Abstract
As a key molecule in the antiviral innate immune response, the activation of TANK-binding kinase 1 (TBK1) is under tight regulation. In this report, we identified phosphatidylserine-specific phospholipase PLA1A as a host factor that modulates the TBK1 activation. Knockdown of PLA1A expression suppressed the innate immune signaling induced by RNA viruses, while PLA1A overexpression enhanced the signaling. PLA1A functioned at the TBK1 level of the signaling pathway, as PLA1A silencing blocked TBK1, but not interferon regulatory factor 3 (IRF3) induced interferon-β (IFN-β) promoter activity. The phosphorylation and kinase activity of TBK1 was reduced in PLA1A knockdown cells. Mechanistically, PLA1A was required in TBK1-mitochondrial antiviral signaling protein (MAVS) interactions but not the interactions of TBK1 with other adaptor proteins. Furthermore, PLA1A knockdown reduced the recruitment of TBK1 and IRF3 to mitochondria, concomitant with altered mitochondria morphology.
Keywords: Antiviral innate immune response, PLA1A, TANK-binding kinase 1, Mitochondria
Introduction
The innate immune response is the first line of host defense against pathogen invasion. Through sensing of pathogen-associated molecular patterns (PAMPs), such as LPS, viral RNA and viral DNA, the pattern-recognition receptors (PRRs) initiate signal transductions through recruiting different adaptors that activate the transcription factors, interferon regulatory factor 3 (IRF3), and nuclear factor κB (NF-κB), which translocate to the nucleus and induce the transcription of type I interferons (IFNs). The rapidly induced and secreted type I IFNs play a key role in the innate immune response by inducing the transcription of a large group of genes that are responsible for the host's resistance to pathogen infection.
The retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) pathway and the stimulator of interferon genes (STING) signaling pathway are 2 important cytosolic pathways responsible for the detection of RNA and DNA patterns respectively. After RNA binding, RIG-I activates mitochondrial antiviral signaling protein (MAVS; also known as VISA, IPS-1 and Cardif) on the mitochondrial membrane, which stimulates the polymerization of MAVS. As a scaffold protein, MAVS recruits various downstream effectors, such as TRAF2, TRAF6 and TRADD to form a “MAVS signalosome,” which activates IκB kinase α and β (IKKα/β) and TANK-binding kinase 1 (TBK1) and IκB kinase ε (TBK1/IKKε) and leads to NF-κB and IRF3 activation [1]. STING (also known as MITA, MPYS, ERIS and TMEM173) was believed to play a central role as a DNA sensor and adaptor in the cytosolic DNA sensing pathways by directly engaging TBK1 to direct IRF3 activation [2]. Diversely in the sensors and adaptors, both RNA- and DNA-sensing pathways converge at the TBK1-IRF3 axis, indicating the important role of TBK1 in innate immunity as an integrator of multiple upstream signals and as a modulator of IFN levels.
TBK1 is a serine/threonine kinase that contains a kinase domain (KD) in its N-terminal region, a ubiquitin-like domain in its middle region and a scaffolding and dimerization domain in its C-terminal region [3]. It is constitutively expressed in many cells and its activity is under tight regulation [4]. Posttranslational modifications of TBK1, such as phosphorylation and ubiquitination, play a key role in regulating the TBK1 function. TBK1 is activated by trans-autophosphorylation at Ser172, which requires compact dimer formation through extensive interactions among the KD, ubiquitin-like domain and scaffolding and dimerization domain [3, 5, 6]. Phosphatases, such as PPM1A, PPM1B, and PP4 have been found to dephosphorylate TBK1 at Ser172 [7, 8, 9]. The modification of TBK1 after dimerization at Lys30 and Lys401 by K63-linked polyubiquitin chains is required for TBK1 kinase activation [5]. Several E3 ubiquitin ligases, such as MIB1, MIB2, Nrdp1, and RNF128 were reported to be required for TBK1 activation by promoting K63-linked polyubiquitination [10, 11, 12]. Additionally, the E3 ubiquitin ligase DTX4 and TRAF-interacting protein have been shown to negatively regulate TBK1 activation by promoting proteasomal degradation of TBK1 [13, 14]. The deubiquitinating enzymes, CYLD and USP2b, inhibit TBK1 activation by cleaving K63-linked ubiquitin chains from TBK1 [15, 16]. Moreover, host factors such as phosphatase Src homology 2 domain-containing protein tyrosine phosphatase 2 bind to the kinase domain of TBK1 and prevent TBK1-mediated downstream substrate phosphorylation [17]. In addition to direct regulation on TBK1, several factors were found to modulate the TBK1 function by influencing the formation of functional TBK1-containing complexes. For example, DOK3 plays a positive role in TLR3 signaling by enabling TRAF3/TBK1 complex formation [18], while FOS-like antigen 1 plays a negative role in the type I IFN response to malaria and viral infection by blocking TBK1 and TRAF3/TRIF interactions [19].
The phosphatidylserine (PS)-specific phospholipase, phospholipase A1 member A (PLA1A), was identified as a host factor involved in HCV assembly by facilitating NS2-E2 and NS2-NS5A complex formation in our previous study [20]. Notably, PLA1A expression was upregulated by dsRNA transfection, which promoted us to analyze the function of PLA1A in the dsRNA-induced type I IFN production. In this study, we found that PLA1A was required for RNA virus-induced type I IFN production, as silencing PLA1A significantly impaired the antiviral response to Sendai virus (SeV). PLA1A targeted the antiviral immune response at the TBK1 level by controlling the phosphorylation and kinase activity of TBK1. Furthermore, PLA1A silencing caused a mitochondrial morphology alteration and reduced the TBK1-associated complexes on mitochondria.
Materials and Methods
Viruses and Cells
SeV and vesicular stomatitis virus (VSV)-green fluorescent protein (GFP) were willingly provided by Prof. Hanzhong Wang and Prof. Hualan Chen respectively. HEK293T and Huh7 cells were maintained in DMEM supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. The cells were cultured under humidified conditions with 5% CO2 at 37°C.
Reagents and Antibodies
The chemical reagents and antibodies used in this study were as follows: poly(I:C) (Sigma-Aldrich, #P9582); mouse anti-β-actin (clone C4) mAb (Santa Cruz Biotechnology [Santa Cruz, CA, USA], sc-47778); mouse anti-HA (clone HA-7) mAb (Sigma-Aldrich, H9658); mouse anti-FLAG (clone M2) mAb (Sigma-Aldrich, F1804); rabbit anti-HA-Tag Ab (C29F4) (#3724), anti-FLAG (#2368), anti-β-Tubulin (9F3; #2128), anti-phospho-TBK1/NAK (Ser172) (D52C2; #5483), anti-TBK1/NAK (#3013), anti-NF-κB p65 (D14E12; #8242), anti-phospho-NF-κB p65 (Ser536; 93H1; #3033), anti-IκBα (L35A5; #4814), anti-phospho-IκBα (Ser32) (14D4; #2859) and anti-phospho-IRF3 (Ser396; D6O1M; #29047) from Cell Signaling Technology (Danvers, MA, USA); IRF3 rabbit polyclonal antibody (#11312-1-AP) and TOM20 rabbit polyclonal antibody (#11802-1-AP) from Proteintech (Wuhan, Hubei, China); Alexa Fluor 561-conjugated secondary Abs and Hoechst 33258 from Invitrogen; and protein G Agarose from Millipore (Darmstadt, Germany). Rabbit polyclonal Ab against human PLA1A was produced by Abmart (Shanghai, China).
Plasmids
The NF-κB, interferon-stimulated response element (ISRE), IRF3 and IFN-β promoter Firefly luciferase reporter plasmids, and pRL-TK Renilla luciferase reporter plasmids, were purchased from Clonetech and Promega respectively. Mammalian expression plasmids for PLA1A, RIG-I, MAVS, STING, TBK1, IKKε, and IRF3 were constructed by standard molecular biology techniques as described previously [20, 21].
Transfection and Dual-Luciferase Reporter Assay
Transient transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were seeded in 24-well plates at 1 × 105 cells per well and transfected the next day with the indicated amount of expression plasmids together with 100 ng of the indicated Firefly luciferase reporter plasmid and 10 ng pRL-TK. The total amount of plasmid DNA per transfection was kept constant by the addition of the empty vector. For some experiments, cells were infected with SeV12 h post-transfection (hpt). Cells were lysed and the luciferase activities were analyzed by the Dual-Glo Luciferase Reporter Assay System (Promega, #E1960). The promoter activity was reported as the ratio of Firefly luciferase activity to Renilla luciferase activity.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA from cultured cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Quantitative real-time polymerase chain reaction (RT-PCR) was performed using the QuantiTect SYBR Green RT-PCR Kit (Qiagen, #204245) on the ABI QuantStudio 6 Flex real-time PCR system (Applied Biosystems). The primers used are listed in Table 1. The 2–ΔΔCT method was used for the calculation of the relative expression level of the genes using the house keeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as the normalizer.
Table 1.
Primers used in the manuscript
| Primer | Primer sequence (5′−3′) |
|---|---|
| PLA1A-F | GGAACTGAGAAACAAGGACACC |
| PLA1A-R | AAACTCGGTTGGAAGACTGAAA |
| ISG15-F | AATGCGACGAACCTCTGAAC |
| ISG15-R | GAAGGTCAGCCAGAACAGGT |
| ISG56-F | GCAGCCAAGTTTTACCGAAG |
| ISG56-R | AGCCCTATCTGGTGATGCAG |
| IFNB1-F | CACGACAGCTCTTTCCATGA |
| IFNB1-R | AGCCAGTGCTCGATGAATCT |
| GAPDH-F | GAAGGTGAAGGTCGGAGTC |
| GAPDH-R | GAAGATGGTGATGGGATTTC |
RNA Interference
To silence endogenous PLA1A expression in HEK293T or Huh7 cells, double-stranded siRNA (Qiagen) targeted the PLA1A mRNA sequence 5′-GGATAGGACTGGTGGAACA-3′ [20]. Cells were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen) for 48 h before further assay. The forward transfection method was used to reach optimal efficiency. siRNA-lipid complexes were formed, added to cells at 60–80% confluence, and mixed gently.
Subcellular Fractionation
Nuclear and cytoplasmic extracts and mitochondria isolation were prepared with a nuclear and cytoplasmic protein extraction kit (Beyotime Biotechnology, #P0027) and a cell mitochondria isolation kit (Beyotime Biotechnology, #C3601) according to the manufacturer's instructions.
Immunoprecipitation, Co-Immunoprecipitation, and Immunoblot Analyses
HEK293T cells were transfected with plasmids encoding Flag- or HA-tagged PLA1A, MAVS, STING, TBK1, or IRF3 as indicated, and lysed using IP buffer (50 mM Tris [pH 7.5], 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 2 mM DTT, 100 mM PMSF, 1 mg/mL proteinase inhibitor). Lysates were quantified with a Bradford assay kit (Bio-Rad) and subjected to immunoprecipitation (IP) using protein G agarose and the corresponding antibody. After 4 washes with IP buffer, adsorbed proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with the indicated antibodies. Cell lysates were also analyzed by SDS-PAGE and immunoblotting to control for protein abundance.
In vitro Kinase Assay
To analyze the TBK1 kinase activity, pFlag-TBK1, pFlag-IRF3, and pHA-PLA1A were transfected into HEK293T cells. The FLAG-TBK1, FLAG-IRF3, or HA-PLA1A proteins were then immunoprecipitated with anti-FLAG or anti-HA antibody 48 hpt. Following 4 washes with IP buffer and 2 washes with kinase assay buffer, immunoprecipitated FLAG-TBK1, FLAG-IRF3, and HA-PLA1A were incubated in kinase assay buffer (20 mM tris-HCl, 1 mM EGTA, 5 mM MgCl2, 0.02% 2-mercaptoethanol, 0.03% Brij-35, BSA [0.2 mg/mL], 20 mM adenosine 5′-triphosphate) at 30°C for 60 min. The reaction was stopped by the addition of 2× SDS loading buffer [7]. The phosphorylation of IRF3 and TBK1 were then detected by western blot.
Results
Impairment of SeV-Induced Type I IFN Response in PLA1A-Knockdown Cells
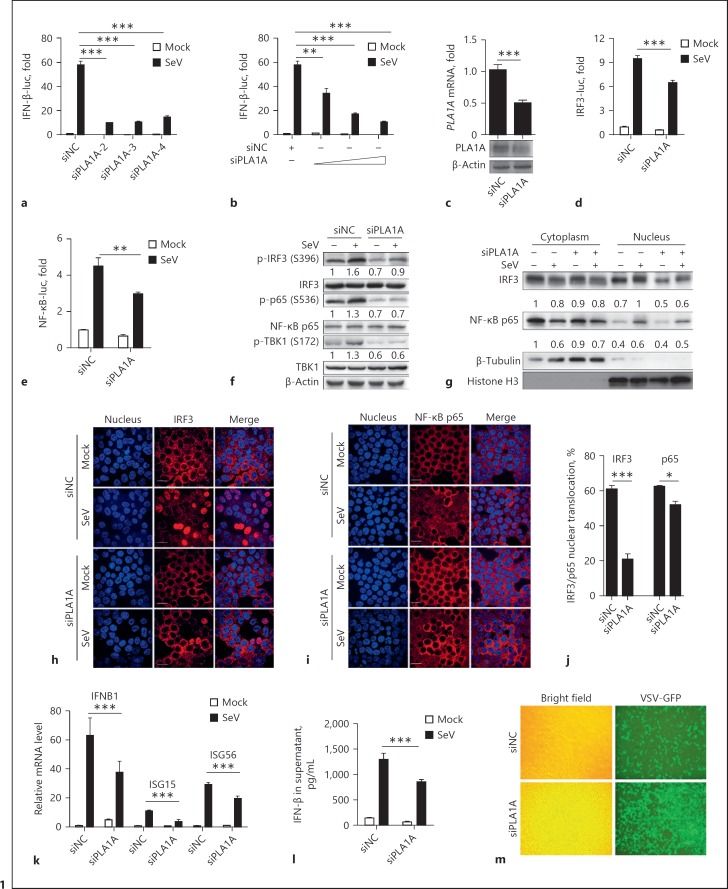
The function of PLA1A in innate immune signaling was analyzed with RNAi experiments. Three siRNAs specific for PLA1A (siPLA1A), which have been verified for their knockdown efficiency [20], were transfected into HEK293T cells together with promoter reporter plasmids. A dramatic decrease in SeV-induced IFN-β promoter activity was observed in all 3 siPLA1A-transfected cells (Fig. 1a), and the siPLA1A functioned in a dose-dependent manner in impairing the SeV-induced IFN-β promoter activity (Fig. 1b). Similarly, the knockdown of PLA1A expression reduced the IFN-β promoter activity stimulated by SeV in Huh7 cells (see online suppl. Fig. S1; for all online suppl. material, see www.karger.com/doi/10.1159/000489832). The knockdown efficiency of selected siPLA1A was shown by the reduced mRNA and protein level of PLA1A (Fig. 1c). Because the transcriptional activation of the IFN-β promoter requires cooperative and coordinative activation of the transcription factors, IRF3 and NF-κB, we then detected whether PLA1A knockdown affected IRF3 and NF-κB reporter promoter activation. SeV-induced IRF3- and NF-κB-dependent promoter activity was reduced in siPLA1A-transfected cells (Fig. 1d, e), indicating that PLA1A affected the activation of IRF3 and NF-κB. Consistently, the phosphorylation levels of IRF3, NF-κB p65, and TBK1 induced by SeV infection were significantly reduced in PLA1A-knockdown cells (Fig. 1f compares lane 2 with lane 4). The localization of IRF3 and NF-κB p65 was then determined by cell fractionation and immuno-fluorescent detection. SeV infection caused a decrease in IRF3 and p65 in the cytoplasm fraction and an increase of IRF3 and p65 in the nuclear fraction (Fig. 1g), indicating the nuclear translocation of both proteins upon SeV infection. Compared with the control, the accumulation of IRF3 and p65 in the nuclear fraction was reduced in PLA1A knockdown cells (Fig. 1g), indicating that knockdown PLA1A expression reduced the nuclear translocation of IRF3 and NF-κB p65. Consistently, immunofluorescence showed that IRF3 was mostly located in the cytoplasm in mock-infected cells, and nuclear localization of IRF3 was found in approximately 61% of SeV-infected cells; however, only 20% of PLA1A knockdown cells showed IRF3 nuclear translocation upon SeV infection (Fig. 1h). Similarly, NF-κB p65 showed cytoplasm localization in most uninfected cells, and SeV infection caused nuclear translocation of NF-κB p65 in approximately 63% cells. When PLA1A was knocked down, SeV-induced nuclear localization of NF-κB p65 was found only in approximately 52% cells (Fig. 1i). The mRNA levels of IFNB1, ISG15, and ISG56 and the IFN-β protein in the supernatant induced by SeV infection were then compared between control siRNA- and siPLA1A-transfected cells. Consistent with the promoter activity assay, the knockdown of PLA1A expression significantly decreased the SeV-induced IFNB1, ISG15, and ISG56 mRNA levels and IFN-β protein in the supernatant (Fig. 1k, l). We further analyzed the effect of PLA1A on viral replication. As shown in Figure 1m, the knockdown of the PLA1A expression increased the replication of vesicular stomatitis virus-GFP, as indicated by more green fluorescence in the siPLA1A-transfected cells, suggesting a reduced antiviral response in the PLA1A knockdown cells. Taken together, these data demonstrated that the knockdown of PLA1A expression impaired the SeV-induced type I IFN production, suggesting that endogenous PLA1A is a positive regulator of innate immune signaling.
Fig. 1.
PLA1A knockdown suppresses the antiviral innate immune response. a, b HEK293T cells were transfected with control siRNA (siNC), different siRNAs targeting PLA1A (siPLA1A; a), or different amounts of siPLA1A-3 (b), together with an interferon (IFN)-β promoter reporter plasmid. Cells were then infected with Sendai virus (SeV) for 12 h. The luciferase activity was detected and presented as fold induction relative to those in uninfected siNC-transfected cells. c HEK293T cells were transfected with siNC or siRNAs specific for PLA1A (siPLA1A). Total RNA was extracted and the mRNA levels of PLA1A were detected by real-time polymerase chain reaction (RT-PCR). Total protein was extracted and the PLA1A was concentrated by IP using an anti-PLA1A antibody. Western blot was performed to detect PLA1A. d, e HEK293T cells were transfected with siNC or siPLA1A together with interferon regulatory factor 3 (IRF3)-luc (d) or nuclear factor κB (NF-κB)-luc (e) reporter plasmids. Cells were then infected with SeV for 12 h, and the luciferase activity was detected and presented as fold induction. f–k HEK293T cells were transfected with siNC or siPLA1A, then infected with SeV for 12 h. f Total protein was extracted and western blot were performed to detect the indicated proteins. g Nuclear and cytoplasmic protein was extracted and subjected in western blot analysis to detect the indicated proteins. h–j The localization of IRF3 and NF-κB p65 was confirmed by immunofluorescence microscopy. Cells were fixed and stained with Hoechst 33258 (blue) and antibodies to IRF3 (red) (h) or NF-κB subunit p65 (red; i). Scale bars, 20 µm. j The percentage of cells with IRF3/p65 nuclear translocation in confocal images were counted (> 100 cells). k Total RNA was extracted and the mRNA levels of IFNB1, ISG15, and ISG56 were detected by real-time polymerase chain reaction (RT-PCR). l Enzyme-linked immunosorbent assay (ELISA) was performed to detect IFN-β in medium. m HEK293T cells were transfected with siNC or siPLA1A, then infected with vesicular stomatitis virus (VSV)-GFP. The replication of VSV-GFP in cells was observed under fluorescence microscopy. Representative results are shown from 3 independent experiments. The 2-tailed t test was used to determine the differences in multiple comparisons (* p < 0.05; ** p < 0.01; *** p < 0.001).
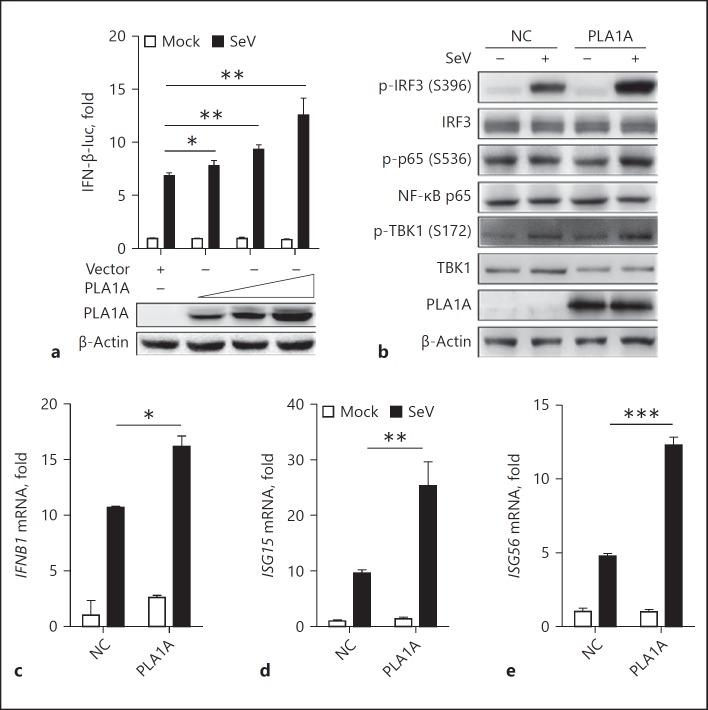
PLA1A Overexpression Enhances the SeV-Induced Type I IFN Response
Endogenously expressed PLA1A is involved in the type I IFN response upon SeV infection. Next, we analyzed the function of exogenously expressed PLA1A in the innate immune response. Using transient transfection and PLA1A stable expression in Huh7 cells (Huh7-PLA1A) that was generated by lentiviral transduction previously, we observed that exogenous expression of PLA1A promoted SeV-induced IFN-β reporter activity in a dose-dependent manner (Fig. 2a) and the phosphorylation of IRF3, NF-κB p65, and TBK1 were slightly stronger in the PLA1A expression cells than that in control cells (Fig. 2b). Notably, high phosphorylation levels of NF-κB p65 were observed in mock-infected cells, which is consistent with our previous findings [22] and published data [23]. Furthermore, the mRNA levels of IFNB1 and several ISGs were detected by real-time RT-PCR, and the results showed that SeV infection induced more IFNB1, ISG15, and ISG56 mRNA expression in the PLA1A stable expressing cells than in control cells (Fig. 2c–e). Collectively, these data suggested that PLA1A overexpression enhanced the SeV-induced phosphorylation of TBK1 and IRF3, and thus enhanced the type I IFN response in Huh7 cells.
Fig. 2.
PLA1A overexpression enhances Sendai virus (SeV)-induced type I interferon (IFN) response in Huh7 cells. a Huh7 cells were transfected with vector or PLA1A expression plasmid together with an IFN-β promoter reporter plasmid, then infected with SeV for 24 h. A luciferase assay was performed and the results are shown as fold induction compared with vector-transfected uninfected cells (upper part). The PLA1A expression levels are shown by western blot (lower part). b–e Huh7-NC or Huh7-PLA1A cells were infected with SeV for 24 h. b Total protein was extracted and subjected to SDS-PAGE. The indicated protein expression levels were detected by western blot. c–e Total cellular RNA was extracted and the mRNA levels of IFNB1 (c), ISG15 (d) and ISG56 (e) were detected by real-time polymerase chain reaction (RT-PCR). The 2-tailed t test was used to determine the differences in multiple comparisons (* p < 0.05; ** p < 0.01; *** p < 0.001).
PLA1A Participates in Type I IFN Signaling at the Level of TBK1
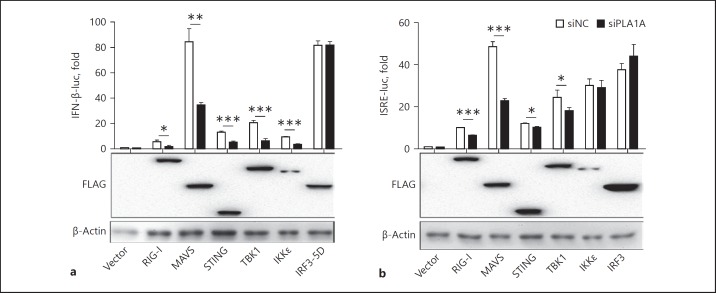
To determine which step of IFN-β induction was influenced by PLA1A, promoter reporter assays were performed in the context of the PLA1A knockdown condition by overexpressing several key components of the RNA virus sensing the RLR signaling pathway, including RIG-I, MAVS, TBK1, IKKε, and IRF3 and the DNA virus sensor/adaptor, STING. The activation of the IFN-β promoter and the ISRE-dependent promoter was determined. The results in Figure 3a and b showed that the knockdown of PLA1A reduced the IFN-β promoter activity stimulated by RIG-I, MAVS, STING, TBK1, and IKKε, as well as the ISRE-dependent promoter activity stimulated by RIG-I, MAVS, STING, and TBK1, but did not influence IRF3–5D (a constitutively active mutant of IRF3) stimulated the IFN-β promoter or ISRE-dependent promoter activation, suggesting that PLA1A regulates type I IFN signaling at the level of TBK1.
Fig. 3.
PLA1A participates in the type I interferon (IFN) signaling at the level of TANK-binding kinase 1 (TBK1). HEK293T cells were transfected with siNC or siRNAs specific for PLA1A (siPLA1A) together with IFN-β promoter (a) or interferon-stimulated response element (ISRE)- (b) reporter plasmids. Cells were then transfected with vector or retinoic acid-inducible gene I (RIG-I), mitochondrial antiviral signaling protein (MAVS), STING, TBK1, IκB kinase (IKK)ε, interferon regulatory factor 3 (IRF3) or IRF3–5D expressing plasmids to stimulate the promoter. A luciferase assay was performed and presented as fold induction relative to siNC and vector-stimulated cells. The 2-tailed t test was used to determine the differences in multiple comparisons (* p < 0.05; ** p < 0.01; *** p < 0.001). The expression levels of the transfected plasmids were confirmed by western blot analysis and shown in the lower part of each plot.
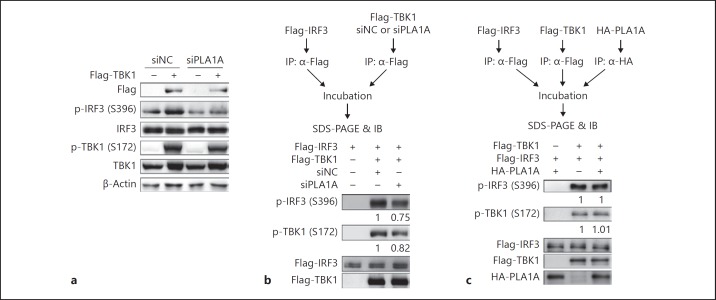
PLA1A Is Required for TBK1 Phosphorylation
As phosphorylation at Ser172 is critical for the kinase activity of TBK1 and overexpression of TBK1 leads to TBK1 autophosphorylation, we then analyzed the phosphorylation levels of TBK1 and IRF3 stimulated by TBK1 overexpression. The results in Figure 4a show that both TBK1 and IRF3 phosphorylation were attenuated in PLA1A knockdown cells. An in vitro kinase assay was then performed using separately IP-purified IRF3 and TBK1, which were purified from siNC or siPLA1A co-transfected cells (Fig. 4b). As expected, no phosphorylation of IRF3 was observed without TBK1 incubation, while IRF3 was robustly phosphorylated by separately purified TBK1. However, the IRF3 phosphorylation level was reduced when incubated with TBK1 purified from PLA1A knockdown cells; meanwhile, a decreased phosphorylation level of TBK1 at Ser172 was found in PLA1A knockdown cells (Fig. 4b), indicating that the kinase activity of TBK1 was diminished in PLA1A knockdown cells due to the reduced TBK1 phosphorylation level. However, the phosphorylation level and kinase activity of TBK1 were not significantly influenced by the PLA1A protein, which was overexpressed and IP-purified separately (Fig. 4c). These results indicated that endogenous PLA1A is required for TBK1 phosphorylation, but PLA1A alone could not modulate the phosphorylation and kinase activity of TBK1.
Fig. 4.
PLA1A is required in TANK-binding kinase 1 (TBK1) phosphorylation. a HEK293T cells were transfected with siNC or siRNAs specific for PLA1A (siPLA1A), then transfected with TBK1 expression plasmid. Total protein was extracted and western blot was performed to detect the indicated protein expression levels. b In vitro kinase assay. HEK293T cells were transfected with pFlag-interferon regulatory factor 3 (IRF3) alone or pFlag-TBK1 with siNC or siRNAs specific for PLA1A (siPLA1A). Flag-IRF3 and Flag-TBK1 were purified separately by IP with an antibody to Flag tag, and then incubated together as indicated. The workflow is shown in the upper part, and the phosphorylation of IRF3 and TBK1 was detected by western blot and shown in the lower part. c In vitro kinase assay. HEK293T cells were transfected with pFlag-IRF3, pFlag-TBK1, or pHA-PLA1A separately. The expressed proteins were immuno-precipitated by the indicated tag antibodies and incubated together as indicated. The workflow is shown in the upper part, and the phosphorylation of IRF3 and TBK1 was confirmed by western blot and shown in the lower part.
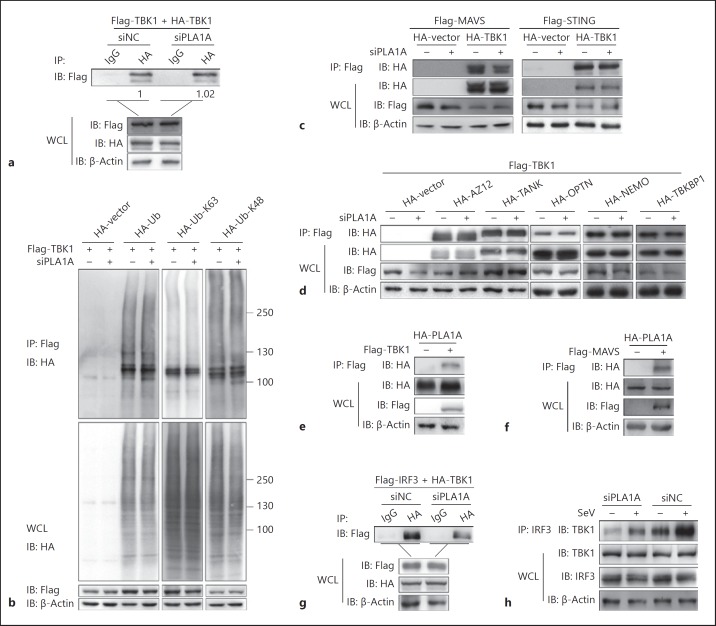
PLA1A Silencing Does Not Affect TBK1 Dimerization or Ubiquitination But Reduces the MAVS-TBK1 Interaction
We then analyzed how TBK1 phosphorylation was modulated by endogenous PLA1A. Increasing evidence suggests that TBK1 dimerization, K63-linked polyubiquitination [3, 5, 6, 24], and the interaction of TBK1 with several adaptors modulate its phosphorylation at Ser172 [25, 26]. We first analyzed the self-interaction and the ubiquitination of TBK1 in the PLA1A knockdown condition. The interaction between Flag-TBK1 and HA-TBK1 was analyzed by co-IP, and results showed that knockdown of PLA1A expression did not affect the interaction between Flag-TBK1 and HA-TBK1, indicating that PLA1A did not interfere in TBK1 dimerization (Fig. 5a). In addition, wild-type ubiquitin, ubiquitin-K63, or ubiquitin-K48 was transfected together with TBK1 into HEK293T cells in which PLA1A expression was knocked down. TBK1 was immunoprecipitated with anti-Flag antibody, then the ubiquitination of TBK1 was determined with anti-HA antibody. The smeared bands of ubiquitinated TBK1 were similar between siNC- and siPLA1A-transfected cells, suggesting no influence of PLA1A on TBK1 ubiquitination (Fig. 5b). Thus, knockdown of PLA1A expression impaired the TBK1 phosphorylation process but not its dimerization or ubiquitination.
Fig. 5.
PLA1A silencing does not affect TANK-binding kinase 1 (TBK1) dimerization or ubiquitination but reduces mitochondrial antiviral signaling protein (MAVS)-TBK1 interactions. a HEK293T cells were transfected with siNC or siRNAs specific for PLA1A (siPLA1A), then transfected with pFlag-TBK1 and pHA-TBK1. Immunoprecipitation (IP) was performed with immunoglobulin G (IgG) or an antibody to HA. Immunoblot (IB) analysis was performed with an antibody to Flag; the corresponding whole cell lysate (WCL) were subject to western blot analysis with antibodies to Flag, HA and β-actin. b HEK293T cells were transfected with siNC or siPLA1A, then transfected with vector, HA-ubiquitin, HA-tagged K63-linked ubiquitin, or an HA-tagged K48-linked ubiquitin expression plasmid together with pFlag-TBK1. IP was performed with Flag antibody, followed by IB analysis with HA antibody. The corresponding WCL was subjected to western blot analysis with antibodies to Flag, HA and β-actin. c–h IP experiments to analyze the protein-protein interactions. c HEK293T cells were transfected with siNC or siPLA1A and then transfected with pFlag-mitochondrial antiviral signaling protein (MAVS) (left) or pFlag-STING (right), plus pHA-TBK1 or empty vector. d HEK293T cells were transfected with siNC or siPLA1A, and then transfected with pFlag-TBK1, plus empty vector or plasmid encoding HA-AZI2, HA-TANK, HA-OPTN (Optineurin), HA-NEMO, and HA-TBKBP1. e, f HEK293T cells were transfected with pHA-PLA1A, together with pFlag-TBK1 (e), pFlag-MAVS (f) or empty vector. g HEK293T cells were transfected with siNC or siPLA1A, then transfected with pFlag-interferon regulatory factor 3 (IRF3) and pHA-TBK1. h HEK293T cells were transfected with siNC or siPLA1A, with or without Sendai virus (SeV) stimulation.
It was reported that the interaction between TBK1 and several adaptors, such as MAVS, MITA, TANK, NEMO, AZI2, TBKBP1, and optineurin (OPTN), can facilitate TBK1 activation [3], which promoted us to analyze the interaction of TBK1 with other adaptor molecules in PLA1A knockdown cells. The co-IP experiments showed that the association between TBK1 and MAVS was depressed in PLA1A knockdown cells, while the association of TBK1 with the other molecules, such as STING, AZI2, TANK, OPTN, NEMO, and TBKBP1 was not affected by PLA1A silencing (Fig. 5c, d). As the MAVS-TBK1 interaction was reduced in PLA1A knockdown cells, we then analyzed whether PLA1A could interact with MAVS and TBK1. Indeed, co-IP experiments showed that PLA1A could interact with MAVS and TBK1 (Fig. 5e, f). Partial co-localization of HA-PLA1A and Flag-TBK1 was observed as shown in online supplementary Figure S3. MAVS was shown to serve as a scaffold to bring TBK1 and IRF3 together and facilitate the phosphorylation of IRF3 by TBK1 [27]. We then analyzed whether PLA1A influences the interaction of TBK1 with its substrate, IRF3. When TBK1 and IRF3 were co-expressed, PLA1A knockdown reduced the amount of IRF3 associated with TBK1 (Fig. 5g). To analyze the interaction between TBK1 and IRF3 in a physiological condition, co-IP was performed in SeV-infected HEK293T cells with anti-IRF3 antibody. More TBK1 was co-immunoprecipitated with IRF3 in the SeV-infected cells than in uninfected cells, indicating the activation of TBK1/IRF3 in these cells, while PLA1A silencing significantly reduced the amount of IRF3 associated with TBK1 (Fig. 5h). Taken together, PLA1A did not affect the TBK1 dimerization or ubiquitination but influenced its interaction with MAVS, thus participating in TBK1 phosphorylation.
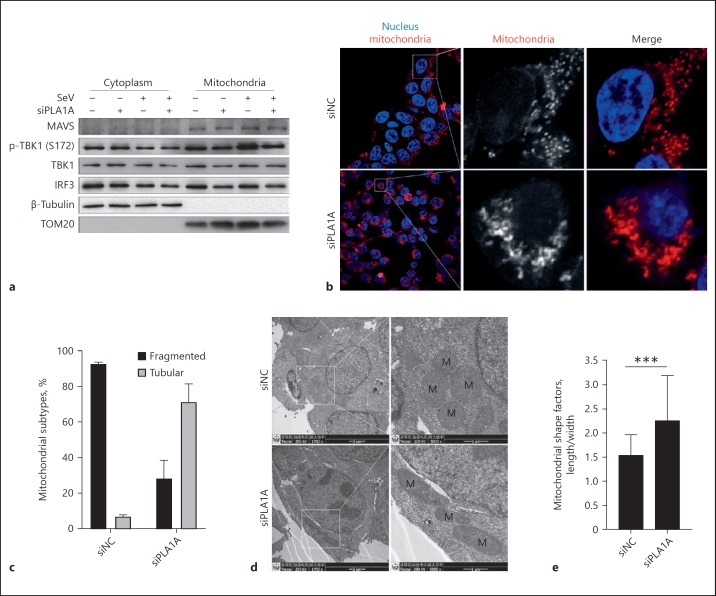
PLA1A Silencing Causes a Mitochondrial Morphology Alteration and Reduces the TBK1-Associated Complexes on Mitochondria
Given that TBK1 is recruited to mitochondria in response to RNA virus infection, where it serves to transduce signals from MAVS and activate IRF3, we then analyzed the localization of TBK1 on mitochondria by cell fractionation. The mitochondria fraction was separated with a cell mitochondria isolation kit from siNC- or siPLA1A-transfected cells and the distribution patterns of MAVS, p-TBK1, TBK1, and IRF3 in the cytoplasm and mitochondria were compared. As shown in Figure 6a, the cytoplasmic protein, β-tubulin, and the mitochondrial protein, TOM20, were only found in the cytoplasm and mitochondrial fractions, respectively, indicating successful mitochondria purification. PLA1A expression knockdown did not change the mitochondrial distribution of MAVS as the expression and mitochondrial localization of MAVS were similar in siNC- and siPLA1A-transfected cells. However, the mitochondrial localized p-TBK1, TBK1, and IRF3 were decreased in siPLA1A transfected cells. SeV infection increased the mitochondrial localized p-TBK1 level in siNC-transfected cells but not in siPLA1A-transfected cells. These results suggest that the recruitment of TBK1 to MAVS and the assembly of the MAVS/TBK1/IRF3 signalosome on mitochondria were impaired in PLA1A-knockdown cells.
Fig. 6.
PLA1A silencing induces mitochondrial elongation. a HEK293T cells were transfected with siNC or siRNAs specific for PLA1A (siPLA1A), then infected with Sendai virus (SeV) for 12 h. Mitochondrial and cytoplasmic protein were extracted and subjected to western blot analysis to detect the indicated proteins. b HEK293T cells were transfected with siNC or siPLA1A. The mitochondria were stained with MitoTracker Red (red) and nuclei were stained with Hoechst 33258 (blue). Representative confocal images were shown. c The percentage of cells with different mitochondria subtypes in confocal images were counted (> 100 cells). d The ultrastructure of HEK293T cells transfected with siNC or siPLA1A were observed under electron microscopy. Scale bar, 1 μm. M, mitochondrion. e The mitochondrial shape factors (length/width) from electron micrographs were calculated (> 500 mitochondria each group).
Interestingly, a difference in the mitochondria morphology between control and PLA1A knockdown cells was observed by MitoTracker Red staining. Dot- or sphere-like clusters of MitoTracker Red, which indicated that fragmented mitochondria, were observed in most of the siNC-transfected cells (approximately 90%), while an elongated morphology was observed in most of the siPLA1A-transfected cells (70%), which indicated tubular mitochondria in these cells (Fig. 6b, c). The mitochondrial morphology was then examined with electron microscopy (Fig. 6d). SiNC-transfected cells contain sphere-shaped mitochondria with a shape factor (ratio of major and minor axes) of 1.54 ± 0.14, while the mitochondria in the siPLA1A-transfected cells were elongated with shape factor of 2.26 ± 0.31 (Fig. 6e). Thus, we concluded that PLA1A expression knockdown caused an alteration in the mitochondrial morphology.
Discussion
Recently, lipid metabolism has been connected to the innate immune response. Here, we report the involvement of PLA1A, a phospholipase, in virus-induced type I IFN production. PLA1A expression knockdown reduced the type I IFN production stimulated by RNA viruses (SeV). PLA1A targeted at TBK1 level, which is a key component of RNA sensing pathways, as the phosphorylation and kinase activity of TBK1 were reduced when PLA1A was silenced. PLA1A expression knockdown reduced the association of TBK1 with mitochondria, concomitant with the mitochondrial morphology change.
TBK1 is a node molecule that participates in many signaling pathways. Increasing evidence suggests that the activation and substrate selection of TBK1 is dependent on its localization, as different adaptor proteins recruit TBK1 to discrete signaling complexes upon different stimulation [25]. Once accumulated, the phosphorylation of TBK1 was thought to be an auto-activation process that requires the dimerization and ubiquitination of TBK1 [3, 5, 6, 24]. Additionally, it is possible that some kinases localized in the same scaffold complex with TBK1 could stimulate TBK1 phosphorylation [28]. In our experiment, PLA1A expression knockdown reduced the phosphorylation of TBK1 stimulated by SeV and TBK1 overexpression. Through interacting with TBK1, PLA1A influenced neither the TBK1 self-interaction nor the ubiquitination of TBK1 (Fig. 5; online suppl. Fig. S2). Moreover, PLA1A did not phosphorylate TBK1 directly, as incubation of PLA1A protein with TBK1 did not enhance the phosphorylation level of TBK1 in an in vitro kinase assay. However, the interactions of MAVS with TBK1 and TBK1 with IRF3 were reduced and the amount of TBK1 that associated with mitochondria was decreased in PLA1A silenced cells; therefore, it is possible that PLA1A modulates TBK1 activation through controlling the recruitment and the local concentration of TBK1. P-TBK1 was reported to localize on the mitochondria and Golgi apparatus in response to RNA virus infections [29, 30], while p-TBK1 shows a cell type-specific subcellular localization upon DNA virus infection, either on mitochondria or co-localized with STING or on other structures [27, 30]. In our research, PLA1A knockdown reduced the distribution of p-TBK1 on mitochondria in SeV-infected cells as well as uninfected cells, suggesting that mitochondrial localized p-TBK1 participates in type I IFN production. However, we still cannot exclude the possibility that PLA1A knockdown may also affect the distribution of p-TBK1 on other organelles or structures.
PLA1A is a phospholipase that hydrolyzes fatty acids at the sn-1 position of PS. Some lipid mediators have been reported to participate in innate immune signaling. For example, 25-hydroxycholesterol potentiates the translocation of IRF3 into the nucleus and the release of IFN-β in poly(I:C)-treated cells [31]. As the fatty acid at the sn-1 position of PS is mostly stearic acid, the main product of PS hydrolysis is lyso-PS and stearic acid. However, we have excluded the involvement of the lipid products of PLA1A in the innate immune response, as the supplementation of lyso-PS and stearic acid did not alter the SeV-induced type I IFN production or restore the type I IFN production in PLA1A knockdown cells (online suppl. Fig. S3).
Mitochondria have been proven to be the signaling platform for the innate immune response induced by both RNA and DNA viruses [32]. In addition to mediating innate immune signaling through the mitochondrial localized protein, MAVS, the mitochondrial dynamics, ROS, membrane potential, mtDNA exposure and oxidative phosphorylation activity were all reported to govern antiviral signaling [32]. Several reports have shown the tight association between mitochondria dynamics with innate immune signaling. For example, the knockdown of genes promoting mitochondrial fission, such as mitochondrial fission protein 1 (Fis1) and dynamin-related protein 1 (Drp1), or overexpression of genes promoting mitochondrial fusion, such as mitochondrial fusion protein 1 (Mfn1), resulted in an enhanced innate immune response, which was accompanied by mitochondria elongation [33]. On the other hand, the knockdown of genes promoting mitochondrial fusion, such as Mfn1 and optic atrophy 1, resulted in reduced RLR signaling, which was accompanied by mitochondria fragmentation [33]. Mfn1 was thought to bind MAVS and be required for the redistribution of MAVS on mitochondria to form speckle-like aggregates [34]. A more recent study demonstrated that fibroblasts deficient in both Mfn1 and Mfn2 exhibited impaired RLR-induced antiviral responses [35]. However, though Mfn1 and Mfn2 have similar mitochondrial dynamic functions, they regulate RLR signaling in a different way. Mfn2 was also thought to interact with MAVS [34], but it produced an inhibitory effect on the RLR signaling downstream of MAVS and upstream of both TRAF6 (the NF-kB activation pathway) and TBK-1 (the IRF-3 activation pathway) [36]. In our experiments, the reduced innate immune signaling in PLA1A knockdown cells was concomitant with elongated mitochondria, while Mfn2 protein was increased in PLA1A knockdown cells (online suppl. Fig. S4). As Mfn2 was shown to have an inhibitory effect on RIG-I signaling, the upregulated Mfn2 might partially explain the impairment of SeV-triggered IFN signaling and the mitochondria elongation in PLA1A knockdown cells [36]. It is also possible that artificially regulated mitochondria dynamics by overexpression or knockdown may change other factors in the mitochondria, such as membrane potential, mitochondrial stress, or the oxidation phosphorylation activity of mitochondria.
It has been demonstrated that many aspects of the mitochondrial function is regulated by the mitochondrial morphology. A recent paper showed that Drp1-dependent mitochondrial dynamic was coupled with oxidative respiration: tubular mitochondria was accompanied with increased oxidative respiration, while fragmented mitochondria achieved the opposite [37]. The oxidative phosphorylation activity was supposed to be required in the RLR innate immune signaling, as SeV-induced IFN production was impaired in cells with oxidative phosphorylation defect, but was restored when the oxidative phosphorylation was recovered [38]. In our study, increased tubular mitochondrial and impaired SeV-induced IFN production was observed in PLA1A knockdown cells. It will be interesting to further analyze how PLA1A contributes to mitochondrial function.
The detailed molecular mechanism how PLA1A modulates mitochondria morphology needs to be studied further. The PS-specific phospholipase, PLA1A, may help maintain the PS balance in cells. As the phosphatidylethanolamine of the mitochondria membrane, which is a kind of phospholipid that plays an important role in the structure and function of mitochondria [39], is mainly produced from PS by mitochondrion-localized PS decarboxylase [40], it is possible that PLA1A expression knockdown may alter the mitochondrial phosphatidylethanolamine level and subsequently induce morphology changes.
In our previous study, PLA1A was upregulated by HCV infection and was supposed to enhance HCV assembly and envelopment by interacting with HCV proteins. As PLA1A is required in RNA virus-induced innate immune signaling, the occupation of PLA1A by HCV proteins might be another explanation for the escape of HCV from the innate immune response. In summary, our current data demonstrate that PLA1A participates in the antiviral innate immune response by targeting TBK1 phosphorylation and mitochondria morphology modulation.
Disclosure Statement
The authors declare that they have no conflicts of interest to disclose.
Author Contribution
X.C. and R.P. conceived and supervised the project. X.G. and D.C. carried out the experiment. X.H. and Y.Z. contributed to the cell culture work. Y.W. provided materials and meaningful suggestions. X.G., R.P., Y.W., C.W., and J.C. analyzed the data. X.G., R.P., and X.C., wrote the manuscript.
Supplementary Material
Supplementary data
Acknowledgments
We are grateful to Dr. Ding Gao, Ms Pei Zhang and Ms Anna Du (The Core Facility and Technical Support, Wuhan Institute of Virology) for their excellent technical support. This work was supported by grants from the National Natural Science Foundation of China (grant numbers 31621061 and 81672021).
References
- 1.Seth RB, Sun LJ, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larabi A, Devos JM, Ng ZL, Nanao MH, Round A, Maniatis T, Panne D. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 2013;3:734–746. doi: 10.1016/j.celrep.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 2013;587:542–548. doi: 10.1016/j.febslet.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, Li Y, Dunn GP, Chan E, Thai T, Yang S, Ficarro SB, Marto JA, Jeon H, Hahn WC, Barbie DA, Eck MJ. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu C, Sankaran B, Chaton CT, Herr AB, Mishra A, Peng J, Li P. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure. 2013;21:1137–1148. doi: 10.1016/j.str.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang W, Zhang Q, Lin X, Wu S, Zhou Y, Meng F, Fan Y, Shen T, Xiao M, Xia Z, Jian Zou, Feng X, Xu P. PPM1A silences cytosolic RNA sensing and antiviral defense through direct dephosphorylation of MAVS and TBK1. Sci Adv. 2016;2:e1501889. doi: 10.1126/sciadv.1501889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Liang L, Fan Y, Sun S, An L, Shi Z, Cheng J, Jia W, Sun W, Mori-Akiyama Y, Zhang H, Fu S, Yang J. PPM1B negatively regulates antiviral response via dephosphorylating TBK1. Cell Signal. 2012;24:2197–2204. doi: 10.1016/j.cellsig.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan Z, Cao H, Xie X, Yang L, Zhang P, Chen Y, Fan H, Liu Z, Liu X. Phosphatase PP4 negatively regulates type I IFN production and antiviral innate immunity by dephosphorylating and deactivating TBK1. J Immunol. 2015;195:3849–3857. doi: 10.4049/jimmunol.1403083. [DOI] [PubMed] [Google Scholar]
- 10.Song G, Liu B, Li Z, Wu H, Wang P, Zhao K, Jiang G, Zhang L, Gao C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol. 2016;17:1342–1351. doi: 10.1038/ni.3588. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 “preferentially” promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Wang L, Zhao X, Zhao K, Meng H, Zhao W, Gao C. TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med. 2012;209:1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Zhao X, Zhang M, Zhao W, Gao C. Ubiquitin-specific protease 2b negatively regulates IFN-β production and antiviral activity by targeting TANK-binding kinase 1. J Immunol. 2014;193:2230–2237. doi: 10.4049/jimmunol.1302634. [DOI] [PubMed] [Google Scholar]
- 16.Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, Xavier R, Ting AT. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An H, Zhao W, Hou J, Zhang Y, Xie Y, Zheng Y, Xu H, Qian C, Zhou J, Yu Y, Liu S, Feng G, Cao X. SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity. 2006;25:919–928. doi: 10.1016/j.immuni.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Kim SS, Lee KG, Chin CS, Ng SK, Pereira NA, Xu S, Lam KP. DOK3 is required for IFN-β production by enabling TRAF3/TBK1 complex formation and IRF3 activation. J Immunol. 2014;193:840–848. doi: 10.4049/jimmunol.1301601. [DOI] [PubMed] [Google Scholar]
- 19.Cai B, Wu J, Yu X, Su XZ, Wang RF. FOSL1 inhibits type I interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. MBio. 2017;8:pii: e02161–16. doi: 10.1128/mBio.02161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo M, Pei R, Yang Q, Cao H, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Lu M, Chen X. Phosphatidylserine-specific phospholipase A1 involved in hepatitis C virus assembly through NS2 complex formation. J Virol. 2015;89:2367–2377. doi: 10.1128/JVI.02982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Pei R, Zhu W, Zeng R, Wang Y, Wang Y, Lu M, Chen X. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J Immunol. 2014;192:1162–1170. doi: 10.4049/jimmunol.1300798. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Zhao K, Su X, Lu L, Zhao H, Zhang X, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Wang Y, Lu M, Chen X, Pei R. MITA/STING and its alternative splicing isoform MRP restrict hepatitis B virus replication. PLoS One. 2017;12:e0169701. doi: 10.1371/journal.pone.0169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo WW, Li S, Li C, Lian H, Yang Q, Zhong B, Shu HB. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 24.Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, Bowman KK, Starovasnik MA, Dueber EC. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc Natl Acad Sci U S A. 2012;109:9378–9383. doi: 10.1073/pnas.1121552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587:1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 26.Goncalves A, Burckstummer T, Dixit E, Scheicher R, Gorna MW, Karayel E, Sugar C, Stukalov A, Berg T, Kralovics R, Planyavsky M, Bennett KL, Colinge J, Superti-Furga G. Functional dissection of the TBK1 molecular network. PLoS One. 2011;6:e23971. doi: 10.1371/journal.pone.0023971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 28.Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, Hough J, McIver EG, Cohen P. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 29.Pourcelot M, Zemirli N, Silva Da Costa L, Loyant R, Garcin D, Vitour D, Munitic I, Vazquez A, Arnoult D. The Golgi apparatus acts as a platform for TBK1 activation after viral RNA sensing. BMC Biol. 2016;14:69. doi: 10.1186/s12915-016-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, Oshiumi H, Miyashita M, Aly HH, Matsumoto M, Seya T. Cell type-specific subcellular localization of phospho-TBK1 in response to cytoplasmic viral DNA. PLoS One. 2013;8:e83639. doi: 10.1371/journal.pone.0083639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koarai A, Yanagisawa S, Sugiura H, Ichikawa T, Kikuchi T, Furukawa K, Akamatsu K, Hirano T, Nakanishi M, Matsunaga K, Minakata Y, Ichinose M. 25-hydroxycholesterol enhances cytokine release and toll-like receptor 3 response in airway epithelial cells. Respir Res. 2012;13:63. doi: 10.1186/1465-9921-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Julkunen I, Namiki H, Yoneyama M, Fujita T. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koshiba T, Onomoto K, Yanagi Y, Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011;4:ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 36.Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, Witzig M, Ishihara N, Mihara K, Ripperger JA, Albrecht U, Frank S, Brown SA, Eckert A. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 2018;27:657–666. doi: 10.1016/j.cmet.2018.01.011. e5. [DOI] [PubMed] [Google Scholar]
- 38.Yoshizumi T, Imamura H, Taku T, Kuroki T, Kawaguchi A, Ishikawa K, Nakada K, Koshiba T. RLR-mediated antiviral innate immunity requires oxidative phosphorylation activity. Sci Rep. 2017;7:5379. doi: 10.1038/s41598-017-05808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borkenhagen LF, Kennedy EP, Fielding L. Enzymatic formation and decarboxylation of phosphatidylserine. J Biol Chem. 1961;236:4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data