Abstract
Signaling by the interleukin-36 receptor (IL-36R) is linked to inflammatory diseases such as psoriasis. However, the regulation of IL-36R signaling is poorly understood. Activation of IL-36R signaling in cultured cells results in an increased polyubiquitination of the receptor subunit, IL-1Rrp2. Treatment with deubiquitinases shows that the receptor subunit of IL-36R, IL-1Rrp2, is primarily polyubiquitinated at the K63 position, which is associated with endocytic trafficking and signal transduction. A minor amount of ubiquitination is at the K48 position that is associated with protein degradation. A focused siRNA screen identified RNF125, an E3 ubiquitin ligase, to ubiquitinate IL-1Rrp2 upon activation of IL-36R signaling while not affecting the activated IL-1 receptor. Knockdown of RNF125 decreases signal transduction by the IL-36R. Overexpression of RNF125 in HEK293T cells activates IL-36R signaling and increases the ubiquitination of IL-1Rrp2 and its subsequent turnover. RNF125 can coimmunoprecipitate with the IL-36R, and it traffics with IL-1Rrp2 from the cell surface to lysosomes. Mutations of Lys568 and Lys569 in the C-terminal tail of IL-1Rrp2 decrease ubiquitination by RNF125 and increase the steady-state levels of IL-1Rrp2. These results demonstrate that RNF125 has multiple regulatory roles in the signaling, trafficking, and turnover of the IL-36R.
Keywords: Ubiquitination, RNF125, Interleukin-36 receptor, Lysosomal trafficking, Protein turnover, Cytokine, Autoimmunity
Introduction
The interleukin (IL)-1 family of receptors has a central role in innate immunity and the inflammatory response for virtually all cells and organs [1]. Dysregulation of signaling by members of the IL-1R family has also been linked to autoinflammatory diseases and degenerative diseases [2]. Perhaps due to the need to carefully regulate signaling, members of the IL-1R family are subject to complex regulations that include receptor antagonists, cytokine decoys, recognition of multiple cytokines, and various signaling inhibitors [1, 2].
Elevated signaling by the IL-36 receptor (IL-36R) has been linked to asthma, inflammatory bowel disease, pustular psoriasis, and plaque psoriasis [3, 4, 5, 6]. Therefore, it is important to better understand the regulation of IL-36R signaling. Many of the requirements for IL-36R signaling are shared with those of the IL-1R [7]. Both the IL-1R and the IL-36R are composed of a heterodimer of the receptor subunit and a receptor accessory protein that are present on the plasma membrane of cells. For the IL-36R, the receptor subunit is named IL-1Rrp2 and the receptor accessor protein is IL-1RAcp. Both proteins contain an extracellular ligand-binding ectodomain, a single pass transmembrane helix, and a Toll-IL-1 (TIR) domain that is located in the cytoplasm. The binding of the IL-36 cytokine by the IL-36R is thought to change the conformation of the receptor and accessory protein subunits to enable their TIR domains to bind adaptor proteins and activate transcription factors to modulate gene expression [8, 9]. Similar to the IL-1R, signal transduction by the IL-36R is coupled to endocytosis and trafficking of the receptor to lysosomes [9].
The covalent attachment of ubiquitins can cause receptors to change cellular locations, modulate protein-protein interactions, and lead to protein degradation [10, 11]. The IL-1R family of proteins are ubiquitinated. The adaptor protein Tollip that affects IL-1R signaling recognizes polyubiquitinated IL-1R, and directs the IL-1R to lysosomes and eventual degradation [12]. Tollip was recently identified to participate in IL-36R signaling and trafficking to lysosomes [7], indicating that the IL-36R will be regulated by ubiquitination. The ubiquitin ligase(s) that act on the IL-36R have not been characterized. In this work, we identified a RING finger ubiquitin ligase, RNF125 (also known as TRAC-1; T-cell RING protein in activation), to ubiquitinate the receptor subunit of the IL-36R, with consequences on IL-36 signaling and degradation.
Materials and Methods
Reagents
The antibody that recognizes the ectodomain of human IL-1Rrp2 was from R&D Systems (catalog No. AF872). Antibodies to recognize human IL-1RAcP (catalog No. PA5-19921) and RNF125 (catalog No. PA5-23155) were from Pierce. Antibodies to Rab11 (catalog No. ab24170) and LAMP1 (catalog No. ab3612) were from Abcam. Antibodies to detect ubiquitin (P4D1; catalog No. sc-8017) and β-actin (catalog No. sc-58679) were from Santa Cruz Biotechnology. The antibody to detect human RIG-I (catalog No. AF4859) was from R&D Systems. The antibody to detect the c-Myc tag was from Santa Cruz Biotechnology (catalog No. 9E10). Secondary antibodies conjugated to Alexa Fluor 488 or 594 were from Life Technologies.
Recombinant human IL-36γ (catalog No. 6835-ILC/CF) and IL-1β (catalog No. 201-LB-005/CF) were from R&D Systems. MG-132 and Bortezomib (Btz) were from Calbiochem (catalog No. 474788) and Santa Cruz Biotechnology (catalog No. sc-217785), respectively. Wortmannin (WTM) was from Invivogen (catalog No. tlrl-wtm).
cDNA Expression Constructs
cDNAs encoding IL-1Rrp2 (NM_003854.2) were cloned into the pcDNA3.1 vector (Invivogen) to generate pIL-1Rrp2. The plasmid that expresses RIG-I was from Ranjith-Kumar et al. [13]. The RNF125 cDNA (OriGene; catalog No. RC204578) was cloned in the PCMV6-entry mammalian vector to generate pRNF125. Mutations in the cDNA encoding IL-1Rrp2 were made by site-directed mutagenesis according to the manufacturer's suggested protocol (Promega). All constructs were sequenced in their entirety to confirm the presence of the mutation and the absence of unintended mutations.
Cells and Culture Media
NCI/ADR-RES (NCI) cells were cultured in RPMI + L-Glu (GIBCO; catalog No. 11875) and 10% fetal bovine serum. BEAS-2B cells were cultured in BEGM media with its supplements (Lonza), as described in Singh et al. [14]. Human dermal fibroblasts (HDF; GIBCO; catalog No. C-013-5C) were grown in Medium 106 (GIBCO; catalog No. M-106-500) with low serum growth supplement (GIBCO; catalog No. S-003-10), gentamicin (10 mg/mL), and amphotericin B (0.25 mg/mL). HEK293T cells were cultured in DMEM high glucose (4 g/L) media (ThermoFisher) with 10% fetal bovine serum. All cells were grown in rat collagen type I-coated flasks (BD Biosciences).
Deubiquitination Assay
The form of ubiquitination of IL-1Rrp2 was analyzed by using the UbiCREST deubiquitinase enzyme kit (BostonBiochem; catalog No. K-400). IL-1Rrp2 was immunoprecipitated from whole cell lysate and treated with the deubiquitinases according to the manufacturer's recommendation. The degree of ubiquitination was determined using Western blots probed to detect ubiquitin.
Western Blot Analysis
Cells were lysed with ice-cold RIPA buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl at pH 7.4, 0.5% Nonidet P-40, and 0.1% SDS) amended with phosphatase inhibitor (Roche; catalog No. 4906837001) and protease inhibitors (Sigma-Aldrich; catalog No. P8340) according to manufacturer's instructions. The lysate was clarified from insoluble materials by centrifugation at 12,000 g for 10 min, suspended in denaturing Laemmli loading buffer [15], and incubated for 5 min at 70°C. Molecules in the lysate were separated by electrophoresis in 4–12% NuPAGE Bis-Tris gel, followed by transfer to PVDF membranes [16]. The Western blot signals were developed with SuperSignal Dura substrate solution (ThermoFisher; catalog No. 34076) and quantified using a ChemiDoc™ XRS+ system and ImageLab software (Bio-Rad).
Screen for Ubiquitin Ligases That Act on IL-1Rrp2
The siRNA library targeting ubiquitin ligases was from Dharmacon Inc. (catalog No. G-005635-025). NCI cells (1.8 × 104 cells/well; 50% confluency) were transfected with 50 nM of either gene-specific siRNAs or a nonspecific control siRNA (catalog No. sc-37007, Santa Cruz Biotechnology) in a 96-well tissue culture plate. Transfection used Lipofectamine RNAiMax following the manufacturer's protocol (Life Technology). The cells were incubated for 48 h prior to addition of the cytokines at 1 ng/mL to activate receptor signaling. Signaling by the receptor was assessed by the amount of IL-6 cytokine secreted into the media using ELISA (Human OptEIATM; BD Biosciences). All ELISA results shown were performed in triplicate and in at least 3 independent experiments.
siRNA Knockdown
HDF (2 × 103 cells/well in 96-well plates) and BEAS-2B cells (2 × 104 cells/well) were transfected at 50% confluency with 50 nM of a mixture of 4 gene-specific siRNAs or a nonspecific control siRNA. The cells were incubated for 48 h after siRNA transfection, and knockdown was confirmed by measuring the abundance of target message using real-time reverse transcription and polymerase chain reaction (RT-PCR). RT used 1 μg of total RNA in a 20-μL reaction mixture that contained 0.5 μM anchored Oligo-dT primers and the M-MuLV reverse transcriptase (NEB; catalog No. M0253S) to produce the cDNAs. PCR was performed with SYBR green to report on the amount of cDNA of interest and gene-specific primers. All RT-PCR data were normalized against the message from GAPDH. The level of target proteins was assessed using Western blots.
Coimmunoprecipitation Assay
Cells grown in 6-well plates to 50% confluence were lysed in RIPA buffer and incubated with primary antibodies followed by incubation with protein A/G Magnetic Beads (ThermoFisher; catalog No. 88803). After 2 washes with Tris-buffered saline (TBS) amended with 0.05% Tween-20 and 0.5 M NaCl, the precipitated materials were solubilized with SDS-PAGE loading buffer for 5 min at 70°C and resolved by electrophoresis on a 4–12% NuPAGE Bis-Tris gel. Western blots were performed as described above.
Luciferase Assay
HEK293T cells were seeded for transfection in CoStar White 96-well plates at 4.4 × 104 cells/mL for transfection. At approximately 70% confluency, the cells were transfected using Lipofectamine 2000 (Invitrogen) with a mixture of plasmids that express the firefly luciferase reporter (pNF-κBLuc; 30 ng; InvivoGen), the Renilla luciferase transfection control (phRL-TK; 5 ng; Promega), and either wild-type or mutant IL-1Rrp2 (1.0 ng). Cells were grown for 24 h prior to the addition of ligands to a final concentration of 1 ng/mL. The cells were assayed 24 h later using the Dual-Glo Luciferase Assay System (Promega). Luminescence was quantified using a plate reader equipped to detect fluorescence (Biotek Inc.).
Flow Cytometry
Immunostaining and flow cytometry experiments were performed as described in Saha et al. [9] and Ranjith-Kumar et al. [17]. NCI cells were treated with fixation buffer (R&D Systems; catalog No. FC004) followed by the permeabilization buffer I (R&D Systems; catalog No. FC005). The cells were stained for 1 h with antibody to detect the IL-1Rrp2 ectodomain. The secondary antibody was a donkey anti-goat immunoglobulin conjugated to Alexa Fluor 488 (Life Technologies). The cells were enumerated using a FACS Calibur flow cytometer (Becton Dickinson), and gated according to forward scatter and side scatter to find viable, single cell events. Background controls were determined using cells stained with only a secondary antibody conjugated to Alexa Fluor® 488. The data were processed using FlowJo software.
Confocal Microscopy
Confocal microscopy used cells grown to 70% confluency on coverslips coated with poly-L-lysine. The cells were treated with the indicated ligand and then fixed with 4% paraformaldehyde for 15 min at room temperature. Cell permeabilization used T buffer composed of phosphate-buffered saline and 0.5% Triton X-100. To reduce nonspecific antibody binding, the cells were incubated for 30 min on ice with 1% normal goat serum in T buffer followed by a 1-h incubation in 2% bovine serum albumin in TBS (TBS-T), pH 7.4, which contains 0.5% Triton X-100. Incubation with the primary antibody was at 4°C in TBS-T amended with 2% bovine serum albumin. The cells were then washed twice with TBS-T and incubated with fluorophore-labeled secondary antibodies for 1 h at room temperature. After 3 additional washes with TBS-T, the coverslips were mounted on glass slides with antifade mounting medium and dried overnight in the dark. Micrographs were acquired with a Nikon A1 scanning confocal microscope with a 60× oil objective. Confocal images were analyzed using the NIS-Elements viewer and ImageJ software. Fluorophore colocalization was quantified using the ImageJ plug-in tool JACoP [18].
Results
IL-36R Signaling Is Associated with Increased Polyubiquitination of IL-1Rrp2
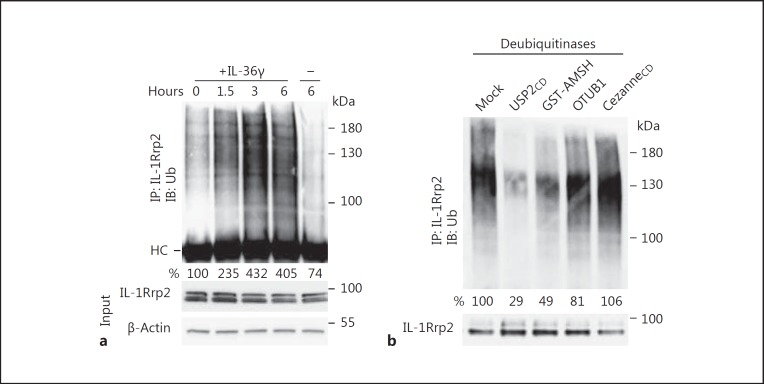
To examine whether IL-36R is ubiquitinated, we immunoprecipitated IL-1Rrp2 from NCI cells and probed the precipitated proteins with an antibody that recognizes ubiquitin. IL-1Rrp2 has a complex pattern of polyubiquitination (Fig. 1a). Furthermore, the amount of polyubiquitination increased over time in cells treated with the IL-36γ cytokine (Fig. 1a). These results confirm that ubiquitination is associated with IL-36R signaling and suggest that activation of receptor signaling is associated with an increase in ubiquitination.
Fig. 1.
Agonist-activated IL-36R is polyubiquitinated. a Polyubiquitination of IL-1Rrp2 after activation of IL-36R by the IL-36γ. NCI cells were not treated or treated with IL-36γ (10 ng/mL) and harvested over time. IL-1Rrp2 polyubiquitination was determined by immunoprecipitation of IL-1Rrp2 and Western blotting probed with an antibody to detect ubiquitin [12]. HC denotes the heavy chain of the immunoglobulin used in the immunoprecipitation assay. b Analysis of the ubiquitination linkages attached to IL-1Rrp2. NCI cells were activated with IL-36γ (10 ng/mL) and incubated for 6 h before the cell lysis with RIPA buffer. IL-1Rrp2 was immunoprecipitated by anti-IL-1Rrp2 antibody to treat with different deubiquitinases. USP2CD removes all ubiquitins, GST-AMSH removes K63-linked ubiquitins, OTUB1 moves K48-linked ubiquitins, and Cezanne removes K6 and K11 ubiquitins. Treated samples were then analyzed by Western blot probed with anti-ubiquitin antibody. The results shown are representative of 3 independent experiments.
The ubiquitin linkage on the target protein is associated with a distinct function [19]. Ubiquitin linked by residue K63 to the target protein can be involved in intracellular trafficking, lysosomal localization, and degradation [20]. Ubiquitin linked by K48 is associated with proteasomal degradation [21]. To identify the form(s) of ubiquitin on IL-1Rrp2, immunoprecipitated IL-1Rrp2 was treated with a series of deubiquitinases that removes linkage-specific ubiquitins. USP-2 that deubiquitinates most ubiquitins was able to largely remove the polyubiquitins on IL-1Rrp2 (Fig. 1b). IL-1Rrp2 polyubiquitination was unaffected by treatment with Cezanne, which removes K6- and K11-linked polyubiquitins (Fig. 1b). Polyubiquitination on IL-1Rrp2 was markedly decreased when treated with AMSH, which remove K63-linked ubiquitins (Fig. 1b). A modest loss of signal was observed with deubiquitinase OTUB1 that removes K48-linked ubiquitins. These results show that activated IL-1Rrp2 had primarily K63 ubiquitination and a minor amount of K48 ubiquitination (Fig. 1b). Given that the different forms of ubiquitin are associated with distinct activities, IL-1Rrp2 ubiquitination is likely to affect signaling, trafficking, and possibly degradation.
RNF125 Participates in IL-36R Signaling
A focused siRNA screen was carried out to identify ubiquitin ligases that act on the IL-36R. The screen was performed in the ovarian tumor cell line, NCI cells, which express endogenous IL-36R and respond to IL-36 cytokines [9]. The cells were transfected with siRNAs to ubiquitin ligases that have been reported to affect inflammatory responses, followed by activation of IL-36R signaling with IL-36γ. The amount of secreted IL-6 detected by ELISA was used as a readout.
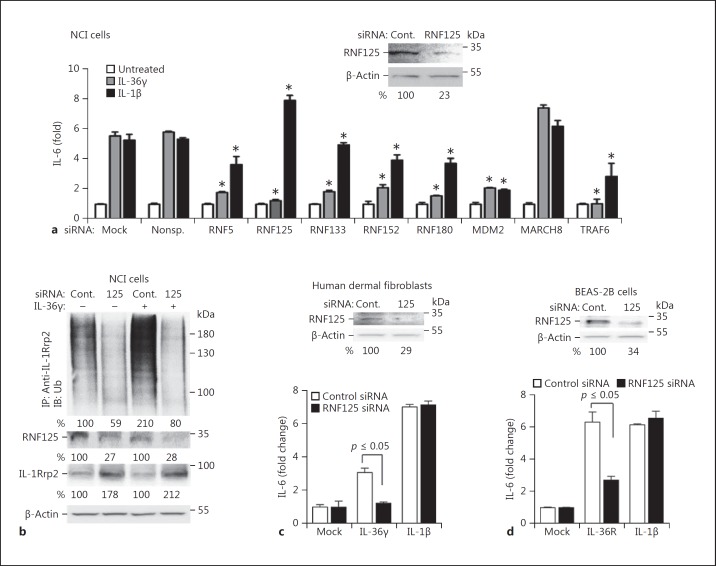
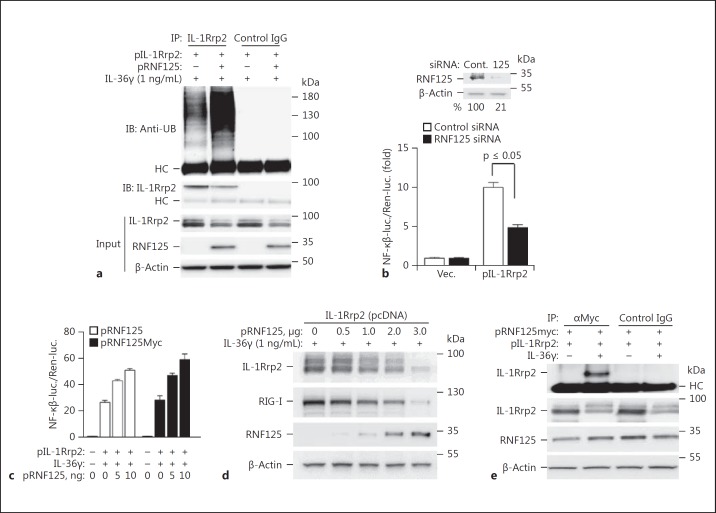
The knockdown of multiple ubiquitin ligases, including RNF5, RNF133, RNF152, and TRAF6, reduced IL-36R signaling. It is likely that some of these ubiquitin ligases affect signal transduction without specifically ubiquitinating the IL-36R. TRAF6 was previously shown to be required for signaling by the IL-1R/TLR superfamily of receptors owing to its adapter function and a role in ubiquitinating the IkB kinase complex (online suppl. Table 1; see www.karger.com/doi/10.1159/000481210 for all online suppl. material) [22]. Interestingly, the knockdown of RNF125 had the most dramatic reduction in IL-36R signaling, but it had only a minimal effect on signaling by the IL-1R (Fig. 2a). RNF125 protein accumulation in these experiments was approximately 25% when compared to that of the cells treated with control siRNAs (Fig. 2a, inset). Knockdown of RNF125 did not affect the proliferation of the cells, but did reduce the abundance of polyubiquitinated IL-1Rrp2 (Fig. 2b; online suppl. Fig. 2). These results demonstrate that RNF125 can ubiquitinate IL-36R and activate its signal transduction, but that it does not activate IL-1R signaling.
Fig. 2.
RNF125 regulates IL-36R signaling. a Sample results of the knockdown of E3 ubiquitin ligases and their effect on IL-36R and IL-1R signaling in NCI cells. Knockdown used 50 nM siRNA specific to E3 ubiquitin ligases or a nonspecific control siRNA. The siRNA specific to RNF125 was a mixture of 4 independently synthesized siRNAs. The cells were incubated for 48 h following transfection of the siRNAs and further treated with IL-36γ (1 ng/mL). Cell media were collected 24 h later for quantification of IL-6 by ELISA. A comparison of the amount of IL-6 produced relative to the samples treated with nonspecific RNA was calculated using the Student t test. * p ≤ 0.05 calculated from 2 experiments with 6 independent samples. Western blots are shown (above) of the accumulation of RNF125 and the β-actin loading controls in NCI cells knocked down with siRNAs specific to RNF125 or with control siRNAs. All Western blot data shown are representative of 3 independent experiments. b RNF125 affects polyubiquitination on IL-1Rrp2 in NCI cells. Immunoprecipitation of IL-1Rrp2 from cell lysate was from cells transfected with RNF125 siRNA. Ubiquitin associated with IL-1Rrp2 was detected using Western blot analysis. The amount of the RNF-125, IL-1Rrp2, and β-actin in the samples used for the analysis of ubiquitinated IL-1Rrp2 were determined by Western blots. c Knockdown of RNF125 results in defective IL-36R signaling in HDF. The mean levels of IL-6 production by IL-36R signaling from 3 independent experiments are shown. Western blots are included (above) of the accumulation of RNF125 and the β-actin loading controls in HDF cells knocked down with siRNAs specific to RNF125 or with control siRNAs. The Western blot data shown are representative of 3 independent experiments. d Effect of RNF125 knockdown on IL-36R signaling in BEAS-2B cells. The results are representative of 3 independent experiments. Western blots are shown (above) of the accumulation of RNF125 and the β-actin loading controls in BEAS-2B cells knocked down with siRNAs specific to RNF125 or with control siRNAs.
Two additional cell lines were examined to determine whether RNF125 plays a role in IL-36R signaling. HDF and human lung epithelial cells (BEAS-2B) both express IL-36R and are functional for IL-36R signaling by the IL-36 cytokines [9]. Knockdown of RNF125 in HDF and in BEAS-2B reduced the level of RNF125 protein to 35% of the level seen in cells treated with control siRNAs (Fig. 2c, d). The knockdown of RNF125 resulted in both cell lines having an approximate 2-fold decrease in IL-36R signaling. Again, signaling by the IL-1R was unaffected in the HDF or in BEAS-2B cells following RNF125 knockdown (Fig. 2c, d). These results confirm that RNF125 has a role in IL-36R signaling.
RNF125 was previously shown to regulate the abundance of the innate immune receptor RIG-I [23]. It also physically interacts with and ubiquitinates the cell cycle checkpoint protein p53, resulting in increased proteasomal degradation of p53 [24, 25]. Li et al. [26] examined various mRNA levels in psoriatic lesional and nonlesional tissues. We examined their data and found that the RNF125 message was decreased in lesional psoriatic tissues by an average of 6.5-fold, while IL-36R and IL-36 cytokine gene expression increased by between 3- and 200-fold (online suppl. Fig. 2). The roles of RNF125 in signaling by the IL-36R, its role in innate immune signaling, stress response, and an inverse relationship between RNF125 and IL-36R expression in lesional psoriatic tissue prompted us to further examine the role of RNF125 in IL-36R signaling.
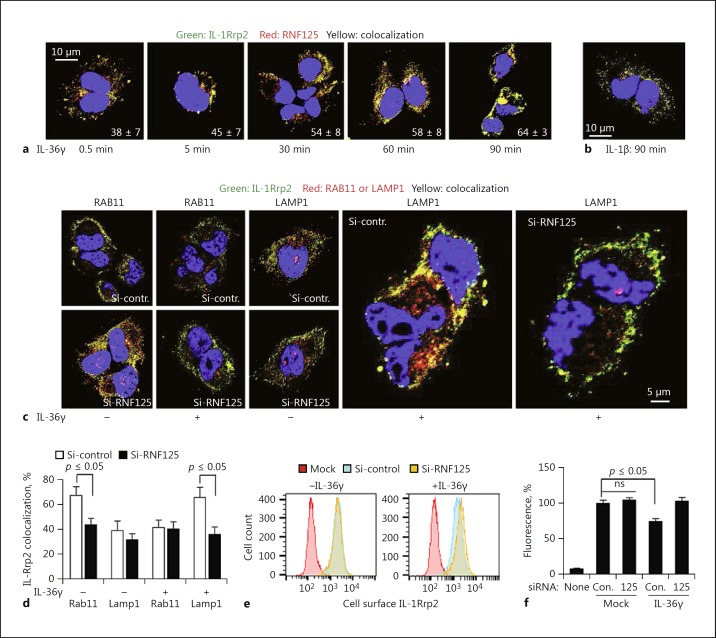
RNF125 Colocalizes with IL-1Rrp2 and Traffics to Lysosomes
The localizations of RNF125 and IL-1Rrp2 were examined in NCI cells that express endogenous proteins. Within minutes of IL-36γ addition, RNF125 colocalized with IL-1Rrp2 (Fig. 3a). The association remained for over a 90-min period when IL-36R trafficked from the plasma membrane, to the cytoplasm, and to the periphery of the nucleus (Fig. 3a). IL-1Rrp2 and RNF125 did not colocalize in cells treated with IL-1β (Fig. 3b), consistent with RNF125 having a specific role in the activation of IL-36R signaling.
Fig. 3.
RNF125 affects IL-36R cellular localization and trafficking to lysosomes. a RNF125 colocalizes with the IL-36R after agonist activation of the receptor. NCI cells were analyzed for colocalization (yellow) of IL-1Rrp2 (green) and RNF125 (red) over a time course following IL-36γ (1 ng/mL) addition to the cell. b IL-36R does not colocalize with RNF125 in the presence of IL-1β in NCI cells. Colocalization (yellow) of IL-1Rrp2 (green) and RNF125 (red) in NCI cells was analyzed following the treatment of IL-1β (1 ng/mL). c NCI cells knocked down for RNF125 show reduced localization of IL-1Rrp2 (green) in LAMP1+ lysosomes (red). Cells transfected with siRNA for 48 h were treated with either IL-36γ (1 ng/mL) or phosphate-buffered saline followed by immune staining and confocal imaging. RAB11 (red) is a marker for recycling endosomes. LAMP1 (red) was used as the lysosomal marker. d Quantification of the colocalization between IL-1Rrp2 and LAMP1 in lysosomes and IL-1Rrp2 and RAB11 in recycling endosomes following RNF125 knockdown. p values were calculated using the Student t test. e Effect of RNF125 knockdown on cell surface expression of IL-36R in NCI cells following IL-36γ (10 ng/mL) treatment by flow cytometry compared to control siRNA-transfected cells. SiRNA transfection to knockdown RNF125 resulted in a higher accumulation of IL-1Rrp2 on the cell surface. f Quantification of the IL-1Rrp2 expression on the cell surface upon RNF125 knockdown. The results were quantified from 3 independent samples that had a minimum of 10,000 cells each. ns, not significant.
Cytokine-activated IL-36R are redirected to traffic to LAMP1+ lysosomes instead of recycling to the plasma membrane [9]. Therefore, we examined whether RNF125 associates with the IL-36R to lysosomes. NCI cells knocked down for RNF125 exhibited reduced IL-1Rrp2 localization to LAMP1+ lysosomes (Fig. 3c, d). There was also a reduction of the IL-1Rrp2 colocalization with Rab11 that marks the recycling endosomes (Fig. 3c, d). In addition, flow cytometry analysis showed that the abundance of the cell surface IL-1Rrp2 decreased by approximately 25% while the intracellular IL-1Rrp2 increased following IL-36γ treatment (Fig. 3e). Cells knocked down for RNF125 had reduced internalization of IL-1Rrp2 upon activation when analyzed by flow cytometry (Fig. 3f). In confocal microscopy, the knockdown of the RNF125 reduced the trafficking of the IL-36R that is normally observed after agonist binding (online suppl. Fig. 3). RNF125 is thus linked to the endocytosis and trafficking of the activated IL-36R to LAMP1+ lysosomes.
Decreased RNF125 Levels Increase IL-1Rrp2 Accumulation
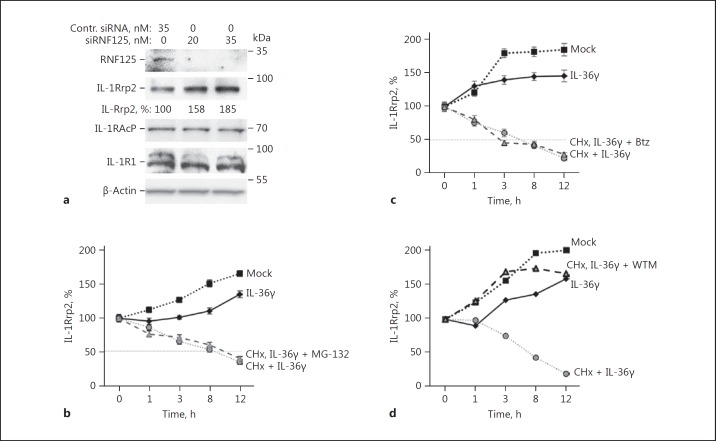
Ubiquitination of RIG-I by RNF125 leads to its degradation [23]. Therefore, we examined whether RNF125 can affect the accumulation of IL-1Rrp2 (Fig. 4a). In these experiments, the knockdown of the RNF125 protein was to approximately 20% of that seen in cells treated with control siRNA. The steady-state level of IL-1Rrp2 protein was increased approximately 2-fold in these experiments, while no obvious effects were observed on the accumulation of IL-1R1 or IL-1RAcp (Fig. 4a). These results suggest that RNF125 has a role in decreasing IL-1Rrp2 accumulation.
Fig. 4.
RNF125 promotes the turnover of IL-1Rrp2. a Knockdown of RNF125 increases the accumulation of IL-1Rrp2 in NCI cells. Knockdown used either 20 or 35 nM of RNF125 siRNA and cells were treated with IL-36γ. The knockdown of RNF125 did not affect the accumulation of the IL-36R accessory protein, IL-1RAcp. The total concentration of siRNA transfected into cells was normalized by the addition of nonspecific control siRNA. Comparable increases in the accumulation of IL-1Rrp2 were observed in 3 independent experiments. b IL-1Rrp2 accumulation in NCI cells was not affected by inhibiting the proteasome. The accumulation of IL-1Rrp2 relative to β-actin was quantified from Western blots following treatment with either IL-36γ (10 ng/mL) alone or in combination with protein synthesis inhibitor cycloheximide (60 μg/mL) and proteasome inhibitor MG-132 (5 μM). Each data point represents the mean and 1 standard error from a sample tested in 3 independent experiments. c The 26S proteasome inhibitor Btz (30 nM final concentration) also did not affect IL-1Rrp2 accumulation after agonist activation of the IL-36R. d Endosome trafficking inhibitor WTM inhibited IL-1Rrp2 turnover. WTM was added to the cells to a final concentration of 100 nM. The results are representative of 2 independent experiments.
Proteins modified with K48 ubiquitin can be subjected to degradation by proteasomes [20, 27]. IL-1 Rrp2 has only a minor amount of K48 ubiquitin, but we wanted to examine whether RNF125 contributed to IL-1Rrp2 turnover by proteasomes. NCI cells activated with IL-36γ that decreased IL-1Rrp2 accumulation were treated with proteasome inhibitors MG-132 or Btz (Fig. 4b, c). The IL-1Rrp2 level in NCI cells increases with cell density after activation by IL-36 [9]. Therefore, the cells were treated with cycloheximide to inhibit the synthesis of new proteins. The addition of MG-132 or Btz did not significantly affected the rate of IL-1Rrp2 turnover (Fig. 4c). These results indicate that the proteasome is not involved in the degradation of IL-1Rrp2 after the cells were treated with the IL-36 cytokine.
Given that the agonist-activated IL-1Rrp2 traffics to lysosomes, it is likely that it is degraded in lysosomes. To examine this, we treated NCI cells activated with IL-36γ with WTM, which inhibits lysosomal trafficking and degradation [28]. We found that the rate of IL-1Rrp2 turnover significantly decreased when compared to cells that were only treated with IL-36γ and cycloheximide (Fig. 4d). We had previously determined that bafilomycin A1 and ammonium chloride that affected the acidification of endosomes did not affect signaling or turnover of the IL-36R [9]. The turnover of agonist-activated IL-1Rrp2 is thus likely to take place in association with lysosomes.
Recombinant RNF125 Ubiquitinates IL-1Rrp2 in HEK293T Cells
We seek to better understand how RNF125 affects IL-36R function in a system where the level and the sequence of both proteins can be manipulated. HEK293T cells lack IL-1Rrp2, but expresses IL-1RAcp. Transient transfection with a plasmid to express IL-1Rrp2 and reconstitute IL-36R signaling in HEK293T cells [29]. Overexpression of RNF125 in HEK293T cells was found to increase the level of polyubiquitinated IL-1Rrp2 only when the cells were induced with IL-36γ, mimicking the property observed in NCI cells that express endogenous IL-36R (Fig. 5a). Furthermore, treatment with deubiquitinases showed that the recombinant IL-1Rrp2 expressed in HEK 293T cells contains mostly ubiquitins linked by the K63 and a smaller proportion were linked by K48 ubiquitins (online suppl. Fig. 4).
Fig. 5.
HEK293T cells reconstituted for IL-36R signalingrespond to RNF125. a RNF125 overexpression in HEK293T cells increased the polyubiquitination of activated IL-1Rrp2. Cells were transfected with plasmids expressing IL-1Rrp2 (1 μg) and RNF125 (1.0 μg) for 36 h, and the degree of IL-1Rrp2 polyubiquitination was determined by immunoprecipitation of IL-1Rrp2 and Western blot to detect ubiquitin. Control IgG was used to determine the degree of nonspecific binding and no polyubiquitinated IL-Rrp2 was observed. All Western blot data shown are representative of 3 independent experiments. b Effect of RNF125 knockdown on IL-36R signaling in HEK293T cells. Cells were transfected with 50 nM of siRNA specific to RNF125 or control siRNAs. Cells were incubated for 48 h following siRNA transfection. All data represent means of 3 independent experiments. p values were calculated using the Student t test. Western blots (above) depict the accumulation of RNF125 and the β-actin loading controls in HEK293T cells knocked down with siRNAs specific to RNF125 or with control siRNAs. c Overexpression of RNF125 or RNF125 with a C-terminal Myc-tag increased IL-36R signaling in HEK293T cells. IL-36R signaling was assessed with the activity of luciferase driven by an NF-κβ promoter element and normalized to the constitutive Renella luciferase expressed from the same cells. All cells were transfected with 1 ng of plasmid expressing IL-1Rrp2 along with an increasing concentration of RNF125-expressing plasmid and incubated for 3 h. The amount of plasmid transfected into cells was normalized using the empty vector plasmids. The cells were treated with IL-36γ (1 ng/mL) for 12 h prior to assessment of the luciferase activity. d RNF125 overexpression decreases IL-1Rrp2 accumulation in HEK293T cells in a concentration-dependent manner. The abundance of RIG-I was assessed to demonstrate that the overexpression had the expected effect, as RNF125 is known to decrease RIG-I abundance. Cells were incubated for 48 h following transfection and the plasmid concentration was normalized by individual empty vector plasmids. e RNF125 forms a coimmunoprecipitable complex with IL-1Rrp2. Coimmunoprecipitation used mAb to the Myc-epitope and control IgG. HC denotes the heavy chain of the IgG used in the immunoprecipitation assay.
HEK293T cells express endogenous RNF125 [23]. Therefore, we used siRNA to knock down RNF125 in HEK293T cells expressing IL-1Rrp2 and examined IL-36R signaling by assessing the level of luciferase expressed from the NF-κβ promoter that could be activated upon IL-36R signaling. The knockdown reduced the RNF125 protein to approximately 20% of the level in cells treated with control siRNA and resulted in a 2-fold reduction in IL-36R signaling (Fig. 5b). Overexpression of increasing amounts of recombinant RNF125 resulted in a dose-dependent increase in IL-36R signaling (Fig. 5c). Interestingly, RNF125 overexpression resulted in a concentration-dependent decrease in the accumulation of the IL-1Rrp2 protein. These results complement those seen in NCI cells where decreased RNF125 levels resulted in an increase in IL-1Rrp2 accumulation (Fig. 5d). Consistent with the report of Arimoto et al. [23], we observed that increased expression of RNF125 decreased the accumulation of RIG-I. These results also demonstrate that HEK293T cells which have been transiently transfected to express recombinant IL-1Rrp2 and/or RNF125 can be used to examine their interaction.
To determine whether RNF125 can interact with IL-1Rrp2, we expressed a version of RNF125 that contains a C-terminal Myc epitope in HEK293T cells. RNF125myc enhanced IL-36R signaling to the same degree as the wild-type RNF125 (Fig. 5c). Immunoprecipitation of RNF125myc resulted in the coprecipitation of IL-1Rrp2. Notably, the coprecipitation of IL-1Rrp2 and RNF125myc occurred only in cells treated with IL-36γ, consistent with the observation from the immunolocalization analysis wherein RNF125 associated with IL-1Rrp2 upon activation of the IL-36R signaling (Fig. 5e). These results suggest that RNF125 binds the agonist-activated IL-36R.
RNF125 Activation of IL-36R Signaling Requires Binding to IL-1Rrp2
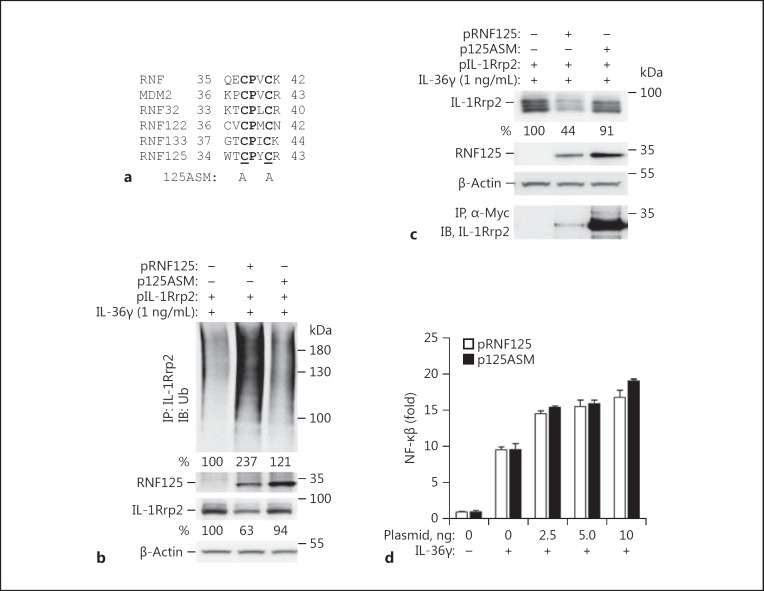
The multiple roles of RNF125 in IL-36R signaling, trafficking, and turnover could be due to either RNF125 ubiquitination of IL-1Rrp2, binding to IL-1Rrp2, or both. To distinguish between these possibilities, we made a version of RNF125 named 125ASM in which highly conserved cysteines in the RNF125 active site were substituted with alanines (Fig. 6a) [30]. Overexpressing 125ASM in HEK293T cells did not increase IL-1Rrp2 polyubiquitination when compared to cells overexpressing wild-type RNF125 (Fig. 6b). In addition, cells overexpressing 125ASM had increased IL-1Rrp2 accumulation when compared to cells overexpressing wild-type RNF125 (Fig. 6b, c). These results suggest that the ubiquitination activity of RNF125 facilitates IL-1Rrp2 degradation. 125ASM did coimmunoprecipitate with IL-1Rrp2, indicating that RNF125 can bind to IL-1Rrp2 without being able to ubiquitinate IL-1Rrp2 (Fig. 6c). Importantly, cells overexpressing 125ASM had comparable signaling to cells overexpressing RNF125 (Fig. 6d). These results suggest that RNF125 had 2 distinguishable activities on IL-1Rrp2. It can bind to IL-36R to activate IL-36R signaling and it can ubiquitinate IL-1Rrp2 to affect the turnover of IL-1Rrp2.
Fig. 6.
IL-36R activation by RNF125 does not require ubiquitination of IL-1Rrp2. a Sequence alignment of the RING finger domain of ubiquitin ligases including RNF125. Bold letters denote conserved residues. b Effect of 125ASM on ubiquitination of IL-1Rrp2 in HEK293T cells. Cells were transfected with plasmids expressing IL-1Rrp2 (1 μg) and RNF125 (1 μg) for 36 h and the degree of IL-1Rrp2 polyubiquitination was determined by immunoprecipitation of IL-1Rrp2 and Western blotting to detect ubiquitin. The abundance of RNF125 was determined by Western blotting probed with antibody specific to RNF125. c Effects of 125ASM on the accumulation of IL-1Rrp2 in HEK293T cells. Coimmunoprecipitation of 125ASM and IL-1Rrp2 used an antibody directed to the Myc tag on the 125ASM and the detection of IL-1Rrp2 in a Western blot probed with antibody specific to IL-1Rrp2. d 125ASM retained the ability to activate IL-36R signaling. HEK293T cells transiently transfected to express the wild-type or 125ASM of RNF125 were assayed for IL-36R signaling upon treatment of IL-36γ for 12 h after 3 h of plasmid transfection using the luciferase reporter assay.
RNF125 Interacts with the C-Terminal Residues of IL-1Rrp2
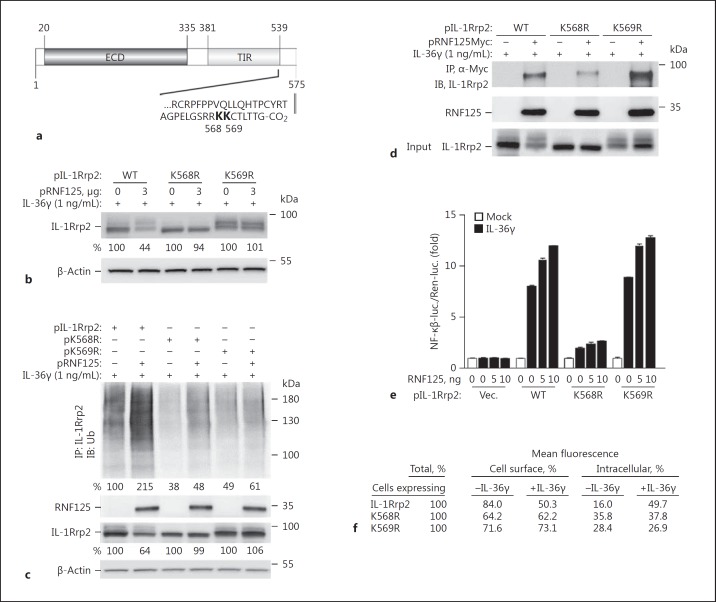
We sought to disable the ubiquitin acceptor site(s) in IL-1Rrp2 to further examine the effects on IL-36R signaling and turnover. Informatics analysis predicts that the 2 most likely ubiquitin-accepting residues in IL-1Rrp2 are in the unstructured tail that is C-terminal to the TIR domain, K568 and K569 (Fig. 7a). Arginine substitution mutants K568R and K569R were constructed and overexpressed in HEK293T cells. The steady-state levels of IL-1Rrp2 in cells expressing either K568R and K569R were increased relative to cells overexpressing wild-type RNF125 (Fig. 7b). In addition, the level of polyubiquitinated IL-1Rrp2 was reduced when compared to cells overexpressing wild-type RNF125 (Fig. 7c). These experiments confirm our previous observation that RNF125-mediated ubiquitination of IL-1Rrp2 led to its decreased accumulation.
Fig. 7.
The C-terminal tail of IL-1Rrp2 mediates RNF125 binding, ubiquitination, and signaling in HEK293T cells. a Location of predicted ubiquitination sites in IL-1Rrp2. The prediction used the IPM algorithm [36]. b Mutants K568R and K569R accumulated to higher levels in HEK293T cells overexpressing RNF125. HEK293T cells were transfected with 1 μg each of plasmids expressing IL-1Rrp2, K568R, or K569R, and 3 μg of pRNF125. c K568R and K569R have decreased polyubiquitination. HEK293T cells were transfected with plasmids expressing IL-1Rrp2 (1 μg) and pRNF125 (0.75 μg) for 36 h. Polyubiquitination was determined by immunoprecipitation of IL-1Rrp2 and Western blots probed with an antibody that recognizes ubiquitin. d IL-1Rrp2 mutants K568R and K569R were also tested for binding to RNF125 upon immunoprecipitation with antibody to the c-Myc sequence followed by Western blots to probe IL-1Rrp2 and RNF125. e IL-36R with the K568R was defective for signal transduction. Signal transduction was performed using the luciferase reporter assay upon treatment of IL-36γ for 12 h after 3 h of plasmid transfection. All data represent the means of 3 independent experiments. WT, wild-type. f Abundance of IL-1Rrp2 and the ubiquitination mutants on the surface of HEK293T cells. Cell surface levels of the proteins were quantified using flow cytometry. The numbers shown are the relative mean fluorescence of the gated cells.
We examined whether K568R and K569R could coimmunoprecipitate with RNF125. K568R was reduced for coimmunoprecipitation with RNF125 while K569R coprecipitated to the same level as wild-type IL-1Rrp2 (Fig. 7d). Since RNF125 binding to IL-1Rrp2 was associated with IL-36R signaling, we examined whether K568R and K569R were affected for signaling. Κ568R, which was reduced in its interaction with RNF125, was also reduced for IL-36R signaling (Fig. 7e). In contrast, K569R, which retained interaction with RNF125, increased its signaling with the RNF125 overexpression. These results demonstrate that the unstructured C-terminal tail of IL-1Rrp2 interacting with RNF125 leads to the activation of signaling. However, ubiquitination of the IL-1Rrp2 C-terminal tail by RNF125 leads to IL-1Rrp2 turnover.
The cellular localizations of mutants K568R and K569R were examined using flow cytometry. The addition of IL-36γ to the cell culture media decreased the cell surface abundance of the wild-type IL-36R, as expected (Fig. 7f). However, both the K568R and the K569R proteins remained on the cell surface even after the addition of IL-36γ (Fig. 7f). Since the K569R mutant was competent for IL-36R signaling (Fig. 7e), these results suggest that signaling by IL-36R does not require endocytosis of the IL-36R. That is, endocytosis and intracellular trafficking of IL-36R to lysosomes can be decoupled from the activation of IL-36R signaling. Altogether, the interaction of RNF125 with IL-1Rrp2 through residues in the IL-1Rrp2 C-terminal tail can affect IL-36R signal transduction, IL-36R trafficking, and IL-1Rrp2 turnover (Fig. 7b-d).
Discussion
In this work, we have demonstrated that the E3 ubiquitin ligase RNF125 contributes to signaling, intracellular trafficking, and turnover of the IL-36R. A reduction in RNF125 expression decreases the signal transduction by the IL-36R (Fig. 2). An increase in IL-36R signaling results from the overexpression of RNF125 (Fig. 5c). RNF125 colocalizes with the IL-1Rrp2 subunit of the IL-36R soon after the agonist activation of the receptor and traffics with IL-1Rrp2 into lysosomes (Fig. 3a, b). Knockdown of RNF125 and a mutation of the residue K568 in the C-terminal tail of IL-1Rrp2 that is recognized by RNF125 resulted in IL-36R remaining at the cell surface (Fig. 7f). Perhaps most interesting is that RNF125 facilitates IL-1Rrp2 turnover by ubiquitinating the receptor (Fig. 5, 6), thus coordinating signaling and the termination of signaling.
The role of RNF125 in both IL-36R signaling and IL-1Rrp2 turnover is likely a mechanism to limit the magnitude of IL-36R signaling to allow the cell to return to the unstimulated state. G-protein-coupled receptors that are activated by agonists also have feedback loops to prevent additional signaling, and these too are regulated by ubiquitination [31, 32]. The Notch signaling pathway that regulates cell proliferation is regulated by proteolytic processing so that the signaling-competent Notch intracellular domain is physically separated from the ligand-binding portion of the Notch receptor to allow only 1 round of signaling [33]. The observation that RNF125 coregulates both signaling and the turnover of the IL-36R subunit suggests that signaling by the IL-36R must be carefully regulated.
Notably, IL-36R turnover does not involve the proteasome (Fig. 4), but is instead associated with IL-1Rrp2 localization to lysosomes. Lysosomes are now recognized to regulate cellular biosynthesis, degradation, and protein exocytosis [34]. Zemoura et al. [20] previously observed that the GABAB receptor undergoes K63-linked ubiquitination followed by internalization and lysosomal degradation. Perhaps IL-1Rrp2 follows the same mechanism after activation by IL-36. The fate of the IL-36R after accumulation in lysosomes remains to be determined.
IL-36R signaling has a number of distinct requirements from those of the IL-1R. While Tollip is required for the proteolytic degradation of the IL-1R [12], Tollip activates signaling and endocytosis of the IL-36R receptor to lysosomes but does not apparently affect IL-36R turnover [9]. RNF125, which plays a role in activating signaling by the IL-36R and also in marking the IL-1Rrp2 for degradation, does not have a demonstrated role in IL-1R signaling or IL-1R accumulation. Both IL-1R and IL-36R play important roles in the inflammatory response. However, the distinguishable requirements for signaling by the 2 receptors suggest that they will have distinct effects on either the level or the timing of the signaling that can impact the resulting inflammatory response. It is possible that the regulation by Tollip and RNF125 will direct different fates for the IL-36R and possibly terminate receptor signaling.
RNF125 likely regulates the molecular crosstalk between the IL-36R and factors that contribute to the inflammatory responses and cell proliferation. RNF125 regulates RIG-I and MDA5, 2 cytoplasmic receptors that can induce interferon production and antipathogen responses when activated by foreign RNAs [23]. RNF125 also negatively regulates p53 function by proteasomal degradation and downmodulate HIV-I replication and inhibits pathogen-induced cytokine production in peripheral mononuclear cells [25, 35]. In psoriatic lesional tissue (online suppl. Fig. 2), RNF125 expression is downregulated while the expression of IL-1Rrp2 and IL-36 are increased. This could affect homeostasis of IL-36R signaling and contribute to the pathologies of psoriasis. RNF125 thus has complex and multifaceted roles in regulating IL-36R signaling and turnover, possibly coordinating the inflammatory response from multiple pathways.
Supplementary Material
Supplementary data
Acknowledgements
We thank D. Singh for making site-directed mutations in IL-1Rrp2. We are grateful to Laura Kao for editing the manuscript and our colleagues at Boehringer-Ingelheim, especially Jay Fine, for support and helpful discussions throughout this work.
References
- 1.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 2.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf J, Ferris LK. Anti-IL-36R antibodies, potentially useful for the treatment of psoriasis: a patent evaluation of WO2013074569. Expert Opin Ther Pat. 2014;24:477–479. doi: 10.1517/13543776.2014.881473. [DOI] [PubMed] [Google Scholar]
- 4.Chustz RT, Nagarkar DR, Poposki JA, Favoreto S, Jr, Avila PC, Schleimer RP, Kato A. Regulation and function of the IL-1 family cytokine IL-1F9 in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2011;45:145–153. doi: 10.1165/rcmb.2010-0075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Towne JE, Sims JE. IL-36 in psoriasis. Curr Opin Pharmacol. 2012;12:486–490. doi: 10.1016/j.coph.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Scheibe K, Backert I, Wirtz S, Hueber A, Schett G, Vieth M, Probst HC, Bopp T, Neurath MF, Neufert C. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2016;66:823–838. doi: 10.1136/gutjnl-2015-310374. [DOI] [PubMed] [Google Scholar]
- 7.Walsh PT, Fallon PG. The emergence of the IL-36 cytokine family as novel targets for inflammatory diseases. Ann NY Acad Sci. 2016 doi: 10.1111/nyas.13280. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 9.Saha SS, Singh D, Raymond EL, Ganesan R, Caviness G, Grimaldi C, Woska JR, Jr, Mennerich D, Brown SE, Mbow ML, Kao CC. Signal transduction and intracellular trafficking by the interleukin 36 receptor. J Biol Chem. 2015;290:23997–24006. doi: 10.1074/jbc.M115.653378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huangfu WC, Fuchs SY. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer. 2010;1:725–734. doi: 10.1177/1947601910382901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 12.Brissoni B, Agostini L, Kropf M, Martinon F, Swoboda V, Lippens S, Everett H, Aebi N, Janssens S, Meylan E, Felberbaum-Corti M, Hirling H, Gruenberg J, Tschopp J, Burns K. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr Biol. 2006;16:2265–2270. doi: 10.1016/j.cub.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 13.Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh D, Vaughan R, Kao CC. LL-37 peptide enhancement of signal transduction by Toll-like receptor 3 is regulated by pH: identification of a peptide antagonist of LL-37. J Biol Chem. 2014;289:27614–27624. doi: 10.1074/jbc.M114.582973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Qi R, Singh D, Kao CC. Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J Biol Chem. 2012;287:32617–32629. doi: 10.1074/jbc.M112.387803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjith-Kumar CT, Miller W, Sun J, Xiong J, Santos J, Yarbrough I, Lamb RJ, Mills J, Duffy KE, Hoose S, Cunningham M, Holzenburg A, Mbow ML, Sarisky RT, Kao CC. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282:17696–17705. doi: 10.1074/jbc.M700209200. [DOI] [PubMed] [Google Scholar]
- 18.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 19.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 20.Zemoura K, Trumpler C, Benke D. Lys-63-linked ubiquitination of γ-aminobutyric acid (GABA), type B1, at multiple sites by the E3 ligase mind bomb-2 targets GABAB receptors to lysosomal degradation. J Biol Chem. 2016;291:21682–21693. doi: 10.1074/jbc.M116.750968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belzile JP, Richard J, Rougeau N, Xiao Y, Cohen EA. HIV-1 Vpr induces the K48-linked polyubiquitination and proteasomal degradation of target cellular proteins to activate ATR and promote G2 arrest. J Virol. 2010;84:3320–3330. doi: 10.1128/JVI.02590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25:1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 23.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Zhou B, Li X, Lu Z, Li W, Huo X, Miao Z. RNF125 is a ubiquitin-protein ligase that promotes p53 degradation. Cell Physiol Biochem. 2015;35:237–245. doi: 10.1159/000369691. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, Voorhees JJ, Kang HM, Nair RP, Abecasis GR, Elder JT. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.d'Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- 28.Martys JL, Wjasow C, Gangi DM, Kielian MC, McGraw TE, Backert JM. Wortmannin-sensitive trafficking pathways in Chinese hamster ovary cells. J Biol Chem. 1996;271:10953–10962. doi: 10.1074/jbc.271.18.10953. [DOI] [PubMed] [Google Scholar]
- 29.Yi G, Ybe JA, Saha SS, Caviness G, Raymond E, Ganesan R, Mbow ML, Kao CC. Structural and functional attributes of the interleukin-36 receptor. J Biol Chem. 2016;291:16597–16609. doi: 10.1074/jbc.M116.723064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, Li CC, Pardo J, Chu PC, Liao CX, Huang J, Dong JG, Zhou X, Huang Q, Huang B, Bennett MK, Molineaux SM, Lu H, Daniel-Issakani S, Payan DG, Masuda ES. A novel E3 ubiquitin ligase TRAC-1 positively regulates T cell activation. J Immunol. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1–24. [PubMed] [Google Scholar]
- 32.Alonso V, Friedman PA. Minireview: ubiquitination-regulated G protein-coupled receptor signaling and trafficking. Mol Endocrinol. 2013;27:558–572. doi: 10.1210/me.2012-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhyay R. Lively lysosomes. ASBMB Today. 2016;5:19–24. [Google Scholar]
- 35.Shoji-Kawata S, Zhong Q, Kameoka M, Iwabu Y, Sapsutthipas S, Luftig RB, Ikuta K. The RING finger ubiquitin ligase RNF125/TRAC-1 down-modulates HIV-1 replication in primary human peripheral blood mononuclear cells. Virology. 2007;368:191–204. doi: 10.1016/j.virol.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data