Abstract
Background
The role of the NFKB1 gene rs28362491 polymorphism and NFKBIA gene rs2233406 polymorphism in the development of head and neck cancer (HNC) remains controversial. This meta-analysis was performed to assess the relationship between the gene polymorphisms and HNC quantitatively.
Methods
PubMed, Embase, Web of Science, WanFang Data, and China National Knowledge databases were used to search for eligible articles. The relationship was evaluated by STATA 11.0.
Results
Eight eligible articles were included in our study. Nine case-control studies from the eight included articles were correlated with rs28362491 polymorphism. Four articles were related to rs2233406 polymorphism. Overall, a significant correlation was observed between the rs28362491 polymorphism and a decreased risk of HNCs (OR = 0.76, 95%CI = 0.60‐0.97 for DD vs. II; OR = 0.80, 95%CI = 0.68‐0.95 for DD vs. DI+II). In subgroup analyses, the rs28362491 polymorphism was associated with the risk of nasopharyngeal carcinoma (NC), but not with oral cancer (OC). In addition, no statistical correlation was found between the polymorphism of rs2233406 and HNCs.
Conclusion
rs28362491 polymorphism was significantly associated with the risk of HNCs, especially with NC. Additionally, our results showed that no association was discovered between rs2233406 polymorphism and HNCs.
1. Introduction
Head and neck cancer (HNC) is the sixth most common cancer [1], which arises in the oral cavity, pharynx, nasal cavity, and larynx. Appropriately 650,000 new cases of HNCs and 350,000 deaths occur each year worldwide according to the statistics [2]. As a cancer with a high morbidity rate and low survival rate, it reduces patient quality of life [3–5]. Although the primary etiology of HNC is related to human papillomavirus (HPV), smoking, alcohol, and Epstein-Barr virus (EBV) infection [6–10], only a small number of subjects exposed to these risk factors will suffer from a HNC, which suggests that individual genetic susceptibility might also be strongly correlated with the development of HNCs [3, 11, 12].
Nuclear factor-κB (NF-κB), which was identified initially in 1986, acts as an essential transcription factor associated with cell survival, cell growth, cell replication, and cell apoptosis [13–15]. NF-κB also plays an important role in the development of carcinogenesis and tumor cell's resistance to radiotherapy and chemotherapy [16]. The NF-κB family consists of five members: p65/Rel A, p50/p105, p52/p100, c-Rel, and Rel B. The heterodimer of the p65/Rel A and p50/p105 subunits, encoded by the NFKB2 and NFKB1 genes, respectively, is the most common form of NF-κB. Moreover, NF-κB activity would be influenced by NF-κB inhibitor alpha (NFKBIA) [17–19]. Inappropriate expression of NF-κB has been reported in certain human tumors, such as colorectal cancer, breast cancer, colon cancer, multiple myeloma, melanoma, prostate cancer, and Hodgkin's lymphoma [20–24].
The NFKB1 and NFKBIA genes, located on chromosome 4q24 and 14q13, respectively, are associated with the development of cancers, including oral cancer (OC) [25], gastric cancer [26], colorectal cancer [27], and Hodgkin's lymphoma [28]. In recent years, it has been reported that some polymorphisms of the NFKB1 gene (rs28362491) and NFKBIA gene (rs2233406) might be associated with HNCs [25, 29–35]. However, the results are conflicting. It might be because of small sample sizes, publication bias, and different ethnic backgrounds. Therefore, it is necessary to perform a comprehensive meta-analysis with a large sample size and high statistical power to assess the association between the polymorphisms of the NFKB1 and NFKBIA genes and HNCs. Additionally, trial sequential analysis (TSA) was also conducted to minimize random errors and strengthen the reliability of our conclusions.
2. Materials and Methods
2.1. Selection of Relevant Studies
The meta-analysis was designed and implemented according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement [36].
PubMed, Embase, Web of Science, WanFang Data, and China National Knowledge databases were searched comprehensively prior to May 1, 2019 with the following terms: “Nuclear factor-κB”, or “Nuclear factor kappa B”, or “NF-κB”, or “NFKB1”, or “rs28362491”, or “NFKBIA”, or “Nuclear factor kappa B inhibitor”, or “rs2233406” AND “carcinoma”, or “cancer”, or “neoplasm”, or “tumor”, or “carcinogenesis” AND “polymorphism”, or “variants”, or “variant”, or “mutation”, or “mutations”, or “polymorphisms” AND “head and neck”, or “HNC”, or “oral”, or “oral cavity”, or “pharyngeal”, or “laryngeal”, or “nasopharyngeal”, or “oropharyngeal”, or “laryngopharyngeal”, or “hypopharyngeal”. Finally, references cited by all the included studies were scanned to identify relevant articles.
2.2. Inclusion and Exclusion Criteria
No limitations were imposed on language or publication date. Articles were enrolled if they met all the following criteria: (1) the paper was case-controlled; (2) the paper assessed the risk of HNC and the rs28362491 polymorphism of the NFKB1 gene and/or the rs2233406 polymorphism of the NFKBIA gene; and (3) information provided by the paper was sufficient to estimate the correlation. Articles which satisfied one of the following criteria were excluded: (1) duplicated papers, reviews, editorials, case reports, commentaries, and nonhuman studies; (2) full-text was not found; and (3) no sufficient data were reported.
2.3. Data Extraction and Quality Assessment
Two writers collected the information of each enrolled article according to the inclusion and exclusion criteria independently. First author, year of publication, country, ethnicity, genotyping method, source of control, distributions of genotype, and cancer type were fetched. Quality assessment of the enrolled papers in the analysis was conducted by the Newcastle-Ottawa Scale (NOS) criteria [37].
2.4. Trial Sequential Analysis (TSA)
Considering that meta-analysis might result in false positive (type-1 error) or false negative (type-2 error) results due to the sparse data [38], TSA was applied to minimize random errors and increase the reliability of our conclusions. In the present study, TSA was conducted with an overall 5% type-1 error risk and 20% type-2 error risk [39]. According to the required information size (RIS) and risk for type-1 error and type-2 error, TSA monitoring boundaries were constructed. If the Z-curve crossed with the TSA monitoring boundary before the RIS was reached or total sample size included in the study reached the RIS, a robust conclusion might have been affirmed and further study would be unnecessary.
2.5. False-Positive Report Probability (FPRP) Analysis
We calculated the false-positive report probability (FPRP) [40] to assess the significant results. We set 0.5 as the FPRP threshold and assigned a prior probability of 0.1 to detect an odds ratio (OR) of 1.50 for an association with genotypes under investigation. Only a significant finding with a FPRP value < 0.5 was considered a noteworthy result.
2.6. Statistical Method
Statistical analysis was performed using STATA 11.0 (StataCorp LP, College Station, TX, United States). The strength of correlation between the gene polymorphisms and HNCs was evaluated through pooled odds ratios (ORs) along with the corresponding 95% confidence intervals (CIs). The ORs and CIs of each gene polymorphism were calculated for the allelic, homozygote, heterozygote, dominant, and recessive genetic models of each article, respectively. The combined ORs were calculated by Z-test, and a value of P < 0.05 was regarded as statistically significant.
The between-article heterogeneity was estimated by Cochran's Q-test and I2 statistics. If the P value in the Q − test > 0.10 and I2 < 50%, the Mantel-Haenszel fixed-effects model was used to estimate the pooled ORs. Otherwise, the DerSimonian and Laird random-effects model was performed to evaluate the correlation between the polymorphisms of the NFKB1 gene (rs28362491), NFKBIA gene (rs2233406), and HNCs. Hardy-Weinberg equilibrium (HWE) in the controls was calculated by the chi-square test [41]. In order to explore the possible heterogeneity among the articles, subgroup analyses according to the genotyping method and tumor type were performed. Sensitivity analysis was conducted by deleting a single article every time in order to assess the reliability of the overall results. Publication bias was detected by Begg's test and Egger's linear regression.
3. Results
3.1. Article Characteristics
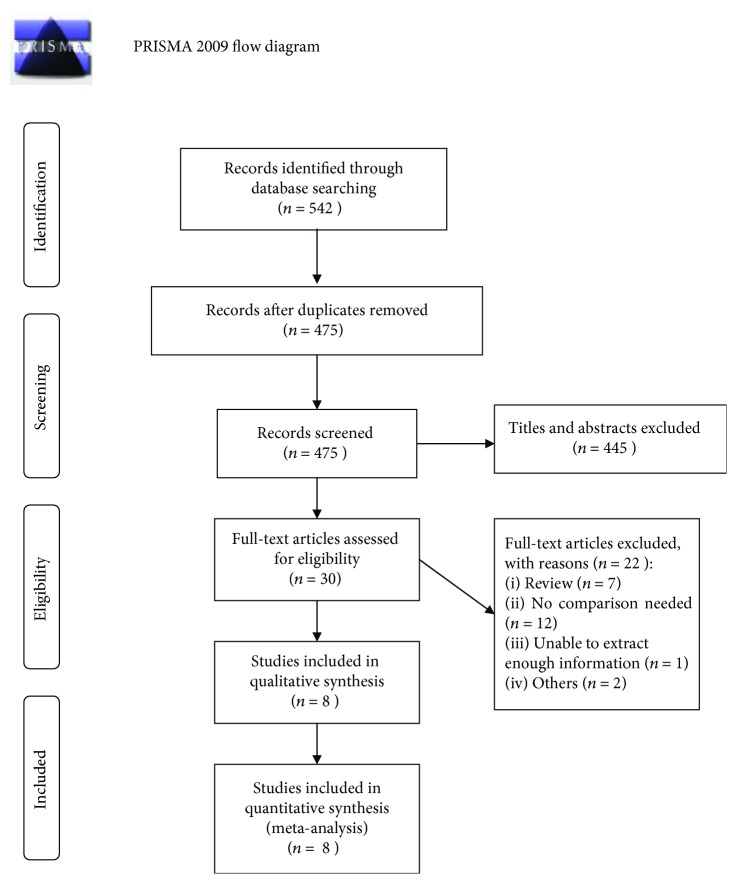
Eight articles [25, 29–35] involving 4434 cases and 4913 controls were finally enrolled in the meta-analysis. The process of study selection is demonstrated in Figure 1.
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram for a study selection process.
Nine case-control studies from eight included articles [25, 29–35] were correlated with rs28362491 polymorphism. Among the eight studies, four studies [25, 30, 34, 35] were related to rs2233406 polymorphism. Overall, the meta-analysis included three OC articles, three nasopharyngeal carcinoma (NC) articles, and two HNC articles. All studies showed that the genotype distribution in the controls was conformed to HWE. The detailed information of these enrolled papers is demonstrated in Table 1.
Table 1.
Characteristics of articles enrolled in the meta-analysis.
| FA (year) | NOS | Ethnicity | Genotyping method | SC | Cases | Controls | Cancer type | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs28362491 | ins/ins | ins/del | del/del | ins/ins | ins/del | del/del | ||||||
| Lin (2006) | 6 | Asian | PCR-PAGE | HB | 59 | 103 | 50 | 43 | 100 | 58 | OC | 0.993 |
| Lehnerdt (2008) | 7 | Caucasian Germany |
PCR-RFLP | HB | 132 | 179 | 53 | 118 | 141 | 48 | HNC | 0.586 |
| Zhou (2009) | 8 | Asian | PCR-PAGE | HB | 74 | 67 | 22 | 71 | 90 | 42 | NC | 0.177 |
| Lin (2012) | 8 | Asian | TaqMan | HB | 116 | 246 | 100 | 81 | 271 | 168 | OC | 0.099 |
| Liu (2012) | 7 | Asian | PCR | PB | 269 | 467 | 170 | 280 | 433 | 193 | NC | 0.289 |
| Liu (2015) | 7 | Asian | TaqMan | HB | 316 | 438 | 152 | 336 | 512 | 224 | NC | 0.262 |
| Liu (2015) | 7 | Asian | TaqMan | HB | 236 | 331 | 117 | 274 | 438 | 195 | NC | 0.169 |
| Chen (2018) | 8 | Asian | MassARRAY | HB | 124 | 197 | 100 | 163 | 230 | 90 | OC | 0.577 |
| Gupta (2017) | 7 | Asian | PCR-RFLP | HB | 132 | 162 | 18 | 144 | 146 | 22 | HNC | 0.064 |
|
| ||||||||||||
| rs2233406 | CC | CT | TT | CC | CT | TT | ||||||
| Lin (2012) | 8 | Asian | TaqMan | HB | 351 | 101 | 10 | 438 | 78 | 4 | OC | 0.797 |
| Liu (2012) | 7 | Asian | PCR | PB | 706 | 185 | 15 | 694 | 201 | 11 | NC | 0.402 |
| Liu (2015) | 7 | Asian | TaqMan | HB | 701 | 188 | 17 | 813 | 244 | 15 | NC | 0.492 |
| Chen (2018) | 8 | Asian | MassARRAY | HB | 308 | 108 | 9 | 365 | 110 | 8 | OC | 0.931 |
FA: first author; SC: source of control; OC: oral cancer; NC: nasopharyngeal carcinoma; HNC: head and neck cancer; HB: hospital-based study; PB: population-based study; HWE: Hardy-Weinberg equilibrium; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PCR: polymerase chain reaction; PCR-PAGE: polymerase chain reaction-polyacrylamide gel electrophoresis.
3.2. Meta-Analysis Results
The results of the overall analyses and subgroup analyses for gene polymorphisms (rs28362491 and rs2233406) with HNCs are shown in Table 2.
Table 2.
Results of overall and subgroup analyses for gene polymorphisms (rs28362491 and rs2233406).
| rs28362491 | No | D versus I | DD versus II | DI versus II | DD+DI versus II | DD versus DI+II | ||||||||||
| OR | 95% CI | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | ||
| Overall | 9 | 0.89 | 0.79-1.00 | 0.059 | 0.76 | 0.60-0.97 | 0.026 | 0.95 | 0.82-1.08 | 0.419 | 0.89 | 0.76-1.05 | 0.165 | 0.80 | 0.68-0.95 | 0.010 |
| PCR-RFLP | 2 | 1.04 | 0.89-1.22 | 0.630 | 0.96 | 0.65-1.40 | 0.813 | 1.17 | 0.93-1.48 | 0.179 | 1.13 | 0.91-1.41 | 0.274 | 0.88 | 0.62-1.26 | 0.495 |
| PCR-PAGE | 2 | 0.74 | 0.61-0.91 | 0.004 | 0.57 | 0.38-0.85 | 0.007 | 0.73 | 0.53-1.02 | 0.063 | 0.67 | 0.49-0.92 | 0.012 | 0.69 | 0.49-0.98 | 0.039 |
| TaqMan | 3 | 0.79 | 0.69-0.91 | 0.001 | 0.61 | 0.45-0.83 | 0.002 | 0.83 | 0.69-1.00 | 0.047 | 0.75 | 0.60-0.94 | 0.014 | 0.70 | 0.60-0.83 | <0.001 |
| OC | 3 | 0.86 | 0.58-1.27 | 0.457 | 0.73 | 0.32-1.65 | 0.446 | 0.82 | 0.56-1.20 | 0.314 | 0.79 | 0.46-1.33 | 0.369 | 0.84 | 0.49-1.46 | 0.543 |
| HNC | 2 | 1.04 | 0.88-1.22 | 0.630 | 0.96 | 0.65-1.40 | 0.813 | 1.17 | 0.93-1.48 | 0.179 | 1.13 | 0.91-1.41 | 0.274 | 0.88 | 0.62-1.26 | 0.495 |
| NC | 4 | 0.87 | 0.78-0.96 | 0.007 | 0.75 | 0.62-0.90 | 0.002 | 0.94 | 0.81-1.09 | 0.400 | 0.87 | 0.74-1.02 | 0.096 | 0.78 | 0.68-0.89 | <0.001 |
|
| ||||||||||||||||
| rs2233406 | No | T versus C | TT versus CC | TC versus CC | TT+TC versus CC | TT versus CC+TC | ||||||||||
| OR | 95% CI | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | OR | (95% CI) | P (z) | ||
| Overall | 4 | 1.13 | 0.90-1.42 | 0.296 | 1.51 | 0.99-2.30 | 0.058 | 1.08 | 0.85-1.39 | 0.523 | 1.12 | 0.86-1.44 | 0.399 | 1.50 | 0.98-2.29 | 0.061 |
| TaqMan | 2 | 1.25 | 0.72-2.16 | 0.431 | 1.68 | 0.93-3.04 | 0.084 | 1.19 | 0.66-2.12 | 0.565 | 1.23 | 0.68-2.23 | 0.497 | 1.67 | 0.92-3.02 | 0.089 |
| OC | 2 | 1.39 | 0.97-1.98 | 0.074 | 1.92 | 0.93-3.98 | 0.080 | 1.36 | 0.99-1.88 | 0.059 | 1.40 | 0.98-2.00 | 0.062 | 1.81 | 0.87-3.75 | 0.111 |
| NC | 2 | 0.96 | 0.83-1.10 | 0.520 | 1.33 | 0.79-2.24 | 0.290 | 0.90 | 0.77-1.05 | 0.180 | 0.92 | 0.79-1.07 | 0.296 | 1.36 | 0.81-2.29 | 0.251 |
3.2.1. Analysis for rs28362491 Polymorphism
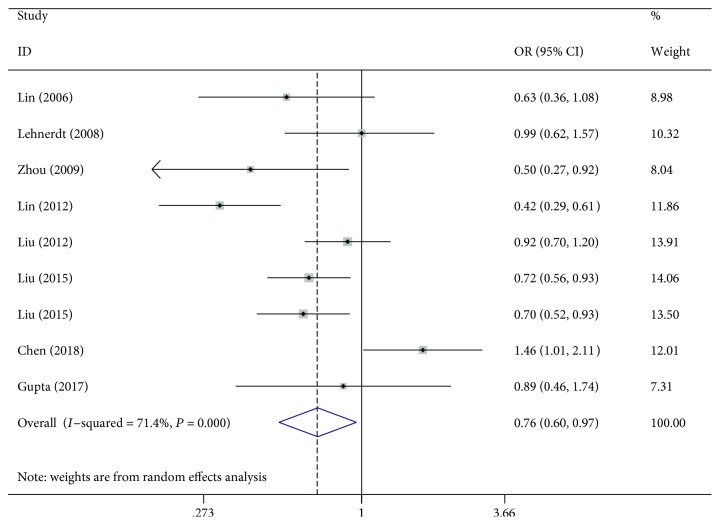
For the existence of significant heterogeneity in the Q-test (value of P in Q − test < 0.10 or I2 > 50%), the random-effects model was performed to analyze the correlation between rs28362491 polymorphism and HNCs under all genetic models. Overall, rs28362491 polymorphism was significantly correlated with a decreased risk of HNCs under homozygote and recessive genetic models (OR = 0.76, 95%CI = 0.60‐0.97 for DD vs. II, Figure 2; OR = 0.80, 95%CI = 0.68‐0.95 for DD vs. DI+II). In subgroup analyses, a significant association was discovered for the polymerase chain reaction-polyacrylamide gel electrophoresis (PCR-PAGE) genotyping method subgroup (OR = 0.74, 95%CI = 0.61‐0.91 for D vs. I; OR = 0.57, 95%CI = 0.38‐0.85 for DD vs. II; OR = 0.67, 95%CI = 0.49‐0.92 for DD+DI vs. II; OR = 0.69, 95%CI = 0.49‐0.98 for DD vs. DI+II), TaqMan genotyping method subgroup (OR = 0.79, 95%CI = 0.69‐0.91 for D vs. I; OR = 0.61, 95%CI = 0.45‐0.83 for DD vs. II; OR = 0.83, 95%CI = 0.69‐1.00 for DI vs. II; OR = 0.75, 95%CI = 0.60‐0.94 for DD+DI vs. II; OR = 0.70, 95%CI = 0.60‐0.83 for DD vs. DI+II), and NC (OR = 0.87, 95%CI = 0.78‐0.96 for D vs. I; OR = 0.75, 95%CI = 0.62‐0.90 for DD vs. II; OR = 0.78, 95%CI = 0.68‐0.89 for DD vs. DI+II).
Figure 2.
Forest plot for the association of rs28362491 polymorphism and HNC risk under a homozygote genetic model.
3.2.2. Analysis for rs2233406 Polymorphism
With respect to rs2233406 polymorphism, the random-effects model was conducted under all genetic models except the recessive model because the statistical heterogeneity between the articles was substantial (value of P in Q − test < 0.10 or I2 > 50%). We discovered that there was no relationship between the rs2233406 polymorphism and the risk of HNCs under all genetic models (OR = 1.07, 95%CI = 0.88‐1.30 for T vs. C; OR = 1.17, 95%CI = 0.65‐2.12 for TT vs. CC; OR = 0.99, 95%CI = 0.76‐1.30 for TC vs. CC; OR = 1.01, 95%CI = 0.77‐1.34 for TT+TC vs. CC; OR = 1.11, 95%CI = 0.84‐1.45 for TT vs. CC+TC). In the stratified analyses by genotyping method and tumor type, no association was observed in all genetic models (P > 0.05).
3.3. Sensitivity Analyses and Publication Bias
In the sensitivity analyses, no substantive change was discovered in the combined ORs after excluding one paper at a time. Egger linear regression tests and Begg's funnel plots were conducted to evaluate publication bias. In all the genetic models, no remarkable publication bias was found by the P value in the Egger test (D vs. I: P = 0.897; DD vs. II: P = 0.788; DI vs. II: P = 0.461; DD+DI vs. II: P = 0.550; DD vs. DI+II: P = 0.996; T vs. C: P = 0.071; TT vs. CC: P = 0.184; TC vs. CC: P = 0.052; TT+TC vs. CC: P = 0.066; TT vs. TC+CC: P = 0.235) and Begg's funnel plot for rs28362491 and rs2233406 polymorphisms.
3.4. TSA and FPRP Analysis Results
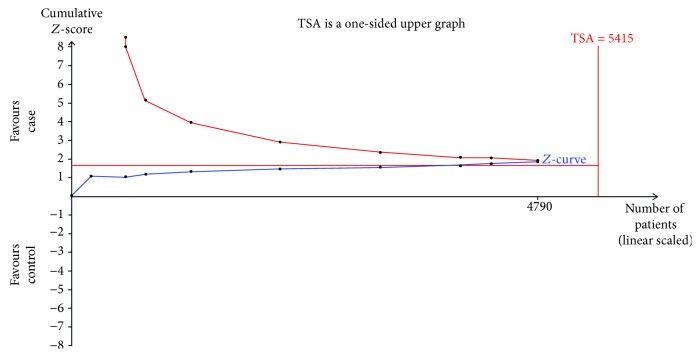
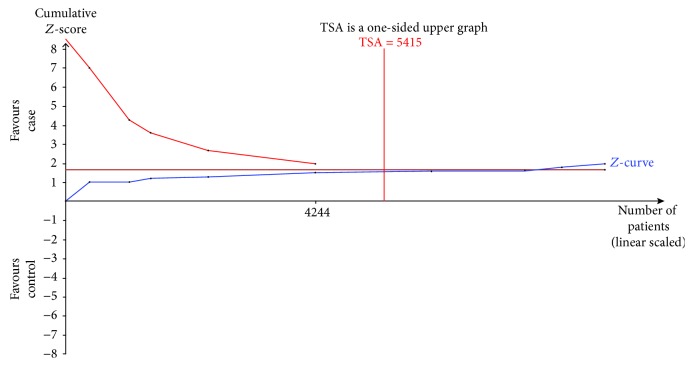
Nine trials were used to evaluate the association between the rs28362491 gene polymorphism and the HNC susceptibility. The results of TSA analysis (taking the homozygous model data for example) showed that the cumulative Z-curve had not crossed the trial monitoring boundary before the RIS was reached (Figure 3), indicating that the cumulative evidence might be insufficient and further studies are needed to strengthen the conclusion. However, the total sample size of another four genetic models included in the study all reached the RIS, suggesting that the cumulative evidence might be sufficient and further studies would be unnecessary (Figure 4 for the recessive model in rs28362491 gene polymorphism). As for rs2233406, the results of TSA analysis demonstrated that the cumulative Z-curve had not crossed the trial monitoring boundary before the RIS was reached under all the genetic models, indicating that further trials are necessary to evaluate the correlation between rs2233406 polymorphism and HNCs (figures were not shown).
Figure 3.
The required information size to demonstrate the association of rs28362491 polymorphism and HNC susceptibility under a homozygote genetic model.
Figure 4.
The required information size to demonstrate the association of rs28362491 polymorphism and HNC susceptibility under a recessive genetic model.
The FPRP values for all the discovered significant results are demonstrated in Table 3. For a prior probability of 0.1, the FPRP values were all less than 0.50 in the significant findings, indicating that these significant correlations were noteworthy.
Table 3.
False-positive report probability values for associations between the rs28362491 polymorphism and the HNC risk.
| Variables | OR (95% CI) | P a | Powerb | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| D versus I | ||||||||
| PCR-PAGE | 0.74 (0.61-0.91) | 0.004 | 0.839 | 0.015 | 0.044 | 0.338 | 0.837 | 0.981 |
| TaqMan | 0.79 (0.69-0.91) | 0.001 | 0.991 | 0.003 | 0.010 | 0.098 | 0.523 | 0.916 |
| NC | 0.87 (0.78-0.96) | 0.006 | 1.000 | 0.016 | 0.048 | 0.355 | 0.847 | 0.982 |
|
| ||||||||
| DD versus II | ||||||||
| Overall | 0.76 (0.60-0.97) | 0.027 | 0.854 | 0.088 | 0.225 | 0.761 | 0.970 | 0.997 |
| PCR-PAGE | 0.57 (0.38-0.85) | 0.006 | 0.221 | 0.073 | 0.192 | 0.723 | 0.963 | 0.996 |
| TaqMan | 0.61 (0.45-0.83) | 0.002 | 0.286 | 0.017 | 0.050 | 0.364 | 0.853 | 0.983 |
| NC | 0.75 (0.62-0.90) | 0.002 | 0.897 | 0.007 | 0.020 | 0.180 | 0.688 | 0.957 |
|
| ||||||||
| DI versus II | ||||||||
| TaqMan | 0.83 (0.69-1.00) | 0.050 | 0.989 | 0.132 | 0.313 | 0.833 | 0.981 | 0.998 |
|
| ||||||||
| DD+DI versus II | ||||||||
| PCR-PAGE | 0.67 (0.49-0.92) | 0.013 | 0.512 | 0.072 | 0.190 | 0.720 | 0.963 | 0.996 |
| TaqMan | 0.75 (0.60-0.94) | 0.013 | 0.847 | 0.042 | 0.117 | 0.594 | 0.937 | 0.993 |
|
| ||||||||
| DD versus DI+II | ||||||||
| Overall | 0.80 (0.68-0.95) | 0.011 | 0.981 | 0.032 | 0.091 | 0.524 | 0.918 | 0.991 |
| PCR-PAGE | 0.69 (0.49-0.98) | 0.038 | 0.576 | 0.166 | 0.374 | 0.868 | 0.985 | 0.998 |
| TaqMan | 0.70 (0.60-0.83) | 0.00004 | 0.713 | <0.001 | 0.001 | 0.006 | 0.054 | 0.363 |
| NC | 0.78 (0.68-0.89) | 0.0002 | 0.990 | 0.001 | 0.002 | 0.022 | 0.184 | 0.693 |
aChi-square test was adopted to calculate the genotype frequency distributions. bStatistical power was calculated using the number of observations in the subgroup and the OR and P values in this table.
4. Discussion
NF-κB plays an essential role in immune response, cell apoptosis, cell proliferation, and the development of cancers [42–44]. Moreover, many inflammatory cytokines, including IL-6, IL-8, and TNF-α would be influenced by NF-κB in regulating their biological effects. Therefore, an abnormality in NF-κB function would disturb these various biological behaviors and eventually lead to tumorigenesis [45].
The -94 ins/del ATTG (rs28362491) promoter polymorphism of the NFKB1 gene encoding p50/p105 subunit of the NF-κB family could modulate the transcription, production, and function of the p50/p105 subunit [45, 46]. NF-κB inhibitor I kappa B (IκB) would inhibit the function of NF-κB by binding to it. NF-κB inhibitor alpha (NFKBIA), which is the most common protein of the IκB family, is encoded by the NFKBIA gene. As a functional polymorphism of the NFKBIA gene, the -826 C/T (rs2233406) polymorphism might play an important role in influencing the function of NF-κB. Recently, various articles have explored the relationship between rs28362491 and rs2233406 polymorphisms and HNCs [25, 29–35]. However, the results were inconclusive.
To the best of our knowledge, this is the first meta-analysis to evaluate the association between rs28362491 and rs2233406 polymorphisms and HNCs. In the present study, nine case-control studies from eight eligible papers were enrolled to assess the correlation between -94 ins/del ATTG polymorphism and HNCs. Overall, the results showed that -94 ins/del ATTG (rs28362491) polymorphism was significantly correlated with a decreased risk of HNCs under homozygote and recessive genetic models (OR = 0.76, 95%CI = 0.60‐0.97 for DD vs. II, Figure 2; OR = 0.80, 95%CI = 0.68‐0.95 for DD vs. DI+II). Although the mechanism remains unknown, considering the essential role of NF-κB in the development of cancer, we hypothesize that the possible mechanisms are as follows. NF-κB plays a critical role in various biological pathways, which brings us to the idea that ATTG deletion results in the loss of binding to nuclear proteins, which causes lower promoter activity [47]. The promoter sequence with -94 del allele leads to decreased transcriptional activity and thereby results in decreased p50/p105 expression levels, which plays an essential role in transactivating antiapoptosis genes and restraining cell apoptosis, thereby promoting cellular proliferation. Besides, compared to the -94 ins allele carriers, the p50 in -94 del carriers could form lesser heterodimers with p65 to regulate the inflammatory pathway. Therefore, the -94 del allele might act as a protective role in HNC risk. However, no association between the rs2233406 polymorphism and the risk of HNCs was observed.
In the subgroup analyses of rs28362491 polymorphism based on the genotyping method, a statistically significant correlation was discovered in the PCR-PAGE genotyping method subgroup under allelic, homozygote, dominant, and recessive genetic models and in the TaqMan genotyping method subgroup under all the genetic models, but not in any other genotyping method subgroups. This might be because various genotyping methods would influence the relationship, indicating that it is necessary to confirm a genotyping method with a high specificity and sensitivity to increase the reliability of the results.
Since HNCs include OC, pharyngeal cancer, NC, and laryngeal neoplasm, further stratification analysis by tumor type was performed. When stratified by tumor type, we found that the rs28362491 polymorphism was significantly correlated with a decreased risk of NC under allelic, homozygote, and recessive genetic models. No association was discovered between the gene polymorphism and OC under all genetic models. This might be caused by various microenvironments, because the same gene could play an individual role in different tumor sites [48]. Surprisingly, our results were different from the results presented by four previous meta-analyses [49–52], which suggested that a significant correlation was seen between the rs28362491 polymorphism and OC. These contradictory results might be because all previous analyses only included two studies associated with OC. Therefore, the results suggested by the previous meta-analyses with limited sample sizes were unreliable.
In the subgroup analyses of rs2233406 polymorphism according to the genotyping method and tumor type, there was no relationship between the rs2233406 polymorphism and HNCs.
However, our study has some inevitable limitations. Firstly, some potential articles that have not been published were not enrolled in the present study, so a publication bias might exist. Secondly, our meta-analysis had a relatively small sample size in each subgroup, so the results of the subgroup analyses might not have enough power to identify the association. Thirdly, the environmental factors, such as smoking and alcohol, also play an essential role in the development of HNCs. Unfortunately, subgroup analyses according to smoking or alcohol consumption could not be conducted since there were no sufficient relevant data from most of the enrolled studies. Finally, the results of TSA analysis (taking the homozygous model data for example) showed that the cumulative evidence might be insufficient and further studies would be needed to strengthen the conclusion of rs2233406 polymorphism and the homozygous model in rs28362491 polymorphism. Therefore, further studies with a larger sample size and more information are required to observe the role of rs28362491 and rs2233406 polymorphisms in HNCs.
Despite these shortcomings, our meta-analysis has several highlights. To our knowledge, this present meta-analysis is the first one to conclude a relationship between rs28362491 polymorphism and HNCs and discover that rs28362491 polymorphism is associated with the development of HNCs, especially NC, although there were four meta-analyses [49–52] which focused on the association between the rs28362491 polymorphism and OC. Compared with these, our meta-analysis has the following advantages: Firstly, nine case-control studies from eight eligible papers were enrolled to assess the correlation between -94 ins/del ATTG polymorphism and HNCs; all the previous analyses only included two studies associated with OC. Secondly, the results presented by the four previous meta-analyses [49–52] were different from our results, which suggested that no significant correlation was seen between the rs28362491 polymorphism and OC. These contradictory results might be because all the previous analyses only included two studies associated with OC. Therefore, the results suggested by our meta-analyses with a higher sample size were more reliable than these previous meta-analyses. Thirdly, our study demonstrated that there was no association between the rs2233406 polymorphism and the risk of HNCs, which was not mentioned in previous articles. Finally, in order to make our conclusion more credible, publication bias analysis and sensitivity analysis were performed. Egger's linear regression tests and Begg's funnel plots demonstrated that no obvious publication bias was found. The sensitivity analysis suggested that our conclusions are reliable. Additionally, we performed FPRP analysis, and the results showed that the significant findings of our study are robust. Besides, the meta-analysis clearly shows that the rs28362491 gene polymorphism could be generally applied as a novel prognostic biomarker for HNCs, especially for NC, and it might play a protective role in populations with HNCs, which provides guidance for precise prediction of prognosis and/or therapeutic response.
5. Conclusions
In conclusion, this meta-analysis explored that the risk of HNCs was significantly correlated with rs28362491 polymorphism, but not with rs2233406 polymorphism. Moreover, the significant correlation between rs28362491 polymorphism and the susceptibility to NC was identified for the first time. However, more gene-environment and gene-gene interaction papers with larger sample sizes should be addressed to evaluate the relationship between rs28362491 and rs2233406 polymorphisms and HNCs.
Acknowledgments
The present article was funded with the following grants: the Clinical Diagnosis and Treatment Capacity Building Project of Liaoning Province (grant no. LNCCC-D17-2015) and the Natural Science Foundation of Liaoning Province, China (grant no. 2015020558).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Argiris A., Karamouzis M. V., Raben D., Ferris R. L. Head and neck cancer. The Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunotto M., Zarate A. M., Bono A., Barra J. L., Berra S. Risk genes in head and neck cancer: a systematic review and meta-analysis of last 5 years. Oral Oncology. 2014;50(3):178–188. doi: 10.1016/j.oraloncology.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Chai R. C., Lambie D., Verma M., Punyadeera C. Current trends in the etiology and diagnosis of HPV-related head and neck cancers. Cancer Medicine. 2015;4(4):596–607. doi: 10.1002/cam4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet. 2015;386(9995):743–800. doi: 10.1016/s0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marron M., Boffetta P., Zhang Z. F., et al. Cessation of alcohol drinking, tobacco smoking and the reversal of head and neck cancer risk. International Journal of Epidemiology. 2010;39(1):182–196. doi: 10.1093/ije/dyp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leemans C. R., Braakhuis B. J. M., Brakenhoff R. H. The molecular biology of head and neck cancer. Nature Reviews Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 8.Kim L., King T., Agulnik M. Head and neck cancer: changing epidemiology and public health implications. Oncology. 2010;24(10):915–919. 924. [PubMed] [Google Scholar]
- 9.Hashibe M., Brennan P., Benhamou S., et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology consortium. Journal of the National Cancer Institute. 2007;99(10):777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 10.Sankaranarayanan R., Masuyer E., Swaminathan R., Ferlay J., Whelan S. Head and neck cancer: a global perspective on epidemiology and prognosis. Anticancer Research. 1998;18(6b):4779–4786. [PubMed] [Google Scholar]
- 11.Lacko M., Braakhuis B. J. M., Sturgis E. M., et al. Genetic susceptibility to head and neck squamous cell carcinoma. International Journal of Radiation Oncology, Biology, Physics. 2014;89(1):38–48. doi: 10.1016/j.ijrobp.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 12.Munshi T., Heckman C. J., Darlow S. Association between tobacco waterpipe smoking and head and neck conditions: a systematic review. Journal of the American Dental Association. 2015;146(10):760–766. doi: 10.1016/j.adaj.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Lin S. C., Lu S. Y., Lee S. Y., Lin C. Y., Chen C. H., Chang K. W. Areca (betel) nut extract activates mitogen-activated protein kinases and NF-κB in oral keratinocytes. International Journal of Cancer. 2005;116(4):526–535. doi: 10.1002/ijc.21104. [DOI] [PubMed] [Google Scholar]
- 14.Hayden M. S., Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Naugler W. E., Karin M. NF-κB and cancer — identifying targets and mechanisms. Current Opinion in Genetics & Development. 2008;18(1):19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koti M., Gooding R. J., Nuin P., et al. Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer. 2013;13(1):p. 549. doi: 10.1186/1471-2407-13-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber S., Nikolaus S., Hampe J. Activation of nuclear factor κB inflammatory bowel disease. Gut. 1998;42(4):477–484. doi: 10.1136/gut.42.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogler G., Brand K., Vogl D., et al. Nuclear factor κB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology. 1998;115(2):357–369. doi: 10.1016/S0016-5085(98)70202-1. [DOI] [PubMed] [Google Scholar]
- 19.Neurath M. F., Fuss I., Schurmann G., et al. Cytokine gene transcription by NF-κB family members in patients with inflammatory bowel disease. Annals of the New York Academy of Sciences. 1998;859(1):149–159. doi: 10.1111/j.1749-6632.1998.tb11119.x. [DOI] [PubMed] [Google Scholar]
- 20.Jana A., Krett N. L., Guzman G., et al. NFkB is essential for activin-induced colorectal cancer migration via upregulation of PI3K-MDM2 pathway. Oncotarget. 2017;8(23):37377–37393. doi: 10.18632/oncotarget.16343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingo-Domenech J., Oliva C., Rovira A., et al. Interleukin 6, a nuclear factor-κB target, predicts resistance to docetaxel in hormone-independent prostate cancer and nuclear factor-κB inhibition by PS-1145 enhances docetaxel antitumor activity. Clinical Cancer Research. 2006;12(18):5578–5586. doi: 10.1158/1078-0432.Ccr-05-2767. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay T., Roth J. A., Maxwell S. A. Altered expression of the p50 subunit of the NF-kappa B transcription factor complex in non-small cell lung carcinoma. Oncogene. 1995;11(5):999–1003. [PubMed] [Google Scholar]
- 23.Bargou R. C., Leng C., Krappmann D., et al. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood. 1996;87(10):4340–4347. [PubMed] [Google Scholar]
- 24.Bours V., Dejardin E., Goujon-Letawe F., Merville M. P., Castronovo V. The NF-κB transcription factor and cancer: high expression of NF-κB- and IκB-related proteins in tumor cell lines. Biochemical Pharmacology. 1994;47(1):145–149. doi: 10.1016/0006-2952(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 25.Lin C. W., Hsieh Y. S., Hsin C. H., et al. Effects of NFKB1 and NFKBIA gene polymorphisms on susceptibility to environmental factors and the clinicopathologic development of oral cancer. PLoS One. 2012;7(4, article e35078) doi: 10.1371/journal.pone.0035078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo S. S., Chen J. H., Wu C. W., Lui W. Y. Functional polymorphism of NFKB1 promoter may correlate to the susceptibility of gastric cancer in aged patients. Surgery. 2009;145(3):280–285. doi: 10.1016/j.surg.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Gao J., Pfeifer D., He L. J., et al. Association of NFKBIA polymorphism with colorectal cancer risk and prognosis in Swedish and Chinese populations. Scandinavian Journal of Gastroenterology. 2007;42(3):345–350. doi: 10.1080/00365520600880856. [DOI] [PubMed] [Google Scholar]
- 28.Chang E. T., Birmann B. M., Kasperzyk J. L., et al. Polymorphic variation in NFKB1 and other aspirin-related genes and risk of Hodgkin lymphoma. Cancer Epidemiology, Biomarkers & Prevention. 2009;18(3):976–986. doi: 10.1158/1055-9965.Epi-08-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S. C., Liu C. J., Yeh W. I., Lui M. T., Chang K. W., Chang C. S. Functional polymorphism in NFKB1 promoter is related to the risks of oral squamous cell carcinoma occurring on older male areca (betel) chewers. Cancer Letters. 2006;243(1):47–54. doi: 10.1016/j.canlet.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Chen F., Liu F., Yan L., et al. A functional haplotype of NFKB1 influence susceptibility to oral cancer: a population-based and in vitro study. Cancer Medicine. 2018;7(5):2211–2218. doi: 10.1002/cam4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B., Rao L., Li Y., et al. A functional insertion/deletion polymorphism in the promoter region of NFKB1 gene increases susceptibility for nasopharyngeal carcinoma. Cancer Letters. 2009;275(1):72–76. doi: 10.1016/j.canlet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Gupta A., Agnihotri V., Kumar R., et al. Effects of tobacco habits on the polymorphism of NFKB1 and NFKB1A gene of head and neck squamous cell carcinoma in Indian population. Asian Pacific Journal of Cancer Prevention. 2017;18(7):1855–1859. doi: 10.22034/apjcp.2017.18.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehnerdt G. F., Bankfalvi A., Grehl S., et al. No association of the NF-κB1 -94ins/delATTG promoter polymorphism with relapse-free and overall survival in patients with squamous cell carcinomas of the head and neck region. International Journal of Immunopathology and Pharmacology. 2008;21(4):827–832. doi: 10.1177/039463200802100407. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Qiu F., Yang L., et al. Polymorphisms of NFκB1 and IκBα and their synergistic effect on nasopharyngeal carcinoma susceptibility. BioMed Research International. 2015;2015:9. doi: 10.1155/2015/362542.362542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y. H. The association between the genetic variants in NFKB1 and NFΚBIA and risk of Nasopharyngeal Carcinoma. Dissertation, Guangzhou Medical University; 2012. [Google Scholar]
- 36.Liberati A., Altman D. G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339(1, article b2700) doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 38.Afshari A., Wetterslev J., Smith A. F. Can systematic reviews with sparse data be trusted? Anaesthesia. 2017;72(1):12–16. doi: 10.1111/anae.13730. [DOI] [PubMed] [Google Scholar]
- 39.Wetterslev J., Thorlund K., Brok J., Gluud C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. Journal of Clinical Epidemiology. 2008;61(1):64–75. doi: 10.1016/j.jclinepi.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. Journal of the National Cancer Institute. 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhagen M., van der Meij A., van Deurzen P. A. M., et al. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Molecular Psychiatry. 2010;15(3):260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- 42.Yu Y., Wan Y., Huang C. The biological functions of NF-κB1 (p50) and its potential as an anti-cancer target. Current Cancer Drug Targets. 2009;9(4):566–571. doi: 10.2174/156800909788486759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 44.Li Q., Verma I. M. NF-κB regulation in the immune system. Nature Reviews Immunology. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 45.Mohd Suzairi M. S., Tan S. C., Ahmad Aizat A. A., et al. The functional -94 insertion/deletion ATTG polymorphism in the promoter region of NFKB1 gene increases the risk of sporadic colorectal cancer. Cancer Epidemiology. 2013;37(5):634–638. doi: 10.1016/j.canep.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Karban A. S., Okazaki T., Panhuysen C. I. M., et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Human Molecular Genetics. 2004;13(1):35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 47.Oltulu Y. M., Coskunpinar E., Ozkan G., et al. Investigation of NF-κB1 and NF-κBIA gene polymorphism in non-small cell lung cancer. BioMed Research International. 2014;2014:6. doi: 10.1155/2014/530381.530381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saidak Z., Mentaverri R., Brown E. M. The role of the calcium-sensing receptor in the development and progression of cancer. Endocrine Reviews. 2009;30(2):178–195. doi: 10.1210/er.2008-0041. [DOI] [PubMed] [Google Scholar]
- 49.Yang X., Li P., Tao J., et al. Association between NFKB1 -94ins/del ATTG promoter polymorphism and cancer susceptibility: an updated meta-analysis. International Journal of Genomics. 2014;2014:8. doi: 10.1155/2014/612972.612972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan W., Wang E., Zhang F., Wang T., You X., Qiao B. Association between the NFKB1-94ins/del ATTG polymorphism and cancer risk: an updated meta-analysis. Cancer Investigation. 2014;32(7):311–320. doi: 10.3109/07357907.2014.911881. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y. Q., Wang D., Gong T., Zhu J. An updated meta-analysis of 37 case-control studies on the association between NFKB1 -94ins/del ATTG promoter polymorphism and cancer susceptibility. Oncotarget. 2016;7(36):58659–58670. doi: 10.18632/oncotarget.10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D., Xie T., Xu J., et al. Genetic association between NFKB1 -94 ins/del ATTG promoter polymorphism and cancer risk: a meta-analysis of 42 case-control studies. Scientific Reports. 2016;6(1, article 30220) doi: 10.1038/srep30220. [DOI] [PMC free article] [PubMed] [Google Scholar]