Abstract
Colorectal cancer (CRC) is one of most common malignancies worldwide. 5-fluorouracil (5-FU) is a mainstay of CRC treatment, particularly in patients with advanced stages of the disease; however, 5-FU-based chemotherapy is not always effective and may result in progression of the disease. The present study investigated several candidate microRNAs (miRs) in parental and 5-FU-resistant HCT116 and HT29 cells, and identified miR-361 as a novel regulator of chemosensitivity. Overexpression of miR-361 enhanced the 5-FU susceptibility of parental and resistant HCT116 and HT29 cells in vitro. Impaired colony formation capacity and increased cell apoptosis (as determined via flow cytometry) was observed in resistant HCT116 and HT29 cells. Furthermore, forkhead box M1 (FOXM1) was identified as a target gene of miR-361 using a dual-luciferase reporter assay, western blotting and reverse transcription-quantitative PCR. Additionally, FOXM1 knockdown improved the cytotoxicity of 5-FU in resistant CRC. ATP binding cassette subfamily C members 5 and 10 (ABCC5/10) were found to be downstream effectors of miR-361. In conclusion, miR-361 increased chemosensitivity, at least in part, via modulation of FOXM1-ABCC5/10. miR-361 may serve as a potential therapeutic target for patients with CRC.
Keywords: microRNA-361, chemosensitivity, forkhead box M1, ATP binding cassette subfamily C, 5-fluorouracil, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignancies of the digestive system and has a worldwide incidence of >1.3 million cases/year (1). Previous studies demonstrated that ~25% of patients were diagnosed at an advanced stage of the disease, and ~50% of patients suffered from liver metastasis, which significantly increased the mortality rates for CRC (2). This highlights the need to identify effective therapeutic agents to treat this malignant disease.
In the past decades, 5-fluorouracil (5-FU) has been a mainstay of CRC treatment, and several clinical trials have revealed that chemotherapy regimens consisting of 5-FU in combination with other cytotoxic agents, such as irinotecan and oxaliplatin, significantly improved the survival of patients with advanced CRC (3–5). However, 5-FU-based chemotherapy is not always effective and the disease may progress, due to the development of chemo-resistance (6). Therefore, it may be beneficial to personalize treatment based on the individual molecular characteristics of the tumor. Development of novel targets to increase the sensitivity of tumors to 5-FU may enhance the cytotoxic effects of 5-FU on tumor cells and decrease the adverse effects of chemotherapy on the immune system. Furthermore, the reduction in treatment costs would have economic benefits.
It has been demonstrated that microRNAs (miRNAs/miRs) play a vital role in the regulation of genes exerting antitumor effects, including genes driving cell apoptosis, inhibiting cell proliferation and regulating drug efflux mechanisms (7,8). miRNAs are a class of small, endogenous, non-coding, regulatory RNA molecules. These small RNAs are 18–24 nucleotides in length and are involved in the regulation of the expression of two-thirds of all human genes through binding to mRNA 3′-untranslated regions (3′UTRs) (9–11). It was previously demonstrated that miRNA-mediated gene regulation is an important process in the occurrence, pathogenesis and progression of several diseases, including gastrointestinal cancer, immune-related diseases and neurodegenerative diseases (10). miRNAs may act as oncogenes or tumor suppressors in tumor occurrence and development. miRNAs may act as oncogenes, referred to as ‘oncomirs’, by inhibiting the effect of tumor suppressor genes or regulating cell apoptosis, promoting the occurrence and development of tumors (8). The identification of miRNAs implicated in the response to antitumor therapy has revealed novel therapeutic approaches to reverse drug resistance (8). For example, overexpression of miR-216b sensitized non-small cell lung cancer cells to cisplatin-induced apoptosis (12). miR-30a overexpression inhibited chemoresistance-associated autophagy in gastric cancer cells (13). Furthermore, overexpression of miR-22 increased the sensitivity of osteosarcoma cells to cisplatin treatment (14). Although several studies have reported that miRNAs can regulate chemosenstivity of CRC cells (7,10,15–19), the molecular mechanism(s) underlying the role of miRNAs in the chemoresistance of CRC has not been fully elucidated.
The present study aimed to identify candidate miRNAs that regulate the susceptibility of CRC cells to 5-FU treatment. Additionally, the downstream targets of the candidate miRNAs associated with the 5-FU response were also investigated. The results obtained in the present study may provide a novel therapeutic strategy to overcome 5-FU resistance in CRC.
Materials and methods
Cell culture and materials
CRC HCT116 and HT29 cell lines were purchased from the Type Culture Collection of the Chinese Academy of Sciences. Cells were grown in high glucose Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The 5-FU-resistant colorectal carcinoma cell lines HCT116-Res and HT29-Res were established as previously described (20). Briefly, the CRC cells were treated with 1 µg/ml of 5-FU (Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2 for 24 h. The spent medium was replaced by fresh culture medium, including DMEM containing 5% fetal bovine serum and 1% penicillin/streptomycin. Upon reaching a confluence of 90%, cells were treated with 2.5 µg/ml 5-FU at 37°C with 5% CO2 for 24 h. This process was repeated using 5 and 10 µg/ml 5-FU. The 5-FU-resistant cell lines HCT116-Res and HT29-Res were therefore established by exposure to gradually increasing concentrations of 5-FU (1, 2.5, 5 and 10 µg/ml).
Cell viability
Cell viability was quantified with a Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.). Cells were plated into 96-well plates at a density of 3×103 cells/well with 100 µl DMEM containing 0, 5, 10 and 20 µg/ml of 5-Fu at 37°C with 5% CO2 for 48 h. Subsequently, 10 µl CCK-8 solution was added into each well, followed by incubation for 2 h at 37°C. The absorbance (A) was read at a wavelength of 450 nm using an absorbance microplate reader (BioTek Instruments, Inc.). The cell viability was expressed as the relative percentage compared with the mean absorbency of the untreated CRC cell lines (control cells). The cell viability percentage was calculated using the following equation: Relative cell viability (% of control)=[(Asample-Ablank)/(Acontrol-Ablank)] ×100. The assay was repeated at least three times.
Transfection of miRNA mimic and siRNA
The miR-361 mimic (sequence 5′-ACCCCUGGAGAUUCUGAUAAUU-3′), miR-361 mimic negative control (NC) (sequence 5′-UUCUCCGAACGUGUCACGUTT-3′), Forkhead box M1 (FOXM1) small interfering (si)RNA (siFOXM1 sequence: Sense: 5′-GGACCACUUUUCCCUACUUUDTDT-3′, antisense: 5′-AAAGUAGGGAAAGUGGUCCDTDT-3′) and siRNA NC (sequence 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′) were designed and synthesized by Shanghai GenePharma Co., Ltd. A total of 2×105 HCT116-Res and HT29-Res cells were seeded in 6-well plates. On reaching ~30% confluence, the cells were transfected with miR-361 mimic or miR NC (both 100 nM) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. siFOXM1 or siRNA NC (both 100 nM) was transfected into HCT116-Res cells or HT29-Res cells by using Lipofectamine® 3000, according to the manufacturer's protocol. RNA extraction and reverse-transcription quantitative PCR (RT-qPCR) experiments were conducted 24 h after transfection, and western blot analysis, colony formation, apoptosis and luciferase assays were carried out 48 h after the transfection.
RNA extraction and RT-qPCR
Total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. miRNA was obtained using an miRcute miRNA extraction kit (Tiangen Biotech Co. Ltd.). RNA and miRNA were reverse transcribed into cDNA using a GoScript™ RT reagent kit (Promega Corporation) according to the manufacturer's protocol. qPCR analysis was subsequently performed using SYBR® Green Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) and an ABI Prism 7900 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Literature review indicated that several miRNAs have been reported to be associated with chemoresistance (7,10,15–19), and were therefore utilized as candidate miRNAs in this study, and their role in 5-Fu resistance was confirmed. The expression of the miRNA candidates by qRCR were identified. The thermocycling conditions were as follow: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec and a final melt curve stage which was 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The following primer pairs were used for qPCR: FOXM1 forward, 5′-CGTCGGCCACTGATTCTCAAA-3′ and reverse, 5′-GGCAGGGGATCTCTTAGGTTC-3′; β-actin forward, 5′-AGAGCTACGAGCTGCCTGAC-3′ and reverse, 5′-AGCACTGTGTTGGCGTACAG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′, and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; miR-361-5p loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTACCCC-3′, forward primer, 5′-ACACTCCAGCTGGGTTATCAGAATCTCCA-3′; miR-21 loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACAT-3′, forward primer, 5′-ACACTCCAGCTGGGUAGCUUAUCAGACUG-3′; miR-224 loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTAAACG-3′, forward primer, 5′-ACACTCCAGCTGGGUCAAGUCACUAGUGGUUC-3′; miR-134 loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCTCCCC-3′, forward primer, 5′-ACACTCCAGCTGGGUGUGACUGGUUGACC-3′; miR-29a loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTAACCGA-3′, forward primer, 5′-ACACTCCAGCTGGGUAGCACCAUCUGAAA-3′; miR-29c loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTAACCGA-3′, forward primer, 5′-ACACTCCAGCTGGGUAGCACCAUUUGAAA-3′; miR-149 loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGGAGTG-3′, forward primer, 5′-ACACTCCAGCTGGGUCUGGCUCCGUGUCUU-3′; miR-200c loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCATCA-3′, forward primer, 5′-ACACTCCAGCTGGGUAAUACUGCCGGGUAA-3′; miR-34a loop reverse primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACCA-3′, forward primer, 5′-ACACTCCAGCTGGGUGGCAGUGUCUUAGC-3′ and the QPCR reverse primer for all miRNA candidates was 5′-TGGTGTCGTGGAGTCG-3′. β-actin and U6 were used for mRNA and miRNA normalization, respectively. Relative quantification of gene expression levels were expressed as fold-change using the 2−ΔΔCq method (21). The relative fold change of miRNAs >1.5 was considered as upregulated miRNAs, while <-1.5 as downregulated miRNAs.
Western blotting
Total protein of HCT116, HT29, HCT116-Res, HT29-Res cells was extracted using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) and a protease inhibitor cocktail (Roche Diagnostics GmbH) according to the manufacturer's protocol. Total protein was quantified using a Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). The cell lysate supernatant was mixed with 6X SDS loading buffer (Beyotime Institute of Biotechnology) and boiled at 100°C for 10 min. Subsequently, 30 µg protein/lane was separated via 10% SDS-PAGE and transferred onto PVDF membranes (Merck KGaA). The membrane was blocked for 1 h at room temperature using 5% non-fat milk dissolved in Tris buffered saline with 0.1% Tween (TBS-T). The membrane was incubated with the following primary antibodies overnight at 4°C: Anti-FOXM1 (1:1,000; cat. no. ab180710 Abcam) and β-actin (1:1,000; cat. no. AA132 Beyotime Institute of Biotechnology), Anti-ABCC5 (1:1,000; cat. no. ab180724; Abcam), Anti-ABCC10 (1:1,000; cat. no. ab91451; Abcam). Membranes were subsequently washed in TBS-T three times and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000; cat. no. A0208 for anti-rabbit and cat. no. A0216 for anti-mouse; Beyotime Institute of Biotechnology) for 1 h at room temperature. The protein bands were visualized using Millipore Immobilon Western Chemiluminescent HRP substrate (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Each reaction was performed in triplicate.
Luciferase assay
Wild-type FOXM1-3′UTR containing a miR-361-5p targeted region or mutant-form 3′UTR was inserted into a pmirGlO Dual-luciferase miRNA Target Expression Vector (cat. no. E1330; Promega Corporation). The HCT116-Res or HT29-Res cells (2×105) were seeded into 24-well plate to reached 70% confluence. Then HCT116-Res or HT29-Res cells were transfected with 2 µg Wild-type FOXM1-3′UTR or mutant-form 3′UTR and 100 nM miR-361-5p mimics using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) Cells were analyzed for luciferase activity was measured 48 h following transfection using the Dual-Glo® Luciferase Assay System (cat. no. E2920; Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Colony formation assay
5-FU-resistant HCT116 or HT29 cells transfected with FOXM1 siRNA, miR-361 mimic or respective NCs were plated in 6-well plates (500 cells/well) and incubated at 37°C for 14 days to allow colony formation. The cell medium was subsequently removed and cells were washed using PBS. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature. The cells were stained with crystal violet kit (cat. no. C0121; Beyotime Institute of Biotechnology) for 15 min at room temperature, according to the manufacturer's protocols. The colonies were washed, imaged by Sony camera (NEX-3N; SONY Corp.) and counted using ImageJ software.
Apoptosis assay
Flow cytometry was performed to detect cell apoptosis by Annexin V and propidium iodide (PI) double staining. 5-FU resistant HCT116 and HT29 cells transfected with the miR-361 mimic and NC were harvested using non-EDTA trypsinase and stained with Annexin V-FITC kit (Miltenyi Biotec, Inc.), according to the manufacturer's protocol. Subsequently, stained cells were detected using a flow cytometer (FACSCanto II; BD Biosciences) and analyzed by BD FACSDiva 8.0.1 software (BD Biosciences). Additionally, a Caspase-Glo 3/7 kit (cat. no. G8090; Promega Corporation) was used to measure caspase 3/7 activity according to the manufacturer's protocols.
miRNA targets prediction
MicroRNA.org (http://www.microrna.org) is a comprehensive resource of microRNA target predictions and expression profiles (22). Target predictions are based on a development of the miRanda algorithm. The algorithm provided a machine learning method named mirSVR to rank microRNA target sites by a downregulation score. By using this online prediction software (August 2010 Release) (23,24), it was found that miR-361 binds to the 3′UTR region of FOXM1 with highest mirSVR score among the predicted targets, and FOXM1 was therefore selected for further study.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 6.0; GraphPad Software, Inc.). Data are presented as the mean ± standard deviation. Differences between treated and untreated control cells were assessed using the unpaired Student's t-test. The Bonferroni test was used to evaluate differences between multiple groups following two-way ANOVA analysis. All experiments were performed at least three times. P<0.05 was used to indicate a statistically significant difference.
Results
Expression level of miR-361 is decreased in 5-FU-resistant HCT116 and HT29 cells
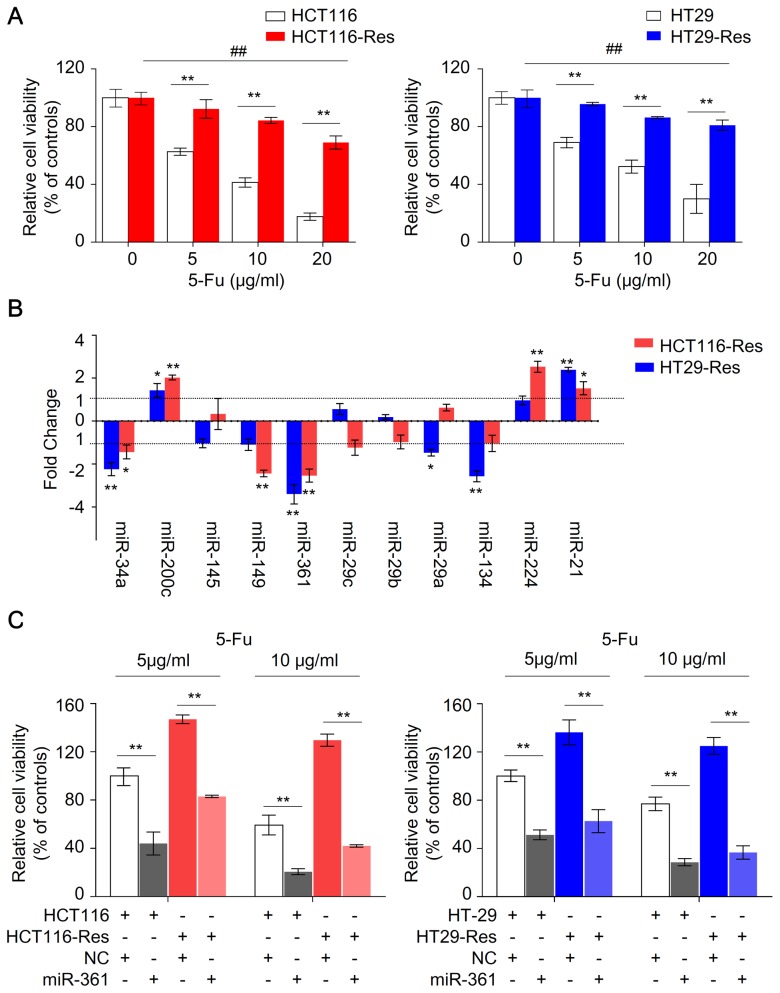
5-FU-resistant HCT116 and HT29 cells were successfully established in the current study using a methodology described in a previous study (25). The resistant and parental cells were treated with 5, 10 and 20 µg/ml 5-FU for 48 h. The viability of parental HCT116 and HT29 cells was significantly decreased compared with the resistant cells (P<0.01; Fig. 1A). The cell viability of resistant HCT116 and HT29 cells at 5 and 10 µg/ml of 5-FU was not significantly inhibited (P>0.05); however, inhibition was observed in resistant cells in the presence of 20 µg/ml 5-FU (P<0.01), suggesting that the resistant cells in the present study exhibited optimal survival in 5 and 10 µg/ml 5-FU.
Figure 1.
miR-361 is upregulated in 5-FU-resistant HCT116 and HT29 cells. (A) Parental and 5-FU-resistant HCT116 and HT29 cells were treated with 5-FU at the indicated concentrations for 48 h and subjected to a CCK-8 assay. Cell viability was presented as the relative value normalized to the 0 µg/ml group (n=6; **P<0.01, parental vs. resistant cells; ##P<0.01, the viability of resistant cells at 20 µg/ml vs. 0 µg/ml). (B) Relative differential expression of candidate miRNAs between parental and 5-FU resistant HCT116 and HT29 cells (n=3; *P<0.05, **P<0.01, vs. parental cells). (C) Overexpression of miR-361 sensitizes parental and resistant HCT116 and HT29 to 5-FU. The parental and 5-FU-resistant HCT116 and HT29 cells were transfected with an miR-361 mimic or NC, exposed to 5-FU at the indicated concentrations for 48 h and subjected to a CCK-8 assay (n=6; **P<0.01). miR/miRNA, microRNA; 5-FU, 5-fluorouracil; CCK-8, Cell Counting Kit-8; NC, negative control; Res, resistant.
To identify novel miRNAs associated with chemoresistance in CRC, the expression levels of several candidate miRNAs in parental and resistant HCT116 and HT29 cells were measured by qPCR (Fig. 1B). miR-21, miR-224 and miR-200c were upregulated, whereas miR-34a, miR-361 miR-134 were downregulated in resistant cells compared with parental cells. Among these miRNAs, the expression level of miR-361 was the most markedly altered, with ~3.5-fold change in resistant HCT116 and HT29 cells compared with parental cells, suggesting that that miR-361 is a potential regulator of 5-FU chemosensitivity in CRC.
Overexpression of miR-361 sensitizes resistant CRC cells to 5-FU, inhibits colony formation and induces apoptosis
To investigate whether miR-361 modulates sensitivity to 5-FU in CRC, parental and resistant cells were transfected with miR-361 mimic or NC, and exposed to 5 or 10 µg/ml 5-FU. Overexpression of miR-361 enhanced the susceptibility of parental HCT116 and HT29 cells to 5-FU and increased 5-FU sensitivity in resistant cells, suggesting that miR-361 may sensitize CRC cells to 5-FU (Fig. 1C). Colony formation and apoptosis assays were subsequently performed to further investigate this effect.
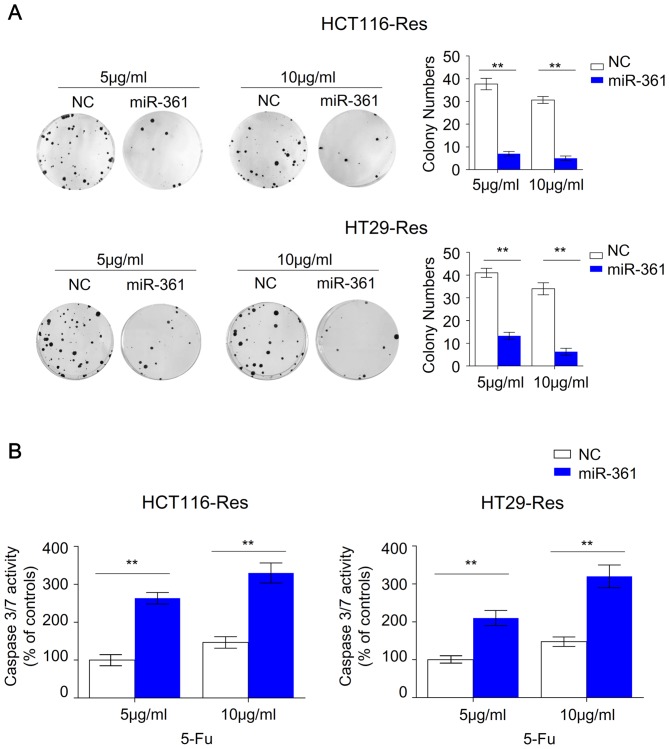
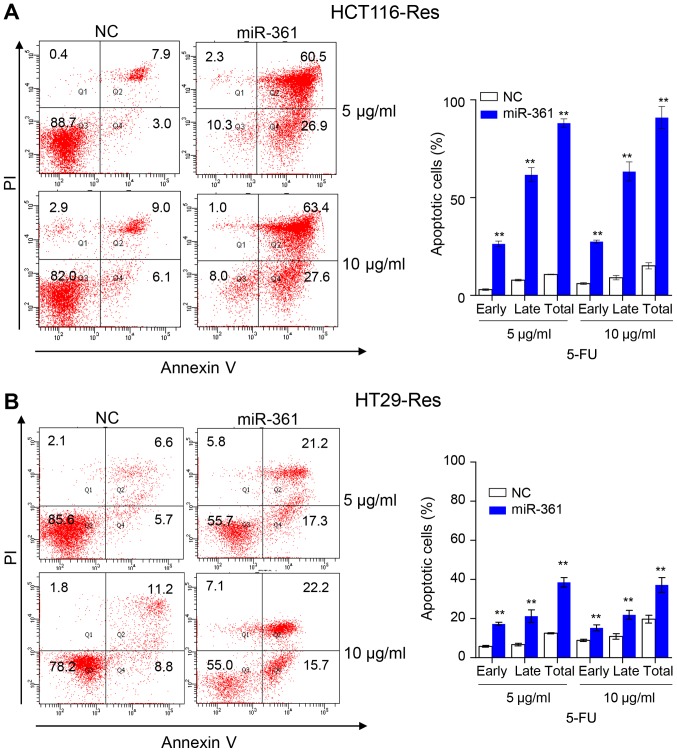
Overexpression of miR-361 in resistant HCT116 and HT29 cells significantly reduced colony numbers at 5 or 10 µg/ml 5-FU, suggesting that miR-361 significantly impaired the ability of resistant cells to grow under 5-FU treatment (Fig. 2A). Previous studies revealed that 5-FU-resistant CRC cells are able to survive under 5-FU treatment with decreased apoptosis compared with parental cells (26–29). The present study subsequently investigated whether the inhibited cell viability and colony formation of the resistant cells was caused by miR-361-induced apoptosis. Overexpression of miR-361 in resistant HCT116 and HT29 cells significantly increased caspase 3/7 activity compared with the NC, suggesting that apoptotic signaling pathways were activated (Fig. 2B). Furthermore, an Annexin V/PI assay revealed that miR-361 overexpression induced apoptosis in resistant HCT116 and HT29 compared with the NC (Fig. 3). Resistant HCT116 cells overexpressing miR-361 exhibited a higher percentage of early apoptotic cells compared with NC cells (26.4±1.4% vs. 2.9±0.4% at 5 µg/ml 5-FU, P<0.001; 27.6±0.8% vs. 6.1±0.5% at 10 µg/ml 5-FU, P<0.001) or a higher percentage of late apoptotic cells compared with NC cells (61.7±3.6% vs. 7.8±0.5% at 5 µg/ml 5-FU, P<0.001; 63.4±4.9% vs. 9.1±1.1% at 10 µg/ml 5-FU, P<0.001). Similarly, resistant HT29 cells overexpressing miR-361 exhibited a higher percentage of early apoptotic cells compared with NC cells (17.3±0.8% vs. 5.8±0.5% at 5 µg/ml 5-FU, P<0.001; 15.3±1.5% vs. 8.9±0.6% at 10 µg/ml 5-FU, P<0.001) or a higher percentage of late apoptotic cells compared with NC cells (21.3±3.2% vs. 6.7±0.7% at 5 µg/ml 5-FU, P<0.001; 21.9±2.3% vs. 10.9±1.4% at 10 µg/ml 5-FU, P<0.001).
Figure 2.
miR-361 inhibits colony formation and activates caspase 3/7 in resistant HCT116 and HT29 cells. (A) Colony formation ability of resistant miR-361-overexpressing cells and NC cells (n=3; **P<0.01). (B) Caspase 3/7 activities of resistant miR-361 overexpressing cells and NC cells were measured via a caspase 3/7 Glo Luc assay (n=3; **P<0.01). miR, microRNA; NC, negative control; Res, resistant; 5-FU, 5-fluorouracil.
Figure 3.
miR-361 induces cell apoptosis in resistant HCT116 and HT29 cells. Annexin V staining of resistant miR-361-overexpressing (A) HCT116 and (B) HT29 cells. Q1: Dead cells, Q2: Late apoptotic cells, Q3: Living cells, Q4: Early apoptotic cells. The histogram indicated the percentage of early, late and total apoptotic cells with different concentrations of 5-FU. The total apoptotic cells include early and late apoptotic cells (n=3, **P<0.01 vs. NC cells). miR, microRNA; 5-FU, 5-fluorouracil; NC, negative control; Res, resistant.
miR-361 is a negative regulator of FOXM1 expression in 5-FU-resistant cells
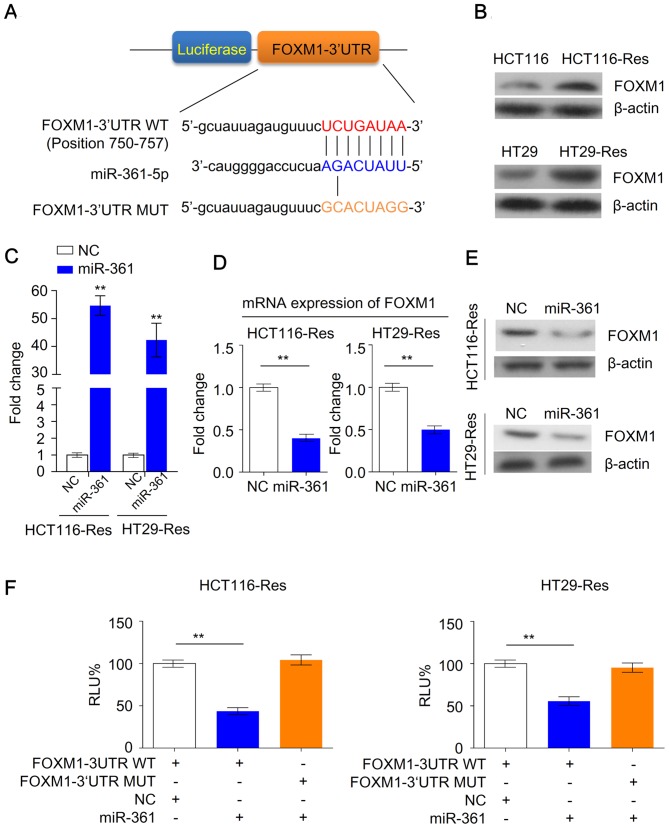
As miR-361 may serve an important role in the regulation of chemosensitivity in CRC cells, the present study investigated the potential targets of miR-361 in resistant cells. Despite the complex interactions between miRNAs and mRNAs, several algorithms are available for exploring such interactions. By using microRNA algorithms (www.microrna.org), FOXM1 was predicted as a potential target of miR-361 (Fig. 4A). Furthermore, western blotting demonstrated that FOXM1 was upregulated in resistant cells compared with parental cells (Fig. 4B). As miR-361 was previously revealed to be downregulated in resistant cells (Fig. 1B), this suggested that FOXM1 may be a potential target of miR-361.
Figure 4.
miR-361 is a direct regulator of FOXM1 in resistant colorectal cancer cells. (A) Schematic representation of the luciferase reporter constructs. The phosphoglycerate kinase promoter in the reporter construct drives the constitutive transcription of a chimeric mRNA containing the firefly luciferase coding sequence fused to the wide-type or mutated FOXM1 3′UTR. (B) FOXM1 expression in parental and 5-FU resistant colorectal cancer cells. (C) Expression levels of miR-361 in resistant HCT116 and HT29 cells transfected with miR-361 mimic or NC (n=3; **P<0.01 vs. NC cells). (D) mRNA and (E) protein levels of FOXM1 in resistant HCT116 and HT29 cells overexpressing miR-361 and NC (n=3; **P<0.01). (F) Relative activity of the luciferase gene fused with the wild-type or mutant FOXM1 3′UTR in resistant HCT116 and HT29 cells. Data were normalized to Renilla luciferase activity (n=3; **P<0.01) miR, microRNA; FOXM1, forkhead box M1; UTR, untranslated region; 5-FU, 5-fluorouracil; NC, negative control; Res, resistant; RLU%, Percentage of relative luminescence; WT, wild type; Mut, mutant.
To further investigate the effects of miR-361 on FOXM1 expression in resistant HCT116 or HT29 cells, miR-361 was overexpressed in CRC cells (Fig. 4C). FOXM1 mRNA and protein levels were significantly decreased following overexpression of miR-361 compared with NC cells, suggesting that FOXM1 may be a direct target of miR-361 in resistant cancer cells (Fig. 4D and E). Luciferase reporter assays were performed to further investigate whether miR-361 regulates the expression of FOXM1 in resistant cells in a direct or indirect manner. miR-361 binding sites of FOXM1 were cloned into luciferase reporter plasmids to establish a wild type FOXM1 3′UTR reporter plasmid. Mutated miR-361 binding sites were cloned to establish a mutant FOXM1 3′UTR reporter plasmid. Resistant HCT116 and HT29 cells were transiently transfected with these reporter constructs along with the NC or the miR-361 mimic. Overexpression of miR-361 reduced the luciferase activity in cells transfected with the wild type FOXM1 3′UTR reporter plasmid; however, no decreased activity was observed in cells transfected the mutated FOXM1 3′UTR reporter plasmid (Fig. 4F). Taken together, these data suggested that miR-361 suppressed FOXM1 expression through direct binding to the putative 3′UTR binding site of FOXM1 mRNA.
Targeting FOXM1 improves the cytotoxicity of 5-FU in resistant CRC cells
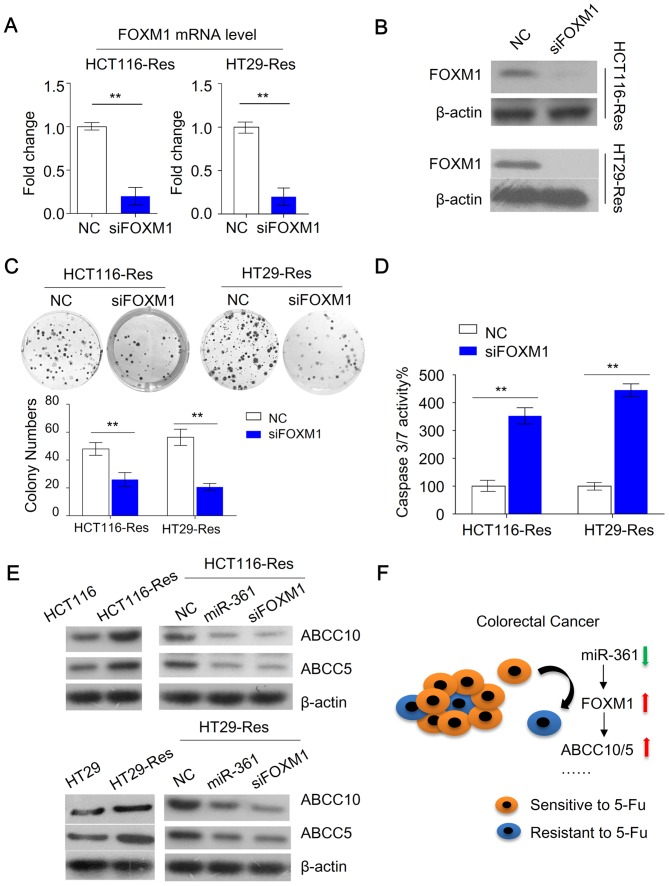
The current study investigated whether changes in FOXM1 expression modulated the chemosensitivity of resistant CRC cells. Knockdown of FOXM1 in resistant HCT116 cells was achieved using FOXM1 siRNA (Fig. 5A and B). The cells were subsequently subjected to colony formation and caspase 3/7 activity assays. FOXM1 knockdown inhibited colony formation compared with NC cells (Fig. 5C). Furthermore, FOXM1 knockdown increased caspase 3/7 activity compared with NC cells (Fig. 5D). Recent studies demonstrated that FOXM1 promotes chemoresistance by upregulating ATP binding cassette subfamily C (ABCC) 10 (30) and ABCC5 (31), which mediate drug efflux leading to acquired drug resistance. The present study investigated whether inhibition of FOXM1 expression affected ABCC10 and ABCC5 expression. Decreased FOXM1 expression resulted in a downregulation of ABCC10 and ABCC5 compared with the NC (Fig. 5E). Furthermore, resistant HCT116 cells overexpressing miR-361 exhibited reduced expression of ABCC10 and ABCC5 compared with the NC (Fig. 5E). Collectively, these data suggested that miR-361 exerted functions in CRC, at least in part, through inhibition of FOXM1, and its downstream targets ABCC10 and ABCC5 (Fig. 5F).
Figure 5.
Inhibition of FOXM1 suppresses colony formation and induces apoptosis through downregulation of ABCC5/10. (A) mRNA and (B) protein levels of FOXM1 were downregulated by siRNA knockdown. (C) Knockdown of FOXM1 resulted in impaired colony formation (D) Knockdown of FOXM1 in resistant cells increased caspase 3/7 activity (n=3; **P<0.01). (E) Protein levels of ABCC5/10 in parental, resistant cells, miR-361-overexpressing and FOXM1 knockdown cells. (F) Reduced expression of miR-361 in colorectal cancer cells contributes to 5-FU chemoresistance by activating the FOXM1-ABCC5/10 signaling pathway. FOXM1, forkhead box M1; ABCC, ATP binding cassette subfamily C; si(RNA), small interfering (RNA); NC, negative control; miR, microRNA; 5-FU, 5-fluorouracil; Res, resistant.
Discussion
CRC is a leading cause of mortality worldwide (1). Patients diagnosed at an early stage typically have a good prognosis; however, a number of patients present with liver metastasis at initial diagnosis, and the prognosis of such patients is usually poor (2). 5-FU-based chemotherapy is a commonly used treatment strategy for patients with advanced CRC. Chemoresistance is associated with the recurrence of CRC during clinical therapy with 5-FU (3–6). The molecular mechanisms underlying chemoresistance in CRC remain largely unknown. It was previously demonstrated that miRNAs regulate diverse biological processes in cancer cells and may serve an important role in chemosenstivity (32). The present study demonstrated that miR-361 is a novel regulator of chemosensitivity in CRC. Furthermore, modulation of miR-361 expression increased the chemosensitivity of resistant CRC cells. Additionally, the present study revealed that miR-361 functions as a chemosensitizer through the FOXM1-ABCC10/5 signaling pathway.
Numerous studies reported aberrant serum or tissue miRNA levels in patients with CRC, and these dysregulated expression profiles are often associated with aggressive clinical phenotypes, drug resistance or poor prognosis (18,33). The current study established in vitro 5-FU-resistant CRC cells to identify 5-FU-associated miRNAs. Several candidate miRNAs, which may function as tumor suppressors or oncogenes, were identified. miR-21, miR-224 and miR-200c were upregulated, whereas miR-34a, miR-361 and miR-134 were downregulated in the resistant cells. Among these miRNAs, the expression levels of miR-361 were the most significantly changed. miR-361 is a tumor suppressor and inhibits cell proliferation and invasion of several types of cancer cells, such as gastric cancer, breast cancer and thyroid cancer (34–36). To the best of our knowledge, the present study is the first to demonstrate that miR-361 restores 5-FU sensitivity in resistant CRC cells, by inhibiting cell viability, colony formation and inducing cell apoptosis, suggesting that miR-361 may serve as a promising therapeutic target to enhance chemosensitivity to 5-FU in CRC.
It has been reported that miRNAs may serve as potential therapeutic agents due to their ability to regulate the expression of multiple target genes (10). While the identification of downstream targets of miRNAs regulating 5-FU sensitivity is challenging, the present study used bioinformatics prediction and several in vitro approaches to identify FOXM1 as an important direct target of miR-361. Previous studies reported that FOXM1 was frequently upregulated in colorectal cancer tissues and serves as a prognostic marker in CRC (9,37). Moreover, overexpression of FOXM1 in CRC cells was shown to correlate with 5-FU resistance (19,30,38). Therefore, 5-FU-based chemotherapy may not be appropriate for patients with CRC with upregulated FOXM1 expression. Moreover, ABCC5 and ABCC10, revealed as downstream targets of FOXM1 in the present study, have been previously suggested as promising therapeutic agents in patients with resistant cancer, such as breast cancer and pancreatic cancer (39–42).
The present study had a number of limitations. The association between miR-361 expression and its target genes FOXM1, ABCC5 and ABCC10 was not evaluated in tissues from 5-FU resistance patients with CRC or animal models with 5-FU resistant xenograft. Future experiments to validate the interaction between miR-361 and its target genes in a large cohort of patients with CRC with complete chemoresponse information are required. Moreover, an miR-361-expressing xenograft animal model to investigate the regulatory effect of miR-361 on its target genes is required.
In summary, the current study investigated the association between miR-361 and FOXM1-ABCC5/10 in 5-FU resistance in CRC. miR-361 may regulate chemosensitivity to 5-FU by targeting FOXM1-ABCC5/10. miR-361-based therapy may serve as a potential strategy to enhance 5-FU sensitivity in patients with resistant CRC. Furthermore, the addition of FOXM1 or ABCC5/10 inhibitors to a chemotherapeutic regimen may substantially reduce the required 5-FU dosage in patients with CRC. The results obtained in the current study may provide novel targets for the treatment of patients with advanced or chemoresistant CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JS conceived and designed the study and participated in data analysis. LZ performed the experiment and drafted the manuscript. BL and BZ performed the data analysis and interpretation. HZ performed the statistical analysis.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser R, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, ESMO Guidelines Working Group Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–iii9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 3.Montagnani F, Chiriatti A, Turrisi G, Francini G, Fiorentini G. A systematic review of FOLFOXIRI chemotherapy for the first-line treatment of metastatic colorectal cancer: Improved efficacy at the cost of increased toxicity. Colorectal Dis. 2011;13:846–852. doi: 10.1111/j.1463-1318.2010.02206.x. [DOI] [PubMed] [Google Scholar]
- 4.Cersosimo RJ. Management of advanced colorectal cancer, Part 1. Am J Health Syst Pharm. 2013;70:395–406. doi: 10.2146/ajhp110532b. [DOI] [PubMed] [Google Scholar]
- 5.Cersosimo RJ. Management of advanced colorectal cancer, Part 2. Am J Health Syst Pharm. 2013;70:491–506. doi: 10.2146/ajhp110532b. [DOI] [PubMed] [Google Scholar]
- 6.Mohelnikova-Duchonova B, Melichar B, Soucek P. FOLFOX/FOLFIRI pharmacogenetics: The call for a personalized approach in colorectal cancer therapy. World J Gastroenterol. 2014;20:10316–10330. doi: 10.3748/wjg.v20.i30.10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu HW, Cho WC. The emerging role of miRNAs in combined cancer therapy. Expert Opin Biol Ther. 2015;15:923–925. doi: 10.1517/14712598.2015.1030390. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 Are promising prognostic biomarkers and novel targets of Tumor-Suppressive miR-342 in human colorectal cancer. Clin Cancer Res. 2016;22:4947–4957. doi: 10.1158/1078-0432.CCR-16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng W, Feng J, Qin H, Ma Y, Goel A. An update on miRNAs as biological and clinical determinants in colorectal cancer: A bench-to-bedside approach. Future Oncol. 2015;11:1791–1808. doi: 10.2217/fon.15.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwandi RA, Vacharaksa A. The role of microRNA in periodontal tissue: A review of the literature. Arch Oral Biol. 2016;72:66–74. doi: 10.1016/j.archoralbio.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Huang G, Pan J, Ye Z, Fang B, Cheng W, Cao Z. Overexpression of miR-216b sensitizes NSCLC cells to cisplatin-induced apoptosis by targeting c-Jun. Oncotarget. 2017;8:104206–104215. doi: 10.18632/oncotarget.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du X, Liu B, Luan X, Cui Q, Li L. miR-30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp Ther Med. 2018;15:599–605. doi: 10.3892/etm.2017.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Natino D, Zhai X, Gao Z, He X. MicroRNA22 inhibits the proliferation and migration, and increases the cisplatin sensitivity, of osteosarcoma cells. Mol Med Rep. 2018;17:7209–7217. doi: 10.3892/mmr.2018.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salendo J, Spitzner M, Kramer F, Zhang X, Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a,-224,-132 and let7g. Radiothe Oncol. 2013;208:451–457. doi: 10.1016/j.radonc.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 16.Ye Q, Su L, Chen D, Zheng W, Liu Y. Astragaloside IV induced miR-134 expression reduces EMT and Increases chemotherapeutic sensitivity by suppressing CREB1 signaling in colorectal cancer cell line SW-480. Cell Physiol Biochem. 2017;43:1617–1626. doi: 10.1159/000482025. [DOI] [PubMed] [Google Scholar]
- 17.Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59:467–474. doi: 10.18388/abp.2012_2079. [DOI] [PubMed] [Google Scholar]
- 18.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Xie T, Mao X, Xue L, Chu X, Chen L. MicroRNA-149 increases the sensitivity of colorectal cancer cells to 5-Fluorouracil by targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem. 2016;39:617–629. doi: 10.1159/000445653. [DOI] [PubMed] [Google Scholar]
- 20.Dabkeviciene D, Jonusiene V, Zitkute V, Zalyte E, Grigaitis P, Kirveliene V, Sasnauskiene A. The role of interleukin-8 (CXCL8) and CXCR2 in acquired chemoresistance of human colorectal carcinoma cells HCT116. Med Oncol. 2015;32:258. doi: 10.1007/s12032-015-0703-y. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource:targets and expression. Nucleic Acids Res. 2008;36((Database Issue)):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, Longley DB, Johnston PG. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–2167. doi: 10.1158/1078-0432.CCR-03-0362. [DOI] [PubMed] [Google Scholar]
- 26.Yao Z, Bhandari A, Wang Y, Pan Y, Yang F, Chen R, Xia E, Wang O. Dihydroartemisinin potentiates antitumor activity of 5-fluorouracil against a resistant colorectal cancer cell line. Biochem Biophys Res Commun. 2018;501:636–642. doi: 10.1016/j.bbrc.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Lai Z, Yan Z, Chen W, Peng J, Feng J, Li Q, Jin Y, Lin J. Hedyotis diffusa Willd suppresses metastasis in 5-fluorouracil-resistant colorectal cancer cells by regulating the TGF-β signaling pathway. Mol Med Rep. 2017;16:7752–7758. doi: 10.3892/mmr.2017.7500. [DOI] [PubMed] [Google Scholar]
- 28.Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, Jia L. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog. 2017;56:2669–2680. doi: 10.1002/mc.22710. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, Zhao Q, Zhang C, Wang G, Yao Y, Huang X, Zhan F, Zhu Y, Shi J, Chen J, et al. miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-KB/XIAP axis. Sci Rep. 2017;7:4194. doi: 10.1038/s41598-017-04172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie T, Geng J, Wang Y, Wang L, Huang M, Chen J, Zhang K, Xue L, Liu X, Mao X, et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget. 2017;8:8574–8589. doi: 10.18632/oncotarget.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou Y, Zhu Q, Li Z, Peng Y, Yu X, Yuan B, Liu Y, Liu Y, Yin L, Peng Y, et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8:e2659. doi: 10.1038/cddis.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian L, Zhao Z, Xie L, Zhu J. MiR-361-5p inhibits the mobility of gastric cancer cells through suppressing epithelial-mesenchymal transition via the Wnt/β-catenin pathway. Gene. 2018;675:102–109. doi: 10.1016/j.gene.2018.06.095. [DOI] [PubMed] [Google Scholar]
- 35.Han J, Yu J, Dai Y, Li J, Guo M, Song J, Zhou X. Overexpression of miR-361-5p in triple-negative breast cancer (TNBC) inhibits migration and invasion by targeting RQCD1 and inhibiting the EGFR/PI3K/Akt pathway. Bosn J Basic Med Sci. 2019;19:52–59. doi: 10.17305/bjbms.2018.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Dong B, Wang Z, Jiang T, Chen G. MicroRNA-361-5p inhibits papillary thyroid carcinoma progression by targeting ROCK1. Biomed Pharmacother. 2018;102:988–995. doi: 10.1016/j.biopha.2018.03.122. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Wu D, Yu Q, Li L, Wu P. Prognostic value of FOXM1 in solid tumors: A systematic review and meta-analysis. Oncotarget. 2017;8:32298–32308. doi: 10.18632/oncotarget.15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2017;9:321–331. doi: 10.18632/oncotarget.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anreddy N, Patel A, Sodani K, Kathawala RJ, Chen EP, Wurpel JN, Chen ZS. PD173074, a selective FGFR inhibitor, reverses MRP7 (ABCC10)-mediated MDR. Acta Pharm Sin B. 2014;4:202–207. doi: 10.1016/j.apsb.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun YL, Chen JJ, Kumar P, Chen K, Sodani K, Patel A, Chen YL, Chen SD, Jiang WQ, Chen ZS. Reversal of MRP7 (ABCC10)-mediated multidrug resistance by tariquidar. PLoS One. 2013;8:e55576. doi: 10.1371/journal.pone.0055576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 42.Hagmann W, Faissner R, Schnölzer M, Löhr M, Jesnowski R. Membrane drug transporters and chemoresistance in human pancreatic carcinoma. Cancers (Basel) 2010;3:106–125. doi: 10.3390/cancers3010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.