Abstract
Invasive aspergillosis mainly occurs in immunocompromised patients and is commonly caused by Aspergillus fumigatus, while A.nidulans is rarely the causative agent. However, in chronic granulomatous disease (CGD) patients, A. nidulans is a frequent cause of invasive aspergillosis and is associated with higher mortality. Immune recognition of A. nidulans was compared to A. fumigatus to offer an insight into why A. nidulans infections are prevalent in CGD. Live cell imaging with J774A.1 macrophage-like cells and LC3-GFP-mCherry bone marrow-derived macrophages (BMDMs) revealed that phagocytosis of A. nidulans was slower compared to A. fumigatus. This difference could be attributed to slower migration of J774A.1 cells and a lower percentage of migrating BMDMs. In addition, delayed phagosome acidification and LC3-associated phagocytosis was observed with A. nidulans. Cytokine and oxidative burst measurements in human peripheral blood mononuclear cells revealed a lower oxidative burst upon challenge with A. nidulans. In contrast, A. nidulans induced significantly higher concentrations of cytokines. Collectively, our data demonstrate that A. nidulans is phagocytosed and processed at a slower rate compared to A. fumigatus, resulting in reduced fungal killing and increased germination of conidia. This slower rate of A. nidulans clearance may be permissive for overgrowth within certain immune settings.
Keywords: Aspergillus fumigatus, Aspergillus nidulans, Phagocytosis, Phagosome acidification, LC3-associated phagocytosis, Chronic granulomatous disease
Introduction
Aspergillus species are environmental molds that play essential roles in the carbon recycling of decaying organic debris. On a daily basis, humans inhale hundreds of conidia, yet these spores are efficiently removed from the lung, which prevents healthy individuals from developing Aspergillus infection.
However, certain patient groups have an elevated risk of developing aspergillosis; this susceptibility strongly depends on the status of the host immune system. Immunocompromised patients are highly susceptible to invasive aspergillosis [1], predominantly due to chemotherapy or treatment with immunosuppressive drugs in the context of malignancies and organ or hematological stem cell transplantation [2, 3]. Such treatments are a major risk factor due to their suppression of the first line of antifungal host defense in the lungs [3]. Although patients with primary immunodeficiency are not usually susceptible to aspergillosis, individuals with dysfunction of the NADPH-oxidase complex, called chronic granulomatous disease (CGD), are highly susceptible to Aspergillus infections [4]. Mutations in the 5 genes (NCF1, NCF2, NCF4, CYBA, and CYBB) encoding the subunits of the NADPH oxidase complex can lead to severely reduced activity of the NADPH oxidase [5, 6]. Defective NADPH oxidase activity in CGD patients (NCF1) and CGD mice (Ncf1−/− and Ncf4−/−) was associated with defective LC3-associated phagocytosis (LAP), leading to reduced fungal killing in the case of NCF1 deficiency [7].
Interestingly, aspergillosis in CGD patients presents as less severe and is associated with a lower mortality of 25–27% compared to 50–60% in hematooncological patients [4]. The epidemiological distribution of Aspergillus species is also significantly different [8]. A. fumigatus is the most commonly isolated species (approx. 62%) in invasive aspergillosis of hematooncological patients, followed by A. flavus (approx. 17%), A. terreus (approx. 10%), and A. niger (approx. 2%) [9, 10, 11, 12]. Infections with A.nidulans are much less common and account for approximately 1% of all invasive aspergillosis [10, 13]. Interestingly, no higher mortality has been attributed to the different species [11]. In contrast, approximately 48% of aspergillosis infections in CGD patients are caused by A. fumigatus, while approximately 33% of infections are caused by A. nidulans[14, 15, 16, 17] (online suppl. Fig. S1A; for all online suppl. material, see www.karger.com/doi/10.1159/000484562). Although A. nidulans is still less prevalent than A. fumigatus in CGD, its disease severity and mortality is significantly higher [15]. Increased mortality is associated with A. nidulans infections compared to A. fumigatus (41% SD ±15 vs. 12% SD ±13; online suppl. Fig. S1B). We hypothesize that in healthy individuals, the immune response against A. nidulans or A. fumigatus may differ. A differential innate response to these Aspergillus species may underpin the capacity of A. nidulans to cause infections in CGD patients. To date, no studies have investigated and compared the initial immune recognition and phagocytosis of A. nidulans versus A. fumigatus. By deciphering the different aspects of the innate immune response under intact NADPH oxidase conditions, we compared the immune recognition of A. nidulans and. A. fumigatus by studying phagocytosis, cytokine induction, and oxidative burst. The differences in these processes might help to explain why A. nidulans infection can occur in CGD patients.
Experimental Procedures
Aspergillus Strains
Aspergillus conidia were cultured and harvested as described in a previous study [18]. Resting conidia of A. fumigatus (strains: AF293/CBS 101355/ATCC MYA-4609, V05–27, ATCC204305, and Ku80) and A. nidulans (CBS 114.63, CBS 119.55, and CBS670.78) were used at a final concentration of 1 × 107/mL, either used as freshly isolated live conidia or heat killed (30 min at 95°C in a water bath) for the peripheral blood mononuclear cell (PBMC) stimulation assays. Live conidia were germinated until the formation of small germ tubes (germlings) by incubation for 4 h at 37°C in 10% human serum and used for the reactive oxygen species (ROS) assay.
LC3-GFP-mCherry Bone Marrow-Derived Macrophages
LC3-GFP-mCherry bone marrow-derived macrophages (BMDMs) were kindly provided by Dr. Fraser P. Coxon (University of Aberdeen) and Dr. Ian Ganley (University of Dundee). The bone marrow was derived from C57Bl/6 mice that constitutively express mCherry-GFP-MAP1LC3b (from rat) from the ROSA26 locus (TaconicArtemis). Differentiation into BMDMs occurred in 5 days at 37°C (5% CO2) in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10 ng/mL M-CSF, 10% fetal bovine serum (heat-inactivated; Invitrogen), 100 U/mL penicillin, and 100 mg/mL streptomycin.
Live Cell Imaging
Murine J774A.1 cells or M-CSF-differentiated BMDMs were seeded in 8-well ibidi imaging dishes at a density of 1 × 105/well or 1.5 × 105/well, respectively, to adhere overnight. Live cell video microscopy phagocytosis assays were carried out at 37°C using an Ultra-VIEW VoX spinning disk microscope (Nikon, Kingston upon Thames, UK). Volocity software was used for data analysis (version 6.3.1; Improvision, PerkinElmer, Coventry, UK). Immediately prior to live cell imaging, DMEM was replaced with 200 μL of prewarmed supplemented CO2-independent medium (Gibco, Invitrogen, Paisley, UK). Live A. fumigatus or A. nidulans cell suspensions of resting conidia were added to cells at an MOI (multiplicity of infection) of 1:1. In experiments with J774A.1 cells, acidic compartments were stained with 1 μM of LysoTracker Red DND-99 (LTR; Invitrogen). Expression and localization was monitored in experiments with mCherry LC3. Volocity software (Improvision) was set to capture images every minute for a 6-h period using an electron-multiplying charge-coupled device (EMCCD) camera. For all conditions, at least 2 independent experiments were carried out, with a minimum of 2 movies per experiment with approximately 20 J774A.1 cells per video or 12 BMDMs per video.
In the video analysis, contact was defined as the moment when the macrophage made visible cell-cell contact with the live conidia. Engulfment was defined as the time frame between contact and complete enclosure. Phagosome acidification was defined as the time until observation of a halo of LTR staining surrounding engulfed conidia, relative to the moment of engulfment. LC3 colocalization was defined as the time until observation of a halo of LC3 surrounding engulfed conidia, relative to the moment of engulfment. Germination of the Aspergillus conidia was defined as the moment when conidia formed a visible germ tube.
Volunteers
Blood was collected from healthy volunteers by venous blood puncture after informed consent was obtained. All experiments were performed and conducted in accordance with good clinical practice, the Declaration of Helsinki, and the approval of the Arnhem-Nijmegen Ethical Committee (No. 2010/104).
PBMC Isolation
Venous blood was drawn in 10-mL EDTA tubes. The blood was diluted 1:1 with phosphate-buffered saline (PBS). Subsequently, PBMCs were isolated using Ficoll-Paque (GE Healthcare, Zeist, The Netherlands) density gradient centrifugation. The PBMCs layer was collected and washed twice in cold PBS. Cells were reconstituted in RPMI+, consisting of RPMI-1640 culture medium (Dutch modification; Gibco, Invitrogen, Breda, The Netherlands) supplemented with 10 µg/mL gentamicin, 2 mM of glutamax and 1 mM of pyruvate (Gibco). The cells were counted with a particle counter (Beckmann Coulter, Woerden, The Netherlands) and the concentration was adjusted to 1 × 107 cells/mL.
PBMC Stimulation and Cytokine Measurements
PBMCs were plated in a 96-well plate (Corning Inc., Corning, NY, USA) at a final concentration of 2.5 × 106/mL, and were stimulated with 1 × 107/mL of A. fumigatus or A. nidulans live or heat-killed conidia in an end volume of 200 μL per well. All PBMC stimulations were performed in the presence of 10% human serum. In several experiments, phagocytosis was blocked by adding 1 ng/mL of cytochalasin D to the stimulations and were compared to a DMSO vehicle control. Cells were incubated at 37°C with 5% CO2. After stimulation for 24 h the supernatants were collected and stored at −20°C. The cytokines IL-1β, IL-6, TNFα, and IL-1Ra were measured in culture supernatants (R&D Systems, Minneapolis, MN, USA) according to the instructions supplied by the manufacturer.
ROS Induction
The induction of ROS was measured by oxidation luminol (5-amino-2,3,dihydro-1,4-phtalazinedione). PBMCs (5 × 105) were resuspended in HBSS and placed in dark 96-well plates. The cells were exposed to HBSS, A. fumigatus heat-inactivated resting conidia, live resting conidia, live germinated conidia (each at 1 × 107/mL), A. nidulans heat-inactivated resting conidia, live resting conidia, live germinated conidia (each at 1 × 107/mL), or zymosan (150 µg/mL), and 20 µL of 1 mM luminol immediately was added. Chemiluminescence was measured in BioTek Synergy HTreader at 37°C for every minute during 1 h.
Fungal Killing Assays
PBMCs (5 × 105/well), J774A.1 cells (1 × 105/well), and BMDMs (1 × 105/well) were plated in 96-well flat-bottom plates and exposed to A. fumigatus or A. nidulans conidia (2 × 106) in a final volume of 200 μL. After 24 h at 37°C all content of the wells was collected and the host cells were lysed with water, pooled with the well contents, and plated in serial dilution on Sabouraud agar plates in order to obtain both conidia associated with cells as well as free conidia. CFUs (colony-forming units) were counted after 24 h at 37°C, and were compared to the inoculum of live conidia in the wells to calculate the percentage killing.
Statistical Analysis
Experimental data were plotted and analyzed using GraphPad Prism v6.0 (GraphPad Software Inc., San Diego, CA, USA). Results are shown as the mean ± standard error of the mean (SEM). The Mann-Whitney U test and 2-way analysis of variance (ANOVA), followed by Bonferroni post hoc tests, were used to test statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Results
Phagocytosis of A. nidulans and A. fumigatus
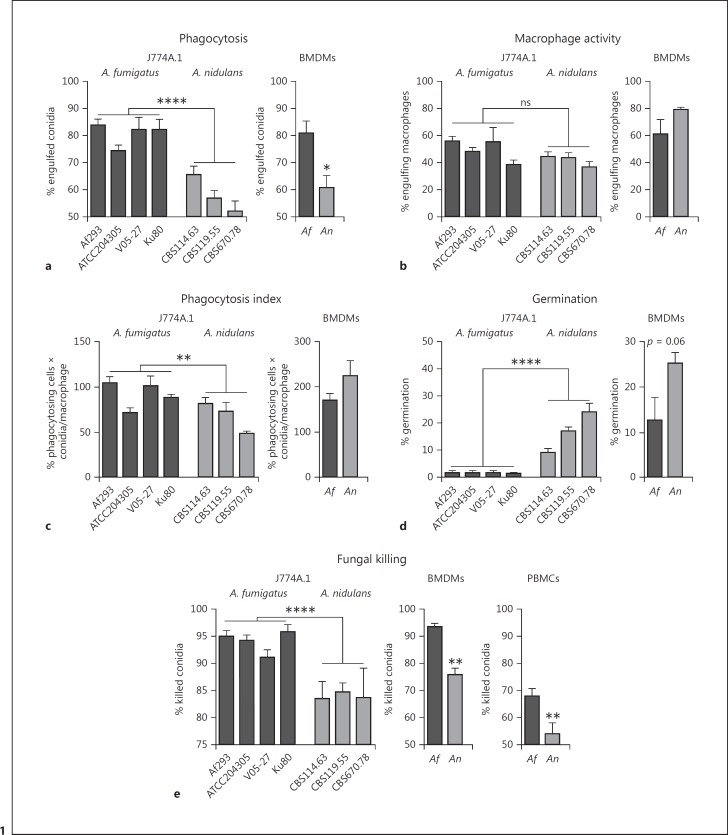
Cells of the murine macrophage-like cell line J774A.1 were exposed to resting conidia isolated from 4 A. fumigatus and 3 A. nidulans strains. Phagocytosis efficiency was assessed after 6 h of exposure to investigate whether there are major differences in overall phagocytosis of A. fumigatus and A. nidulans. After the 6-h time period, J774A.1 cells had engulfed a significantly higher percentage of A. fumigatus conidia compared to A. nidulans conidia (Fig. 1a). However, no significant difference was observed in the percentage of J774A.1 cells that were engulfing conidia (Fig. 1b). The phagocytic index of J774A.1 cells engulfing A. fumigatus strains was significantly higher, however, in a strain-to-strain comparison, not all A. fumigatus strains were engulfed with a higher phagocytic index compared to the A. nidulans strains. The reduced engulfment of A. nidulans strains led to significantly more conidia germinating and forming hyphae (Fig. 1d). Fungal killing assays were performed to assess whether the reduced capacity to engulf A. nidulans translated into a reduced capacity to kill A. nidulans conidia. After 24 h of exposure of J774A.1 cells to live conidia, the cells demonstrated a reduced capacity to kill conidia of A. nidulans strains compared to A. fumigatus strains (Fig. 1e).
Fig. 1.
Comparison of phagocytosis and killing of A. fumigatus and A. nidulans by J774A.1 murine cells and BMDMs. a Percentage of engulfed live resting A. fumigatus (Af293, ATCC204305, V05–27, and Ku80) and A. nidulans (CBS114.63, CBS119.55, and CBS670.78) conidia after 6 h of exposure to J774A.1 cells or A. fumigatus (Af293) and A. nidulans (CBS114.63) in M-CSF-differentiated BMDMs at an MOI of 1:1. b Percentage of J774A.1 cells or BMDMs with engulfed conidia after 6 h. c Phagocytic index as calculated by the percentage of phagocytosing cells multiplied by the number of conidia per J774A.1 cell or BMDM. d Percentage of germinating conidia within the total population of conidia after 6 h of exposure to J774A.1 cells or BMDMs. e Percentage of conidia killed after 6 h of exposure to J774A.1 cells, BMDMs, or human PBMCs measured by CFUs remaining after plating on Sabouraud agar. Data are presented as the mean ± SEM derived from at least 4 independent experiments for J774A.1 cells with a total of 14 frames for Af293, 6 for V05–27, 6 for ATCC204305, 6 for Ku80, 14 for CBS114.63, 6 for CBS119.55, and 6 for CBS670.78. For BMDMs, 2 independent experiments are shown with 2 and 4 videos. Means were compared for significance using the Mann-Whitney U test; * p < 0.05, **p < 0.01, ****p < 0.0001.
For the remaining experiments, a representative strain of each of the fungal species was used (Af293 for A. fumigatus and CBS114.63 for A. nidulans).
To validate the difference in phagocytosis and killing of A. nidulans spores, M-CSF-differentiated BMDMs were assessed for their phagocytosis and killing capacity. BMDMs also engulfed more A. fumigatus conidia after 6 h (Fig. 1a). Although no significant differences were observed in the percentage of engulfing macrophages and the phagocytic index (Fig. 1b, c), a trend towards more germination of A. nidulans conidia was observed (Fig. 1d). As with J774A.1 cells, the BMDMs and human PBMCs also demonstrated a reduced capacity to kill A. nidulans in comparison to A. fumigatus (Fig. 1e). A full characterization of the growth dynamics of the 2 strains AF293/CBS 101355/ATCC MYA-4609 (A. fumigatus) and CBS 114.63 (A. nidulans) was performed to exclude that the differences in phagocytosis were due to differences in growth of the 2 fungal species (online suppl. Fig. S2).
J774A.1 Cells and BMDMs Engulf A. nidulans at a Slower Rate than A. fumigatus
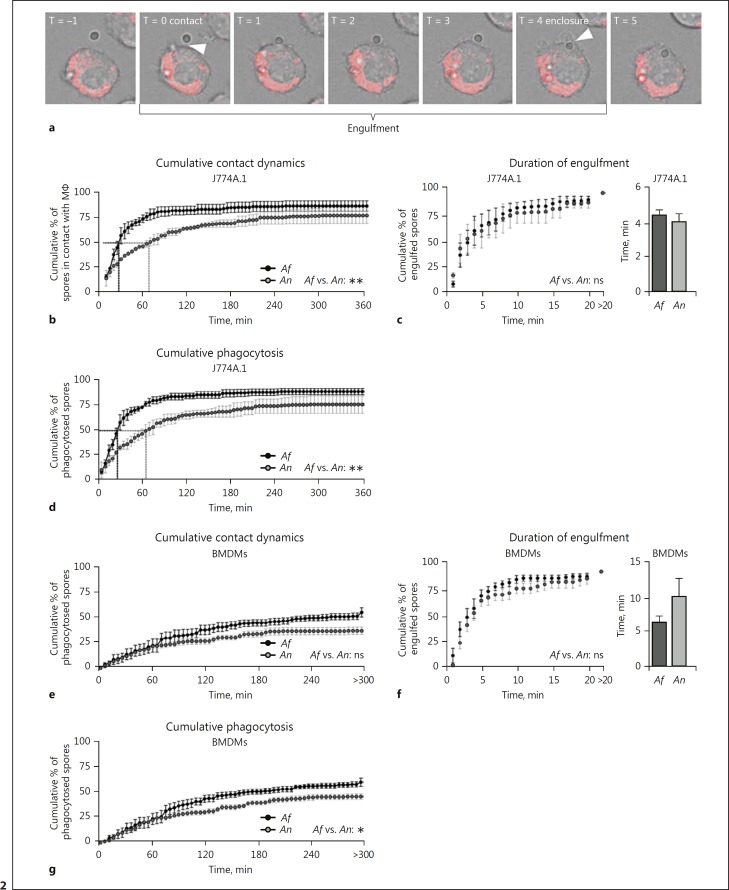
To dissect the dynamics of recognition and phagocytosis of live resting A. fumigatus and A. nidulans conidia in more detail, imaging videos were analyzed (see online suppl. Video 1 for a representative live cell-imaging video). The moment of contact (timepoint at which contact occurs between phagocyte and conidium) was established from 1-min interval time lapse videos (Fig. 2a). A significant difference was observed in the moment of contact between J774A.1 cells and conidia of A. fumigatus versus A. nidulans (Fig. 2b). Fifty percent of all A. fumigatus conidia were in contact with cells after 30 min, while this took over 1 h for A. nidulans (Fig. 2b). Subsequently, the time of engulfment was assessed that was defined as the time between contact and complete enclosure (Fig. 2c). However, no significant difference in the mean time of engulfment was observed (Fig. 2d). BMDMs exhibited similar interactions, although the dynamics of contact between macrophage and spore (Fig. 2e) and speed of engulfment (Fig. 2f) was not significantly slower. Collectively, these observations demonstrate that A. nidulans conidia are phagocytosed more slowly than A. fumigatus conidia (Fig. 2g).
Fig. 2.
J774A.1 cells and BMDMs engulf A. nidulans at a slower rate compared to A. fumigatus. a Representative frame series illustrating the definition of contact and full enclosure (phagocytosis). b, e Cumulative percentage of A. fumigatus and A. nidulans conidia in contact with J774A.1 cells (b) or BMDMs (e). Contact is defined as the timepoint where the cell makes visible physical contact with the spore. c, f Cumulative percentage of A. fumigatus and A. nidulans conidia engulfed by J774A.1 cells (c) or BMDMs (f) relative to the time of contact between spore and macrophage and mean time of duration between contact and engulfment. d, g Cumulative percentage of A. fumigatus and A. nidulans conidia engulfed by J774A.1 cells (d) or BMDMs (g) relative to the time of the start of the experiment. All bars and dot plots represent the mean ± SEM derived from 2 independent experiments with 4 videos per experiment for J774A.1 cells and 2 independent experiments with 2 and 4 videos for BMDMs. Statistical analysis was performed with 2-way ANOVA for cumulative graphs and means were compared for significance using the Mann-Whitney U test; * p < 0.05, **p < 0.01.
Enhanced Migration of J774A.1 Cells in Response to A. fumigatus Conidia
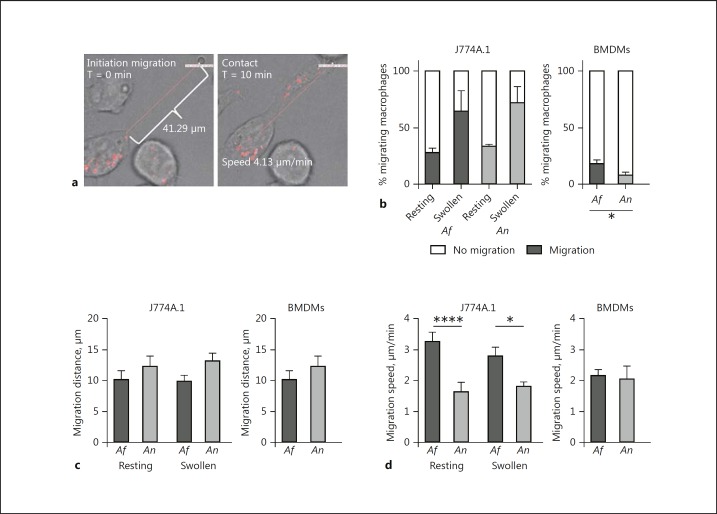
Phagocyte migration was analyzed during exposure to each fungal species to understand whether this was a component of slower A. nidulans engulfment (Fig. 3a). For individual engulfed conidia, we investigated whether cells actively migrated to engulf the spore. No significance difference was observed in the number of J774A.1 cells migrating towards conidia. Although cells migrated significantly more towards conidia that became swollen during the course of the experiment, no significant differences in the percentage of cells that migrated were observed between A. fumigatus and A. nidulans (Fig. 3b). In addition, no significant differences were observed in the distance that cells migrated to engulf conidia (Fig. 3c). However, the J774A.1 cells migrated with significantly greater velocity towards A. fumigatus resting and swollen conidia than to A. nidulans (Fig. 3d). In contrast, BMDMs did not show a faster migration towards A. fumigatus conidia (Fig. 3d). However, significantly more cells were migrating to engulf A. fumigatus conidia when compared to A. nidulans conidia (Fig. 3b).
Fig. 3.
A. fumigatus is more efficiently recognized and phagocytosed than A. nidulans. a Two representative frames from live cell-imaging videos illustrating the measurement of migration distance (left panel) and definition of contact between an A. fumigatus conidium and a J774A.1 cell (right panel). b Percentage of J774A.1 cells (left panel) and BMDMs (right panel) that show active migration to A. fumigatus or A. nidulans resting conidia or swollen conidia. c Mean distance (μm) measured using Volocity software from the moment of initiation of migration by J774A.1 cells (left panel) and BMDMs (right panel). d Migration speed (μm/min) measured by Volocity software from the moment of movement initiation to the moment of contact between J774A.1 cells (left panel) and BMDMs (right panel) and conidia. All bars and dot plots represent the mean ± SEM, derived from 2 independent experiments with 4 videos per experiment for J774A.1 cells and 2 independent experiments with 2 and 4 videos for BMDMs. Means were compared for significance using the Mann-Whitney U test; * p < 0.05, ****p < 0.0001
LC3-Associated Phagocytosis Is Less Efficient for A. nidulans
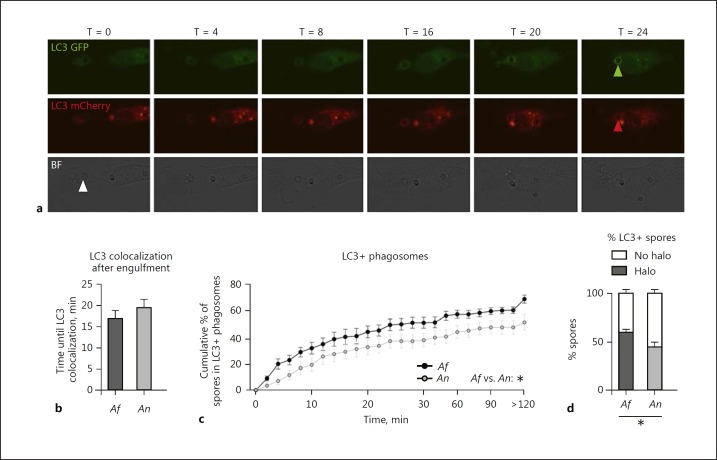
Phagosomes containing Aspergillus conidia mature into a phagolysosome, a process that is characterized by fusion with endosomes and lysosomes. Over the past years it has been demonstrated that a fully functional LAP is required for inducing an optimal phagosome maturation of phagosomes containing A. fumigatus by monocyte/macrophages [19, 20, 21, 22]. Moreover, cells from CGD patients recruit LC3 to Aspergillus-containing phagosomes less efficiently [7, 20]. We therefore investigated whether A. nidulans delayed LAP, which could be a contributing factor in the enhanced susceptibility of CGD patients to A. nidulans. BMDMs that constitutively express LC3-GFP-mCherry were exposed to A. fumigatus and A. nidulans live resting conidia, and the time until formation of an LC3 halo was assessed relative to the moment of phagocytosis of the conidia (Fig. 4a; online suppl. Video 2, 3). Although the time until colocalization of an LC3 halo was not significantly different for A. nidulans compared to A. fumigatus (Fig. 4b), the cumulative percentage of engulfed conidia that acquire an LC3 halo over time was significantly delayed for A. nidulans (Fig. 4c), and significantly fewer A. nidulans conidia recruited LC3 following phagocytosis, when compared to A. fumigatus conidia.
Fig. 4.
A. nidulans-containing phagosomes show delayed LC3 recruitment. a Representative image series of GFP-mCherry LC3 expressing M-CSF-differentiated BMDMs showing the LC3 GFP-mCherry signal and appearance of an LC3 halo surrounding the conidia at 24 min after engulfment. b Mean time until an LC3 halo was visible relative to the moment of engulfment. c Cumulative dynamics of LC3 halo colocalization. d Percentage of engulfed conidia that become surrounded by an LC3 halo (halo) and the percentage of conidia that are not enclosed by a halo of LC3 (no halo). All bars and dot plots represent the mean ± SEM derived from 2 independent experiments with 2 and 4 videos, respectively. Statistical analysis was performed with 2-way ANOVA for cumulative graphs and the means were compared for significance using the Mann-Whitney U test; * p < 0.05.
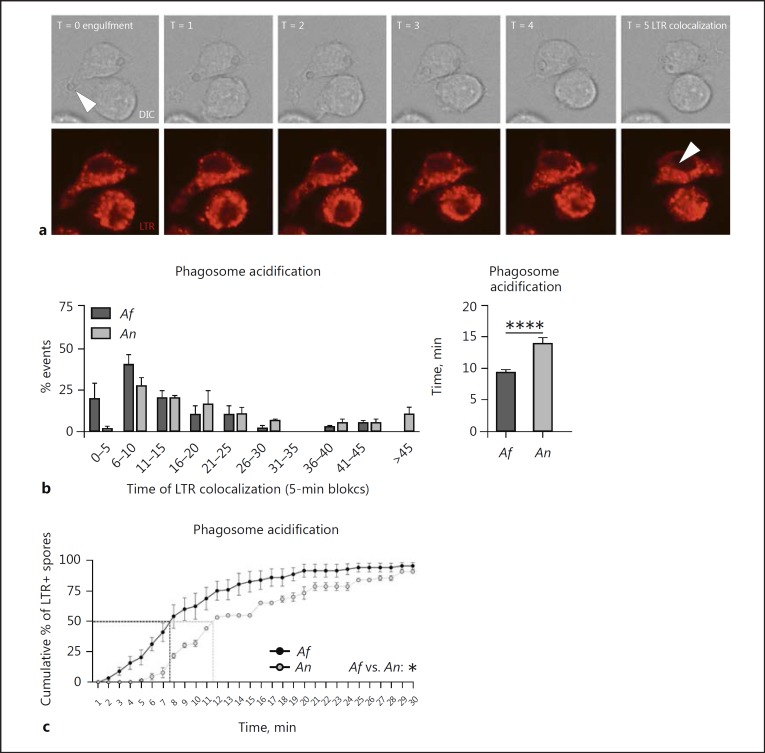
Phagosome Acidification Is Slower for Phagosomes Containing A. nidulans
Phagosome acidification has previously been shown to be crucial for the killing of A. fumigatus [23]. LTR was used to stain the acidic compartments of J774A.1 cells. Upon exposure of J774A.1 cells to live resting conidia, the onset of phagosome acidification was assessed by localization of LTR as a halo around the engulfed spore, relative to the moment of complete enclosure of the particle (Fig. 5a). Phagosome acidification of A. nidulans-containing phagosomes in J774A.1 cells was significantly slower than phagosome acidification of A. fumigatus-containing phagosomes (Fig. 5b). Analysis of the dynamics reveal that 50% of engulfed A. fumigatus conidia were in acidified compartments of the J774A.1 cells by 8 min postengulfment, while for A. nidulans this took over 11 min (Fig. 5c).
Fig. 5.
A. nidulans-containing phagosomes show delayed phagolysosomal fusion. a Representative image series showing LTR staining of acidic compartments in J774A.1 cells and the appearance of an LTR halo surrounding the conidia at 5 min after engulfment. b Histogram illustrating the distribution of time in which the LTR halo colocalized with the engulfed conidia relative to the time of engulfment and a plot of the average time of LTR-halo colocalization relative to the time of engulfment compared between A. fumigatus and A. nidulans engulfed conidia in J774A.1 cells. c Cumulative dynamics of LTR halo colocalization relative to the moment of engulfment in J774A.1 cells. All bars and dot plots represent the mean ± SEM derived from 2 independent experiments with 4 videos. Statistical analysis was performed with 2-way ANOVA for cumulative graphs and means were compared for significance using the Mann-Whitney U test; * p < 0.05, ****p < 0.0001.
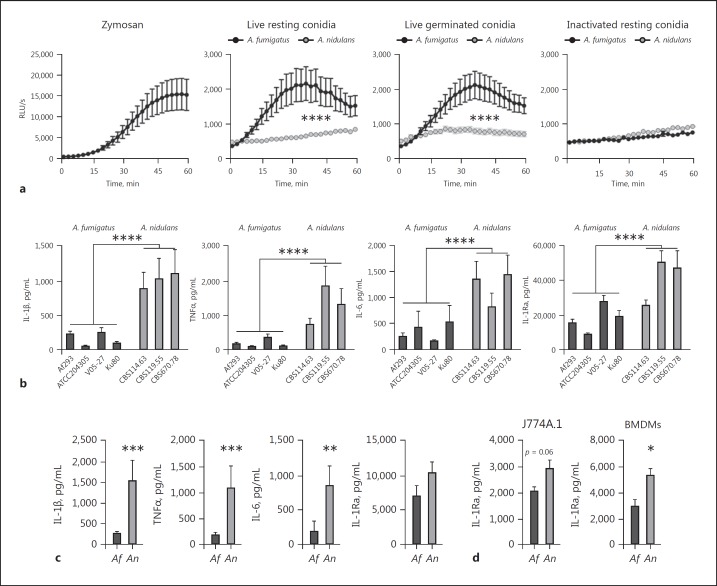
A. fumigatus Induces a Significantly Stronger Oxidative Burst than A. nidulans
The oxidative burst is thought to be essential for successful killing of Aspergillus conidia, since NADPH-induced ROS was found to regulate LAP [7, 20], a mechanism that is required for fungal killing and antigen presentation [20, 24]. Therefore, the capacity to induce an oxidative burst was compared between A. fumigatus and A. nidulans using live resting conidia, live germinated conidia (germlings), and heat-inactivated resting conidia (Fig. 6a-d). In line with previous studies [25], we confirmed that A. nidulans induced a significantly reduced oxidative burst in human PBMCs (Fig. 6c, d). The induction of oxidative burst was below the detection limit of the assay for J774A.1 and BMDMs (data not shown).
Fig. 6.
A. fumigatus induces higher ROS but lower cytokine levels in human PBMCs compared to A. nidulans. a Oxidative burst assayed by luminol oxidation by human PBMCs (5 × 105 cells/well) from healthy volunteers (n = 6) that were incubated with Zymosan, live resting conidia (1 × 107/mL), live germinated conidia (1 × 107/mL), or inactivated conidia (1 × 107/mL; panels from left to right). b Cytokine release by human PBMCs measured in the cell culture supernatant by ELISA were stimulated with live conidia of various strains of A. fumigatus (Af293, n = 16; ATCC204305, n = 10; V05–27, n = 10, and Ku80, n = 8) and A. nidulans (CBS114.63, n = 16; CBS119.55, n = 10, and CBS670.78, n = 5). c Cytokine release by human PBMCs induced by heat-inactivated conidia of A. fumigatus (Af293, n = 13) or A. nidulans (CBS114.63, n = 13). d IL-1Ra release measured in the cell culture supernatants of J774A.1 cells (n = 10) and BMDMs (n = 6) after stimulation with A. fumigatus (Af293) or A. nidulans (CBS114.63). All bars and dot plots represent the mean ± SEM; * p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Statistical analysis was performed with 2-way ANOVA for ROS curves and the Mann-Whitney U test for cytokine data.
A. nidulans-Induced Cytokine Levels Are Significantly Higher Compared to A. fumigatus
Defective ROS production has been associated with a higher cytokine production in CGD patients [26]. Therefore, we investigated whether the reduced oxidative burst elicited by A. nidulans correlated with an increased cytokine response by stimulating human PBMCs of healthy volunteers with heat-killed and live resting conidia of A. fumigatus or A. nidulans. Cytokine production in human PBMCs induced by live resting conidia of the 3 A. nidulans strains was significantly higher for the innate cytokines IL-6, TNFα, IL-β, and IL-1Ra compared to stimulation with 4 A. fumigatus strains (Fig. 6b). Killed resting conidia of A. nidulans induced significantly more IL-1β, TNFα, and IL-6 (Fig. 6c). Although no induction of IL-1β, TNFα, and IL-6 could be detected in J774A.1 cells and BMDMs in response to A. nidulans (strain CBS114.63) and A. fumigatus (strain Af293), a trend towards higher IL-1Ra release was observed in both cell types (Fig. 6d).
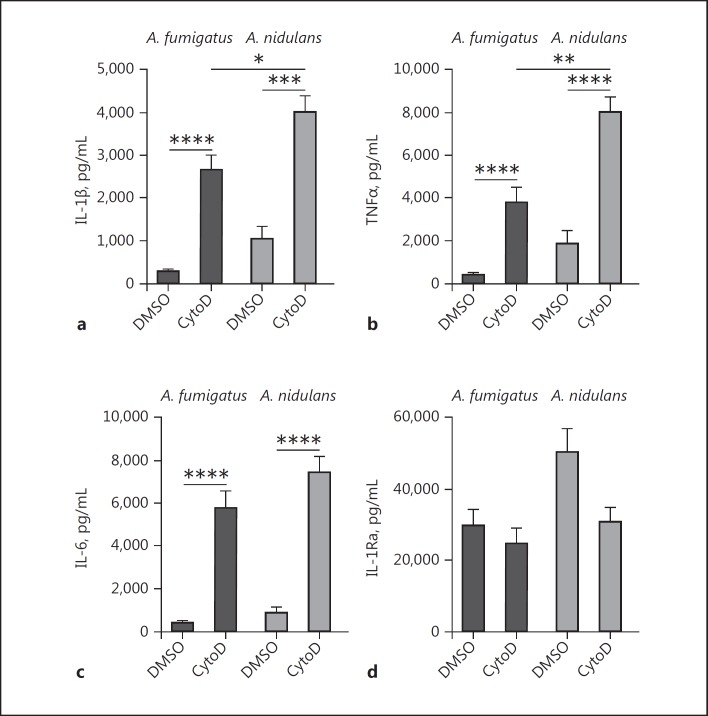
Inhibition of Phagocytosis Increases A. fumigatus- and A. nidulans-Induced Cytokine Responses
In order to assess whether the differences in cytokine release could be attributed to differences in phagocytosis, the capacity of human PBMCs to engulf particles was inhibited by cytochalasin D. Inhibition of actin-polymerization [27], and thus phagocytosis, resulted in a significant increase of IL-1β, TNFα, and IL-6 induction in response to both heat-inactivated resting conidia of the A. nidulans (strain CBS114.63) and A. fumigatus (strain Af293; Fig. 7a-c). The capacity to induce IL-1Ra, however, did not increase after the blockade of phagocytosis with cytochalasin D (Fig. 7d). Despite inhibition of phagocytosis, A. nidulans still induced significantly more IL-1β and TNFα than A. fumigatus.
Fig. 7.
Influence of phagocytosis inhibition on cytokine release induced by A. fumigatus and A. nidulans. IL-1β (a), TNFα (b), IL-6 (c), and IL-1Ra (d) release measured by ELISA in cell culture supernatants of human PBMCs (n = 10 donors) after stimulation with heat-inactivated A. fumigatus (Af293) or A. nidulans (CBS114.63) conidia, in the presence of 1 ng/mL of cytochalasin D or vehicle control. All bars represent the mean ± SEM; * p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Statistical analysis was performed with the Mann-Whitney U test.
Discussion
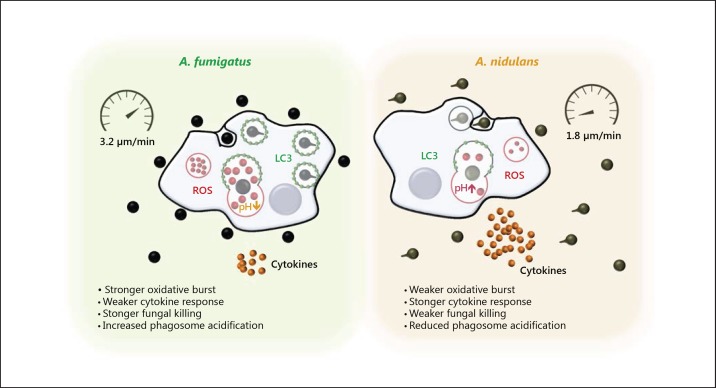
In this study, various aspects of the innate immune response, in particular the phagocytosis of A. fumigatus and A. nidulans, were compared. Differences in the phagocytic and immune-stimulatory capacities of these 2 distinct opportunistic pathogens were observed, which might help to explain their differential pathogenicity. By 6 h phagocytes are less able to control A. nidulans, which continue to grow and germinate, resulting in reduced killing of A. nidulans compared to A. fumigatus. Both species differed markedly in their engulfment and uptake kinetics during interactions with host cells: A. fumigatus was recognized and taken up faster compared to A. nidulans. Strikingly, LC3 appeared more rapidly in A. fumigatus-containing phagosomes and phagosomes containing A. fumigatus were acidified significantly faster than those containing conidia of A. nidulans. Human PBMCs exposed to A. nidulans produced increased concentrations of proinflammatory cytokines compared to exposure to A. fumigatus conidia and oxidative burst was diminished in the response to A. nidulans. These findings are summarized in Figure 8.
Fig. 8.
Schematic comparison of the innate immune response of A. fumigatus and A. nidulans. The innate immune response against A. nidulans differs from the immune response against A. fumigatus in delayed recognition, delayed phagocytosis, slower cell migration, slower phagosome acidification, a less pronounced oxidative burst, and enhanced cytokine production.
Phagosome maturation leading to phagosome acidification is crucial for the killing of A. fumigatus[23]. LAP utilizes autophagy machinery for more efficient killing of engulfed pathogens [19, 20] and MHCII-mediated antigen presentation [28]. Specifically, LAP affects the downstream maturation of Aspergillus-containing phagosomes, including acidification [19, 20, 21]. Therefore, delayed LAP dynamics could indicate differential processing of the 2 Aspergillus species in the phagolysosomes.
We observed a difference in the kinetics of phagosome acidification using LTR-stained J774A.1 cells and LC3 halos in LC3-GFP-mCherry transgenic BMDMs. Phagosome acidification in J774A.1 occurred earlier than the appearance of LC3 halos in BMDMs. This difference in kinetics may be explained by the use of 2 different cell types for each of the observations. However, an alternative explanation is that LC3 is present in low quantity which is visible by microscopy much earlier than appearance of the halo. A relatively weak LC3 signal could be observed in the videos during engulfment (online suppl. Video 2). Further investigations will determine whether early localization of LC3 concurrently with engulfment regulates phagosome acidification. Since the early weak LC3 signal was impractical for quantification, and previous reports focused on LC3+ halos surrounding engulfed conidia as a measurement of LAP, in the current study LAP was also quantified as LC3 halos surrounding engulfed conidia.
Using live cell imaging, we conducted detailed comparisons of the phagocytosis dynamics of A. nidulans and A. fumigatus; these observations may have relevance in the setting of CGD. We observed that phagocytes migrated poorly to resting conidia of either species, but upon the end stage of conidial swelling and start of germ tube formation of the conidia, both J774A.1 cells (online suppl. Video 4) and BMDMs migrated actively. Interestingly, this is different from another fungus, Candida albicans, which elicits active migration of phagocytes in both yeast and hyphal forms [29]. Delayed phagocytosis of resting Aspergillus conidia might be due to PAMP masking by spore surface hydrophobins [30], until germination reveals underlying immune-stimulatory molecules following the shedding of the hydrophobin layer. Phagocytes appear to sense germination over a substantial distance (online suppl. Video 4), perhaps by the sensing of secreted fungal molecules, such as galactomannan or galactosaminogalactan (GAG). Alternatively, macrophages might contact-sense distant germinating conidia using superfine projections, as previously seen with Candida interactions by scanning electron microscopy [31]. Nevertheless, the factors that trigger the migration of phagocytes towards germinating conidia warrants further investigation.
C. albicans forms hyphae inside macrophages, facilitating lysis and escape [32]. This phenomenon was not observed during the phagocytosis of A. fumigatus and A. nidulans; however, hyphae were expulsed from the cell following intracellular germination (online suppl. Video 5), as was previously described for Candida and Cryptococcus [33, 34]. These observations provide new insights into the interplay between macrophages and Aspergillus, and highlight the potential of live cell imaging as a tool to study the dynamics of phagocytosis.
Previously, phagocytosis of A. fumigatus by J774A.1 macrophage-like cells was evaluated to be similar in terms of morphology and efficiency to the phagocytosis by alveolar macrophages [35]. Nevertheless, a potential limitation of the study is the use of macrophage-like cells (J774A.1 cells) and nontypical macrophages (BMDMs). Although these cell types have been extensively used to study the dynamics of phagocytosis of fungi [29, 32, 33, 34, 36], the findings may not be directly translatable to bona fide tissue macrophages, such as alveolar macrophages, and further studies are warranted using primary macrophages, and in particular primary human alveolar macrophages. Protocols for the isolation of primary human macrophages are becoming available [37, 38]; however, it remains challenging to obtain large quantities of cells for experiments without activating them and under sterile conditions.
The findings of this study may help us understand why the susceptibility to A. nidulans in CGD patients is much higher compared to hematooncology patients. The differences in the prevalence of cases associated with fungal species suggest that factors other than neutrophils may determine the pathogenicity of A. nidulans, since A. nidulans infections are rarely seen in neutropenic patients. The residual cells of hematooncology patients, such as residential alveolar macrophages, have normal antifungal killing activity, while CGD patients have defects in neutrophils, macrophages, monocytes, and other immune cells. Although the primary defect in CGD patients is an inability to produce NADPH-dependent ROS, killing of A. nidulans is still possible in NADPH oxidase-deficient monocytes/macrophages [17]. However, the defect in the NADPH complex in CGD patients has also been associated with a reduced efficiency of LAP [20, 39]. Restoration of LAP in CGD mice was successful for controlling invasive aspergillosis caused by A. fumigatus [7].
The epidemiology of Aspergillus infections in CGD patients implies that A. nidulans has an advantage in colonizing patients within this particular disease background. Although extensive studies are lacking, several studies support the assumption that A. fumigatus is the more abundant pathogen in the environment [40, 41]. In addition, A. fumigatus conidia are more efficiently dispersed in the environment due to their higher hydrophobicity [42]. The emergence of infections from less environmentally abundant A. nidulans in CGD patients but not in other immunocompromised groups points towards a selective advantage for this fungus.
Here, we demonstrated that healthy macrophages phagocytose and control A. nidulans with reduced efficiency. Crucial antifungal processes, including LAP and phagosome acidification, were impaired in response to A. nidulans. Phagocytes of CGD patients have significantly reduced LAP [7, 20], an impairment that may be compounded upon further repression of LC3 recruitment and phagosome acidification associated with A. nidulans. Exposure to these spores may thus result in increased susceptibility and higher mortality of A. nidulans infection in CGD [15, 16, 17].
Hematooncology patients experience a relatively short time period of risk for Aspergillus infections, while CGD patients are exposed to environmental Aspergillus conidia over years. Therefore, it is tempting to speculate that the net result of a chronic defective Aspergillus phagocytic machinery characteristic for CGD in combination with the slower phagocytic rate, delayed phagosome acidification, and less efficient LAP of A. nidulans might explain why A. nidulans infections are only seen in CGD patients and not in other disease settings, where this severe threshold of defective host defense against A. nidulans is not reached.
The higher immunostimulatory capacity of A. nidulans compared to A. fumigatus is in agreement with earlier studies using different strains than those presented here [26, 43]. Interestingly, CGD patients, also exhibit a heightened proinflammatory cytokine profile [44]. This response in CGD patients especially for interleukin-1 production [7, 26, 43] has been associated with colitis [7]. Failure to control excessive inflammatory responses has been associated with a poor clinical outcome and increased mortality from aspergillosis [45], and the higher mortality observed with A. nidulans could also be a result of excessive inflammatory reactions leading to immunopathology.
Differences in cell wall architecture between A. nidulans and A. fumigatus may explain how they elicit differential cytokine responses. GAG is an important anti-inflammatory polysaccharide of the cell wall of A. fumigatus that induces the anti-inflammatory cytokine IL-1Ra [46]. The absence of GAG in the cell wall of A. nidulans [47] may contribute to its proinflammatory capacity, yet this does not explain the additional high induction of an IL-1Ra response. It should be noted, however, that other cell wall components including chitin [48] and β-glucan [49] are known to induce high levels of IL-1Ra, but in contrast to GAG also have the capacity to induce strong proinflammatory responses. High IL-1Ra levels could also be a result of a feedback loop of proinflammatory cytokines such as IL-1β, which are known to induce IL-1Ra [50]. Furthermore, a high immunostimulatory capacity associated with high levels of the proinflammatory IL-1β or TNFα can be the result of continuous extracellular signaling via pattern recognition receptors and activation of the subsequent signaling pathways. Frustrated phagocytosis of long filaments is a known phenomenon associated with the production of high levels of proinflammatory cytokines [51]. Although these filaments were much bigger than the conidia used in this study, we observed differential phagocytosis between the fungi, and a continuous presence of extracellular filaments might be one of the reasons for the increased proinflammatory response associated with A. nidulans. In line with this, we observed that the blockade of phagocytosis using cytochalasin D enhanced the induction of IL-1β, TNFα, and IL-6 by both A. nidulans and A. fumigatus.
Collectively, our data provide a novel insight to Aspergillus phagocytosis and illustrate clear differences between Aspergillus species. These findings offer a possible means to understanding the pathogenicity of A. nidulans in the CGD patient.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
The authors would like to acknowledge Fraser P. Coxon and Ian Ganley for providing LC3-GFP-mCherry BMDMs. M.S.G. was supported by an FEMS research grant and F.L.v.d.V. was supported by ZonMW under the name EURO-CMC frame of E-Rare-2, the ERA-Net for Research on Rare Diseases.
References
- 1.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW, Brown JM, Brumble LM, Freifeld AG, Hadley S, Herwaldt LA, Kauffman CA, Knapp K, Lyon GM, Morrison VA, Papanicolaou G, Patterson TF, Perl TM, Schuster MG, Walker R, Wannemuehler KA, Wingard JR, Chiller TM, Pappas PG. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis. 2010;50:1091–1100. doi: 10.1086/651263. [DOI] [PubMed] [Google Scholar]
- 3.Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood. 2002;100:4358–4366. doi: 10.1182/blood-2002-05-1496. [DOI] [PubMed] [Google Scholar]
- 4.Henriet SS, Verweij PE, Warris A. Aspergillus nidulans and chronic granulomatous disease: a unique host-pathogen interaction. J Infect Dis. 2012;206:1128–1137. doi: 10.1093/infdis/jis473. [DOI] [PubMed] [Google Scholar]
- 5.Leiding JW, Holland SM. Chronic granulomatous disease. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mefford HC, Stephens K, Amemiya A, Ledbetter N, editors. Seattle: University of Washington; 1993. GeneReviews. [PubMed] [Google Scholar]
- 6.Roos D, de Boer M. Molecular diagnosis of chronic granulomatous disease. Clin Exp Immunol. 2014;175:139–149. doi: 10.1111/cei.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, Begun J, Plantinga TS, Joosten LA, van der Meer JW, Chamilos G, Netea MG, Xavier RJ, Dinarello CA, Romani L, van de Veerdonk FL. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci USA. 2014;111:3526–3531. doi: 10.1073/pnas.1322831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregg KS, Kauffman CA. Invasive aspergillosis: epidemiology, clinical aspects, and treatment. Semin Respir Crit Care Med. 2015;36:662–672. doi: 10.1055/s-0035-1562893. [DOI] [PubMed] [Google Scholar]
- 9.Baddley JW, Stroud TP, Salzman D, Pappas PG. Invasive mold infections in allogeneic bone marrow transplant recipients. Clin Infect Dis. 2001;32:1319–1324. doi: 10.1086/319985. [DOI] [PubMed] [Google Scholar]
- 10.Hsiue HC, Wu TH, Chang TC, Hsiue YC, Huang YT, Lee PI, Hsueh PR. Culture-positive invasive aspergillosis in a medical center in Taiwan, 2000–2009. Eur J Clin Microbiol Infect Dis. 2012;31:1319–1326. doi: 10.1007/s10096-011-1445-1. [DOI] [PubMed] [Google Scholar]
- 11.Perfect JR, Cox GM, Lee JY, Kauffman CA, de Repentigny L, Chapman SW, Morrison VA, Pappas P, Hiemenz JW, Stevens DA, Mycoses Study Group The impact of culture isolation of Aspergillus species: a hospital-based survey of aspergillosis. Clin Infect Dis. 2001;33:1824–1833. doi: 10.1086/323900. [DOI] [PubMed] [Google Scholar]
- 12.Wald A, Leisenring W, van Burik JA, Bowden RA. Epidemiology of Aspergillus infections in a large cohort of patients undergoing bone marrow transplantation. J Infect Dis. 1997;175:1459–1466. doi: 10.1086/516480. [DOI] [PubMed] [Google Scholar]
- 13.Simon-Nobbe B, Denk U, Poll V, Rid R, Breitenbach M. The spectrum of fungal allergy. Int Arch Allergy Immunol. 2008;145:58–86. doi: 10.1159/000107578. [DOI] [PubMed] [Google Scholar]
- 14.Henriet S, Verweij PE, Holland SM, Warris A. Invasive fungal infections in patients with chronic granulomatous disease. Adv Exp Med Biol. 2013;764:27–55. doi: 10.1007/978-1-4614-4726-9_3. [DOI] [PubMed] [Google Scholar]
- 15.Blumental S, Mouy R, Mahlaoui N, Bougnoux ME, Debre M, Beaute J, Lortholary O, Blanche S, Fischer A. Invasive mold infections in chronic granulomatous disease: a 25-year retrospective survey. Clin Infect Dis. 2011;53:e159–e169. doi: 10.1093/cid/cir731. [DOI] [PubMed] [Google Scholar]
- 16.Dotis J, Roilides E. Osteomyelitis due to Aspergillus species in chronic granulomatous disease: an update of the literature. Mycoses. 2011;54:e686–e696. doi: 10.1111/j.1439-0507.2010.02001.x. [DOI] [PubMed] [Google Scholar]
- 17.Segal BH, DeCarlo ES, Kwon-Chung KJ, Malech HL, Gallin JI, Holland SM. Aspergillus nidulans infection in chronic granulomatous disease. Medicine (Baltimore) 1998;77:345–354. doi: 10.1097/00005792-199809000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Netea MG, Warris A, van der Meer JW, Fenton MJ, Verver-Janssen TJ, Jacobs LE, Andresen T, Verweij PE, Kullberg BJ. Aspergillus fumigatus evades immune recognition during germination through loss of toll-like receptor-4-mediated signal transduction. J Infect Dis. 2003;188:320–326. doi: 10.1086/376456. [DOI] [PubMed] [Google Scholar]
- 19.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Kyrmizi I, Gresnigt MS, Akoumianaki T, Samonis G, Sidiropoulos P, Boumpas D, Netea MG, van de Veerdonk FL, Kontoyiannis DP, Chamilos G. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J Immunol. 2013;191:1287–1299. doi: 10.4049/jimmunol.1300132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akoumianaki T, Kyrmizi I, Valsecchi I, Gresnigt MS, Samonis G, Drakos E, Boumpas D, Muszkieta L, Prevost MC, Kontoyiannis DP, Chavakis T, Netea MG, van de Veerdonk FL, Brakhage AA, El-Benna J, Beauvais A, Latge JP, Chamilos G. Aspergillus cell wall melanin blocks LC3-associated phagocytosis to promote pathogenicity. Cell Host Microbe. 2016;19:79–90. doi: 10.1016/j.chom.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Sprenkeler EG, Gresnigt MS, van de Veerdonk FL. LC3-associated phagocytosis: a crucial mechanism for antifungal host defence against Aspergillus fumigatus. Cell Microbiol. 2016;18:1208–1216. doi: 10.1111/cmi.12616. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim-Granet O, Philippe B, Boleti H, Boisvieux-Ulrich E, Grenet D, Stern M, Latge JP. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect Immun. 2003;71:891–903. doi: 10.1128/IAI.71.2.891-903.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, Ziegler U, Roesler J, Dengjel J, Reichenbach J, Munz C. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol. 2013;203:757–766. doi: 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriet SS, Hermans PW, Verweij PE, Simonetti E, Holland SM, Sugui JA, Kwon-Chung KJ, Warris A. Human leukocytes kill Aspergillus nidulans by reactive oxygen species-independent mechanisms. Infect Immun. 2011;79:767–773. doi: 10.1128/IAI.00921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smeekens SP, Henriet SS, Gresnigt MS, Joosten LA, Hermans PW, Netea MG, Warris A, van de Veerdonk FL. Low interleukin-17A production in response to fungal pathogens in patients with chronic granulomatous disease. J Interferon Cytokine Res. 2012;32:159–168. doi: 10.1089/jir.2011.0046. [DOI] [PubMed] [Google Scholar]
- 27.Wang E, Michl J, Pfeffer LM, Silverstein SC, Tamm I. Interferon suppresses pinocytosis but stimulates phagocytosis in mouse peritoneal macrophages: related changes in cytoskeletal organization. J Cell Biol. 1984;98:1328–1341. doi: 10.1083/jcb.98.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012;287:34149–34156. doi: 10.1074/jbc.M112.382812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NA, Erwig LP. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012;8:e1002578. doi: 10.1371/journal.ppat.1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latge JP. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 31.Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- 32.McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, Gow NA, Erwig LP. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect Immun. 2010;78:1650–1658. doi: 10.1128/IAI.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bain JM, Lewis LE, Okai B, Quinn J, Gow NA, Erwig LP. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet Biol. 2012;49:677–678. doi: 10.1016/j.fgb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston SA, May RC. Cryptococcus interactions with macrophages: evasion and manipulation of the phagosome by a fungal pathogen. Cell Microbiol. 2013;15:403–411. doi: 10.1111/cmi.12067. [DOI] [PubMed] [Google Scholar]
- 35.Behnsen J, Narang P, Hasenberg M, Gunzer F, Bilitewski U, Klippel N, Rohde M, Brock M, Brakhage AA, Gunzer M. Environmental dimensionality controls the interaction of phagocytes with the pathogenic fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 2007;3:e13. doi: 10.1371/journal.ppat.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bain JM, Louw J, Lewis LE, Okai B, Walls CA, Ballou ER, Walker LA, Reid D, Munro CA, Brown AJ, Brown GD, Gow NA, Erwig LP. Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio. 2014;5:e01874. doi: 10.1128/mBio.01874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexis N, Soukup J, Ghio A, Becker S. Sputum phagocytes from healthy individuals are functional and activated: a flow cytometric comparison with cells in bronchoalveolar lavage and peripheral blood. Clin Immunol. 2000;97:21–32. doi: 10.1006/clim.2000.4911. [DOI] [PubMed] [Google Scholar]
- 38.Efthimiadis A, Spanevello A, Hamid Q, Kelly MM, Linden M, Louis R, Pizzichini MM, Pizzichini E, Ronchi C, van Overvel F, Djukanovic R. Methods of sputum processing for cell counts, immunocytochemistry and in situ hybridisation. Eur Respir J Suppl. 2002;37:19s–23s. doi: 10.1183/09031936.02.00001902. [DOI] [PubMed] [Google Scholar]
- 39.De Luca A, Zelante T, D'Angelo C, Zagarella S, Fallarino F, Spreca A, Iannitti RG, Bonifazi P, Renauld JC, Bistoni F, Puccetti P, Romani L. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3:361–373. doi: 10.1038/mi.2010.22. [DOI] [PubMed] [Google Scholar]
- 40.Guinea J, Pelaez T, Alcala L, Bouza E. Outdoor environmental levels of Aspergillus spp. conidia over a wide geographical area. Med Mycol. 2006;44:349–356. doi: 10.1080/13693780500488939. [DOI] [PubMed] [Google Scholar]
- 41.Sharma R, Deval R, Priyadarshi V, Gaur SN, Singh VP, Singh AB. Indoor fungal concentration in the homes of allergic/asthmatic children in Delhi, India. Allergy Rhinol. 2011;2:21–32. doi: 10.2500/ar.2011.2.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus - what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013;9:e1003743. doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henriet SS, van de Sande WW, Lee MJ, Simonetti E, Momany M, Verweij PE, Rijs AJ, Ferwerda G, Sheppard DC, de Jonge MI, Warris A. Decreased cell wall galactosaminogalactan in Aspergillus nidulans mediates dysregulated inflammation in the chronic granulomatous disease host. J Interferon Cytokine Res. 2016;36:488–498. doi: 10.1089/jir.2015.0095. [DOI] [PubMed] [Google Scholar]
- 44.Weisser M, Demel UM, Stein S, Chen-Wichmann L, Touzot F, Santilli G, Sujer S, Brendel C, Siler U, Cavazzana M, Thrasher AJ, Reichenbach J, Essers MA, Schwable J, Grez M. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J Allergy Clin Immunol. 2016;138:219–228. doi: 10.1016/j.jaci.2015.11.028. e9. [DOI] [PubMed] [Google Scholar]
- 45.Montagnoli C, Fallarino F, Gaziano R, Bozza S, Bellocchio S, Zelante T, Kurup WP, Pitzurra L, Puccetti P, Romani L. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J Immunol. 2006;176:1712–1723. doi: 10.4049/jimmunol.176.3.1712. [DOI] [PubMed] [Google Scholar]
- 46.Gresnigt MS, Bozza S, Becker KL, Joosten LA, Abdollahi-Roodsaz S, van der Berg WB, Dinarello CA, Netea MG, Fontaine T, De Luca A, Moretti S, Romani L, Latge JP, van de Veerdonk FL. A polysaccharide virulence factor from Aspergillus fumigatus elicits anti-inflammatory effects through induction of interleukin-1 receptor antagonist. PLoS Pathog. 2014;10:e1003936. doi: 10.1371/journal.ppat.1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee MJ, Liu H, Barker BM, Snarr BD, Gravelat FN, Al Abdallah Q, Gavino C, Baistrocchi SR, Ostapska H, Xiao T, Ralph B, Solis NV, Lehoux M, Baptista SD, Thammahong A, Cerone RP, Kaminskyj SG, Guiot MC, Latge JP, Fontaine T, Vinh DC, Filler SG, Sheppard DC. The fungal exopolysaccharide galactosaminogalactan mediates virulence by enhancing resistance to neutrophil extracellular traps. PLoS Pathog. 2015;11:e1005187. doi: 10.1371/journal.ppat.1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker KL, Aimanianda V, Wang X, Gresnigt MS, Ammerdorffer A, Jacobs CW, Gazendam RP, Joosten LA, Netea MG, Latge JP, van de Veerdonk FL. Aspergillus cell wall chitin induces anti- and proinflammatory cytokines in human PBMCs via the Fc-γ receptor/Syk/PI3K pathway. mBio. 2016;7:e01823–15. doi: 10.1128/mBio.01823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smeekens SP, Gresnigt MS, Becker KL, Cheng SC, Netea SA, Jacobs L, Jansen T, van de Veerdonk FL, Williams DL, Joosten LA, Dinarello CA, Netea MG. An anti-inflammatory property of Candida albicans β-glucan: induction of high levels of interleukin-1 receptor antagonist via a Dectin-1/CR3 independent mechanism. Cytokine. 2014;71:215–222. doi: 10.1016/j.cyto.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyles MS, Young L, Brown DM, MacCalman L, Cowie H, Moisala A, Smail F, Smith PJ, Proudfoot L, Windle AH, Stone V. Multi-walled carbon nanotube induced frustrated phagocytosis, cytotoxicity and pro-inflammatory conditions in macrophages are length dependent and greater than that of asbestos. Toxicol In Vitro. 2015;29:1513–1528. doi: 10.1016/j.tiv.2015.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data