Abstract
After infection or invasion is recognized, biochemical mediators act in signaling insect immune functions. These include biogenic amines, insect cytokines, eicosanoids, and nitric oxide (NO). Treating insects or isolated hemocyte populations with different mediators often leads to similar results. Separate treatments with an insect cytokine, 2 biogenic amines, and an eicosanoid lead to a single result, hemocyte spreading, understood in terms of intracellular cross-talk among these signaling systems. This study focuses on the cross-talk between NO and eicosanoid signaling in our model insect, Spodoptera exigua. Bacterial injection increased NO concentrations in the larval hemocytes and fat body, and RNA interference (RNAi) of the S. exigua NO synthase (NOS) gene suppressed NO concentrations. RNAi treatment also led to a significant reduction in hemocyte nodulation following bacterial injection. Similar RNAi treatments led to significantly reduced PLA2 activities in the hemocytes and fat body compared to control larvae. Injection of L-NAME also prevented the induction of PLA2 activity following bacterial challenge. An injected NO donor, S-nitroso-N-acetyl-DL-penicillamine, increased PLA2 activity in a dose-dependent manner. However, eicosanoids did not influence NO concentrations in immune-challenged larvae. We infer that NO and eicosanoid signaling operate via cross-talk mechanisms in which the elevated NO concentrations activate PLA2 and eicosanoid biosynthesis, which finally mediates various immune responses.
Keywords: Nitric oxide, Nitric oxide synthase, Eicosanoid, Immune response, Spodoptera exigua
Introduction
Insects defend themselves from pathogens and parasites with their complex innate immune system, usually assorted into hemocytic (phagocytosis, nodulation, and encapsulation) and humoral (the synthesis of anti-microbial peptides [AMPs] and prophenol oxidase activation) functions. Hemocytes and the larval fat body are the main immunity-conferring tissues [1], although several epithelia also produce and secrete AMPs. Upon immune challenge, non-self-recognition mechanisms initiate immune signaling, which propagates to nearby hemocytes and to the fat body via immune mediators [2]. These include biogenic monoamines [3, 4], the ENF family of cytokines [5], and eicosanoids such as prostaglandins (PGs), leukotrienes (LTs), and epoxyeicosatrienoic acids (EETs) [6, 7].
Nitric oxide (NO) is a small, membrane-permeable signal molecule that acts in the nervous and immune systems as a mediator in insects and vertebrates [8]. NO is synthesized from L-arginine by NO synthase (NOS) which, in mammals, occurs in 3 forms, eNOS, constitutively expressed in endothelial cells, nNOS constitutively expressed mostly in neurons, and inducible NOS (iNOS) which can generate very large amounts of NO [9]. NO is associated with neuronal processing and long-term memory of chemo- and visual signals in insects [10]. It also plays a crucial role in producing light in fireflies by inhibiting oxygen usage of mitochondria to supply enough oxygen to photocytes to oxidize luciferin [11]. In blood-sucking insects, NO is secreted from the salivary gland and transferred to the host to act as vasodilator or platelet-aggregation inhibitor [12]. In mosquitoes transmitting malarial protozoa, NOS expression is rapidly induced after blood feeding and it elevates NO concentrations [13], which directly limits the development of the protozoan parasites [14, 15]. NO also induces both cellular and humoral immune responses via the Toll/IMD signal pathways in Drosophila [16, 17]. Regulation of NOS expression is directly associated with the immune response in Manduca sexta[18]. An ENF-family cytokine that triggers AMP gene expression elevates NO concentration in Bombyx mori by inducing NOS expression, suggesting a cross-talk mechanism between cytokine and NO signaling in insects [19].
Eicosanoids are a group of oxygenated C20 polyunsaturated fatty acids. Eicosanoid biosynthesis begins with hydrolysis of arachidonic acid (AA) from cellular phospholipids by phospholipase A2 (PLA2). The AA is oxygenated into PGs by cyclooxygenase (or POX in insects), into LTs by lipoxygenase, or into EETs by epoxidase [20]. Eicosanoids mediate cellular and humoral immune responses in insects [7]. Both PGs and LTs mediate the activation of hemocyte-spreading behavior and nodule formation [21]. PGs mobilize the sessile hemocytes in hemopoietic organs and attached to the body wall into the hemolymph circulation [22], and mediate the release of inactive prophenol oxidase from hemocytes (i.e., oenocytoids) into the plasma by activating oenocytoid cell lysis through a specific membrane receptor [23, 24]. Inhibition of eicosanoid biosynthesis by RNA interference of PLA2 suppresses both the IMD and Toll signaling pathways in flies [25, 26]. In mosquitoes, EETs are synthesized in vitro from AA by tissue lysate, and have also been detected in larval and adult tissues [27, 28]. Inhibition of EET degradation by amending blood meals with the urea-based epoxyhydrolase inhibitor 12-(3-adamantan-1-yl-ureido)dodecanoic acid (AUDA) led to the increased hemolymph EET concentrations and the increased expression of genes encoding some, but not all, tested AMPs [29]. These treatments did not influence mosquito fecundity, fertility or longevity, indicating that they specifically inhibited soluble epoxy hydrolase. These findings indicate that all 3 major eicosanoid groups play crucial signaling roles in insect immunity.
Eicosanoid signaling is induced by cytokines, eicosanoids, and biogenic monoamines via a small protease, Rac1, which mediates cross-talk between these signaling pathways [30]. The cross-talk between cytokine and NO signals indicates that NO may influence eicosanoid signaling in insects. Inhibition of PLA2 activity results in the significant immunosuppression of cellular and humoral immune responses [31]. Some entomopathogenic bacteria in the genera Xenorhabdus and Photorhabdus target PLA2 to induce host immunosuppression [32]. These findings led us to pose a hypothesis that NO signaling is an upstream component of eicosanoid signaling in response to immune challenge. Here, we report on the outcomes of experiments designed to test our hypothesis.
Material and Methods
Insect Rearing and Bacterial Culture
S. exigua larvae were reared on an artificial diet [33] under standard laboratory conditions of 25 ± 1°C, a 16L:8D photoperiod, and 60 ± 10% relative humidity. The adults were fed 10% sucrose. Identity of larval instars (L1-L5) was determined by head width [33]. An entomopathogenic bacterium, X. hominickii ANU101 was isolated from the nematode, Steinernema monticolum[34], and its primary form was preserved on 25% glycerol stock at −80°C. The glycerol stock was cultured on Luria-Bertani (LB) broth for 48 h at 28°C with shaking at 180 rpm. A hemocytometer (Neubauer, Marienfeld, Germany) was used to quantify the bacterial cells under a phase contrast microscope (BX41, Olympus, Tokyo, Japan) at ×200 magnification, recorded in cells/mL. Bacterial culture broth was mixed with ethyl acetate to extract bacterial metabolites according to the method of Sadekuzzaman et al. [31].
Chemicals
NOS inhibitor L-NAME (Nω-nitro-L-arginine methyl ester hydrochloride) and its inactive enantiomer D-NAME (Nω-nitro-D-arginine methyl ester hydrochloride), NO donor SNAP (S-nitroso-N-acetyl-DL-penicillamine), and 2 PLA2 inhibitors of dexamethasone (DEX: [11β, 16α]-9-fluoro-11, 17, 21-trihydroxy-16-methylpregna-1, 4-diene-3) and oxindole (C8H9NO) were purchased from Sigma-Aldrich Korea (Seoul, Korea). PLA2 substrate, 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycerol-3-phos- phatidylcholine, was purchased from Molecular Probes (Eugene, OR, USA).
Determination of NO Concentration
The NO concentration was indirectly quantified by measuring its nitrate form using the nitrate/nitrite colorimetric assay kit (Cayman Chemical, Ann Arbor, MI, USA). Heat-killed (95°C for 20 min) X. hominickii (1 × 105 cells/larva) was injected with a microsyringe (Hamilton, Reno, NV, USA) through the abdominal proleg into L5 larval hemocoels. At times post-injection (PI) (see Results), the abdominal proleg was cut and the exuded hemolymph was collected into tubes containing a few granules of phenylthiourea to block melanization. The hemolymph was centrifuged at 3,000 g for 5 min to separate hemocytes and plasma. The fat body was collected into 100 mM phosphate-buffered saline (PBS, pH 7.4). Hemocytes and the fat body were homogenized in PBS by using a homogenizer (Ultra-Turrax T8, Ika Laboratory, Funkentstort, Germany). Hemolymph (20 larvae/sample) and fat body samples (9 larvae/sample) were collected per replication. Each treatment was replicated 3 times with independent biological sample collections. The homogenized tissues were centrifuged at 14,000 g for 20 min, and the supernatants were used to measure nitrate concentrations using the kit. The protein concentrations in hemocyte, plasma, or fat body samples were determined by Bradford assay [35]. To quantify the nitrate concentrations of the samples, a nitrate standard curve was prepared with the final concentrations of 0, 5, 10, 15, 20, 25, 30, and 35 μM in a 200-μL reaction volume. The absorbance at 540 nm was recorded on a microplate reader (SpectraMax® M2, Molecular Devices, Sunnyvale, CA, USA).
Quantitative PCR Analysis of S. exigua NOS
Total RNA was isolated from the indicated developmental stages and larval tissues using Trizol reagent (Life Technologies, Carlsbad, CA, USA). For developmental stages, 300 eggs, 100 larvae at L1-L2, 20 larvae at L3-L4, 1 larva of L5, 1 pupa, and 1 adult were used for each RNA extraction. For the larval tissues, the hemocytes, fat body, midgut, and epidermis were isolated from L5 larvae. cDNA was synthesized with Maxime RT PreMix (Intron Biotechnology, Seoul, Korea) according to the manufacturer's instruction. In each cDNA reaction, 1 μg of RNA was used. PCR was conducted with S. exigua NOS (SeNOS)-specific primers (5′′-GCGAAGGACCTGTACACTATG-3′′ and 5′′-CAAGGCCAAACGAGCTTTATG-3′′) at 94°C for 1 min for denaturation, followed by 35 cycles at 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min.
For quantitative (q)PCR, 20 μL of reaction mixture was run with gene-specific primers (5′′-GAGGGCGATCATAGGAAATACG-3′′ and 5′′- ATTCCGTAAGCTGGCGATAAA-3′′) and SYBR® Green real-time PCR master mix (Toyobo, Osaka, Japan) according to Bustin et al. [36]. qPCR was run with 40 cycles at 95°C for 25 s, 52°C for 45 s, and 72°C for 35 s, using a thermocycler (CFX Connect™ real-time PCR detection system, Bio-Rad, Hercules, CA, USA). A reference gene, RL32, was used as a constitutively expressed gene [37] for quantification of SeNOS mRNA accumulation with gene-specific primers (5′′-ATGCCCAACATTGGTTACGG-3′′ and 5′′-TTCGTTCTCCTGGCTGCGGA-3′′). After the PCR reactions, a melting curve analysis from 60 to 95°C was applied to all of the reactions to ensure the consistency and specificity of the amplified product. The quantification analysis of SeNOS expression followed the comparative CT (ΔΔCT) method of Livak and Schmittgen [38].
RNA Interference Silencing of SeNOS
Double-stranded RNA was synthesized by using MEGAscript RNAi kit (Ambion, Austin, TX, USA) according to the manufacturer's instruction. To generate the template, gene-specific primers were prepared by adding T7 primer (underlined) at the 5′′ end of the forward (TAATACGACTCACTATAGGGAGAGCGAAGGACCTGTACACTATG) and reverse (TAATACGACTCACTATAGGGAGACAAGGCCAAACGAGCTTTATG) primers, respectively. PCR conditions were 95°C for 5 min for denaturation, followed by 35 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. The PCR product (355 bp) was diluted 2× with deionized distilled water, and 1 μL was used for a template and mixed with nucleotide substrate and T7 RNA polymerase in a final volume of 20 μL. After the RNA synthesis was performed at 37°C for 4 h, the double-stranded (ds)RNA mixture was digested with DNase I (4 units/reaction) to degrade the DNA template. The resulting mixture was mixed with binding buffer (5.5 M guanidine hydrochloride, 20 mM Tris-HCl, pH 6.6) and dsRNA was cleaned through a silica column in the kit with washing buffer (20 mM NaCl, 80% ethanol, 2 mM Tris-HCl, pH 7.5). The dsRNA was eluted with 100 μL of elution buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0), and quantified using a spectrophotometer (GeneQuant pro, Amersham Biosciences, Science Park, Singapore).
To evaluate RNA interference (RNAi) efficiency against SeNOS, 1 μL (0.6 μg) of dsRNA was injected into each L5 larva, and the fat body was collected at 0, 24, 48, and 72 h PI. After cDNA preparation as described above, SeNOS mRNA levels were quantified by qPCR. Each treatment was analyzed in 3 biologically independent replicates. For control, dsRNA specific to a viral gene, CpBV-ORF302[39], was prepared as just described.
Nodulation Assay
Hemocyte nodule formation was assessed in L5 larvae using heat-killed X. hominickii as described [31]. Each L5 larva was injected with 1 × 105 bacterial cells at 48 h PI. After 8 h of incubation at 25°C, the larvae were dissected in PBS under a stereomicroscope (Stemi SV11, Zeiss, Jena, Germany). Brown or black nodules were counted on the fat body, gut, and trachea at ×100 magnification. Each treatment used 10 larvae and data are presented as numbers of nodules/larva.
Influence of NOS Inhibitor or NO Donor on PLA2 Activity and NO Concentration
L5 larvae were challenged with bacteria as just described. For the NOS inhibitor assay, each larva was coinjected with 50 μg of L-NAME or D-NAME (negative control) along with the bacteria. The NO donor, SNAP, was injected into L5 larvae at concentrations indicated in the Results section. Each treatment was replicated 3 times, with biologically independent groups of 20 larvae for hemocyte collection and 9 larvae for fat-body collection. At 8 h PI, the hemocytes and fat body were collected as described, and homogenized in PBS using a homogenizer (Ultra-Turrax T8). Following centrifugation at 10,000 g for 10 min, the supernatant was used as the enzyme preparation.
PLA2 activity measurement followed the method of Radvanyi et al. [40]. Briefly, a total reaction volume (150 μL) consisted of 136.5 μL of 50 mM Tris (pH 7.0), 1.5 μL of 10% bovine serum albumin (BSA), 1 μL of CaCl2, 10 μL of enzyme source and 1 μL of pyrene-labeled substrate (10 mM in ethanol). A spectrofluorometer (SpectraMax M2) was used to measure the fluorescence intensity at Ex345 and Em398. The enzyme activity was calculated by changes in fluorescence per minute. The specific enzyme activity was calculated by dividing the fluorescence change by the protein amount in the reaction (data presented as ΔFLU/min/mg). Protein concentration in each enzyme source was determined by Bradford assay using BSA as a standard. Each treatment was replicated with 3 biologically independent enzyme preparations using different larval samples. NO levels were monitored by estimating nitrate concentrations in the samples.
Bioinformatics Analysis
Similarities of SeNOS with other NOSs were analyzed using the online Basic Local Alignment Search Tool program (BLASTX) (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi?PROGRAM = blastx&PAGE_TYPE = BlastSearch&LINK_LOC = blasthome). Prediction of protein domain structure was performed using the NCBI Conserved Domains program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the SMART program (http://smart.embl-heidelberg.de/). MEGA v6.0 was used to construct the phylogenetic tree using the neighbor-joining method.
Statistical Analysis
All assays were analyzed in one-way ANOVA by PROC GLM for continuous variables using an SAS program [41]. Mortality data were subjected to arcsin transformation and used for ANOVA. The means were compared by the least-squared difference (LSD) test at type I error = 0.05.
Results
Bacterial Challenge Upregulates NO Concentration
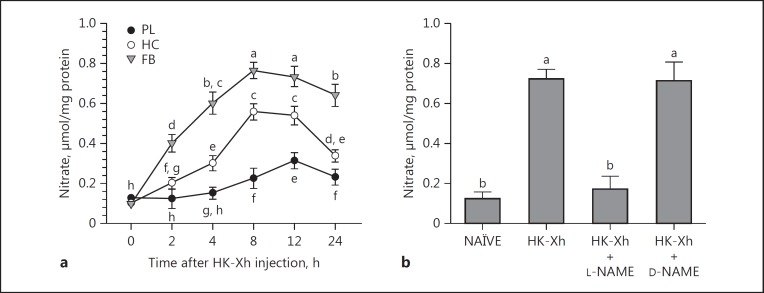
We recorded significant increases in NO (determined as nitrate) concentrations in the fat body and hemocytes and, to a lesser degree, in the plasma, following bacterial challenge, beginning at about 2 h PI (Fig. 1a). These tissues had constitutive NO concentrations of about 0.1 μmol/mg protein, which increased by 5- to 8-fold within 8 h PI in the fat body and hemocytes, and, to a lesser degree, in the plasma. The NO-enhancing effect of the bacterial challenge was reversed by L-NAME-bacterial (but not by D-NAME [inactive enantiomer]-bacterial) coinjections (Fig. 1b).
Fig. 1.
Bacterial challenge leads to increased NO concentrations (measured in nitrate form using the Griess reagent). a Nitrate concentrations in hemocytes (HC), fat body (FB), and plasma (PL). L5 larvae were injected with heat-killed X. hominickii (HK-Xh; 1 × 105 per larva). At indicated times post-infection, tissues were isolated and processed for nitrate amounts. Data points report nitrate concentrations and error bars indicate 1 SD, n = 3 biologically independent replicates using 9 larvae/replicate. Points annotated with the same lower-case letters are not significantly different at type I error = 0.05 (LSD test). b The NOS-specific inhibitor, L-NAME, but not inactive D-NAME, decreased FB NO concentrations at 8 h following the indicated treatments. Histogram bars report mean NO concentrations and error bars show 1 SD, n = 3 biologically independent replicates using 9 larvae/replicate. Bars annotated with the same lower-case letters are not significantly different at type I error = 0.05 (LSD test). NAÏVE, PBS-injected controls.
Identification of SeNOS
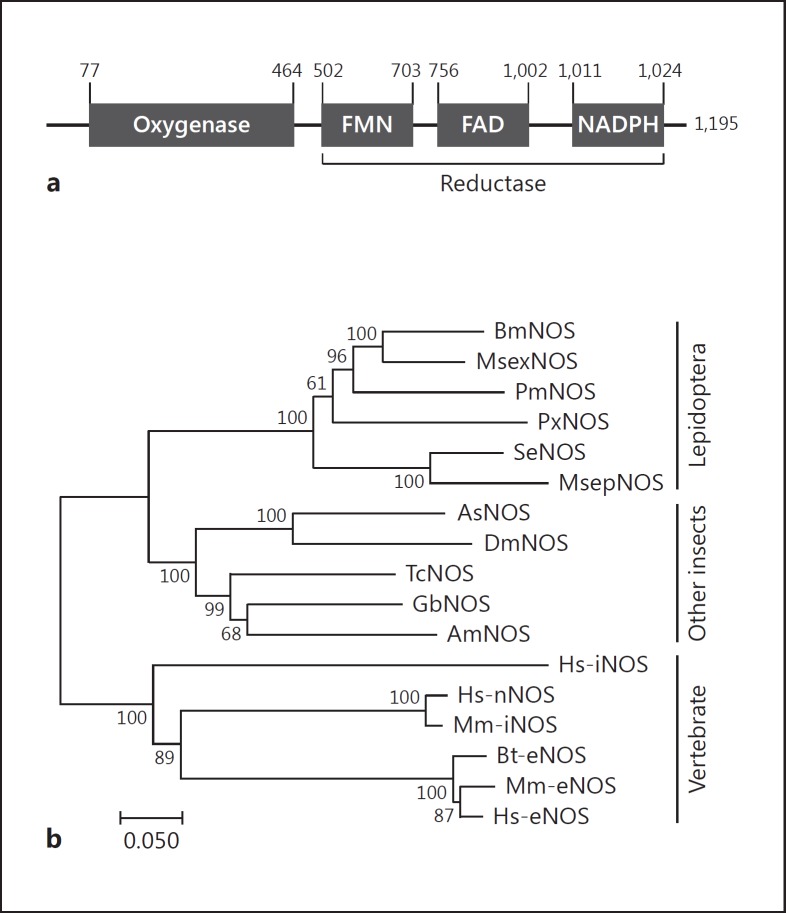
We identified a NOS-like open reading frame (ORF) encoding a 1,195 amino acid protein. Its molecular weight is predicted to be about 136.1 kDa and pI value about 6.6. Domain analysis indicated that SeNOS contained oxygenase and reductase domains containing the cofactor-binding sites, flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and NADPH (Fig. 2a). Phylogenetic analysis with the predicted amino acid sequence indicated that SeNOS was clustered with other lepidopteran NOSs and that all insect NOSs are distinct from their vertebrate counterparts (Fig. 2b).
Fig. 2.
Prediction of S. exigua NOS (SeNOS) candidate gene (GenBank accession No. MF162295). a Domain and binding motifs of SeNOS. Two domains were predicted to be oxygenase and reductase. The reductase domain consists of 3 binding motifs: flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and NADPH. b Phylogenetic analysis of SeNOS with other NOS genes. Phylogenetic tree was constructed by the neighbor-joining method using MEGA v6.0 software. Bootstrap values were obtained after 1,000 repetitions and are denoted on each branch. GenBank accession Nos. are Q8T8C0 for Bombyx mori NOS (BmNOS), O77104 for Manduca sexta (MsexNOS), A0A194QQW5 for Papilio machaon (PmNOS), XP_011558120.1 for Plutella xylostella (PxNOS), H9TN28 for Mythimna separata (MsepNOS), O61608 for Anopheles stephensi (AsNOS), NP_523541.2 for Drosophila melanogaster (DmNOS), D6WZB5 for Tribolium castaneum (TcNOS), B7XH76 for Gryllus bimaculatus (GbNOS), Q5FAN1 for Apis mellifera (AmNOS), AAC19133.1 for Homo sapiens (Hs-iNOS), AAB60654.1 for Homo sapiens (Hs-nNOS), NP_032738.1 for Mus musculus (Mm-nNOS), P29473.3 for Bos taurus (BtNOS), P70313 for Mus musculus (Mm-eNOS), and X76303.1 for Homo sapiens (Hs-eNOS).
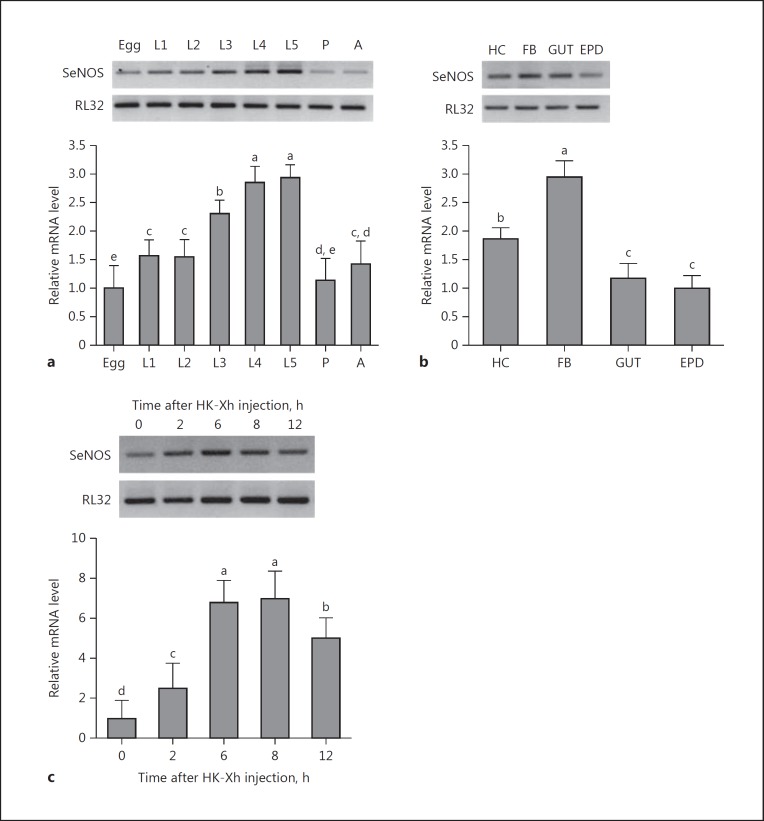
SeNOS was expressed in all developmental stages from egg to adult (Fig. 3a). Late larval instars had higher levels compared to other stages. In L5 larvae, mRNAs encoding SeNOS are present in all tested tissues with the highest accumulation in the fat body (Fig. 3b). SeNOS expression was inducible following bacterial challenge (Fig. 3c). The induction was rapid, appearing within 2 h PI.
Fig. 3.
SeNOS expression profile. a Variation of SeNOS expression in all developmental stages: egg, larval instars (L1-L5), pupa (P), and adult (A). b Variation of SeNOS expression in 4 larval tissues, hemocyte (HC), fat body (FB), midgut (GUT), and epidermis (EPD). c Inducible expression of SeNOS following bacterial injection. Heat-killed X. hominickii (HK-Xh; 1 × 105 bacterial cells per larva) were injected into L5 larva. After the indicated time intervals, FB was collected and processed for measuring mRNA levels. Each treatment had 3 biologically independent replicates. Different letters above the SD bars indicate significant difference among means at type I error = 0.05 (LSD test). A ribosomal gene, RL32, was used as the stably expressed reference gene for RT-PCR (upper panel) and qPCR (lower panel).
SeNOS Influences NO Concentration and Cellular Immunity
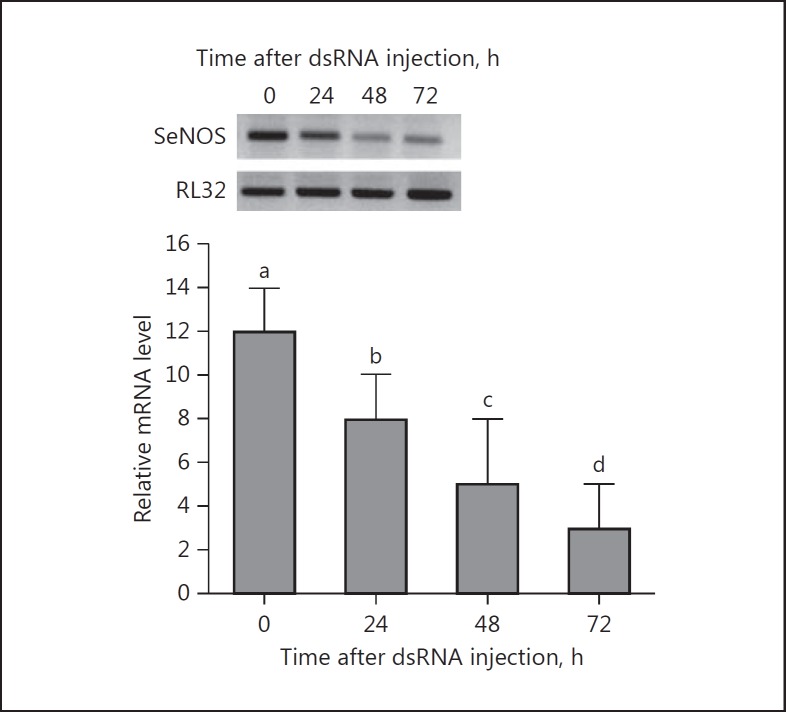
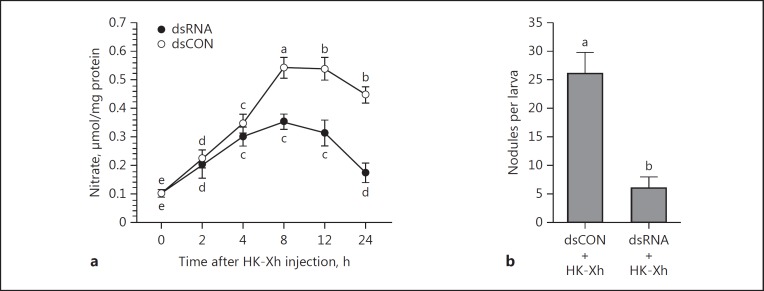
Double-stranded SeNOS injection led to significant, time-related reductions in mRNA encoding SeNOS, with an approximately 4-fold reduction by 72 h PI (Fig. 4). Similar injections significantly suppressed NO concentrations (down by approx. 35% by 6 h PI and approx. 65% by 24 h PI; Fig. 5a). The reduced NO concentrations resulted in a significant reduction in hemocyte nodule formation in response to bacterial challenge (down by approx. 75% at 8 h PI; Fig. 5b).
Fig. 4.
dsSeNOS treatments led to reduced accumulation of mRNA encoding SeNOS. One microliter (2 μg) of dsRNA was injected into each L5 larva. At the indicated times post-injection, fat bodies were collected and processed for mRNA levels. The histogram bars indicate mean relative mRNA accumulation, n = 3 biologically independent determinations. Histogram bars annotated with different lower-case letters are significantly different at type I error = 0.05 (LSD test). A ribosomal gene, RL32, was used as the stably expressed reference gene for RT-PCR (upper panel) and qPCR (lower panel).
Fig. 5.
dsSeNOS treatments led to reductions in NO concentrations and hemocyte nodule formation in S. exigua. a dsSeNOS injections suppressed bacterial-stimulated NO production in L5 fat body. One microliter (2 μg) of dsRNA was injected into each larva. At 48 h post-injection (PI), heat-killed X. hominickii (HK-Xh; 1 × 105 bacterial cells per larva) were injected into the larvae. At the indicated times PI, fat bodies were isolated and processed for nitrate determinations. The data points represent mean NO concentrations and the error bars represent 1 SD. Data points annotated with the same lower-case letters are not significantly different at type I error = 0.05 (LSD test). b dsSeNOS suppressed bacterial-stimulated nodule formation. dsRNA was injected into L5 larvae, and at 48 h PI, HK-Xh (1 × 105 bacterial cells per larva) were injected into the treated larvae. For dsRNA control (dsCON), dsRNA specific to a viral gene, CpBV-ORF302[34], was injected into the larvae (the same amount of dsSeNOS). At 8 h PI, nodules were counted by dissection. Each treatment included 5 biologically independent replicates. Histogram bars represent mean numbers of nodules/larva and the error bars represent 1 SD. Different letters above the SD bars indicate significant difference among means at type I error = 0.05 (LSD test).
Bacterial Challenge Led to NO-Mediated Upregulation of PLA2 Activity
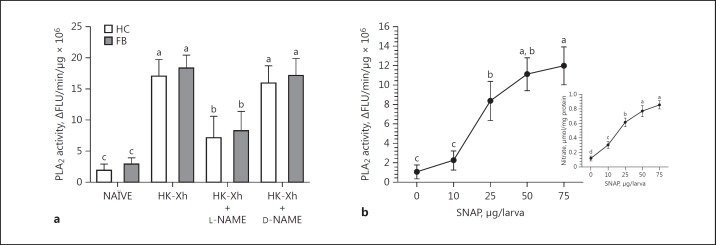
Bacterial challenge led to dose-dependent increases (approx. 6-fold) in PLA2 activities in the hemocytes and fat body (Fig. 6a), which was prevented in larvae coinjected with the NOS-specific inhibitor, L-NAME, and bacteria. The negative control experiment with coinjection with bacteria and the inactive enantiomer, D-NAME, did not prevent PLA2 activity induction. Treating L5 larvae with SNAP (an NO-releaser) injections led to dose-dependent increases in fat-body PLA2 activity (Fig. 6b) and nitrate concentrations by 8 h PI (Fig. 6b, inset).
Fig. 6.
Bacterial challenge and increased internal NO concentrations activate PLA2. a PLA2 activity is increased in bacteria-infected (heat-killed X. hominickii (HK-Xh; 1 × 105 bacterial cells per larva) larvae relative to naïve controls. The increased activity is blocked by cotreatments with the bacteria plus the NOS-specific inhibitor, L-NAME. Similar coinjections with the inactive D-NAME did not influence PLA2 activity. At 8 h post-injection (PI), the fat body was collected to measure PLA2 activity. Each treatment included 3 biologically independent replicates, 9 larvae/replicate. The histogram bars show mean PLA2 activity and the error bars show 1 SD. b Influence of NO increase on PLA2 activity. Each L5 larva was injected with the indicated concentrations of an NO donor, SNAP. At 8 h PI, fat body was collected to measure nitrate concentration (inset) and PLA2 activity. Each treatment included 3 biologically independent replicates. Each point represents mean PLA2 activity. Different letters above the SD bars indicate significant difference among means at type I error = 0.05 (LSD test). NAÏVE, PBS-injected controls.
A Change in PLA2 Activity Did Not Modulate NO Concentration
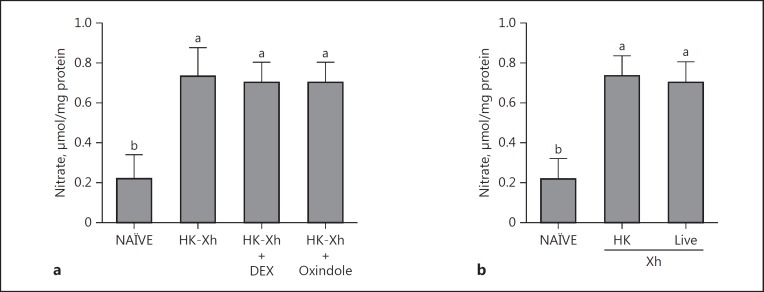
From the data in Figure 6, we inferred that NO activates eicosanoid signaling by activating PLA2. Another possibility is that eicosanoid signaling modulates NO concentrations. We tested this idea by coinjecting L5 larvae with bacterial and 1 of 2 pharmaceutical PLA2 inhibitors, DEX or oxindole. Neither treatment prevented the bacteria-driven elevation of NO concentrations. Another test is based on the report that live X. hominickii inhibits S. exigua PLA2 activity by synthesizing and releasing heat-sensitive PLA2 inhibitor(s) [31]. The heat-killed bacteria lose PLA2 inhibitory activity. Figure 7b shows that injecting heat-killed, and, separately, live X. hominickii led to significantly increased NO concentrations in L5 larvae.
Fig. 7.
Inhibiting PLA2 activity did not influence NO concentrations in S. exigua larvae. a Influence of PLA2-specific inhibitors, dexamethasone (DEX) and oxindole, on NO concentrations. NO concentration was measured in nitrate form using Griess reagent. Heat-killed X. hominickii (HK-Xh; 1 × 105 cells per larva) was injected into L5 larva. For inhibition assay, each S. exigua L5 larva was coinjected with HK-Xh and 10 μg of DEX or oxindole. At 8 h post-injection (PI), fat body was collected to measure nitrate concentrations. b Influence of an entomopathogenic live bacterium, X. hominickii, on NO concentrations. Live X. hominickii (Xh; 1 × 105 cfu per larva) or HK-Xh (1 × 105 bacterial cells per larva) were injected to L5 larva. At 8 h PI, fat body was collected to measure nitrate concentration. Each treatment was replicated 3 biologically independent times, 9 larvae/replicate. Histogram bars show mean NO concentrations. Different letters above the SD bars indicate significant difference among means at type I error = 0.05 (LSD test). NAÏVE, PBS-injected controls.
Discussion
The data reported here potently support our hypothesis that NO signaling is an upstream component of eicosanoid signaling in response to immune challenge. The argument is composed of several germane points. First, bacterial injections led to increases in NO concentrations in the fat body, hemocytes, and plasma, indicating that NO is produced in response to bacterial challenge. The increases were inhibited in larvae coinjected with bacteria and the NOS inhibitor, L-NAME, showing that bacteria-stimulated increases in NO concentrations follow from SeNOS activity. Second, mRNA encoding SeNOS is present in all S. exigua life stages and in 4 tested tissues: the hemocytes, the fat body, the gut, and the epidermis. Third, bacterial challenge led to time-dependent increases in the accumulation of fat body mRNA encoding SeNOS, from which we infer that NO production follows the induction of a specific gene, SeNOS. Fourth, injections of dsSeNOS led to time-dependent reductions in fat body mRNA encoding SeNOS, to reductions in bacteria-stimulated fat-body NO concentrations, and to steep reductions in a well-established immune function parameter, hemocyte nodulation. Fifth, bacterial challenge led to substantial increases in PLA2 activity, which were sharply reduced by about 2-fold in larvae coinjected with the NOS inhibitor, L-NAME, indicating to us that increased PLA2 activity occurs downstream of increased SeNOS activity. This point is reinforced by the demonstration that injecting the NO donor, SNAP, led to increased fat body NO concentrations and increased PLA2 activity. Finally, if PLA2 activity was increased upstream of SeNOS activity, we would expect to see the inhibition of PLA2 activity leading to reductions in tissue NO concentrations, which we directly ruled out. Taken together, these points amount to a persuasive demonstration that NO mediates hemocytic immune reactions via increasing PLA2 activity and eicosanoid biosynthesis.
Relative to identifying an S. exigua gene encoding SeNOS, screening S. exigua transcriptomes predicted a SeNOS construct, which contained a full ORF. The SeNOS amino acid sequence has oxygenase and reductase domains, which requires several cofactors, including NADPH, FMN, and FAD [42]. Our in silico analysis shows that SeNOS has binding sites for these cofactors. Our phylogenetic analysis shows that SeNOS clusters with other lepidopteran NOSs. A single NOS is encoded in each known insect genome. Drosophila encodes single NOS that shares 47% sequence similarity to the mammalian nNOS[43]. Also, a single Anopheles stephensi NOS is highly inducible in response to feeding on malaria parasite-containing blood [15]. We infer that SeNOS is the sole NOS gene in this - and possibly all - insect genomes.
SeNOS was expressed in all developmental stages and larval tissues. Bacterial challenge significantly increased SeNOS expression, indicating that SeNOS is an iNOS, as seen in other lepidopterans, including M. sexta[18] and B. mori[19].
NO mediates various physiological processes in insects [8]. NO, per se, is cytotoxic at high concentrations and exhibits antimicrobial/antiparasitic activities in mammalian immunity [44]. A rapid increase (of 100- to 1,000-fold) in NO concentrations follows the expression of iNOS, which the constitutive eNOSs and nNOSs cannot do [45]. At low concentrations, NO mediates cellular and humoral immune responses [46]. Our indirect measurement indicated an almost 8-fold increase in fat body NO after bacterial injection. We infer from this relatively slight increase that NO acts in mediating immune signals to the hemocytes and fat body. Indeed, we show that the increased NO concentration led to hemocyte nodule formation in response to bacterial challenge, which was blocked by L-NAME treatments. Our interpretation is that NO mediates the cellular immune responses of S. exigua and possibly all insect species. For example, D. melanogaster, a strain resistant to the endoparasitoid wasp, Leptoplina boulardi, produces a significant increase in NO concentrations from differentiated lamellocytes [47], and the elevated NO level mediates hemocyte encapsulation of the parasitoid eggs [16]. NO also acts in humoral immunity by inducing genes encoding AMPs in insects. For example, NO directly induces the expression of genes encoding AMPs in D. melanogaster[17]. The expression of 5 genes encoding AMPs was influenced by L-NAME, from which it was inferred that NO mediates humoral immunity in B. mori [19]. Because eicosanoids mediate humoral immune reactions [21, 48], we propose that NO mediates humoral as well as hemocytic immune responses in S. exigua.
dsSeNOS treatments suppressed the bacteria-stimulated upregulation of NO concentrations, which led to immunosuppression. This is consistent with the idea that NO generally acts in signaling insect immune functions, as seen in mosquitoes [15], an endoparasitoid wasp [16], and bacteria [17]. Foley and O'Farrell [17] and Carton et al. [49] found that the injection of L-NAME into the Drosophila hemocoel prior to infection by gram-negative bacteria decreased the survival of the flies, thereby suggesting that NO is a component of insect immune response signaling. However, NO is a Janus-faced molecule. Excessive NO production may reduce cell and host survival [50]. For example, 1-octen-3-ol, a volatile organic compound secreted from most fungi, is harmful due to generating NO and its subsequent reaction with reactive oxygen species (ROS) to generate toxic peroxynitrile (ONOO−). This leads to a shortened Drosophila life span [51]. In Galleria mellonella, hemocytes produce NO to mediate immune responses, but exhibit differential production of NO between 2 developmental stages, in which sessile hemocytes at the prepupal pharate stage produce more NO than circulating hemocytes at the larval stage [52]. Overproduction of NO at the prepupal stage was cytotoxic in response to bacterial challenge. Thus, the expression of all NOSs, including SeNOS, should be tightly regulated. The slight increase in NO concentration following bacterial challenge may favor S. exigua by minimizing the detrimental hazard of NO-driven toxic ROS. More to the point, the inducible expression of NOS is site-specific to the foci of pathogen infection in M. sexta. Eleftherianos et al. [18] demonstrated this by infecting hornworms with a bacterial pathogen, Photorhabdus luminescens, at different sites. M. sexta expressed NOS only in gut tissue following oral infection and only in the fat body and hemocytes following infection by hemocelic injection. We infer that the situation in insects is similar to the vertebrate CNS [50]; on the one hand, Janus-face NO is neuroprotective at physiological concentrations and, on the other, it is cytotoxic at higher concentrations.
Eicosanoids mediate cellular and humoral immune responses in insects [7]. Eicosanoid immune signals act as a common downstream signal for the cytokine plasmatocyte-spreading peptide (PSP) and 2 biogenic monoamines in S. exigua [4, 53]. We proposed cross-talk mechanisms to account for the coordinated cellular immune responses recorded in S. exigua[53]. Here, we propose an additional element in the intracellular signaling in S. exigua, in which increasing NO concentrations, following bacterial challenge or SNAP treatment, led to enhanced PLA2 activity. The upregulated PLA2 activity would, in turn, drive upregulated eicosanoid biosynthesis. The situation appears to be complex, however, because the inhibition of PLA2 activity with specific inhibitors did not reduce the upregulation of the NO concentration in response to bacterial challenge. Our suggestion is that eicosanoids act as a downstream component of the NO signal pathway. In B. mori, another cytokine, paralytic peptide, acts upstream relative to NO immune signaling [19]. Eicosanoids also mediate nodulation reactions to bacterial infection in B. mori[54], and we propose cross-talk mechanisms between paralytic peptide and eicosanoid signaling in B. mori. Because paralytic peptide acts upstream to NO signaling in B. mori, we propose the presence of influential interactions between NO and eicosanoid signals.
The concept of cross-talk between NO and eicosanoid signaling was initially reported for a mouse macrophage cell line, RAW264.7 [55]. The cells were treated with lipopolysaccharide which induced iNOS and the inducible form of cyclooxygenase (COX-2), and thereby significantly elevated NO and PG levels. We note that gene sequences encoding an insect COX have not been found in the published insect genomes. The COX functions of mammals are carried out by various forms of peroxidase (POX) in insects [37]. The biosynthesis of both NO and PGE2 was blocked by treating cells with 1 of 2 NOS inhibitors, NG-monomethyl-L-arginine or aminoguanidine. The authors also stimulated human fetal fibroblasts with interleukin 1β, and then treated the cells with exogenous NO, either as a gaseous solution or with an NO donor, sodium nitroprusside. This treatment significantly induced COX-2 activity [56, 57]. They concluded that NO interacts directly with COX-2 to simulate the production of proinflammatory PGs; this point has not yet been settled [58], however, because later work showed that NO stimulates COX-2 expression via various transcriptional activators in cancer cells [59, 60]. Our view, based on the work with mammalian cell lines, is that the cross-talk between NO and eicosanoids occurs in multiple cell-specific patterns in mammals. The work on B. mori and S. exigua discussed here indicates that multiple patterns occur in insects and, likely, in most eukaryotic cells in general.
Disclosure Statement
The authors have declared that no conflict of interests exists.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2017R1A2B3009815). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. All programs and services of the US Department of Agriculture are offered on a nondiscriminatory basis without regard to race, color, national origin, religion, sex, age, marital status, or handicap.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Gillespie JP, Trenczek T, Kanost MR. Biological mediators of insect immunity. Annu Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 3.Baines D, Downer RG. Octopamine enhances phagocytosis in cockroach hemocytes: involvement of inositol trisphosphate. Arch Insect Biochem Physiol. 1994;26:249–261. doi: 10.1002/arch.940260402. [DOI] [PubMed] [Google Scholar]
- 4.Kim GS, Nalini M, Lee DW, Kim Y. Octopamine and 5-hydroxytryptamine mediate hemocytic phagocytosis and nodule formation via eicosanoids in the beet armyworm, Spodoptera exigua. Arch Insect Biochem Physiol. 2009;70:162–176. doi: 10.1002/arch.20286. [DOI] [PubMed] [Google Scholar]
- 5.Strand MR, Hayakawa Y, Clark KD. Plasmatocyte spreading peptide (PSP1) and growth blocking peptide (GBP) are multifunctional homologs. J Insect Physiol. 2000;46:817–824. doi: 10.1016/s0022-1910(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 6.Stanley D, Miller J, Tunaz H. Eicosanoid actions in insect immunity. J Innate Immun. 2009;1:282–290. doi: 10.1159/000210371. [DOI] [PubMed] [Google Scholar]
- 7.Stanley DW, Kim Y. Eicosanoid signaling in insects: from discovery to plant protection. Crit Rev Plant Sci. 2014;33:20–63. [Google Scholar]
- 8.Rivero A. Nitric oxide: an antiparasitic molecule of invertebrates. Trends Parasitol. 2006;22:219–225. doi: 10.1016/j.pt.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Colasanti M, Gradoni L, Mattu M, Persichini T, Salvati L, Venturini G, Ascenzi P. Molecular bases for the anti-parasitic effect of NO. Int J Mol Med. 2002;9:131–134. [PubMed] [Google Scholar]
- 10.Müller U. The nitric oxide system in insects. Prog Neurobiol. 1997;51:363–381. doi: 10.1016/s0301-0082(96)00067-6. [DOI] [PubMed] [Google Scholar]
- 11.Trimmer BA, Aprille JR, Dudzinski DM, Lagace CJ, Lewis SM, Michel T, Qazi S, Zayas RM. Nitric oxide and the control of firefly flashing. Science. 2001;292:2486–2488. doi: 10.1126/science.1059833. [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro JMC. Reversible binding of nitric oxide by a salivary heme protein from a blood sucking insect. Science. 1993;260:539–541. doi: 10.1126/science.8386393. [DOI] [PubMed] [Google Scholar]
- 13.Lim J, Gowda DC, Krishnegowda G, Luckhart S. Induction of nitric oxide synthase in Anopheles stephensi by Plasmodium falciparum: mechanism of signaling and the role of parasite glycosylphosphatidylinositols. Infect Immun. 2005;73:2778–2789. doi: 10.1128/IAI.73.5.2778-2789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luckhart S, Vodovotz Y, Cui L, Rosenberg R. The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc Natl Acad Sci USA. 1998;95:5700–5705. doi: 10.1073/pnas.95.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nappi AJ, Vass E, Frey F, Carton Y. Nitric oxide involvement in Drosophila immunity. Nitric Oxide. 2000;4:423–430. doi: 10.1006/niox.2000.0294. [DOI] [PubMed] [Google Scholar]
- 17.Foley E, O'Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 2003;17:115–125. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eleftherianos I, Felföldi G, ffrench-Constant RH, Reynolds SE. Induced nitric oxide synthesis in the gut of Manduca sexta protects against oral infection by the bacterial pathogen Photorhabdus luminescens. Insect Mol Biol. 2009;18:507–516. doi: 10.1111/j.1365-2583.2009.00899.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishii K, Adachi T, Hamamoto H, Oonishi T, Kamimura M, Imamura K, Sekimizu K. Insect cytokine paralytic peptide activates innate immunity via nitric oxide production in the silkworm Bombyx mori. Dev Comp Immunol. 2013;39:147–153. doi: 10.1016/j.dci.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50:S237–S242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrestha S, Kim Y. Various eicosanoids modulate the cellular and humoral immune responses of the beet armyworm, Spodoptera exigua. Biosci Biotech Biochem. 2009;73:2077–2084. doi: 10.1271/bbb.90272. [DOI] [PubMed] [Google Scholar]
- 22.Park J, Kim Y. Change in hemocyte populations of the beet armyworm, Spodoptera exigua, in response to bacterial infection and eicosanoid mediation. Korean J Appl Entomol. 2012;51:349–356. [Google Scholar]
- 23.Shrestha S, Kim Y. Eicosanoids mediate prophenoloxidase release from oenocytoids in the beet armyworm, Spodoptera exigua. Insect Biochem Mol Biol. 2008;38:99–112. doi: 10.1016/j.ibmb.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Shrestha S, Kim Y, Stanley D. PGE2 induces oenocytoid cell lysis via a G protein-coupled receptor in the beet armyworm, Spodoptera exigua. J Insect Physiol. 2011;57:1568–1576. doi: 10.1016/j.jinsphys.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Hyršl P, Dobes P, Wang Z, Hauling T, Wilhelmsson C, Theopold U. Clotting factors and eicosanoids protect against nematode infections. J Innate Immun. 2011;3:65–70. doi: 10.1159/000320634. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Dong X, Zheng W, Zhang H. The PLA2 gene mediates the humoral immune responses in Bactrocera dorsalis (Hendel) Dev Comp Immunol. 2017;67:293–299. doi: 10.1016/j.dci.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Morisseau C, Hammock BD. Expression and characterization of an epoxide hydrolase from Anopheles gambiae with high activity on epoxy fatty acids. Insect Biochem Mol Biol. 2014;54:42–52. doi: 10.1016/j.ibmb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Morisseau C, Yang J, Mamatha DM, Hammock BD. Epoxide hydrolase activities and epoxy fatty acids in the mosquito Culex quinquefasciatus. Insect Biochem Mol Biol. 2015;59:41–49. doi: 10.1016/j.ibmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Morisseau C, Yang J, Lee KS, Kamita SG, Hammock BD. Ingestion of the epoxide hydrolase inhibitor AUDA modulates immune responses of the mosquito, Culex quinquefasciatus during blood feeding. Insect Biochem Mol Biol. 2016;76:62–69. doi: 10.1016/j.ibmb.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J, Stanley D, Kim Y. Rac1 mediates cytokine-stimulated hemocyte spreading via prostaglandin biosynthesis in the beet armyworm, Spodoptera exigua. J Insect Physiol. 2013;59:682–689. doi: 10.1016/j.jinsphys.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Sadekuzzaman M, Park Y, Lee S, Kim K, Jung JK, Kim Y. An entomopathogenic bacterium, Xenorhabdus hominickii ANU101, produces oxindole and suppresses host insect immune response by inhibiting eicosanoid biosynthesis. J Invertebr Pathol. 2017;145:13–22. doi: 10.1016/j.jip.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Ji D, Cho S, Park Y. Two groups of entomopathogenic bacteria, Photorhabdus and Xenorhabdus, share an inhibitory action against phospholipase A2 to induce host immunodepression. J Invertebr Pathol. 2005;89:258–264. doi: 10.1016/j.jip.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Goh HG, Lee SG, Lee BP, Choi KM, Kim JH. Simple mass-rearing of beet armyworm, Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae), on an artificial diet. Korean J Appl Entomol. 1990;29:180–183. [Google Scholar]
- 34.Park Y, Kang S, Sadekuzzaman M, Kim H, Jung JK, Kim Y. Identification and bacterial characteristics of Xenorhabdus hominickii ANU101 from an entomopathogenic nematode, Steinernema monticolum. J Invertebr Pathol. 2017;144:74–87. doi: 10.1016/j.jip.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 36.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:4. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 37.Park J, Stanley D, Kim Y. Roles of peroxinectin in PGE2-mediated cellular immunity in Spodoptera exigua. PLoS One. 2014;9:e105717. doi: 10.1371/journal.pone.0105717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Park B, Kim Y. Transient transcription of a putative RNase-containing BEN domain encoded in Cotesia plutellae bracovirus induces an immunosuppression of the diamondback moth, Plutella xylostella. J Invertebr Pathol. 2010;105:156–163. doi: 10.1016/j.jip.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Radvanyi F, Jordan L, Russo-Marie F, Bon C. A sensitive and continuous fluorometric assay for phospholipase A2 using pyrene-labeled phospholipids in the presence of serum albumin. Anal Biochem. 1989;177:103–109. doi: 10.1016/0003-2697(89)90022-5. [DOI] [PubMed] [Google Scholar]
- 41.SAS Institute. SAS/STAT User's Guide. SAS Institute, Inc., Cary, NC. 1989 [Google Scholar]
- 42.Mayer B. Regulation of nitric oxide synthase and soluble guanylyl cyclase. Cell Biochem Funct. 1994;12:167–177. doi: 10.1002/cbf.290120304. [DOI] [PubMed] [Google Scholar]
- 43.Regulski M, Tully T. Molecular and biochemical characterization of dNOS: a Drosophila Ca2+/calmodulin-dependent nitric oxide synthase. Proc Natl Acad Sci USA. 1995;92:9072–9076. doi: 10.1073/pnas.92.20.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 45.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guzik TJ, Korbut R, Adamek-Guzik T. Nitric oxide and superoxide in inflammation and immune regulation. J Physiol Pharmacol. 2003;54:469–487. [PubMed] [Google Scholar]
- 47.Arefin B, Kucerova L, Krautz R, Kranenburg H, Parvin F, Theopold U. Apoptosis in hemocytes induces a shift in effector mechanisms in the Drosophila immune system and leads to a pro-inflammatory state. PLoS One. 2015;10:e0136593. doi: 10.1371/journal.pone.0136593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C, Dai L, Wang L, Qian C, Wei G, Li J, Zhu B, Liu C. Inhibitors of eicosanoid biosynthesis influencing the transcripts level of sHSP21.4 gene induced by pathogen infections, in Antheraea pernyi. PLoS One. 2015;10:e0121296. doi: 10.1371/journal.pone.0121296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carton Y, Frey F, Nappi AJ. Parasite-induced changes in nitric oxide levels in Drosophila paramelanica. J Parasitol. 2009;95:1134–1141. doi: 10.1645/GE-2091.1. [DOI] [PubMed] [Google Scholar]
- 50.Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 51.Inamdar AA, Bennett JW. A common fungal volatile organic compound induces a nitric oxide mediated inflammatory response in Drosophila melanogaster. Sci Rep. 2014;4:3833. doi: 10.1038/srep03833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnan N, Hyršl P, Šimek V. Nitric oxide production by hemocytes of larva and pharate prepupa of Galleria mellonella in response to bacterial lipopolysaccharide: cytoprotective or cytotoxic? Comp Biochem Physiol C. 2006;142:103–110. doi: 10.1016/j.cbpc.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Srikanth K, Park J, Stanley DW, Kim Y. Plasmatocyte-spreading peptide influences hemocyte behavior via eicosanoids. Arch Insect Biochem Physiol. 2011;78:145–160. doi: 10.1002/arch.20450. [DOI] [PubMed] [Google Scholar]
- 54.Stanley-Samuelson D, Pedibhotla VK, Rana RL, Nor Aliza AR, Hoback WW, Miller JS. Eicosanoids mediate nodulation responses to bacterial infections in larvae of the silkmoth, Bombyx mori. Comp Biochem Physiol. 1997;118A:93–100. [Google Scholar]
- 55.Salvemini D, Misko TP, Masferrer JL, Seibert K, Currie MG, Needleman P. Nitric oxide activates cyclooxygenase enzymes. Proc Natl Acad Sci USA. 1993;90:7240–7244. doi: 10.1073/pnas.90.15.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salvemini D, Seibert K, Masferrer JL, Misko TP, Currie MG, Needleman P. Endogenous nitric oxide enhances prostaglandin production in a model of renal inflammation. J Clin Invest. 1994;93:1940–1947. doi: 10.1172/JCI117185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salvemini D, Masferrer JL. Interactions of nitric oxide with cyclooxygenase: in vitro, ex vivo, and in vivo studies. Methods Enzymol. 1996;269:12–25. doi: 10.1016/s0076-6879(96)69005-3. [DOI] [PubMed] [Google Scholar]
- 58.Kim SF. The role of nitric oxide in prostaglandin biology; update. Nitric Oxide. 2011;25:255–264. doi: 10.1016/j.niox.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Borchert GL, Phang JM. Polyoma enhancer activator 3, an ETS transcription factor, mediates the induction of cyclooxygenase-2 by nitric oxide in colorectal cancer cells. J Biol Chem. 2004;279:18694–18700. doi: 10.1074/jbc.M308136200. [DOI] [PubMed] [Google Scholar]
- 60.Park SW, Sung MW, Heo DS, Inoue H, Shim SH, Kim KH. Nitric oxide upregulates the cyclooxygenase-2 expression through the cAMP-response element in its promoter in several cancer cell lines. Oncogene. 2005;24:6689–6698. doi: 10.1038/sj.onc.1208816. [DOI] [PubMed] [Google Scholar]