Abstract
The present study was to identify the drug resistance, resistance mechanism and the extended-spectrum β-lactamase (ESBLs) genotypes of Shigella flexneri (S. flexneri) in Jinan. Susceptibility tests were performed by MIC-determination. The genotypes of β-lactamase were identified using PCR and DNA sequencing. The resistance transfer ability of the ESBL-producing strains was examined by conjugation tests. A total of 105 S. flexneri isolates were collected, and 34 (32.4%) were ESBL-producing isolates. All ESBL-producing isolates were susceptible to cefoxitin and imipenem, and 35.3% isolates were resistant to ciprofloxacin. ESBL-producing isolates showed high level resistant to ampicillin (100%), cefotaxime (100%), tetracycline (100%), chloramphenicol (100%), trimethoprim/sulfamethoxazole (100%), ceftazidime (73.5%) and cefepime (73.5%). Three types of β-lactamase genes (blaTEM, blaOXA and blaCTX-M) were identified in all ESBL-producing isolates, and the genotypes were confirmed as blaTEM-1 (23/34), blaOXA-30 (34/34), blaCTX-M-14 (9/34) and blaCTX-M-15 (25/34) by sequencing. In conclusion, the Shigella strains isolated in Jinan are cross-resistant and multi-drug resistant. The main genotypes of ESBLs are CTX-M-14 and CTX-M-15.
Keywords: antimicrobial resistance, ESBLs, genotype, Shigella flexneri
Introduction
Shigella is a highly infectious intestinal bacteria that can cause serious harm. According to DuPont et al. [1], ingestion of 10–100 Shigella dysentery can make healthy individuals ill. Shigella’s endotoxin, exotoxin, enterotoxin and temperature regulation genes can cause fever, bloody purulent stool, abdominal cramps in infected individuals. Some patients can have very severe symptoms including hemolytic uremic syndrome, hypoglycemia, hyponatremia, intestinal perforation, seizures, encephalopathy and even death while others experience self-limiting illness. According to WHO in 1999, there are 165 million dysentery patients around the world every year, with about 162 million in the developing world. Dysentery was the cause of death for 1.1 million people, mainly children under the age of 5 [2]. Shigella was named by WHO in 1996 as a life threatening bacteria due to its growing resistance to treatment [3] and the morbidity associated with economic conditions, public health, living habits and epidemic Shigella serotypes. Bacillary dysentery is a serious public health problem in China [4], considered the third most dangerous among infectious diseases. Bacterial dysentery must be controlled through eliminating the source of infection, cutting off the route of transmission and protecting the vulnerable groups in time. However, in recent years, Shigella has become more drug resistant, causing inefficient treatment. Those Shigella patients who protracted course of disease are a primary source of infection in others, causing great difficulties for disease control and prevention and clinical treatment.
Bacterial drug resistance seriously impedes the efficacy of clinical treatment, increasing the cost of treatment, shortening the application period for new drugs and increasing the cost of drug development. In the 1950s, sulfonamides were effective for treatment of shigellosis, but soon produced drug resistance. Research indicated that the gene cassettes dfrA1, sat1 and aadA1 carried by Shigella encode resistance to sulfonamides, streptomycin and aminoglycoside antibiotics [5,6]. Shigella has been resistant to tetracycline, chloramphenicol and ampicillin since the 1970s; studies [7] show that in the R plasmid-mediated resistance of Shigella to these antibiotics, the transmissible plasmid carried the corresponding resistance genes. Shigella developed resistance to quinolones at the beginning of the 1980s, which has been attributed to its gyrase A subunit (gyra) mutation and topoisomerase gene (parc) mutation. The third-generation cephalosporins have been highly effective against Shigella in the clinic up until 1999, when Ahamed & Kundul [8] first observed in India that Shigella developed SHV-11 type extended-spectrum β-lactamase (ESBLs), leading to cross resistance to penicillin and first-generation, second-generation and third-generation cephalosporins and monoamide antibiotics. Researchers in France then found SHV-2 type ESBLs, those in Japan found TOHO-1 type ESBLs, and those studying populations in Argentina found PER-2 type ESBLs. South Korea, Turkey, Lebanon and other countries reported CTX-M type ESBLs, which the main genotypes were being CTX-M-2, CTX-M-3, CTX-M-14 and CTX-M-15. The main genotypes in China were CTX-M-3, CTX-M-14 and CTX-M-15 [8–10]. The current paper presents results of research conducted in China between 2011 and 2016, analyzing the resistance, popular genotypes and resistant gene dissemination of clinical sporadic ESBLs-producing Shigella and the difference between CTX-M-14 and CTX-M-15 in affecting the capability of antibiotics.
Materials and methods
Bacterial isolates
All isolates were obtained from clinic patients with diarrhea in China from 2011 to 2016 and identified by ID 32E and serotyping as Shigella flexneri (S. flexneri). Quality control strains of Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603 and E. coli K12RifRLac- were presented by Doctor Xu Yuanhong of the First Affiliated Hospital of Anhui Medical University. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki*, and that all subjects provided written informed consent.
ESBLs screening and confirmation
ESBLs screening and confirmation were done to the standard of CLSI recommendations for K. pneumoniae, Klebsiella oxytoca and E. coli. Highly suspicious strains were identified by the bacteriostatic ring (≤27 mm to cefotaxime and ≤22 mm to ceftazidime) and confirmed by the double disk synergy test. In those drugs with clavulanic acid, as opposed to those without clavulanic acid, an inhibition zone diameter of ≥5 mm may be a preliminary indication of ESBL-producing strains.
Antimicrobial susceptibility testing
The Kirby–Bauer (K–B) disk diffusion method and MIC determination were utilized as the method and criterion for antimicrobial susceptibility testing, respectively, according to the CLSI 2010 version of M100-S20 regulations.
Extraction of DNA
Genomic DNA was extracted from the isolates using TIANamp Bacteria DNA kit (Tiangen Biotech (Beijing) Co., Ltd.), All operations are carried out in accordance with the Kit specification.
Primer synthesis
According to the primer sequence described in Table 1, TEM, SHV, OXA, the CTX-M-1 group, the CTX-M-2 group, the CTX-M-8 group, the CTX-M-9 group and the CTX-M-25 group-encoding gene primers were synthesized.
Table 1. Sequence and annealing temperature of β-lactamase gene primers.
| Primer | Sequence (5′→3′) | Product size | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| TEM | FP: CCCTGGTAAATGCTTC | 919 | 45 | [1] |
| RP: GAGTAAACTTGGTCTG | ||||
| SHV | FP: GGTTATGCGTTATATTCGCC | 864 | 58 | [3] |
| RP: TTAGCGTTGCCAGTGCTC | ||||
| OXA | FP: ACACAATACATATCAACTTCGC | 885 | 50 | [3] |
| RP: AGTGTGTTTAGAATGGTGATC | ||||
| CTX-M-1 group | FP: CGT CAC GCT GTT GTT AGG AA | 780 | 50 | [4] |
| RP: ACG GCT TTC TGC CTT AGG TT | ||||
| CTX-M-2 group | FP: TTA ATG ATG ACT CAG AGC ATT C | 902 | 49 | [5] |
| RP: GAT ACC TCG CTC CAT TTA TTT | ||||
| CTX-M-8 group | FP: ACT TCA GCC ACA CGG ATT CA | 948 | 50 | [5] |
| RP: AAG TGG AGC GAC AGA GC | ||||
| CTX-M-9 group | FP: CGG AAGCAGTCTAAATTC TTCGTGAAATAG | 1160 | 54 | [1] |
| RP: CGG GCC AGT TGG TGA TTT GA | ||||
| CTX-M-25 group | FP: GTA AGG CGG GCG ATG TTA AT | 856 | 50 | [5] |
| RP: AAC CGT CGG TGA CAA TTC TG |
PCR amplification and DNA sequencing
The PCR reaction system was a mixture of 0.15 μl of Taq DNA polymerase (5 U/μl), 3 μl of 10× PCR buffer (Mg2+) and 2.4 μl of dNTP mixture (2.5 mM). Next, 1.5 μl of forward and reverse primers (10 μM), 3 μl of DNA template and sterile double-distilled water were added to give a total reaction volume of 30 μl. The PCR amplification conditions included pre-denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 s, annealing (at respective annealing temp) for 30 s and 72°C for 40 s; and final extension at 72°C for 5 min. The PCR products were analyzed by 1% agarose gel electrophoresis, and the positive PCR products were sent to be sequenced.
Conjugation analysis
The ESBL-producing strains were specified as the donor bacteria, and E. coli K12RifRLac- was specified as the recipient bacteria. An individual colony of donor and recipient bacteria were inoculated in 1 ml of common broth. After incubation at 35°C for 6 h, 100 μl of donor bacteria and 100 μl of recipient bacteria were placed into a 500 μl sterile ordinary broth mix, conjugated for 2 h at 35°C, then the positive colony was picked up on a China blue plate containing cefotaxime (1 mg/l). The red colony could be used for further pure culture and identified by ID 32E. The genotype and MIC of the conjugation were determined as previously described.
Relative hydrolysis rate determination
The CTX-M-14 and CTX-M-15 Shigella genotypes were in nutrient broth overnight then centrifuged at 4000 r/min for 15 min. The precipitate was washed three times with physiological brine and finally suspended in 2 ml of normal saline. The precipitate in saline was frozen and thawed eight times then centrifuged at 8000 r/min for 30 min. The supernatant was a ESBLs crude extract. Thirty microliters of the extract was then added to 3 ml of cephalosporin, diluting it to a final concentration of 0.1 mmol/l, and the OD value was determined by ultraviolet spectrophotometry. After a 15 min water bath at 37°C, the OD value was determined again. The enzymatic hydrolysis of 100% cefazolin was used as a benchmark to calculate the relative hydrolysis rate of each kind of cephalosporin (%) (the optimum wavelength of each would be: cefazolin, 265 nm; cefuroxime, 265 nm; cefotaxime, 257 nm; ceftazidime, 257 nm; aztreonam, 277 nm; cefepime, 261 nm; and cefoxitin, 260 nm). The relative hydrolysis rate = (ΔOD of other cephalosporins/ΔOD of cefazolin) × 100%.
Results
The isolated rate of ESBL-producing Shigella
In 105 strains of S. flexneri, 40 suspicious strains were screened by the K–B method, and a preliminary determination indicated that 34 strains were ESBL-producing. The genotyping method confirmed 34 strains (32.4%) were ESBL-producing.
Antimicrobial susceptibility analysis
All 34 ESBL-producing Shigella strains were resistant to ampicillin, cefotaxime, tetracycline, chloramphenicol, sulfamethoxazole and trimethoprim; 28 strains were resistant to cefepime and ceftazidime, and 13 to ciprofloxacin. All 34 ESBL-producing Shigella strains were sensitive to cefoxitin, imipenem and gentamicin. The results are shown in Table 2.
Table 2. MIC of 34 ESBL-producing S. flexneri and corresponding conjugon (μg/ml).
| Antimicrobial agents | Range of MIC | Crit. conc. | MIC of 34 ESBLs- producing S. flexneri (quantity) | MIC of 34 conjugon (quantity) | ||
|---|---|---|---|---|---|---|
| CTX-M-14type(9) | CTX-M-15type(25) | CTX-M-14type(9) | CTX-M-15type(25) | |||
| Ampicillin | 0.016–256 | 16 | >256(9) | >256(25) | >256(9) | >256(25) |
| Cefotaxime | 0.016–256 | 2 | 32(7), >256(2) | >256(25) | 16(2), 32(7) | >256(25) |
| Ceftazidime | 0.016–256 | 8 | 1(7), 2(2) | 16(2), 32(23) | 0.5(1), 1(1), 2(7) | 8(2), 16(23) |
| Cefoxitin | 0.016–256 | 16 | 2(2), 4(7) | 2(2), 4(23) | 2(1), 4(8) | 2(1), 4(24) |
| Cefepime | 0.016–256 | 16 | 4(5), 8(4) | 64(8), 128(9), 256(8) | 1(3), 2(4), 4(2) | 16(8), 64(14), 256(3) |
| Imipenem | 0.06–32 | 8 | 0.25(7), 0.5(2) | 0.25(23), 0.5(2) | 0.125(7), 0.25(2) | <0.06(10), 0.125(15) |
| Ciprofloxacin | 0.02–32 | 2 | 0.125(2), 4(7) | 0.25(20), 4(5) | <0.02(9) | <0.02(25) |
| Gentamicin | 0.016–128 | 8 | 1(5), 2(3), 32(1) | 1(21), 2(4) | 0.25(8), 0.5(1), 8(1) | 0.25(15), 0.5(6), 1(4) |
| Chloramphenicol | 0.016–256 | 16 | 128(3), >256(6) | 64(11), 128(5), 256(9) | 4(5), 8(4) | 4(23), 8(2) |
| Tetracycline | 0.016–256 | 8 | 128(1), 256(8) | 128(2), 256(23) | 1(8), 2(1) | 1(23), 128(2) |
| Sulfamethoxazole | 2–1024 | 64 | >1024(9) | >1024(25) | 4(2), 8(7) | 4(7), 8(16), 16(2) |
| Trimethoprim | 0.125–64 | 4 | >64(9) | >64(25) | <0.125(9) | <0.125(25) |
Results of PCR amplification
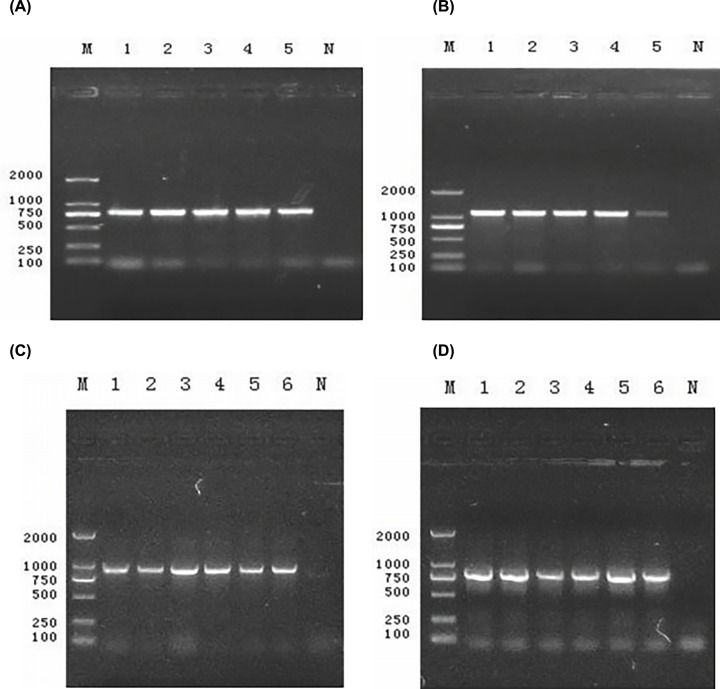
All 34 ESBL-producing Shigella strains were positive for the blaCTX-M gene, including 25 in the CTX-M-1 group (Figure 1A) and nine in the CTX-M-9 group (Figure 1B). Twenty-three strains were positive for blaTEM (Figure 1C) and 34 were positive for blaOXA (Figure 1D) but were not detected in SHV, CTX-M-2, CTX-M-8 or CTX-M-25 positive strains. Positive amplification products by DNA sequencing confirmed that the CTX-M-1 group was the blaCTX-M-15 gene, and the CTX-M-9 group was the blaCTX-M-14 gene that encoded ESBLs. The blaTEM and blaOXA genes were blaTEM-1 and blaOXA-30, respectively, encoding β-lactamase instead of ESBLs.
Figure 1. Electrophoretogram of blaCTX-M gene.
M: Marker D2000; 1, 2, 3, 4, 5 and 6: Random strain; N: E. coli ATCC 25922. (A,B,C,D): Electrophoretogram of CTX-M-1, CTX-M-9, TEM and OXA gene amplification product.
Conjugation analysis
All 34 ESBL-producing Shigella conjugative transfer tests were positive, and the biochemical identification results of conjugation were the same as the recipient bacterium E. coli K12RifRLac−. PCR amplification confirmed that all strains transferred blaCTX-M and that the genotype was consistent with the donor bacteria. MIC results and conjugons are presented in Table 3.
Table 3. Relative hydrolysis rate determination of CTX-M Type S. flexneri (%).
| Antimicrobial agents | CTX-M-14 | CTX-M-15 |
|---|---|---|
| Cefazolin | 100 | 100 |
| Cefuroxime | 85.8 | 88.0 |
| Cefotaxime | 70.7 | 65.9 |
| Ceftazidime | 2.6 | 7.7 |
| Aztreonam | 0 | 7.2 |
| Cefepime | 51.9 | 73.7 |
| Cefoxitin | 0 | 0 |
Discussion
With the wide application of third-generation cephalosporins, the ESBL-producing Shigella detection rate increased gradually [11]. In the present study, 32.4% (34/105) of S. flexneri strains produced ESBLs, which was more than the separation rate of 10.3% in this area during 2003–2006 [12]. These statistics indicate that there is an increasing trend of ESBL-producing Shigella in the district. One hand, with third-generation cephalosporins being used in clinical experience widely against the increasing multi-drug resistance of Shigella, the selective pressure caused resistance to grow more gradually. On the other hand, in the ESBL-producing Enterobacteriaceae (such as E. coli and K. pneumoniae) family, plasmids transferred the ESBLs resistance genes through transformation, transduction and conjugation. Conjugation analysis also confirmed the presence of conjugative plasmid in ESBL-producing Shigella, which carried the ESBLs resistance genes that had completely transferred to the receptor bacteria; three strains were also carrying the gentamicin and tetracycline resistance gene. Nicolas [13] found that S. flexneri has SHV-2 ESBLs, located in an 80-Kb plasmid; and Kim et al. [14] found that the ESBL-encoding gene of Shigella sonnei was located in the conjugative plasmid, with individual strains of two or three types of ESBLs. All of these results reveal that conjugative plasmid plays a key role in the transmission of ESBLs and other resistance genes, leading to horizontal transfer of the drug resistance genes and communication between the homologous and heterologous bacteria.
The first ESBLs of the CTX-M type were isolated in 1989 from E. coli [15]. The name CTX-M was given because these ESBLs more efficiently hydrolyze cefotaxime compared with ceftazidime, and the homology between this enzyme and the TEM and SHV enzymes is only 40%. At present, CTX-M- type ESBLs have become the most popular worldwide, and more than 90 CTX-M-type enzymes have been discovered [16]. In the present study, 34 strains of ESBL-producing Shigella were identified as CTX-M-type ESBLs with two distinct subtypes.
Through the analysis of drug susceptibility of these two groups of ESBL-producing S. flexneri, we found that there was a conspicuous difference in resistance to cefepime, ceftazidime and aztreonam. The CTX-M-14 type strains were more sensitive, while the CTX-M-15 type strains appeared to have a different degree of resistance. Relative hydrolysis rate determination also showed that the CTX-M-14-type enzyme cannot hydrolyze these three drugs. This inability to hydrolyze may be related to the existence of only Ser in the 237 position [17] with a lack of Lys or Arg active groups in the 240 position.
The drug susceptibility results showed that 34 strains of ESBL-producing S. flexneri exhibited serious cross resistance and multidrug resistance phenomena—35.3% resistant to ciprofloxacin, 73.5% resistant to ceftazidime and cefepime, while 100% sensitive to cefoxitin, imipenem and gentamicin (patients with diarrhea are often given oral treatment in China). These results mean that there are a very limited number of antibiotics that one can choose to treat bacterial dysentery caused by ESBL-producing Shigella.
ESBLs are widely distributed in several bacteria. The Enterobacteriaceae bacteria is particularly beneficial for the production of resistance to antibiotics in Shigella because of its plasmid-mediated and integron-captured properties [18–20], which make it difficult to implement interventions that can successfully control and treat the disease. Therefore, to strengthen the monitoring and molecular epidemiological study of resistance phenotype and genotype of ESBLs-producing strains, find out the epidemic characteristics of ESBL-producing Shigella in the region, and provide data support for clinicians, which is of great significance for delaying the production of bacterial resistance and controlling the spread and prevalence of resistant strains in a timely and effectively.
Abbreviations
- ESBL
extended-spectrum β-lactamase
- S. flexneri
Shigella flexneri
Author Contribution
Guarantor of integrity of the entire study and manuscript preparation: F.Z.B. and M.X.Y. Study concepts: H.M.F. Study design: F.Z.B., G.Y.Y. and S.Z.W. Definition of intellectual content: F.Z.B. Data analysis: F.Z.B. and S.Z.W. Manuscript editing: S.Z.W. and Y.G.S.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
References
- 1.DuPont H.L., Levine M.M., Hornick R.B. and Formal S.B. (1989) Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 159, 1126–1128 10.1093/infdis/159.6.1126 [DOI] [PubMed] [Google Scholar]
- 2.Navia M.M., Capitano L., Ruiz J., Vargas M., Urassa H., Schellemberg D.. et al. (1999) Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara. Tanzania J. Clin. Microbiol. 37, 3113–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J. (1996) Bacteria on the rampage. Nature 383, 219–220 10.1038/383219a0 [DOI] [PubMed] [Google Scholar]
- 4.Yan W., Xu Y., Yang X. and Zhou Y. (2010) A hybrid model for short-term bacillary dysentery prediction in Yichang City, China. Jpn. J. Infect. Dis. 63, 264–270 [PubMed] [Google Scholar]
- 5.DeLappe N., O’Halloran F., Fanning S., Corbett-Feeney G., Cheasty T. and Cormican M. (2003) Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J. Clin. Microbiol. 41, 1919–1924 10.1128/JCM.41.5.1919-1924.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan J.C., Ye R., Meng D.M., Zhang W., Wang H.Q. and Liu K.Z. (2006) Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J. Antimicrob. Chemother. 58, 288–296 10.1093/jac/dkl228 [DOI] [PubMed] [Google Scholar]
- 7.Farrar W.E. Jr and Eidson M. (1971) Antibiotic resistance in Shigella mediated by R factors. J. Infect. Dis. 123, 477–484 10.1093/infdis/123.5.477 [DOI] [PubMed] [Google Scholar]
- 8.Ahamed J. and Kundu M. (1999) Molecular characterization of the SHV-11 beta-lactamase of Shigella dysenteriae. Antimicrob. Agents Chemother. 43, 2081–2083 10.1128/AAC.43.8.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Z., Li T., Xu Y. and Li J. (2007) Detection of CTX-M-14 extended-spectrum beta-lactamase in Shigella sonnei isolates from China. J. Infect. 55, e125–e128 10.1016/j.jinf.2007.07.017 [DOI] [PubMed] [Google Scholar]
- 10.Zhang R., Zhou H.W., Cai J.C., Zhang J., Chen G.X., Nasu M.. et al. (2011) Serotypes and extended-spectrum beta-lactamase types of clinical isolates of Shigella spp. from the Zhejiang province of China. Diagn. Microbiol. Infect. Dis. 69, 98–104 10.1016/j.diagmicrobio.2010.08.027 [DOI] [PubMed] [Google Scholar]
- 11.Sreenivasan S., Kali A. and Pradeep J. (2016) Multidrug resistant Shigella flexneri infection simulating intestinal intussusception. J. Lab. Physicians 8, 55–57 10.4103/0974-2727.176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan G.Y., Bian F.Z., Zhu J.M., Zheng X.F., Zhang Y.F. and Zhang L. (2008) Study on plasmid-mediated extended spectrum β-lactamases and their resistance phenotypes in Shigella. Chin. J. Lab. Med. 31, 1245–1248 [Google Scholar]
- 13.Nicolas X., Granier H. and Le Guen P. (2007) Shigellosis or bacillary dysentery. Presse. Med. 36, 1606–1618 10.1016/j.lpm.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 14.Kim S., Kim J., Kang Y., Park Y. and Lee B. (2004) Occurrence of extended-spectrum beta-lactamases in members of the genus Shigella in the Republic of Korea. J. Clin. Microbiol. 42, 5264–5269 10.1128/JCM.42.11.5264-5269.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauernfeind A., Grimm H. and Schweighart S. (1990) A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18, 294–298 10.1007/BF01647010 [DOI] [PubMed] [Google Scholar]
- 16.Rubtsova M.Y., Ulyashova M.M., Bachmann T.T., Schmid R.D. and Egorov A.M. (2010) Multiparametric determination of genes and their point mutations for identification of beta-lactamases. Biochemistry (Mosc). 75, 1628–1649 10.1134/S0006297910130080 [DOI] [PubMed] [Google Scholar]
- 17.Bradford P.A. (2001) Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madiyarov R.S., Bektemirov A.M., Ibadova G.A., Abdukhalilova G.K., Khodiev A.V., Bodhidatta L.. et al. (2010) Antimicrobial resistance patterns and prevalence of class 1 and 2 integrons in Shigella flexneri and Shigella sonnei isolated in Uzbekistan. Gut Pathog. 2, 18 10.1186/1757-4749-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang C.Y., Lu P.L., Lin C.C., Lee T.M., Tsai M.Y. and Chang L.L. (2011) Integron types, gene cassettes, antimicrobial resistance genes and plasmids of Shigella sonnei isolates from outbreaks and sporadic cases in Taiwan. J. Med. Microbiol. 60, 197–204 10.1099/jmm.0.022517-0 [DOI] [PubMed] [Google Scholar]
- 20.Gassama Sow A., Aidara-Kane A., Barraud O., Gatet M., Denis F. and Ploy M.C. (2010) High prevalence of trimethoprim-resistance cassettes in class 1 and 2 integrons in Senegalese Shigella spp isolates. J. Infect. Dev. Ctries. 4, 207–212 10.3855/jidc.583 [DOI] [PubMed] [Google Scholar]