM‐1 concentrations than sd‐SP and bd‐SP groups at both 4 and 8 hours, similar at 12 hours. For early infection, lower sICAM‐1 concentration is infaust for the accumulation of leukocytes in infectious sites so as to aid in S pneumoniae's surviving and progressing in CSF. The concentrations of such interleukins and adhesin were equal between sd‐SP and bd‐SP groups, while our previous study proved sd‐SP is less virulent than bd‐SP.8, 9 The data in this study were some different from those in our previous study,33 which was due to the different concentrations of S pneumoniae and THP‐1 cells. In this study, the ratio of S pneumoniae and THP‐1 cells was much lower, which resulted in the faster clearance of S pneumoniae. The 16S rRNA cycle threshold >32.0 in qRT‐PCR also suggested much poorer survival of S pneumoniae (data not shown). And Figure 6 also confirms the scarcity of S pneumoniae and more healthy conditions of THP‐1 cells. Rapid accumulation of THP‐1 cells was captured after S pneumoniae was added. With the stimulation time progressing, THP‐1 cells tended to show similar morphisms while S pneumoniae kept decreasing. In this study, THP‐1 cells showed rapid clearance ability of S pneumoniae, in agreement with another report.34
Our study had some limitations. Many types of cell lines are involved in S pneumoniae‐host interactions, the amount and concentrations of which are hard to determine. However, only one fixed concentration of THP‐1 cells was used to simulate actual interactions. Furthermore, THP‐1 cell line derived from leukemia cells, which is capable of self‐proliferation and owns different ability of phagocytosing and killing S pneumoniae from normal monocytes.
In summary, our data demonstrated lower secretions of sICAM‐1 from monocytes may facilitate S pneumoniae's invading CSF. And further investigations need to be conducted.
AUTHORS’ CONTRIBUTIONS
Ping Ren and Dakang Hu conceived of the study and were involved in the stimulation test. Lianhua Yu and Chunyan Gao were dedicated to strains’ collection and identification. Jin Zhang carried out PCR and ELISA tests. Ying Qu and Xinyu Jiang analyzed the data. Jin Zhang and Chunyan Gao wrote the manuscript, which was revised by Dakang Hu and Yixia Zhou. All authors read and approved the final manuscript.
ETHICAL APPROVAL
This work was approved by the Ethics Committee of Taizhou Municipal Hospital.
ACKNOWLEDGMENTS
This work was supported in part by Zhejiang Natural Science Foundation (LQ19H20002), Zhejiang Medical and Health Research Program (2018KY901), Jiaojiang Science and Technology Development Program, Taizhou City (10275 and 153050), Research Program of Taizhou College (2015PY033 and 2017PY055) and National Natural Science Foundation of China (81860089).
Ren P, Zhang J, Yu L, et al. Impact of different Streptococcus pneumoniae on the secretion of interleukin and adhesin from THP‐1 monocytes. J Clin Lab Anal. 2019;33:e22927 10.1002/jcla.22927
Ping Ren and Jin Zhang equally contributed.
REFERENCES
- 1. O'Brien KL, Wolfson LJ, Watt JP, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374(9693):893‐902. [DOI] [PubMed] [Google Scholar]
- 2. Koedel U, Scheld WM, Pfister HW. Pathogenesis and pathophysiology of pneumococcal meningitis. Lancet Infect Dis. 2002;2(12):721‐736. [DOI] [PubMed] [Google Scholar]
- 3. Weisfelt M, van de Beek D, Spanjaard L, Reitsma JB, de Gans J. Clinical features, complications, and outcome in adults with pneumococcal meningitis: a prospective case series. Lancet Neurol. 2006;5(2):123‐129. [DOI] [PubMed] [Google Scholar]
- 4. Suzuki H, Ikeda K. Mode of action of long‐term low‐dose macrolide therapy for chronic sinusitis in the light of neutrophil recruitment. Curr Drug Targets Inflamm Allergy. 2002;1(1):117‐126. [DOI] [PubMed] [Google Scholar]
- 5. Khan MN, Pichichero ME. The host immune dynamics of pneumococcal colonization: implications for novel vaccine development. Hum Vaccin Immunother. 2014;10(12):3688‐3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nelson JC, Jackson M, Yu O, et al. Impact of the introduction of pneumococcal conjugate vaccine on rates of community acquired pneumonia in children and adults. Vaccine. 2008;26(38):4947‐4954. [DOI] [PubMed] [Google Scholar]
- 7. Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819‐826. [DOI] [PubMed] [Google Scholar]
- 8. Hu DK, Liu Y, Li XY, Qu Y. In vitro expression of Streptococcus pneumoniae ply gene in human monocytes and pneumocytes. Eur J Med Res. 2015;20:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu DK, Wang DG, Liu Y, et al. Roles of virulence genes (PsaA and CpsA) on the invasion of Streptococcus pneumoniae into blood system. Eur J Med Res. 2013;18:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robson RL, Reed NA, Horvat RT. Differential activation of inflammatory pathways in A549 type II pneumocytes by Streptococcus pneumoniae strains with different adherence properties. BMC Infect Dis. 2006;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thornton J, McDaniel LS. THP‐1 monocytes up‐regulate intercellular adhesion molecule 1 in response to pneumolysin from Streptococcus pneumoniae . Infect Immun. 2005;73(10):6493‐6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jambo KC, Sepako E, Heyderman RS, Gordon SB. Potential role for mucosally active vaccines against pneumococcal pneumonia. Trends Microbiol. 2010;18(2):81‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knapp S, Leemans JC, Florquin S, et al. Alveolar macrophages have a protective antiinflammatory role during murine pneumococcal pneumonia. Am J Respir Crit Care Med. 2003;167(2):171‐179. [DOI] [PubMed] [Google Scholar]
- 14. du Plessis M, Smith AM, Klugman KP. Rapid detection of penicillin‐resistant Streptococcus pneumoniae in cerebrospinal fluid by a seminested‐PCR strategy. J Clin Microbiol. 1998;36(2):453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dinarello CA. Immunological and inflammatory functions of the interleukin‐1 family. Annu Rev Immunol. 2009;27:519‐550. [DOI] [PubMed] [Google Scholar]
- 17. Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin‐1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11(8):633‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karmakar M, Katsnelson M, Malak HA, et al. Neutrophil IL‐1beta processing induced by pneumolysin is mediated by the NLRP3/ASC inflammasome and caspase‐1 activation and is dependent on K+ efflux. J Immunol. 2015;194(4):1763‐1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albrecht LJ, Tauber SC, Merres J, et al. Lack of proinflammatory cytokine interleukin‐6 or tumor necrosis factor receptor‐1 results in a failure of the innate immune response after bacterial meningitis. Mediators Inflamm. 2016;2016:7678542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duell BL, Tan CK, Carey AJ, Wu F, Cripps AW, Ulett GC. Recent insights into microbial triggers of interleukin‐10 production in the host and the impact on infectious disease pathogenesis. FEMS Immunol Med Microbiol. 2012;64(3):295‐313. [DOI] [PubMed] [Google Scholar]
- 21. Mege J‐L, Meghari S, Honstettre A, Capo C, Raoult D. The two faces of interleukin 10 in human infectious diseases. Lancet Infect Dis. 2006;6(9):557‐569. [DOI] [PubMed] [Google Scholar]
- 22. Cyktor JC, Turner J. Interleukin‐10 and immunity against prokaryotic and eukaryotic intracellular pathogens. Infect Immun. 2011;79(8):2964‐2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Penaloza HF, Nieto PA, Muñoz‐Durango N, et al. Interleukin‐10 plays a key role in the modulation of neutrophils recruitment and lung inflammation during infection by Streptococcus pneumoniae . Immunology. 2015;146(1):100‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warner N, Burberry A, Pliakas M, McDonald C, Núñez G. A genome‐wide small interfering RNA (siRNA) screen reveals nuclear factor‐kappaB (NF‐kappaB)‐independent regulators of NOD2‐induced interleukin‐8 (IL‐8) secretion. J Biol Chem. 2014;289(41):28213‐28224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Sullivan MJ, Hirota N, Martin JG. Sphingosine 1‐phosphate (S1P) induced interleukin‐8 (IL‐8) release is mediated by S1P receptor 2 and nuclear factor kappaB in BEAS‐2B cells. PLoS ONE. 2014;9(4):e95566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang JM, An J. Cytokines, inflammation, and pain. Int Anesthesiol Clin. 2007;45(2):27‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morona JK, Morona R, Paton JC. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci USA. 2006;103(22):8505‐8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Etzioni A. Adhesion molecules in leukocyte endothelial interaction. Adv Exp Med Biol. 1996;408:151‐157. [DOI] [PubMed] [Google Scholar]
- 29. Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76(2):301‐314. [DOI] [PubMed] [Google Scholar]
- 30. Rijneveld A, de Vos A, Florquin S, Verbeek J, van der Poll T. CD11b limits bacterial outgrowth and dissemination during murine pneumococcal pneumonia. J Infect Dis. 2005;191(10):1755‐1760. [DOI] [PubMed] [Google Scholar]
- 31. van Zoelen M, Schouten M, de Vos AF, et al. The receptor for advanced glycation end products impairs host defense in pneumococcal pneumonia. J Immunol. 2009;182(7):4349‐4356. [DOI] [PubMed] [Google Scholar]
- 32. Weber SE, Tian H, Pirofski LA. CD8+ cells enhance resistance to pulmonary serotype 3 Streptococcus pneumoniae infection in mice. J Immunol. 2011;186(1):432‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Hu D‐K, Wang D‐G, et al. Effects of clinical isolates of Streptococcus pneumoniae on THP‐1 human monocytic cells. Mol Med Rep. 2013;8(5):1570‐1574. [DOI] [PubMed] [Google Scholar]
- 34. Broadley S, Plaumann A, Coletti R, et al. Dual‐track clearance of circulating bacteria balances rapid restoration of blood sterility with induction of adaptive immunity. Cell Host Microbe. 2016;20(1):36‐48. [DOI] [PubMed] [Google Scholar]


