Abstract
Melanoma differentiation-associated gene 5 (MDA5) is a pattern recognition receptor that recognizes cytoplasmic viral double-stranded RNA (dsRNA) and initiates rapid innate antiviral responses. MDA5 forms a filament-like multimer along the dsRNA leading to oligomerization, which in turn activates the adaptor protein mitochondrial antiviral signaling protein (MAVS) to provide a signal platform for the induction of type I interferon (IFN) and proinflammatory cytokines. The conformational switch of MDA5 causes antiviral defense, but excessive activation of the MDA5-MAVS pathway may result in autoimmune diseases. The regulatory mechanisms of MDA5 activation remain largely unknown. By yeast 2-hybrid, we identified DNAJB1, a member of the HSP40 (heat shock protein 40) family, as an MDA5-binding protein. HSP40s usually cowork with HSP70s. We found that dsRNA stimulation with physiological conditions upregulated the expression levels of DNAJB1 and HSP70; then the proteins were coupled and translocated into the stress granules, where MDA5 encounters dsRNA. DNAJB1 disrupted MDA5 multimer formation, resulting in the suppression of type I IFN induction. The disruption of endogenous DNAJB1 increased MDA5- and MAVS-mediated IFN promoter activation and rendered cells virus resistant. HSP70 inhibitor also enhanced the IFN-inducing function of MDA5 and MAVS. These results suggest that the DNAJB1-HSP70 complex functions for the natural maintenance of RNA sensing by interacting with MDA5/MAVS.
Keywords: Melanoma differentiation-associated gene 5, Type I interferon, Pattern recognition receptors
Introduction
The innate immune mechanism is the first line of defense against viral infections. After viral infection, the pathogen components are recognized by pattern recognition receptors, leading to the induction of rapid innate immune responses [1]. Among them, retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) work as cytoplasmic viral RNA sensors that sense viral double-stranded RNA (dsRNA). RLRs are composed of melanoma differentiation-associated gene 5 (MDA5), RIG-I, and laboratory of genetics and physiology 2 [2, 3]. RIG-I recognizes relatively short dsRNA and 5′-triphosphate RNA, whereas MDA5 recognizes longer dsRNA (>1 kbp) [4, 5, 6]. MDA5 and RIG-I comprise 2 N-terminal caspase activation and recruitment domains (CARDs), a DEAD/H box RNA helicase domain, and a C-terminal domain (CTD). Following dsRNA recognition, MDA5 and RIG-I change conformation and bind the adaptor protein mitochondrial antiviral signaling protein (MAVS [7]; also referred to as IPS-1, VISA, and Cardif) via their CARD domains on the mitochondrial outer membrane. This RLR-MAVS interaction induces MAVS activation, resulting in the production of type I interferon (IFN) and other inflammatory cytokines [2].
MDA5 recognizes measles virus (MV), picornavirus, such as poliovirus, encephalomyocarditis virus [8, 9, 10], and so on. The MDA5 CTD binds viral dsRNA [11], allowing MDA5 proteins to assemble along the dsRNA, leading to MDA5 filament formation [12, 13, 14]. Recent studies have shown that MDA5 is activated by protein phosphatase 1α/γ-mediated dephosphorylation of the MDA5 CARDs in response to viral infection [15]. Thus, preceding phosphorylation of de novo MDA5 prevents excessive activation of MDA5. We found the core MDA5 oligomerization strictly regulated by right open reading frame kinase 3 (RIOK3)-mediated phosphorylation of the CTD of MDA5 [16]. These findings suggest that switching between phosphorylation and dephosphorylation is a key factor in MDA5 functional regulation. The core MDA5 filament is necessary for the oligomerization of the 2 MDA5 CARDs, which promotes MAVS prion-like aggregation [17]. This MAVS multimer formation is also essential for the activation of MAVS and works as a signaling platform for activating downstream molecules of MAVS, for example TNF receptor-associated factor (TRAF) 2 and TRAF6 [18]. Thus, the MDA5-MAVS pathway plays an essential role in antiviral defense, while recent studies have suggested that excess activation of MDA5-MAVS signaling causes various autoimmune responses [19, 20, 21, 22]. Therefore, the molecular mechanisms regulating MDA5-MAVS signaling may be implicated in autoimmune disorders.
Here, we identified DNAJB1 (DnaJ homolog subfamily B member 1), one of the HSP40 (heat shock protein 40) family members, as a negative regulator of the MDA5-MAVS pathway. HSP40s (also referred to as J proteins) work as cofactors of HSP70s that are involved in various biological processes, such as protein refolding, protein interaction, and protein transport, by supporting protein conformational changes in an ATP-dependent manner [23]. DNAJB1 thus may interfere with MDA5 oligomerization by coupling with HSP70. We found here that DNAJB1 first bound MDA5 and formed a complex with HSP70, leading to disaggregation of MDA5. A similar reaction occurred on MAVS but not RIG-I. HSP40s stimulate HSP70 ATPase activity, resulting in protein refolding and the prevention of protein aggregation [24]. Through maintaining protein homeostasis, HSP70 works for stress tolerance, largely resulting in the cooling of inflammation [25, 26]. Our data also revealed that the DNAJB1-HSP70 complex disrupts the formation of MDA5 multimers, resulting in the natural attenuation of IFN induction by MDA5/MAVS.
Materials and Methods
Cells, Viruses, and Reagents
Vero SLAM and HEK293 cells were cultured in DMEM low glucose (Gibco) with 10% heat-inactivated fetal calf serum (FCS). HEK293FT cells were cultured in DMEM high glucose with 10% FCS. HeLa cells were cultured in MEM with 10% FCS and 2 mM of L-Gln. The MV Nagahata strains were amplified in Vero SLAM cells, and viral titers were determined by plaque assay as reported [27]. The yeast AH109 strain was purchased from Clontech. Anti-FLAG M2 monoclonal antibody (mAb), anti-hemagglutinin (HA) polyclonal antibody (pAb), anti-β-actin mAb, and HSP70 inhibitor VER-155008 were purchased from Sigma. Anti-DNAJB1 pAb, anti-HSP70 mAb (3A3), and anti-G3BP mAb were purchased from Abcam. Short polyinosinic:polycytidylic acid (polyI:C), also referred to as polyI:C LMW, was purchased from Invivogen.
Plasmid
cDNA fragments encoding the DNAJB1 partial C-terminal region were isolated by screening of the human lung cDNA library using a yeast 2-hybrid system with a MDA5-C-terminal fragment (731–1,025 aa) as bait. Full-length DNAJB1 cDNA was cloned from total RNA derived from HepG2 cells by RT-PCR, and we sequenced the cDNA fragment and confirmed that there was no PCR error. The obtained DNAJB1 cDNA was inserted into pEF-BOS and fused to HA-tag at the C-terminal end. The DNAJB1 point mutant construct was constructed by PCR-mediated site direct mutagenesis, and we confirmed that there was no PCR error. Full-length MDA5, MDA5-ΔCARD (296–1025 aa), and MDA5-CTD fragment (731–1025 aa) were inserted into pEF-BOS and fused to FLAG-tag at the N-terminal end. Full-length MAVS fragments were inserted into the pEF-BOS and fused to the HA-tag at the C-terminal end, or into the pcDNA3.1 vector and fused to 3× FLAG-tag at the N-terminal end.
Yeast 2-Hybrid System
The AH109 yeast strain was transformed with a plasmid encoding the Gal4 DNA binding domain fused to an MDA5-CTD fragment (BD-MDA5-C) and a plasmid carrying the Gal4 activation domain-fused DNAJB1 (AD-DNAJB1) or the AD alone as a negative control [28]. The transformed cells were plated onto a selective medium (SD-WLHA). The yeast cells transformed with BD-MDA5-C and AD-DNAJB1 but not AD alone grew on the selective plate.
Immunoprecipitation and Western Blotting
Indicated expression plasmids were transfected into HEK293FT cells using Lipofectamine 2000 (Invitrogen). At 24 h posttransfection, cell lysates were prepared with a lysis buffer (20 mM Tris-HCl, pH 7.5, 10% glycerol, 125 mM NaCl, 1 mM EDTA, 1% NP-40, 20 mM IAA, and 2 mM PMSF, 30 mM NaF, and 5 mM Na3VO4). The lysates were precleared with protein G-Sepharose (GE Healthcare) for 1 h at 4°C, and then incubated with anti-FLAG M2 mAb for 2 h at 4°C. Protein G-Sepharose were added to the lysates. Cell lysates were further incubated for 2 h at 4°C. After 4 washes with the above buffer, the precipitated samples were resuspended in a sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 35% glycerol, and BPB). The samples were resolved on SDS-PAGE (7.5 or 10% gel), blotted onto PVDF membranes (Millipore), and labeled with anti-FLAG M2 mAb, anti-HA pAb, or anti-β-actin mAb.
Immunofluorescence Analysis
HeLa cells were cultured on glass coverslips in 12-well plates, and transfected with indicated expression vectors using Lipofectamine 2000. After 24 h, the cells were fixed in PBS containing 3% formaldehyde for 30 min, and then permeabilized with PBS that contains 0.2% Triton X-100 for 15 min. In the case of stimulation with polyI:C, HeLa cells were transfected with polyI:C for 4 h, and then the cells were fixed and permeabilized. Blocking of permeabilized cells was performed with PBS containing 1% of BSA for 30 min. Cells were subsequently incubated with anti-FLAG M2 mAb, anti-HA pAb, anti-DNAJB1 pAb, anti-HSP70 mAb, anti-G3BP mAb, or Mitotracker Red (Invitrogen) in PBS containing 1% BSA overnight at 4°C. After 4 washes, the cells were treated with Alexa 488- or 568-conjugated 2nd antibodies (1/1,000) for 30 min at room temperature. Finally, cover glass was mounted onto slide glass with Prolong Gold for staining the nuclei. The stained cells were observed using ×63 magnification under a LSM510 META microscope (Zeiss, Jena, Germany).
Proximity Ligation Assay
Transfected HeLa cells were fixed and permeabilized. Cells were incubated with anti-HA pAb and anti-FLAG M2 mAb in PBS containing 1% BSA overnight at 4°C. Thereafter, we detected the interaction between DNAJB1 and RLRs or adaptors using a Duolink in situ Proximity Ligation Assay (PLA) kit (Sigma) according to the manufacturer's protocol. In situ PLA probe anti-rabbit MINUS and anti-mouse PLUS bound to primary Abs. After ligation between oligonucleotides with the probes, PL positive (PL+) signals were amplified and visualized by Duolink in situ Detection Reagents Green. PL+ signals were observed by confocal microscopy and analyzed usingy the Duolink Image Tool.
Luciferase Reporter Assay
HEK293 cells cultured in 24-well plates were transfected with the expression vectors with the p125 luc reporter plasmid and a Renilla luciferase vector as an internal control. The p125 luc reporter plasmid encoding the human IFN-β promoter was donated by Dr. T. Taniguchi (University of Tokyo, Tokyo, Japan). Upon polyI:C stimulation, at 24 h posttransfection, cells were transfected with polyI:C for 20 h. The cells were collected and lysed by passive lysis buffer (Promega). Subsequently, luciferase activities were determined by the dual luciferase assay kit (Promega), and were normalized to Renilla luciferase activities to determine the relative luciferase activities.
RNA Interference
Knockdown of DNAJB1 was performed by small interfering RNA (siRNA). Each siRNA (DNAJB1, No. s7008; negative control, No. AM4635) was purchased from Ambion. Cells cultured in 24-well plates were transfected with siRNA (6 pmol) using Lipofectamine iMAX (Invitrogen). Forty-eight hours after transfection, the knockdown efficiency was determined by RT-qPCR using RNA extracted from cells, or by Western blotting using the cell lysate.
Generation of DNAJB1 Knockout Cells
We designed a guide RNA targeting a DNAJB1 exon1 (TAA AGA CTA CTA CCA GAC GTT GG) manually. This guide RNA oligonucleotide was inserted into the pX330-PGK-Puro3F vector expressing CAS9 and Puromycin resistance gene. HEK293 cells were transfected with the pX330-PGK-Puro3F vectors containing a guide RNA sequence for DNAJB1 or mock control, using Lipofectamine 2000. At 48 h posttransfection, we selected the positive cells using 3 μg/mL of puromycin for 48 h, and confirmed DNAJB1 knockout (KO) by Sanger sequencing and Western blotting.
RT-qPCR
Total RNAs were extracted from cells using TRIzol (Invitrogen), and 400 ng of RNA was treated with DNase I, and then rcDNA was prepared by reverse-transcription with random primers using the High Capacity cDNA Transcription Kit (ABI) according to the manufacturer's instructions. We performed qPCR using the Step One Real-Time PCR system (ABI). The RNA expression levels were normalized to β-actin. The primers used for qPCR are shown in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000480740) [16].
Native PAGE
Cell lysates were prepared from approximately 1 × 106 HEK293 FT cells with sample buffer containing Tris-HCl, pH 6.8, 15% glycerol, and BPB. Samples were resolved using Mini-PROTEAN TGX™ Precast Gels (Bio-Rad). Western blotting was then performed with anti-FLAG M2 mAb.
Statistical Analysis
p values were calculated with the Student t test. Error bars represent the SD or SEM between samples.
Results
Identification of DNAJB1 as a Novel MDA5 Binding Factor
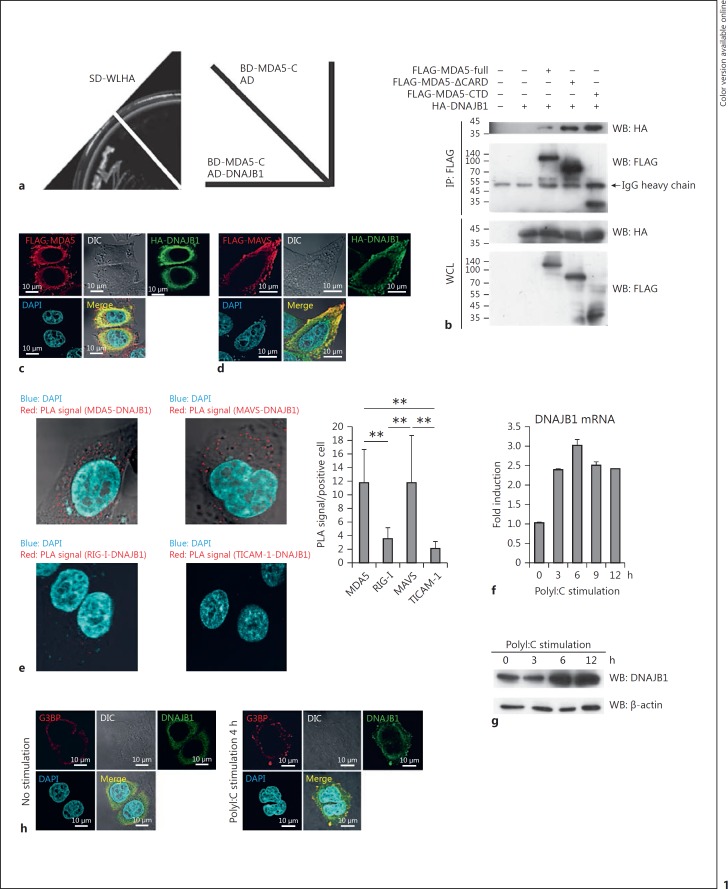
To isolate a novel MDA5 binding factor, we screened a human lung cDNA library, using a yeast 2-hybrid system. Using the MDA5 CTD as bait, we isolated cDNA clones that encoded partial C-terminal regions of DNAJB1 (Fig. 1a; online Fig. S1a). To confirm the interaction of MDA5 and DNAJB1, we performed immunoprecipitation assays. Ectopically expressed HA-tagged DNAJB1 was coimmunoprecipitated with FLAG-tagged full-length MDA5 (Fig. 1b). DNAJB1 merged with MDA5 in the cytoplasm on confocal analysis (Fig. 1c), indicating colocalization. Furthermore, immunofluorescence analysis showed that DNAJB1 also colocalized with MAVS but not RIG-I (Fig. 1d; online Fig. S1b). To confirm the interaction between DNAJB1 and RLRs/MAVS, we performed PLA. PLA signals were detected as high when DNAJB1 was ectopically expressed with MDA5 or MAVS. Under similar conditions in which RIG-I or toll-interleukin 1 receptor domain-containing adaptor molecule (TICAM)-1 was overexpressed, PLA signals were barely detected (Fig. 1e). These data support the finding that DNAJB1 interacts with MDA5 and MAVS, but not RIG-I.
Fig. 1.
DNAJB1 was identified as a novel MDA5 binding factor. a A plasmid carrying Gal4 DNA binding domain fused to an MDA5-C fragment (BD-MDA5-C) was transfected into AH109 yeast cells together with a plasmid carrying Gal4 activation domain-fused DNAJB1 (AD-DNAJB1) or AD only. The transfected cells were plated onto a selective medium (SD-WLHA). The yeast cells with BD-MDA5-C and AD-DNAJB1 but not AD only grew on the selective plate, showing the interaction between MDA5-C and DNAJB1. b FLAG-tagged MDA5-expressing vector was transfected into HEK293FT cells with HA-tagged DNAJB1-expressing vector. Twenty-four hours after transfection, cells were lysed with a lysis buffer. The proteins were immunoprecipitated with anti-FLAG M2 mAb. The immunoprecipitates were analyzed by SDS-PAGE and then immunoblotted using anti-FLAG M2 mAb and anti-HA pAb. c, d Plasmids carrying FLAG-tagged MDA5 or FLAG-tagged MAVS were transfected into HeLa cells together with HA-tagged DNAJB1-expressing plasmid. At 24 h posttransfection, cells were fixed with formaldehyde and permeabilized with Triton X-100. After labeling with anti-FLAG M2 mAb and anti-HA pAb, cells were stained with second antibodies. The stained cells were observed by confocal microscopy. e Plasmids expressing FLAG-tagged MDA5 (upper left panel), MAVS (upper right panel), RIG-I (lower left panel), or TICAM-1 (lower right panel) were transfected into HeLa cells together with HA-tagged DNAJB1- expressing plasmid. Twenty-four hours after plasmid transfection, cells were fixed and permeabilized. After labeling with anti-FLAG M2 mAb and anti-HA pAb, protein interaction was detected using a Duolink in situ PLA kit. PLA signal were observed by confocal microscopy. The numbers of PLA signals in each positive cell (each >170 cells) were counted with the Duolink Image Tool (right panel). ** p < 0.01 (Student t test). f HEK293 cells were transfected with polyI:C, and then total RNA was extracted at the indicated times after transfection. The mRNA expressions of DNAJB1 were determined by RT-qPCR and normalized to β-actin. g HEK293FT cells were transfected with polyI:C, and then cells were harvested and lysed at the indicated times after stimulation. The protein expression levels of DNAJB1 were determined by Western blotting with anti-DNAJB1/HSP40 pAb. h HeLa cells were transfected with polyI:C for 4 h, and cells were subsequently fixed and permeabilized. The cells were labeled with anti-DNAJB1/HSP40 pAb and anti-G3BP mAb, and subsequently stained with second antibodies. The stained cells were observed by confocal microscopy.
PolyI:C stimulation increased the levels of DNAJB1 mRNA (Fig. 1f) and protein (Fig. 1g) in HEK293 cells. Next, we examined the subcellular distribution of DNAJB1 in the presence or absence of polyI:C stimulation. dsRNA was detected in the cytoplasm of HeLa cells 4 h after polyI:C stimulation (online Fig. S1c). Confocal microscopic analysis demonstrated that DNAJB1 was colocalized with a stress granule component Ras-GAP SH3 domain-binding protein (G3BP) after polyI:C stimulation (Fig. 1h). Stress granules represent a site where RLRs interact with dsRNA, and MAVS is needed for their formation. We also confirmed that DNAJB1 was partially translocated to mitochondrial outer membrane in response to polyI:C stimulation (online Fig. S1d). Therefore, our data suggest that DNAJB1 is translocated into the stress granules and mitochondrial membrane in response to dsRNA and interacts with MDA5 and MAVS.
DNAJB1 Negatively Regulates MDA5-MAVS-Mediated IFN-β Signaling
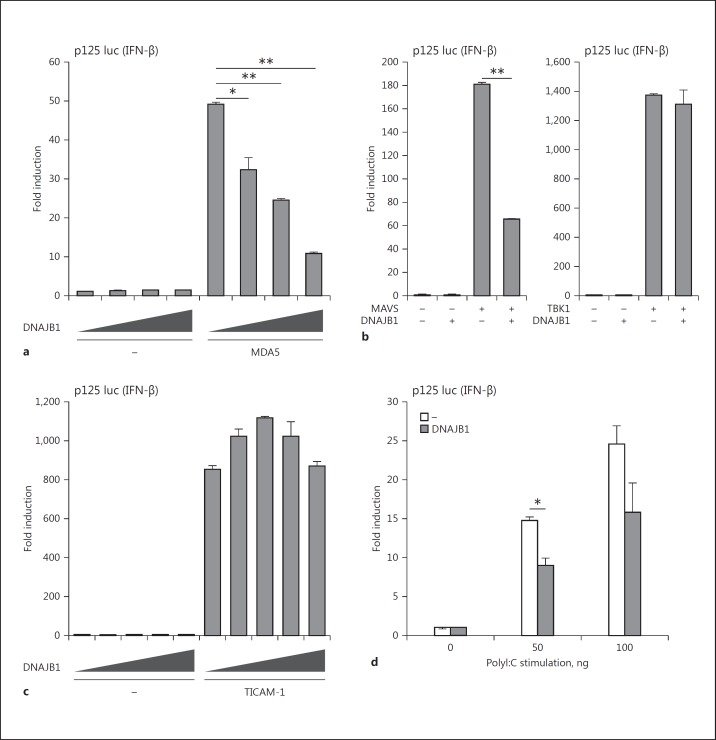
Next, we investigated the effect of DNAJB1 on MDA5-mediated IFN-β induction. DNAJB1 overexpression in HEK293 cells suppressed MDA5-mediated IFN-β promoter activation in a dose-dependent manner (Fig. 2a). Overexpression of DNAJB1 also inhibited IFN induction and NF-κB promoter activation via overexpressed MAVS (Fig. 2b; online Fig. S2a). However, TANK-binding kinase 1 (TBK1)-mediated IFN-β promoter activation was not affected by DNAJB1 overexpression judged by reporter output (Fig. 2b). MAVS is an adaptor molecule for MDA5 as well as RIG-I, and RIG-I CARDs binds MAVS to activate TBK1 and IRF3 [29], suggesting that DNAJB1 inhibits IFN-β induction upstream of TBK1. This is consistent with our observation that DNAJB1 colocalized with not only MDA5 but also MAVS (Fig. 1c, d). Although DNAJB1 failed to interact with RIG-I, DNAJB1 partially suppressed RIG-I-mediated IFN-β promoter activation (online Fig. S2b), which reflects the fact that DNAJB1 has some effect on MAVS function. In contrast, no regulation was observed with DNAJB1 in IFN-β promoter activation mediated by TICAM-1 (Fig. 2c), which is an adaptor for TLR3 [28]. No effect of DNAJB1 on TICAM-1 interaction was confirmed by PLA assay (Fig. 1e). Moreover, overexpressed DNAJB1 inhibited polyI:C-induced IFN-β promoter activation (Fig. 2d). These results suggest that DNAJB1 happens to interact with MDA5 and MAVS, with the MAVS signaling thereby being selectively dampened.
Fig. 2.
DNAJB1 suppresses MDA5-MAVS-mediated IFN-β signaling. a HEK293 cells were transfected with MDA5 vector (100 ng) together with DNAJB1-encoding vector (0, 100, 200, or 400 ng), IFN-β promoter reporter plasmid (p125 luc), and Renilla luciferase vector (internal control). b HEK293 cells were transfected with vectors carrying MAVS (100 ng) or TBK1 (100 ng) together with DNAJB1 (100 ng)-expressing vector, p125 luc, and Renilla luciferase vector. c HEK293 cells were transfected with plasmid carrying TICAM-1 (10 ng) together with DNAJB1 (0, 10, 20, 50, or 100 ng)-expressing plasmid, p125 luc, and Renilla luciferase plasmid. After 24 h posttransfection, luciferase activities were determined and normalized to Renilla luciferase activities. d DNAJB1 (100 ng)-expressing plasmid was transfected into HEK293 cells together with p125 luc and internal control plasmid for 24 h, and then stimulated by polyI:C transfection (0, 50, or 100 ng) for 24 h. Subsequently, luciferase activities were measured. * p < 0.05, ** p < 0.01 (Student t test).
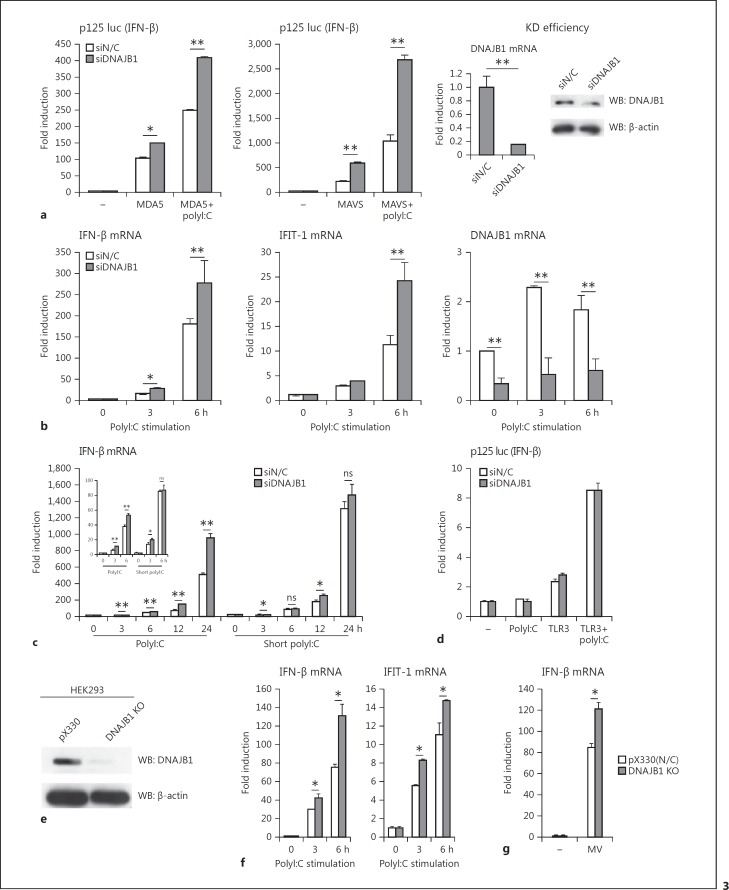
To examine the role of endogenous DNAJB1 in the MDA5-MAVS pathway, we performed knockdown analysis using siRNA. Under overexpression of MDA5 or MAVS, DNAJB1 knockdown increased IFN-β promoter activation, irrespective of polyI:C stimulation (Fig. 3a). Furthermore, DNAJB1 knockdown enhanced polyI:C- induced IFN-β and IFN-induced protein with tetratricopeptide repeats (IFIT)-1 mRNA expression (Fig. 3b). Notably, DNAJB1 knockdown barely enhanced RIG-I-mediated IFN-β induction (Fig. 3c), where we used short polyI:C, a RIG-I-dominant ligand in the cytoplasm [4, 30]. The effect of DNAJB1 knockdown was less prominent in short polyI:C than polyI:C (Fig. 3c), suggesting that preferential suppression for IFN-β production occurs in MDA5 rather than RIG-I through DNAJB1. Knockdown of DNAJB1 also did not affect Toll-like receptor 3 (TLR3)-mediated IFN-β promoter activation in response to polyI:C (Fig. 3d). Thus, DNAJB1 mainly modulates the function of MDA5 protein. To further confirm the data, we generated DNAJB1 KO cells using CRISPR-Cas9 system, wherein endogenous DNAJB1 harbored a frame-shift mutation. In DNAJB1 KO HEK293 cells, the endogenous DNAJB1 protein was not detected by Western blotting (Fig. 3e). Predictably, DNAJB1 KO increased polyI:C-induced IFN-β and IFIT-1 mRNA expression (Fig. 3f). Moreover, to investigate the virus-induced innate immune response in DNAJB1 KO cells, we used the MV Nagahata strain which is preferentially recognized by MDA5, leading to the induction of type I IFN [31]. DNAJB1 KO enhanced MV Nagahata-induced IFN-β mRNA expression (Fig. 3g). These knockdown data are reciprocal to the overexpression results, indicating that endogenous DNAJB1 negatively regulates the function of the MDA5 and MAVS axis upon dsRNA stimulation.
Fig. 3.
Knockdown of DNAJB1 enhances MDA5- and MAVS-mediated IFN-β induction. a HEK293 cells were transfected with plasmids expressing MDA5 (left panel) or MAVS (middle panel), and siRNA for negative control or DNAJB1, together with p125 luc and Renilla luciferase vector. Twenty-four hours after transfection, cells were transfected with polyI:C (0 or 100 ng) for 24 h, and then luciferase activities were measured. Knockdown efficiency of DNAJB1 was determined 48 h after siRNA transfection by RT-qPCR at the mRNA level. The knockdown efficiency of DNAJB1 at the protein levels was assessed by immunoblotting with anti-DNAJB1 pAb and anti-β-actin mAb (right panel). b Knockdown assay was performed with siRNA for negative control or DNAJB1 siRNAs were transfected into HEK293 cells for 48 h. Cells were stimulated by polyI:C transfection. Total RNA was extracted at the indicated times after stimulation. IFN-β, IFIT-1, and DNAJB1 mRNA expressions were analyzed by RT-qPCR and normalized to β-actin. c Negative control or DNAJB1 siRNA were transfected into HEK293 cells for 48 h. Cells were subsequently transfected with polyI:C or short polyI:C. Total RNA was extracted at indicated times after ligand stimulation. IFN-β mRNA expression was determined by RT-qPCR and normalized to β-actin. d HEK293 cells were transfected with siRNA for negative control or DNAJB1. At 24 h posttransfection, TLR3 (500 ng)-expressing vector was transfected to cells together with p125 luc and Renilla luciferase vector. Twenty-four hours after plasmid transfection, the culture medium was removed and polyI:C (0 or 10 μg) was added with fresh medium. The luciferase activities were determined 6 h after stimulation. e Cell lysate was prepared from control (pX330) and DNAJB1 KO HEK293 cells, and then immunoblotted with anti-DNAJB1 pAb and anti-β-actin mAb. f Control (pX330) and DNAJB1 KO HEK293 cells were transfected with polyI:C and harvested at the indicated times after transfection. IFN-β and IFIT-1 mRNA expressions were determined by RT-qPCR and normalized to β-actin. g Control (pX330) and DNAJB1 KO HEK293 cells were infected with MV Nagahata strain for 18 h at MOI = 0.1. IFN-β mRNA expression was determined by RT-qPCR and normalized to β-actin. ns, not significant. * p < 0.05, ** p < 0.01 (Student t test).
DNAJB1 Inhibits MDA5 Multimer Formation
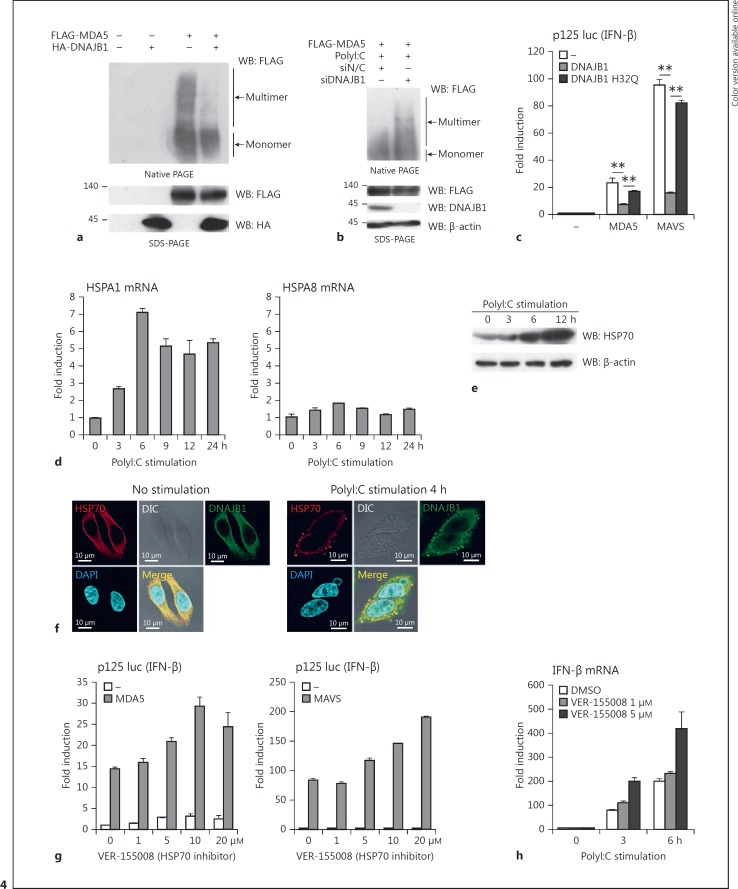
MDA5 multimer formation is essential for its activation. To reveal the molecular mechanism for DNAJB1-mediated inhibition of the MDA5 function, we investigated whether DNAJB1 affected MDA5 multimer formation by native PAGE. Ectopically expressed DNAJB1 severely reduced the multimerization of FLAG-MDA5, whereas the protein expression levels of MDA5 were unaffected by DNAJB1 coexpression (Fig. 4a). Conversely, DNAJB1 knockdown increased FLAG-MDA5 multimer formation upon polyI:C stimulation (Fig. 4b). Therefore, these biochemical data indicate that DNAJB1 interferes with MDA5 oligomerization. Members of the HSP40 family, including DNAJB1, couple with HSP70 protein and increase its ATPase activity to regulate multiple functions of HSP70, including protein refolding, protein import, protein-protein interaction, and prevention of protein aggregation [24, 32]. Therefore, to investigate whether the DNAJB1-HSP70 complex regulates the MDA5/MAVS-mediated type I IFN induction, we used the DNAJB1 H32Q mutant as a control that is unable to interact with HSP70 [33, 34]. Ectopic expression of the H32Q mutant failed to reduce MDA5- and MAVS-mediated IFN-β promoter activation (Fig. 4c). These data suggest that DNAJB1-HSP70 interaction is required for regulation of MDA5 multimer formation and MDA5-MAVS function.
Fig. 4.
DNAJB1 and HSP70 form a negative feedback mechanism for the MDA5-MAVS pathway. a FLAG-tagged MDA5-expressing plasmid was transfected into HEK293FT cells together with HA-tagged DNAJB1-expressing plasmid for 24 h. Cell lysates were subjected to native PAGE or SDS-PAGE, and then immunoblotted using anti-FLAG M2 mAb and anti-HA pAb. b Negative control and DNAJB1 siRNAs were transfected into HEK293FT cells for 48 h, and then MDA5 (400 ng)-expressing vector was transfected. At 24 h after transfection, cells were stimulated by polyI:C (400 ng) transfection for 12 h. Cells were lysed, and the cell lysates were subjected to native PAGE or SDS-PAGE. The proteins were detected by Western blotting using anti-FLAG M2 mAb, anti-DNAJB1 pAb, and anti-β-actin mAb. c Plasmids carrying MDA5 or MAVS, and plasmids carrying DNAJB1 WT or DNAJB1 H32Q, were transfected into HEK293 cells together with p125 luc and Renilla luciferase vector for 24 h, and then the luciferase activities were determined. ** p < 0.01 (Student t test). d HEK293 cells were transfected with polyI:C. Total RNA was extracted at the indicated times after stimulation. HSPA1 and HSPA8 mRNA expression were determined by RT-qPCR and normalized to β-actin. e HEK293FT cells were transfected with polyI:C, and then cells were harvested and lysed at the indicated times after stimulation. The protein expression levels of total HSP70 were determined by Western blotting with anti-HSP70 pAb. f HeLa cells were transfected with polyI:C for 0 h (left panel) or 4 h (right panel). The cells were fixed and permeabilized. The fixed cells were labeled with anti-DNAJB1 pAb and anti-HSP70 mAb, and subsequently stained with second antibodies. The stained cells were observed by confocal microscopy. g HEK293 cells were pretreated with the HSP70 inhibitor VER-155008 (0, 1, 5, 10, or 20 μM) for 24 h. The pretreated cells were transfected with plasmids expressing MDA5 (left panel) or MAVS (right panel), together with p125 luc and Renilla luciferase vector. Twenty-four hours after plasmid transfection, the luciferase activities were determined. h HEK293 cells were pretreated with VER-155008 (0, 1, or 5 μM) for 24 h. The pretreated cells were transfected with polyI:C (100 ng) for the indicated times. IFN-β mRNA expression was determined by RT-qPCR and normalized to β-actin.
HSP70 Is Induced by dsRNA Stimulation and Inhibits the MDA5-MAVS Pathway
The HSP70 family contains stress-inducible members (HSPA1) and constitutively expressed members (HSPA8) [23]. We found that polyI:C stimulation induced HSPA1 mRNA but not HSPA8 mRNA (Fig. 4d). The total protein expression levels of HSP70 were also increased in response to polyI:C (Fig. 4e). Although lipofection itself induced transient HSP induction, the cells stimulated with dsRNA maintained high expression levels of HSPs for over 6 h (online Fig. S3). These data reflect the fact that DNAJB1 and HSP70 are both induced by polyI:C stimulation. A confocal microscopic analysis demonstrated that HSP70 and DNAJB1 exhibited diffuse cytoplasmic colocalization in the absence of stimulation (Fig. 4f, left panel). In response to polyI:C, however, HSP70 and DNAJB1 accumulated in cytoplasmic granules (Fig. 4f, right panel). Protein levels of DNAJB1 were increased in response to polyI:C (Fig. 1g). Because DNAJB1 localized to the G3BP-positive stress granules after polyI:C stimulation (Fig. 1h), HSP70 and DNAJB1 are recruited to the stress granules secondary to polyI:C stimulation.
Next, we investigated the role of HSP70 in the MDA5-MAVS function using the HSP70 chemical inhibitor VER-155008 [35]. In the presence of overexpressed MDA5 or MAVS, VER-155008 enhanced IFN-β promoter activation in a dose-dependent manner (Fig. 4g). Moreover, the pretreatment of cells with VER-155008 increased polyI:C-induced IFN-β mRNA expression (Fig. 4h). Collectively, these data indicate that HSP70 interferes with the MDA5-MAVS function.
Discussion
Our study indicates that DNAJB1 (an HSP40 family member) is a novel MDA5-binding protein. In the cytosol, DNAJB1 interacts with MDA5 but not RIG-I. In response to dsRNA stimulation, DNAJB1 translocates to stress granules, where RLRs encounter viral dsRNA. Furthermore, DNAJB1 partially accumulates in mitochondria membrane after dsRNA stimulation. DNAJB1 also colocalizes with MAVS, which is required to form dsRNA-induced stress granules in the cytoplasm. Thus, DNAJB1 may selectively affect the function of MDA5 and MAVS in stress granules. Ectopic expression of DNAJB1 inhibits MDA5- and MAVS-mediated IFN-β induction, whereas DNAJB1 knockdown or knockout enhances MDA5- and MAVS-mediated IFN-β production. DNAJB1 does not affect RIG-I- or TLR3-mediated IFN-β induction. The physiological relevance of this protein is shown in MV-infected cells, where DNAJB1 disruption renders the cells resistant to viruses by inducing high levels of type I IFNs. Thus, DNAJB1 functionally serves as a natural regulator of the MDA5-MAVS pathway.
Following the recognition of dsRNA, MDA5 proteins form protein-protein contacts to assemble along dsRNA in a head to tail arrangement, leading to MDA5 filament formation. Previously, we have shown that the core MDA5 multimer formation is regulated by RIOK3-mediated phosphorylation of the MDA5 CTD. The core MDA5 oligomer is essential for the multimerization of the MDA5 CARDs, which promotes MAVS prion-like aggregation. Here, our data indicate that DNAJB1 abolishes MDA5 multimer formation.
HSP40s usually couple with HSP70s, suggesting that the disruption of MDA5 oligomerization is attributable to the complex of DNAJB1-HSP70. We show evidence that the DNAJB1-HSP70 complex negatively affects MAVS aggregation because the H32Q mutation of DNAJB1 (unable to interact with HSP70) lost the inhibitory activity of DNAJB1 against MAVS-mediated IFN-β induction.
Many HSPs are induced by various types of cell stress, for example, heat shock, radiation, oxidation, and infection. Our data indicate that dsRNA stimulation induces DNAJB1 (Fig. 1f, g) and HSPA1 (an HSP70) expression (Fig. 4d). Although HSPs were transiently induced by sensing the lipofection reagent, the cells transfected with polyI:C maintained expression levels of HSPs greater than cells treated with Lipofectamine 2000 reagent alone (online Fig. S3a, b). Thus, dsRNA stimulation or its byproducts enhance the expression levels of these HSPs. In response to dsRNA stimulation, stress granules are formed in a MAVS- and protein kinase R (PKR)-dependent manner [36]. Moreover, MDA5 translocates to stress granules. Immunofluorescence analysis demonstrates that DNAJB1 and HSP70 congregate in stress granules in response to polyI:C. Furthermore, DNAJB1 partially translocates to mitochondrial outer membrane after polyI:C stimulation. These data suggest that DNAJB1 and HSP70 are likely to exist in close proximity to MDA5 and MAVS upon dsRNA stimulation. MAVS aggregates like the prion protein [18], and a recent report suggests that HSPs block the self-aggregation of prion proteins [37]. It is likely that HSPs may control the aggregation of MDA5 and/or MAVS in response to cellular stress even in natural conditions, similar to that for the disaggregation of prion. However, we have not obtained direct evidence that DNAJB1 merges with endogenous MDA5 or MAVS and disrupts interaction between MDA5 and MAVS. The mechanistic analysis for MDA5 functional regulation by DNAJB1 remains to be settled.
A recent report has shown that the dsRNA sensor PKR works downstream of MDA5 and upstream of MAVS, leading to MAVS-dependent type I IFN induction [38]. Our data suggest that DNAJB1 is involved in the MDA5-MAVS axis, but not RIG-I or TICAM-1. PKR does not interact with RIG-I but is involved in MDA5-MAVS signaling [38]. Furthermore, dsRNA-mediated PKR signaling is dependent on MAVS, but not TICAM-1 [39]. DNAJB1 interacts with P58IPK (also called DNAJC3), which regulates the activity of PKR [40], and the dissociation of P58IPK from DNAJB1 inhibits the PKR phosphorylation. Thus, we speculate that free DNAJB1 per se may affect MDA5-PKR-MAVS signaling. Further studies are required to unravel the regulation mechanisms of this protein cascade.
In conclusion, our studies identified the DNAJB1-HSP70 complex as a novel negative regulator of the MDA5-MAVS axis. Excessive inflammation has an adverse impact on our body. Aberrant activation of MDA5-MAVS signaling triggers an SLE-like autoimmune response [19]. The involvement of the DNAJB1-HSP70 complex in homeostasis of the MDA5-MAVS pathway may contribute to the avoidance of excessive inflammation, and help us search for strategies controlling immunological disorders.
Disclosure Statement
The authors declare no financial or commercial conflicts of interest.
Author Contributions
K.T. performed the experiments. K.T., H.O., M.M., and T.S. designed the experiments and analyzed the data. H.O., M.M., and T.S. supervised the research. K.T., M.M., and T.S. wrote the manuscript.
Supplementary Material
Supplementary data
Acknowledgements
We thank N. Inoue (Osaka Medical Center for Cancer and Cardiovascular Diseases, Osaka, Japan) and T. Taniguchi (University of Tokyo, Tokyo, Japan) for providing the pX330-PGK-Puro3F vector and p125 luc reporter plasmid, respectively. We also wish to express gratitude to our laboratory members, particularly Dr. K. Funami, for many helpful discussions.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 4.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 9.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe Y, et al. The toll-like receptor 3-mediated antiviral response is important for protection against poliovirus infection in poliovirus receptor transgenic mice. J Virol. 2012;86:185–194. doi: 10.1128/JVI.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahasi K, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peisley A, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 15.Wies E, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takashima K, Oshiumi H, Takaki H, Matsumoto M, Seya T. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11:192–200. doi: 10.1016/j.celrep.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Wu B, Hur S. How RIG-I like receptors activate MAVS. Curr Opin Virol. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funabiki M, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lincez PJ, Shanina I, Horwitz MS. Reduced expression of the MDA5 gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 22.Shao WH, et al. Prion-like MAVS aggregation in lupus patients associates with increased interferon-I. Arthritis Rheumatol. 2016;68:2697–2707. doi: 10.1002/art.39733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, Shibazaki A, Ono R, Kaisho T. HSP70 mediates degradation of the p65 subunit of nuclear factor κB to inhibit inflammatory signaling. Sci Signal. 2014;7:ra119. doi: 10.1126/scisignal.2005533. [DOI] [PubMed] [Google Scholar]
- 26.Borges TJ, et al. The anti-inflammatory mechanisms of Hsp70. Front Immun. 2012;3:95. doi: 10.3389/fimmu.2012.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shingai M, et al. Wild-type measles virus infection in human CD46/CD150-transgenic mice: CD11c-positive dendritic cells establish systemic viral infection. J Immunol. 2005;175:3252–3261. doi: 10.4049/jimmunol.175.5.3252. [DOI] [PubMed] [Google Scholar]
- 28.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 29.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 30.Lang X, et al. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med. 2017;214:459–473. doi: 10.1084/jem.20160592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shingai M, et al. Differential type I IFN-inducing abilities of wild-type versus vaccine strains of measles virus. J Immunol. 2007;179:6123–6133. doi: 10.4049/jimmunol.179.9.6123. [DOI] [PubMed] [Google Scholar]
- 32.Hageman J, van Waarde MAWH, Zylicz A, Walerych D, Kampinga HH. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J. 2011;435:127–142. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- 33.Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michels AA, Kanon B, Bensaude O, Kampinga HH. Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J Biol Chem. 1999;274:36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- 35.Schlecht R, et al. Functional analysis of Hsp70 inhibitors. PLoS One. 2013;8:e78443. doi: 10.1371/journal.pone.0078443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, et al. IPS-1 plays an essential role in dsRNA-induced stress granule formation by interacting with PKR and promoting its activation. J Cell Sci. 2014;127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- 37.Reidy M, Sharma R, Roberts B-L, Masison DC. Human J-protein DnaJB6b cures a subset of Saccharomyces cerevisiae prions and selectively blocks assembly of structurally related amyloids. J Biol Chem. 2016;291:4035–4047. doi: 10.1074/jbc.M115.700393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham AM, et al. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog. 2016;12:e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P, Samuel CE. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J Biol Chem. 2008;283:34580–34587. doi: 10.1074/jbc.M807029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melville MW, Hansen WJ, Freeman BC, Welch WJ, Katze MG. The molecular chaperone hsp40 regulates the activity of P58IPK, the cellular inhibitor of PKR. Proc Natl Acad Sci. 1997;94:97–102. doi: 10.1073/pnas.94.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data