Abstract
Different liver cell types are endowed with immunological properties, including cell-intrinsic innate immune functions that are important to initially control pathogen infections. However, a full landscape of expression and functionality of the innate immune signaling pathways in the major human liver cells is still missing. In order to comparatively characterize these pathways, we purified primary human hepatocytes, hepatic stellate cells, liver sinusoidal endothelial cells (LSEC), and Kupffer cells (KC) from human liver resections. We assessed mRNA and protein expression level of the major innate immune sensors, as well as checkpoint-inhibitor ligands in the purified cells, and found Toll-like receptors (TLR), RIG-I-like receptors, as well as several DNA cytosolic sensors to be expressed in the liver microenvironment. Amongst the cells tested, KC were shown to be most broadly active upon stimulation with PRR ligands emphasizing their predominant role in innate immune sensing the liver microenvironment. By KC immortalization, we generated a cell line that retained higher innate immune functionality as compared to THP1 cells, which are routinely used to study monocyte/macrophages functions. Our findings and the establishment of the KC line will help to understand immune mechanisms behind antiviral effects of TLR agonists or checkpoint inhibitors, which are in current preclinical or clinical development.
Keywords: Checkpoint inhibitors, Hepatic stellate cell, Host defense, Kupffer cell, Liver sinusoidal endothelial cells, Macrophages, Pathogen-associated molecular patterns, Pattern recognition receptors, Primary cells, Primary human hepatocytes
Introduction
Infection by microorganisms leads to the activation of the host immune response through a sensing mediated by innate pattern recognition receptors (PRRs). PRRs include Toll-like receptors (TLRs), C-type lectin receptors, RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), and DNA sensors, such as IFI16 (Gamma-interferon-inducible protein 16), cGAS or AIM2 (absent in melanoma 2) [1]. Each PRR detects specific pathogen-associated molecular patterns (PAMPs) derived from viruses, bacteria, mycobacteria, fungi, and parasites that initiate the recruitment of distinct sets of adaptor molecules such as Myd88 (Myeloid differentiation primary response gene 88), TRIF (TIR-domain-containing adapter-inducing interferon-β), MAVS (Mitochondrial antiviral-signaling protein), and STING (Stimulator of interferon genes), among others [1]. Activation of those signaling pathways leads to the secretion of many inflammatory cytokines, including interferons (IFN), but also different chemokines and antimicrobial peptides.
The liver is located at the crossroads of the systemic and enteric circulations and carries out important metabolic functions such as detoxification and glucose and lipid metabolism. In addition, the liver performs many essential immune tasks and is considered a secondary lymphoid organ due to the number of flowing-through, infiltrating, and resident immune cells it contains [2].
Mechanisms of physiologic tolerogenicity are in place in the liver in order to prevent a persistent inflammation in reaction to permanent exposure to gut-derived “microbial degradation products” or even live bacteria, which can pass-through gut mucosa [2, 3]. Among tolerogenic mechanisms, there are checkpoint ligand-receptor systems (i.e., PD-1/PD-L1, CTLA-4/B7–1 or 2…) that modulate T-cell receptor-mediated T cell activity [4, 5]. This physiologic tolerogenicity represents a sort of Achilles' heel of the liver, which consequently can be the target of various pathogens establishing chronic infections [6].
The liver is, however, also capable of mounting a potent antimicrobial response. The liver tissue environment is composed of highly specialized cell types, including parenchymal and a number of nonparenchymal cells that play a key role in regulating hepatic immune functions. Parenchymal cells, called hepatocytes, account for 80% of liver mass and respond to different type of stimuli [2, 3, 7]. LSEC are also well known to participate in liver immune response by secreting cytokines upon pathogenic stimuli [8]. These cells also play a key role upon danger signal leading to fibrosis since, upon shear stress, they will undergo cytoskeletal remodeling, leading to a loss of fenestration [8, 9]. Hepatic stellate cell (HSC), the liver fibroblasts, and producers of extracellular matrix are localized in the space of Disse, an area between hepatocytes and sinusoids, and thus are not directly exposed to the bloodstream. These cells normally represent 5–8% of the total number of the liver cells. However, upon chronic inflammation, HSC undergo transformation to become myofibroblasts, the activated state of HSC [9, 10, 11]. Once activated, these cells proliferate and start secreting numerous components of the extracellular matrix creating a scar-like tissue [12]. However, during uncontrolled inflammation and scarring/healing process, the overproduction of extracellular matrix induces fibrosis, which can ultimately lead to cirrhosis and favor the development of hepatocellular carcinoma (HCC) [9, 12]. Finally, Kupffer cells (KC), the liver resident macrophages, represent 80% of total macrophages population within the body [13]. As macrophages, they form the first line of defense against pathogens and are specialized in pathogen recognition [14]. In response to stimulation, they produce a large spectrum of cytokines and chemokines that attract other immune cells such as neutrophils or infiltrating monocytes that will differentiate into macrophages upon entering the liver. KCs have a high phagocytic capacity and are implicated in the elimination of aging blood cells and pathogens. KC can subsequently present associated antigens to lymphocytes to reactivate them at the site of injury or infection [13].
Successful liver pathogens evolved strategies to either passively or actively evade innate and adaptive immunity. Indeed, hepatitis B virus (HBV) and hepatitis C virus (HCV) can persistently infect the hepatocytes. HBV and HCV, chronically infect around 70 and 250 million people worldwide respectively, leading to more than 1.2 million deaths per year (WHO 2017). While curative treatments have been recently developed for HCV [15], leading to viral clearance in more than 95% of cases, HBV cure is not achievable yet, as available treatments (i.e., nucleoside analogues) only allow the control of viral replication and require lifelong administration [16]. New treatments that could lead to a “functional cure” are currently under evaluation [16]. Among them, TLR7 agonists are evaluated in clinical phase II study after showing promising antiviral effects in preclinical models of HBV infections [17, 18]. Moreover, PD-1/PD-L1 inhibitors are considered to combat the development of several types of cancers [19, 20], including HCC in chronic HBV infection [21, 22]. Interestingly, they also are currently in clinical trial to treat CHB patients without signs of HCC. In this context, a better knowledge of the expression and functionality of innate sensors in liver cells would help developing novel PRR agonists, with antimicrobial or anticancer properties, or other strategies to revert immune inhibitory processes. This led us to examine the expression and functionality of some PRR in isolated primary liver cells, as well as in cell lines derived thereof.
Material and Methods
Liver Primary Cells Purification and Cells Culture
Peripheral blood mononuclear cell (PBMC) from at least 3 blood donors were isolated by Ficoll gradient cultured in RPMI medium supplemented with 10% of decomplemented FBS and 50 U/mL of penicillin/streptomycin (macrophage medium) at a density of 250,000 cells/cm2. Liver cells were isolated from liver resections (online suppl. Fig. S1A and S1B; for all online suppl. material, see www.karger.com/doi/10.1159/000489966) obtained from three surgical departments in Lyon (Centre Léon Bérard, Hôpital de la Croix Rousse and Centre Hospitalier Lyon-Sud) with the French ministerial authorizations (AC 2013-1871, DC 2013-1870, AFNOR NF 96 900 sept 2011). After collagenase treatment, the liver extract is filtered and centrifuged, as previously described [23]. Pellets contain primary human hepatocytes (PHH), whereas supernatants contain remaining nonparenchymal and other liver resident cells. PHH were cultured on collagen layer and maintained in PHH medium (Williams medium supplemented with 5% of fetal clone II serum, 50 U/mL of penicillin/streptomycin, 1× glutamax, 5 µg/mL of bovine insulin, 5 × 10–5 M of hydrocortisone, and 2% of DMSO) at a density of 250,000 cells/cm2. The supernatants (containing all liver cells except PHH) were either used to purify total liver mononuclear cells (LMNC) by Ficoll gradient or used to purify hepatic stellate cells (HSC), KC and LSEC, by a two-phase iodixanol gradient [24] (online suppl. Fig. S1A). LMNC were further cultured in macrophage medium at a density of 250,000 cells/cm2. HSC were further cultured in Williams medium supplemented with 10% of fetal clone II serum, 50 U/mL of penicillin/streptomycin, 1× glutamax, 5 µg/mL of bovine insulin, and 5 × 10–5 M of hydrocortisone at a density of 125,000 cells/cm2. LSEC were purified by positive selection with CD146 microbeads (Miltenyi Biotec) and culture in LSEC medium (MCDB131 supplemented with 20% of fetal clone II serum, 50 U/mL of penicillin/streptomycin, 5× glutamax, 35 mM of hydrocortisone, and 10 mg/mL of cAMP) at a density of 125,000 cells/cm2. KC were isolated by negative selection using pan monocyte isolation kit (Miltenyi Biotec) and cultured in macrophage medium at a density of 125,000 cells/cm2. At least 3 batches of each primary cells were used to perform the experiments. The photos of the liver resections and the yield of purified cells for each donor are presented in the online supplementary Figure 1B. THP1 (obtained from Thierry Walzer's laboratory, CIRI, Lyon) were cultured and differentiated as previously described [25]. Immortalized Kupffer cells (iKC) were immortalized by transduction with lentiviruses expressing the HPV E6E7 proteins, cultured in macrophage medium and differentiated by a 48-h treatment with 2% DMSO to stop proliferation at a density of 125,000 cells/cm2.
RNA Extraction, Reverse Transcription, dPCR, and qPCR
Total RNA was extracted with Nucleospin RNA II (Macherey-Nagel) and cDNA synthetized using the SuperScript®III Reverse Transcriptase (Life technologies) according to the manufacturer's instructions. cDNA was first analyzed by digital PCR (dPCR), on a 96 × 96 Biomark HD system (Fluidigm) using EvaGreen® dye according to the manufacturer's instructions. Primers sequences are presented in online supplementary Table S1. qPCR analyses were performed using “Express SYBR GreenERTM qPCR SuperMix Universal” (Invitrogen). mRNA expression was assessed by comparative cycle threshold (Ct) method, by normalizing the amount of target cDNA on housekeeping genes: RPLP0 and GUS for dPCR and RPLP0 for qPCR (2–ΔCt).
Heat Map Construction and Analysis
Primary results obtain with Biomark HD system were analyzed with manufacturers software (Fluidigm Real-Time PCR Analysis). Construction of the Heat map was performed on MEV software.
Immunoblot Analysis
Cells were lysed in RIPA buffer supplemented with protease inhibitors for 30 min on ice, sonicated and boiled in Laemmli buffer complemented with DTT. Protein concentration was measured by BCA protein assay kit (Thermo Scientific). Equal amounts of protein from total cell lysates (30–40 μg) were loaded onto SDS-PAGE Stain-Free precast gels (BioRad) and transferred onto nitrocellulose membranes (BioRad). The membranes were blocked with TBST (1× TBS with 0.1% Tween 20) +5% milk at room temperature for 1 h and incubated overnight at 4°C with primary antibodies. Primary antibodies and their corresponding positive controls used in this study are described in online supplementary Table S2. Membranes were washed and incubated with horseradish peroxidase-coupled secondary antibody (SIGMA) for 1 h at room temperature. Activity was visualized by chemiluminescence, and the signal was quantified by ImageLab software. Stain-free analysis was performed following manufacturer's instructions. Western blot analyses were performed on a pool of protein samples obtained from at least three different donors. The nature of positive control samples is specified in online supplementary Table S2.
PRR Stimulation and ELISA
TLR or RLR ligands from Invivogen or Riboxx life science are listed in online supplementary Table S3. It is worth noting that a low concentration of RIGI/MDA5 agonist (Poly(I/C)-(LMW)- LyoVec from Invivogen) was used (i.e., 0.025 μg/mL) to stimulate cells, as this concentration was found to be not toxic in repetitive administration in HepaRG cells [26]. Cells were exposed to the ligands, supernatants were collected 24 h later, and IL-6/IP-10 concentration were assed using Duoset ELISA (R&D systems) according to the manufacturer's instructions.
Phagocytosis Assay
THP1, IKC, and KC phagocytosis capacity was assessed using pHrodoTM Green E. coli BioparticlesTM conjugates for phagocytosis. Cells were exposed for 1 h to the bioparticles at either 4 or 37°C. After 1 h, cells were collected by scrapping, and phagocytosis was assessed by flow cytometry using FACScalibur flow cytometer and analyzed using CellQuestPro software (BD Biosciences).
Results and Discussion
Innate Immunity Gene RNA Expression in Various Primary Liver Cells
PHH, LSEC, HSC, and KC were purified from different liver resections (online suppl. Fig. S1A and S1B). RT-qPCR analyses using specific markers (i.e., HNF4α for PHH, L-SIGN for LSEC, α-SMA for HSC, and CD68 for KC) revealed a high enrichment for each cell type (online suppl. Fig. S1C and S1D). The expression level of 53 mRNAs that are known to be involved in pathogens sensing and immune cell regulation was assessed in nonstimulated cells (i.e., basal state) by microfluidic high-throughput quantitative RT-dPCR (Fluidigm, Biomark) assays. As controls, we used total PBMC and total nonparenchymal liver mononuclear cells (LMNC) from different donors that were not infected with HBV, HCV, or HIV.
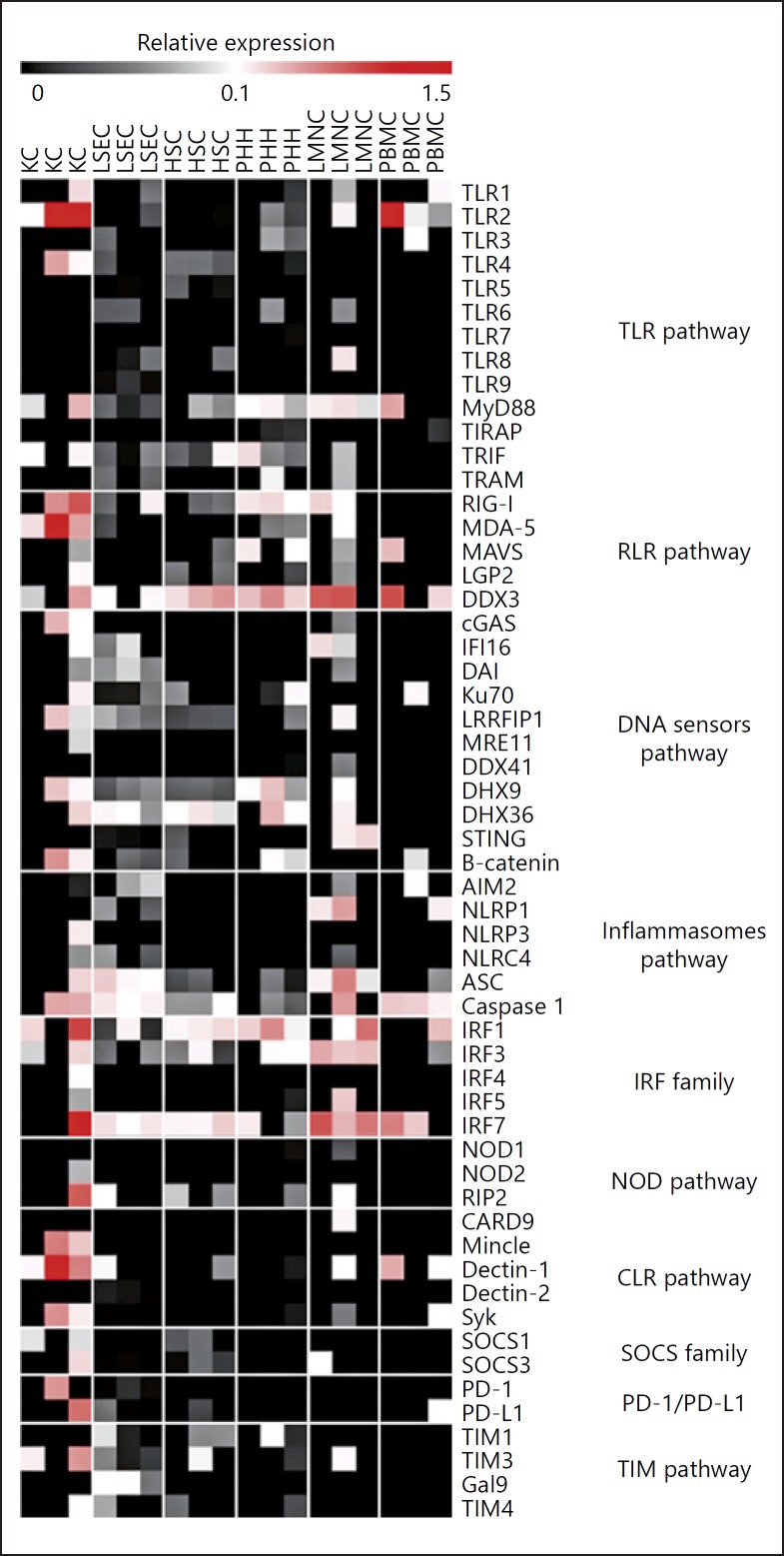
Interestingly, resident LMNCs showed, overall, higher expressions of the tested genes at basal state compared to PBMC supporting the fact that the liver is important for host immunity [2]. Nonstimulated liver cells showed high expression of cytosolic DNA (i.e., DDX3, Ku70, DHX9, or DHX36) and RNA sensors (i.e., RLR, RIG-I, and MDA-5) compared to PBMC (Fig. 1). Except for TLR2 and TLR4, which are highly expressed at basal level in KCs, TLR mRNA levels were relatively low in liver cells. In contrast, MyD88, TRIF, and TRAM, the main TLR adaptors, showed high basal expression level in all the different primary liver cells (Fig. 1) and IRF1, 3, and 7, the transcription factors involved in TLR signaling, were detected in almost all tested cells. Similarly, expression of NOD1 and NOD2 (previously described to be functional in hepatocytes [27, 28]) was barely detectable at basal level, but RIP2, a NOD adaptor protein was detected in almost all cells tested (Fig. 1). Inflammasome receptor mRNA (NLRP1, NLRP3, NLRC4, AIM2) were also barely detected, whereas mRNA coding for their signaling proteins (ASC and caspase 1) were readily detected in all liver cells. Finally, C-type lectin receptors, SOCS family, PD-1 and PD-L1, and TIM pathways were barely detected in the liver at basal state using microfluidic high-throughput quantitative RT-PCR (Fig. 1).
Fig. 1.
Liver cells expression of innate sensors and related molecules. PHH, LSEC, HSC, KC, LMNC, and PBMC were purified from different donors and cultured for 24 h. RNAs were extracted, and the expression level of 53 mRNAs was assessed by microfluidic high-throughput quantitative RT-PCR (Fluidigm, Biomark) assays. Data are presented as relative expression to two housekeeping genes (RPLP0 and GUS). Black square indicates an absence of expression, nuances of grey a low expression, and nuances of red a higher expression. TLR, toll-like receptor; RLR, retinoic acid inducible gene (RIG)-like receptor; IRF, interferon regulatory factor; NOD, nucleotide-binding oligomerization domain (NOD)-like receptor; CLR, C-type lectin receptor; SOCS, suppressor of cytokine signaling protein; PD-1, programmed cell death one; PD-L1, programmed cell death ligand one; TIM, T-cell immunoglobulin and mucin domain.
Innate Immunity Gene Protein Expression and Functionality in Various Primary Liver Cells
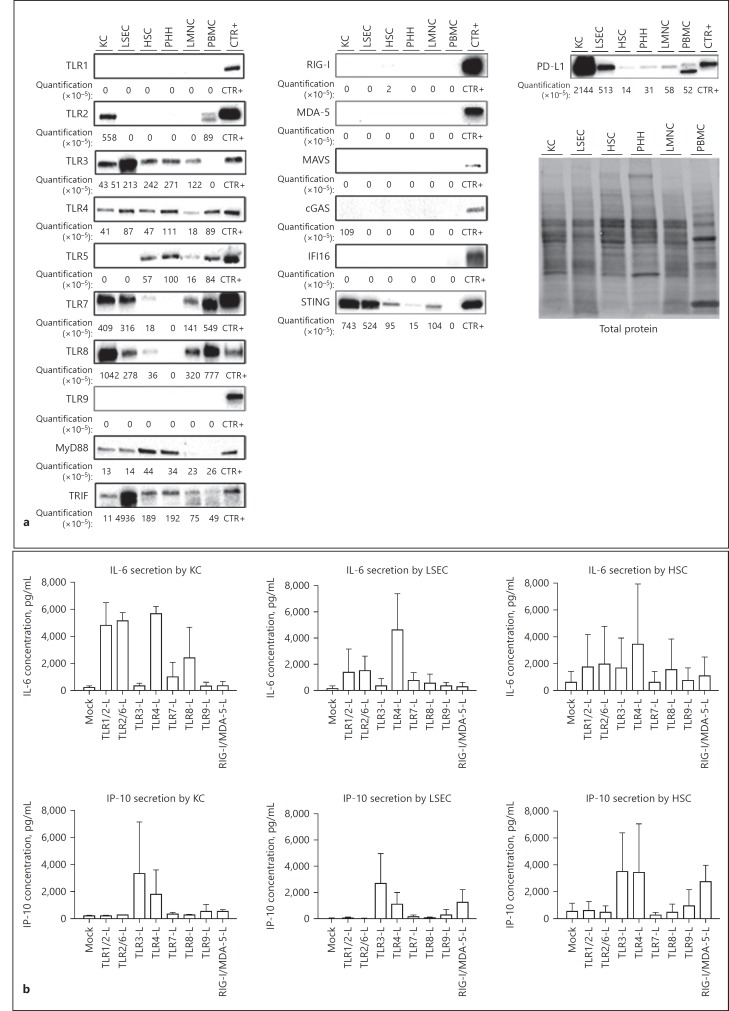
By Western blot analyses, we assessed more specifically the expression patterns of PD-L1 as well as a subset of proteins belonging to the TLR, RLR, and DNA cytosolic sensor families in liver cells. TLR1, TLR9, members of the RLR family (RIG-I, MDA-5, and MAVS), and the DNA sensor IFI16 and cGAS proteins could not be detected in any of the tested cells (Fig. 2a). To confirm whether the mRNAs of these different proteins were indeed negative, we performed standard RT-qPCR analyses, which are more sensitive than Fluidigm ones (used in Fig. 1) to detect them. With this method, we were able to detect the presence of these different mRNAs (online suppl. Fig. S2), suggesting that those proteins are expressed in liver cells at very low levels at basal state but could be upregulated upon specific stimulation, as already described [29, 30]. Surprisingly, we could not detect TLR9 proteins either in PMBCs or in LMNCs (Fig. 2a) that should contain pDCs and B cells known to express TLR9 [31], and as confirmed with pDC purified from PBMC (Fig. 2a, TLR9, CTR+). This might be due to the heterogeneity of cell proportion between donors. TLR4, TLR5, TLR7, TLR8, and TRIF proteins were detected (although at various levels) in both LMNC and PBMC (Fig. 2a), whereas TLR3 and STING proteins were only detected in LMNC, suggesting that percentage of the different immune cell populations are slightly different in blood compared to the liver and/or that the liver microenvironment may influence the phenotype of immune liver cells. TLR3, TLR4, MyD88, TRIF, and STING are ubiquitously expressed by KC, LSEC, HSC, and PHH (Fig. 2a) implying that the liver is particularly well equipped to detect lipopolysaccharides as well as cytosolic RNA and DNA [32]. Whereas TLR2 mRNAs were present in all cell types, their level was the highest in KCs (online suppl. Fig. S2), which translated at the protein levels as TLR2 protein being only detectable in KCs. In accordance with their phagocytic capacities [3], KC and LSEC express TLR7 and TLR8 on protein level (Fig. 2a), both TLRs involved in intracellular ssRNA detection [33]. Interestingly, TLR5 protein was only detected in HSC and PHH (Fig. 2a). Since these cells are not directly exposed to blood streams, this specific basal expression could be an evolutionary way to detect infiltrating pathogens and avoid persistent inflammation due to the exposition of bacterial components coming from the enteric circulation. Finally, KC [1] but also LSEC highly express PD-L1 protein (Fig. 2a), highlighting those cells as the main drivers of liver tolerogenic activity [34]. Altogether, we observed that all mRNAs from the tested TLR, RLR, DNA sensors and their respective adaptors are at least expressed by one of the liver resident cell types. While adaptors (Myd88, TRIF, and STING) are detected at the protein level in all cells, a large number of proteins from these pathways are not detected. These results suggest the majority of sensors might be expressed at relatively low levels probably to prevent chronic inflammatory stimulation at basal state [35]. However, in the presence of pathogens, parenchymal and nonparenchymal liver cells might mount an efficient immune response since all the adaptor molecules and the transcription factors are highly expressed already at basal state (Fig. 1, 2a). For instance, we previously showed that PHH express a number of PRR and that their agonization can efficiently inhibit HBV replication [36]. PHH responded to TLR1/2, TLR3, TLR4, TLR5, TLR2/6, and RIG-I/MDA-5 stimulation [36] despite the undetectable basal level of expression of several of those PRRs, such as TLR1/2 and RLR proteins (Fig. 2a). Functionality of TLR and RLR were also assessed in KC, LSEC, and HSC by stimulating cells from three to four different donors with the respective ligands. As expected from the protein expression data (Fig. 2a), KC produced IL-6 and/or IP-10 in response to TLR1/2, TLR3, TLR4, TLR7, and TLR8 ligands (Fig. 2b). Similarly, LSEC did also respond to TLR3 and TLR4 but not to TLR7 or TLR8 stimulations (Fig. 2b) suggesting that the amount of those proteins detected by Western blot analysis (Fig. 2a) was too low to derive a measurable response upon ex vivo stimulation with agonists. In contrast, LSEC produced IL-6 and IP-10 in response to TLR2 and RIG-I/MDA-5 ligands respectively (Fig. 2b), despite the fact that TLR2, RIG-I, or MDA-5 proteins were not detected (Fig. 2a) and only low amounts of mRNAs were detected in those cells (online suppl. Fig. S2). HSC mostly produced IP-10 in response to ligands for TLR3, TLR4, and RIGI/MDA5, whereas IL-6 production was very variable from one donor to the other (Fig. 2b).
Fig. 2.
TLR, RLR, cytosolic DNA sensors and PD-L1 expression in nonstimulated primary liver resident cells and in responses to PRR stimulations. a PHH, LSEC, HSC, KC, LMNC, and PBMC were purified from at least 3 different donors and cultured for 24 h. Proteins were extracted, pooled, and TLR1 to TLR9, MyD88, TRIF, RIG-I, MDA-5, MAVS, cGAS, IFI16, STING, and PD-L1 protein expression was assessed by Western blot analysis. Target protein levels were normalized to total protein quantification assessed by stain-free gels. Stimulated cells were used as controls (CTR+) for primary antibody efficiencies as indicated in Table S2. b KC, LSEC, and HSC from different donors were exposed to the indicated ligands at the concentration indicated in online supplementary Table S3 for 24 h. Supernatants were collected, and IL-6 and IP-10 secretions were analyzed by ELISA. Data are presented as mean ± standard deviation of three biological replicates.
Generation and Characterization of an iKC Line
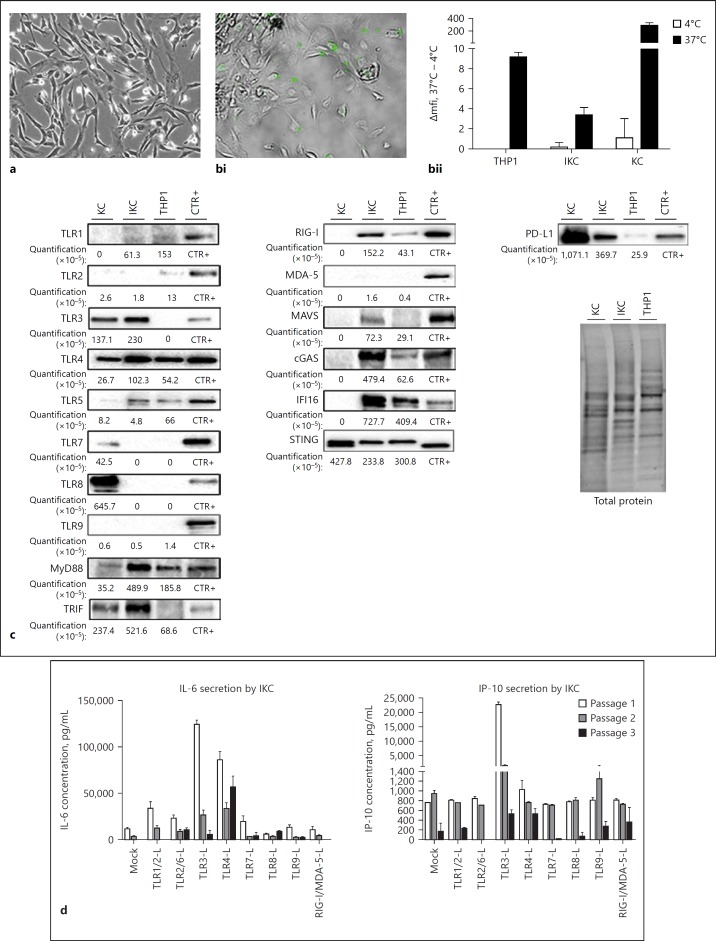
Access to liver resection and therefore primary liver cells is limited. To facilitate the study of liver cells, several models have been implemented over the years as surrogate models of PHH (Huh7 [37], HepG2 cells [38], and dHepaRG cells [39]), LSEC (TRP3 [40]), and HSC (LX-1 and LX-2 [41]). However, to our knowledge, no human KC derivate cell line has been described so far. KCs are a unique macrophage population differentiated from myeloid progenitor during embryogenesis and have specific functions, such as self-renewing capacity, which distinguish them from circulating monocytes [42]. We therefore immortalized KC by expressing the E6 and E7 proteins of the papilloma virus [43] to derive a polyclonal cell line (Fig. 3a). iKC acquired proliferative capacity, far beyond the self-renewing capacity of KC ex vivo. To stop iKC proliferation, cells were treated with 2% DMSO during 48 h for the following experiments. Although to a lower extend compared to primary KC, iKC retained their ability to phagocyte bacteria (Fig. 3bi, bii). KC, iKC, and THP1 (i.e., the most commonly used monocyte/macrophage-like cell line) [44, 45, 46] had a similar pattern of protein expression for TLR1, TLR4, MDA-5, and STING (Fig. 3c). Similar to the abolishment of TLR9 expression by HPV [47], TLR7 and TLR8 proteins were not detected in iKC (and THP1) as compared to KC, whereas higher amounts of TLR5, MyD88, RIG-I, cGAS, and IFI16 proteins were found in both cell lines (Fig. 3c). MAVS protein was only detected in iKC (Fig. 3c). Even though, KC and iKC express different amounts of TLR7, TLR8, Myd88, RIG-I, and MAVS proteins, their responses to the respected stimulations were overall similar (Fig. 3d). Whereas a low basal signal was detected in both KC and THP1, TLR2 protein was not detected in iKC (Fig. 3c) and response to TLR1/2 agonization was much lower in IKC than in KC (Fig. 3d, to be compared to Fig. 2b). Interestingly, TLR3, TRIF, and PD-L1 protein expression pattern in IKC was closer to KC than THP1 (Fig. 3c). Furthermore, KC and iKC strongly responded to TLR3 and TLR4 stimulations (Fig. 3d). Of note, amounts of secreted IL-6 upon IKC stimulations were 10 times higher than those detected in KC, suggesting that immortalization processes potentiate these pathways (compare Fig. 2b and 3d). Of note, IL-6 and IP-10 secreted amounts (Fig. 3d), as well as proliferating capacity (data not shown), decreased with cell passaging of IKC highlight the importance to perform experiments on low passage cells.
Fig. 3.
Characterization of immortalized Kupffer cells (iKC). iKC from three different passages were seeded and cultured for 48 h with 2% DMSO. KC purified from three different donors were seeded and cultured for 24 h. THP1 were seeded and cultured for 48 h with PMA. Cells were then left untreated (a, c), exposed to pHrodoTM bacteria for 1 h at 4°C or 37°C (b) or exposed to the indicated ligands at the concentration indicated in online supplementary Table S3 for 24 h (d). a, bi Microscopic analyses of cells (×20 magnification) were performed. bii Bacteria phagocytosis was assessed by flow cytometry analysis. c Proteins were extracted, pooled and TLR1 to TLR9, MyD88, TRIF, RIG-I, MDA-5, MAVS, cGAS, IFI16, STING, and PD-L1 protein expression was assessed by Western blot analyses. Target protein levels are normalized to total protein quantification assessed by stain-free staining. Stimulated cells were used as controls (CTR+) for primary antibody efficiencies as indicated in online supplementary Table S2. d Supernatants were collected, and IL-6 and IP-10 secretions were analyzed by ELISA. Data are presented as mean ± standard deviation of three biological replicates.
To summarize, we provided here a comprehensive analysis of innate immune signaling capacity of the most important liver cell populations and demonstrate their ability to respond to pathogens or PRR ligand stimulation. This knowledge will be useful to understand mechanisms behind antiviral effects of TLR agonists or check-point inhibitors [16, 19, 20, 21, 48, 49], whose development as direct effectors or “adjuvant” in more complex immune-therapeutic strategies are currently investigated to treat cancer and chronic viral infection. For instance, TLR7, TLR8, RIGI, and STING agonists are currently developed to fight chronic HBV infections [16] but our work highlights other ligands such as TLR2 and TLR3 agonists have the best direct anti-HBV properties in HBV-replicating PHH [50]. In addition, in the liver microenvironment, the agonization of TLR2 and TLR3 might trigger the production of antiviral secretion by KC and/or LSEC.
Disclosure Statement
The authors declare that they have no conflicts of interest.
Author Contributions
J.L., S.F.-D., S.V., and D.D. designed the study; S.F.-D., S.V., J.L., L.A., L.D., and M.Bo. performed and/or analyzed experiments; S.F.-D., K.E., M.Br., U.P., and N.B.-V. contributed to the establishment of protocols for liver cells isolation; M.R., G.P., M.L., J.-Y.M., and C.D. provided liver resections. F.Z. and A.S. provided material. J.L., S.F.-D., S.V., and D.D. wrote the manuscript. U.P., F.Z., and N.B.-V. did a critical reading of the manuscript.
Supplementary Material
Supplementary data
Acknowledgments
The work was supported by grants from INSERM, ANRS (French national agency for research on AIDS and viral hepatitis; grants from CSS4 and CSS7 study committees), the DEVweCAN LABEX (ANR-10-LABX-0061), and the FINOVI foundation. The authors would like to thank Maud Michelet, Jennifer Molle, Loïc Peyrot, Anaelle Dubois, Océane Floriot, and Marie-Laure Plissonnier for their help in the isolation of primary human hepatocytes, as well as Prof. Michel Rivoire's, Prof. Jean-Yves Mabrut's, and Dr. Guillaume Passot's staff for providing liver resections. The authors would also like to thank Maëlle Locatelli, Léa Monnier, Brieux Chardès, and Hélène Chabrolles for providing the hepatocyte cell lines. Finally, the authors thank the EFS (French establishment for blood donation) for providing the blood bags.
References
- 1.Pandey S, Kawai T, Akira S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol. 2014;7:a016246. doi: 10.1101/cshperspect.a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 3.Crispe IN. Liver antigen-presenting cells. J Hepatol. 2011;54:357–365. doi: 10.1016/j.jhep.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dal Bello MG, Alama A, Coco S, Vanni I, Grossi F. Understanding the checkpoint blockade in lung cancer immunotherapy. Drug Discov Today. 2017;22:1266–1273. doi: 10.1016/j.drudis.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Arasanz H, et al. PD1 signal transduction pathways in T cells. Oncotarget. 2017;8:51936–51945. doi: 10.18632/oncotarget.17232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 7.Kmieć Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161:III–XIII, 1–151. doi: 10.1007/978-3-642-56553-3. [DOI] [PubMed] [Google Scholar]
- 8.Sørensen KK, Simon-Santamaria J, McCuskey RS, Smedsrød B. Liver sinusoidal endothelial cells. Compr Physiol. 2015;5:1751–1774. doi: 10.1002/cphy.c140078. [DOI] [PubMed] [Google Scholar]
- 9.Eggert T, Greten TF. Tumor regulation of the tissue environment in the liver. Pharmacol Ther. 2017;173:47–57. doi: 10.1016/j.pharmthera.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva BO, Ramos LF, Moraes KCM. Molecular interplays in hepatic stellate cells: apoptosis, senescence and phenotype reversion as cellular connections that modulates liver fibrosis. Cell Biol Int. 2017;41:946–959. doi: 10.1002/cbin.10790. [DOI] [PubMed] [Google Scholar]
- 11.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 12.Moran-Salvador E, Mann J. Epigenetics and liver fibrosis. Cell Mol Gastroenterol Hepatol. 2017;4:125–134. doi: 10.1016/j.jcmgh.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 14.Seki S, et al. The liver as a crucial organ in the first line of host defense: the roles of Kupffer cells, natural killer (NK) cells and NK1.1 Ag+ T cells in T helper 1 immune responses. Immunol Rev. 2000;174:35–46. doi: 10.1034/j.1600-0528.2002.017404.x. [DOI] [PubMed] [Google Scholar]
- 15.Nehra V, Rizza SA, Temesgen Z. Sofosbuvir/velpatasvir fixed-dose combination for the treatment of chronic hepatitis C virus infection. Drugs Today. 2017;53:177–189. doi: 10.1358/dot.2017.53.3.2604176. [DOI] [PubMed] [Google Scholar]
- 16.Durantel D, Zoulim F. New antiviral targets for innovative treatment concepts for hepatitis B virus and hepatitis delta virus. J Hepatol. 2016;64((suppl)):S117–S131. doi: 10.1016/j.jhep.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Lawitz E, et al. Safety, pharmacokinetics and pharmacodynamics of the oral toll-like receptor 7 agonist GS-9620 in treatment-naive patients with chronic hepatitis C. Antivir Ther (Lond) 2015;20:699–708. doi: 10.3851/IMP2845. [DOI] [PubMed] [Google Scholar]
- 18.Gane EJ, et al. The oral toll-like receptor-7 agonist GS-9620 in patients with chronic hepatitis B virus infection. J Hepatol. 2015;63:320–328. doi: 10.1016/j.jhep.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 19.Bertucci F, Finetti P, Birnbaum D, Mamessier E. The PD1/PDL1 axis, a promising therapeutic target in aggressive breast cancers. Oncoimmunology. 2016;5:e1085148. doi: 10.1080/2162402X.2015.1085148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagasaka M, et al. PD1/PD-L1 inhibition as a potential radiosensitizer in head and neck squamous cell carcinoma: a case report. J Immunother Cancer. 2016;4:83. doi: 10.1186/s40425-016-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Z, Zhou Q, Lu M, Tan D, Xu X. A programmed cell death-1 haplotype is associated with clearance of hepatitis B virus. Ann Clin Lab Sci. 2017;47:334–343. [PubMed] [Google Scholar]
- 22.Wenjin Z, Chuanhui P, Yunle W, Lateef SA, Shusen Z. Longitudinal fluctuations in PD1 and PD-L1 expression in association with changes in anti-viral immune response in chronic hepatitis B. BMC Gastroenterol. 2012;12:109. doi: 10.1186/1471-230X-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecluyse EL, Alexandre E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol Biol. 2010;640:57–82. doi: 10.1007/978-1-60761-688-7_3. [DOI] [PubMed] [Google Scholar]
- 24.Hösel M, et al. Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors. Hepatology. 2012;55:287–297. doi: 10.1002/hep.24625. [DOI] [PubMed] [Google Scholar]
- 25.Schütze N, et al. The selenoprotein thioredoxin reductase is expressed in peripheral blood monocytes and THP1 human myeloid leukemia cells - regulation by 1,25-dihydroxyvitamin D3 and selenite. Biofactors. 1999;10:329–338. doi: 10.1002/biof.5520100403. [DOI] [PubMed] [Google Scholar]
- 26.Luangsay S, et al. Expression and functionality of Toll- and RIG-like receptors in HepaRG cells. J Hepatol. 2015;63:1077–1085. doi: 10.1016/j.jhep.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Vegna S, et al. NOD1 participates in the innate immune response triggered by hepatitis C virus polymerase. J Virol. 2016;90:6022–6035. doi: 10.1128/JVI.03230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott MJ, Chen C, Sun Q, Billiar TR. Hepatocytes express functional NOD1 and NOD2 receptors: a role for NOD1 in hepatocyte CC and CXC chemokine production. J Hepatol. 2010;53:693–701. doi: 10.1016/j.jhep.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matikainen S, et al. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J Virol. 2006;80:3515–3522. doi: 10.1128/JVI.80.7.3515-3522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Goffic R, et al. Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 31.Mathan TSM, Figdor CG, Buschow SI. Human plasmacytoid dendritic cells: from molecules to intercellular communication network. Front Immunol. 2013;4:372–372. doi: 10.3389/fimmu.2013.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 33.Adzavon YM, et al. TLR7 and TLR8 agonist resiquimod (R848) differently regulates MIF expression in cells and organs. Cytokine. 2017;97:156–166. doi: 10.1016/j.cyto.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Folkl A, Bienzle D. Structure and function of programmed death (PD) molecules. Vet Immunol Immunopathol. 2010;134:33–38. doi: 10.1016/j.vetimm.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Leifer CA, Medvedev AE. Molecular mechanisms of regulation of Toll-like receptor signaling. J Leukoc Biol. 2016;100:927–941. doi: 10.1189/jlb.2MR0316-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luangsay S, et al. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol. 2015;63:1314–1322. doi: 10.1016/j.jhep.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Nakabayashi H, Taketa K, Yamane T, Miyazaki M, Miyano K, Sato J. Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gan. 1984;75:151–158. [PubMed] [Google Scholar]
- 38.Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 39.Hantz O, Parent R, Durantel D, Gripon P, Guguen-Guillouzo C, Zoulim F. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J Gen Virol. 2009;90:127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- 40.Parent R, et al. An immortalized human liver endothelial sinusoidal cell line for the study of the pathobiology of the liver endothelium. Biochem Biophys Res Commun. 2014;450:7–12. doi: 10.1016/j.bbrc.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 41.Xu L, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 43.Schütze DM, et al. Immortalization capacity of HPV types is inversely related to chromosomal instability. Oncotarget. 2016;7:37608–37621. doi: 10.18632/oncotarget.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mantecca P, et al. Airborne nanoparticle release and toxicological risk from metal oxide-coated textiles: toward a multi-scale safe-by-design approach. Environ Sci Technol. 2017;51:9305–9317. doi: 10.1021/acs.est.7b02390. [DOI] [PubMed] [Google Scholar]
- 45.Hernandez-Sánchez F, et al. High glucose induces O-GlcNAc glycosylation of the vitamin D receptor (VDR) in THP1 cells and in human macrophages derived from monocytes. Cell Biol Int. 2017;41:1065–1074. doi: 10.1002/cbin.10827. [DOI] [PubMed] [Google Scholar]
- 46.Xu B, Gao Y, Zhan S, Ge W. Quantitative proteomic profiling for clarification of the crucial roles of lysosomes in microbial infections. Mol Immunol. 2017;87:122–131. doi: 10.1016/j.molimm.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Hasan UA, et al. TLR9 expression and function is abolished by the cervical cancer-associated human papillomavirus type 16. J Immunol. 2007;178:3186–3197. doi: 10.4049/jimmunol.178.5.3186. [DOI] [PubMed] [Google Scholar]
- 48.Nowicki TS, Anderson JL, Federman N. Prospective immunotherapies in childhood sarcomas: PD1/PDL1 blockade in combination with tumor vaccines. Pediatr Res. 2016;79:371–377. doi: 10.1038/pr.2015.246. [DOI] [PubMed] [Google Scholar]
- 49.Tsukahara T, et al. The future of immunotherapy for sarcoma. Expert Opin Biol Ther. 2016;16:1049–1057. doi: 10.1080/14712598.2016.1188075. [DOI] [PubMed] [Google Scholar]
- 50.Lucifora J, et al. Direct antiviral effects of various pattern recognition receptor (PRR) agonists in HBV-replicating hepatocytes. J Hepatol. 2015;62:S515–S516. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data