Abstract
Lung cancer has high incidence and mortality rates, and lung squamous cell carcinoma (LUSC) is a common form of non-small-cell lung carcinoma (NSCLC). The aim of our study was to discover long non-coding RNAs (lncRNAs) associated with LUSC prognosis. RNA-sequencing data obtained from LUSC samples were extracted from The Cancer Genome Atlas database. Using the limma package, differentially expressed genes (DEGs; including differentially expressed lncRNA genes (DELs), coding genes (DECs), and other genes (DEOs)) between LUSC and control samples were analyzed. Using Kaplan-Meier survival analysis, prognosis-associated lncRNAs were further selected. Following the calculation of Pearson's correlation coefficients between DELs and other DEGs, the DEL-DEG co-expression network was visualized using Cytoscape software. Using the clusterProfiler package, potential functions for DECs co-expressed with DELs were predicted. There were 1,305 DEGs in LUSC samples, including 153 DELs, 1,109 DECs, and 43 DEOs. Based on survival analysis, 22 prognosis-associated lncRNAs (including surfactant associated 1, pseudogene (SFTA1P), long intergenic non-protein coding RNA 968 (LINC00968), GATA6 antisense RNA 1, (GATA6-AS1) TBX5 antisense RNA 1 (TBX5-AS1) and FEZF1 antisense RNA 1 (FEZF1-AS1)) in LUSC were selected from these DELs, and the associated abnormal expression levels were also verified in LUSC clinical samples. A DEL-DEG co-expression network was constructed, which involved 93 DELs. Co-expressed DECs were enriched for only 8 prognosis-associated DELs, including LINC00968, SFTA1P, and TBX5-AS1. Specifically, mitogen-activated protein kinase (MAPK) signaling pathway-associated genes were enriched in DECs co-expressed with LINC00968, SFTA1P, GATA6-AS1, TBX5-AS1 and FEZF1-AS1, which may be prognosis-associated lncRNAs in LUSC. In addition, LINC00968 may affect the outcome of patients with LUSC via the MAPK signaling pathway.
Keywords: lung squamous cell carcinoma, long non-coding RNAs, survival analysis, co-expression network, enrichment analysis
Introduction
Lung cancer is classified as small-cell lung carcinoma (SCLC) or non-small-cell lung carcinoma (NSCLC) (1). Patients with lung cancer are frequently effected by weight loss, shortness of breath, coughing and chest pains (2). Lung cancer has a high rate of incidence and mortality; in 2012, 1.8 million people were effected, resulting in 1.6 million deaths globally (3). NSCLC accounts for ~85 % of all lung cancer cases, and is less sensitive to chemotherapy than SCLC (4,5). As a type of NSCLC, lung squamous cell carcinoma (LUSC) has a higher incidence in males compared with females, and is closely associated with a history of smoking (6). It was therefore necessary to investigate the pathogenesis of LUSC in order to improve the clinical outcomes of patients, and approaches to treatment.
Long non-coding RNAs (lncRNAs) are non-coding RNAs longer than 200 nucleotides, which serve roles in various biological phenomena, including tumorigenesis and cancer progression (7,8). A number of lncRNAs associated with the outcome of patients with NSCLC have been reported; downregulation of HMlincRNA717 (also known as gastric cancer associated transcript 2) is associated with NSCLC progression, and HMlincRNA717 expression serves as an independent marker for predicting the survival of patients with NSCLC (9). Expression of TatD DNase domain containing 1 (TATDN1) is associated with the increased invasion and metastatic potential of NSCLC cells, indicating that TATDN1 may function as a promising prognostic biomarker and therapeutic target for the disease (10). Overexpression of SBF2 antisense RNA 1 (SBF2-AS1) is associated with lymph node metastasis, histological grade and poor overall survival, indicating that SBF2-AS1 is an independent prognostic marker and promising therapeutic target for patients with NSCLC (11). Decreased expression of tubulin alpha 4b has been reported in NSCLC cell lines and tissues, where it mediates cell proliferation and is an adverse prognostic predictor for NSCLC (12). The Pvt1 oncogene may be a candidate marker for the diagnosis and prognosis of patients with NSCLC, and may accelerate NSCLC cell proliferation by decreasing the expression of p15 and p21 (13,14). B-cell chronic lymphocytic leukemia (CLL)/lymphoma 2 is implicated in the favorable prognosis of patients with localized NSCLC via its interaction with the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (15). However, elucidation of the roles of lncRNAs associated with the prognosis of patients with LUSC requires further investigation.
In order to determine the mechanisms of multiple underlying diseases, gene expression profiles of human tissues are increasingly being analyzed using bioinformatics tools (16). In the present study, RNA-sequencing data obtained from LUSC samples was downloaded from public databases. This was followed by differential expression analysis, survival analysis, co-expression network analysis and enrichment analysis to determine key lncRNAs effecting the prognosis of patients with LUSC. This may provide a basis for prognostic prediction and targeted therapy in LUSC.
Materials and methods
Data extraction and pre-processing
Using the R package of The Cancer Genome Atlas (TCGA) Biolinks package (http://www.bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html) (17), RNA-sequencing and clinical data for LUSC were extracted from TCGA http://cancergenome.nih.gov/) database. The raw data was pre-processed, and genes with no expression in >10% samples were defined as those with low expression, and subsequently filtered out. Based on the human reference genome GENCODE (hg 19) (18), ensemble gene IDs were mapped to gene symbols and the corresponding gene types were characterized. The GENCODE database classified the following types of genes as lncRNA genes: 3prime_overlapping_ncRNA, antisense_RNA, bidirectional_promoter_lncRNA, lincRNA, macro_lncRNA, non_coding, proessed_transcript, sense_intronic, sense_overlaping and TEC (18). According to this typing standard, genes from the RNA-sequencing data were grouped as lncRNA genes, coding genes and other genes.
Differential expression analysis
Data were normalized using the R package limma (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (19), and differential expression analysis for LUSC and the control samples was performed. From the genes obtained upon analysis, those with thresholds of false discovery rate (FDR) <0.01, and |log fold change (FC)| ≥3, differentially expressed genes (DEGs; including differentially expressed lncRNA genes (DELs), coding genes (DECs), and other genes (DEOs)) were selected.
Survival analysis for DELs
To analyze the prognostic features of the DELs, samples were classified into high and low expression groups with the median expression of DELs as the threshold. Kaplan-Meier (KM) survival analysis (20), was performed for clinical data to compare the survival differences between high and low expression groups. P<0.05 was considered to indicate a statistically significant difference.
Co-expression network and enrichment analysis
Co-expression network analysis is a valuable method of studying the biological functions of lncRNAs, and its theoretical basis is that the expression profiles of genes involved in the same biological function may be correlated (21). Pearson's correlation coefficients were determined between the DELs and DEOs, with the correlation coefficient |r|≥0.6 as the threshold for screening co-expression pairs. The DEL-DEG co-expression network was constructed using Cytoscape software v3.6.0 (http://www.cytoscape.org/) (22). Based on the results of the co-expression network, potential functions of prognosis-associated DELs were analyzed. Using the R package cluster Profiler (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (23), Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were predicted for those DECs co-expressed with DELs. P<0.05 was considered to indicate a statistically significant difference.
Patient sample collection
LUSC tissues and adjacent non-tumorous tissues were collected from 37 patients (age range, 40–80 years; mean age, 56 years; with 23 males and 14 females) with NSCLC who underwent surgical treatment at The Second Affiliated Hospital of Nanchang University (Nanchang, China) between May 2013 and September 2016. Following surgical resection, tissue samples were immediately frozen in liquid nitrogen and stored at −80°C. The study protocols conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and were approved by the Institutional Ethical Review Committee of The Second Affiliated Hospital of Nanchang University. All patients enrolled in the study gave written informed consent.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from patient tissue samples using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and treated with DNase I (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's protocol. RT-qPCR was performed using the TaqMan High-Capacity cDNA Reverse Transcription Kit and the TaqMan Fast PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the corresponding primers, according to the manufacturer's protocol. GAPDH was used as an internal control. The RT-qPCR protocol included an initial denaturation step (95°C for 5 min) and 40 cycles of denaturation (95°C for 10 sec), annealing (60°C for 20 sec) and extension (72°C for 10 sec). The relative expression levels were calculated using the 2−ΔΔCq method as previously described. The primer sequences for surfactant associated 1, pseudogene (SFTA1P), long intergenic non-protein coding RNA 968 (LINC00968), GATA6 antisense RNA 1, (GATA6-AS1) TBX5 antisense RNA 1 (TBX5-AS1) and FEZF1 antisense RNA 1 (FEZF1-AS1) were as follows: SFTA1P forward, 5′-CAGGAGGTCACAGGGAAG-3′, and reverse, 5′-AAGCCGAATACAGTTGCC-3′; LINC00968 forward, 5′-GGCAGTTTTATTGTGGTGATT-3′, and reverse, 5′-AATGGAAGTTGACGGGATAG-3′; GATA6-AS1 forward, 5′-ACGGTTTCTGGACCTTCTC-3′, and reverse, 5′-TTGTGAACTTGTGGCTCCT-3′; TBX5-AS1 forward, 5′-GCAAAAGAAAGGGTTGGTC-3′, and reverse, 5′-AAAAGTGGGGAAACCAGAA-3′; FEZF1-AS1 forward, 5′-CTGGAGCCTTACCTGCCTT-3′, and reverse, 5′-CTGGAGGGACACACCTCAC-3′; MKP-1 forward, 5′-GGACATTTGGGCTGTGTG-3′, and reverse, 5′-CCGCTTTTGGACTGAGAGA-3′; TGF-β receptor type-2 (TGFBR2) forward, 5′-CCCCAGGTAAGGATAGCAG-3′, and reverse, 5′-CCAGGTAGGCAGTGGAAA-3′; and GAPDH forward, 5′-TCCTCTGACTTCAACAGCGACAC-3′, and reverse, 5′-CACCCTGTTGCTGTAGCCAAATTC-3′.
Statistical analysis
Statistical analysis was performed using GraphPad 6.0 statistical software (GraphPad Software, Inc., La Jolla, CA, USA). Data is shown as mean ± standard deviation. Data from two groups were analyzed using unpaired t-test and >2 groups were analyzed using one-way analysis of variance with Tukey's post hoc test. Spearman's correlation analysis was used to determine the correlations between the expression levels of LINC00968 and TGFBR2/MKP-1 in LUSC tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
Differential expression analysis
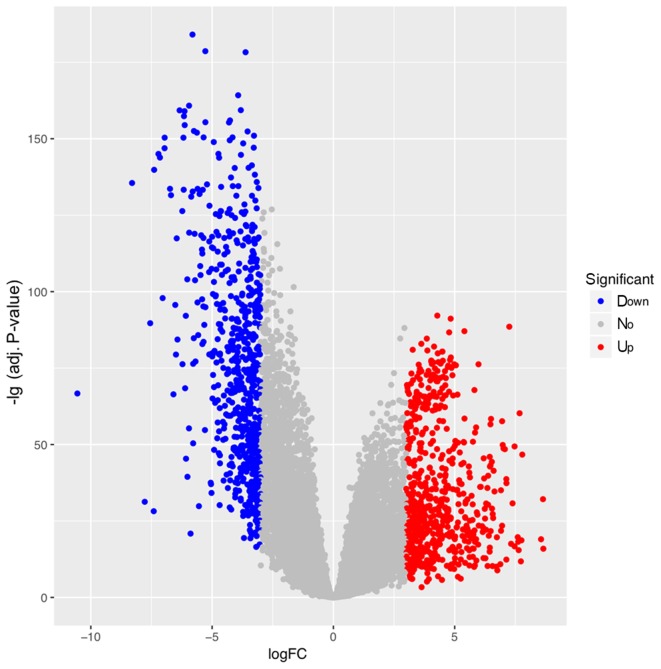
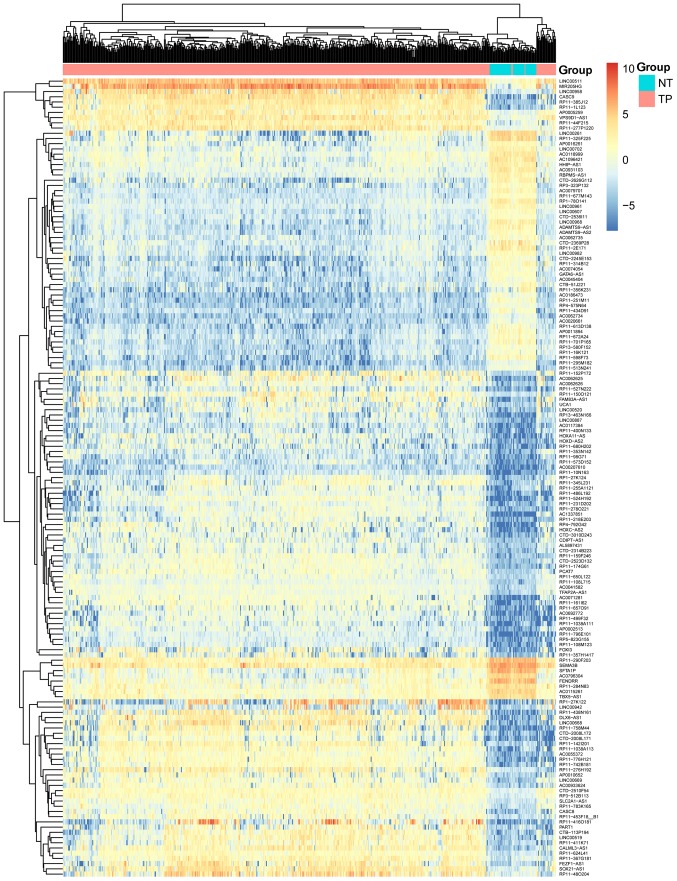
A total of 22,271 genes (including 3,685 lncRNAs, 16,045 coding genes and 2,541 other genes) from 551 samples (including 50 LUSC and 49 control samples) were obtained. Under FDR <0.01 and |logFC| ≥3, a total of 1,305 DEGs were identified (including 631 upregulated, and 674 downregulated genes). Among these DEGs, there were 153 DELs (99 upregulated and 54 downregulated), 1,109 DECs (504 upregulated and 605 downregulated) and 43 DEOs (28 upregulated and 15 downregulated). The volcano plot for the DEGs is displayed in Fig. 1. The clustering heat map suggested that the identified DELs may be used to distinguish between LUSC samples and controls (Fig. 2).
Figure 1.
Volcano plot of DEGs associated with lung squamous cell carcinoma. Red, blue and grey represent upregulated, downregulated and non-DEGs, respectively. DEG, differentially expressed gene.
Figure 2.
Heat map of differentially expressed long non-coding RNAs associated with lung squamous cell carcinoma. In the sample strip, red and blue represent tumor samples and non-tumor samples, respectively.
Survival analysis for identified DELs
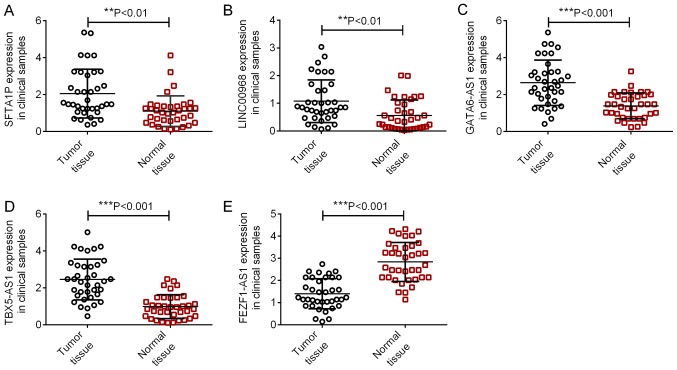
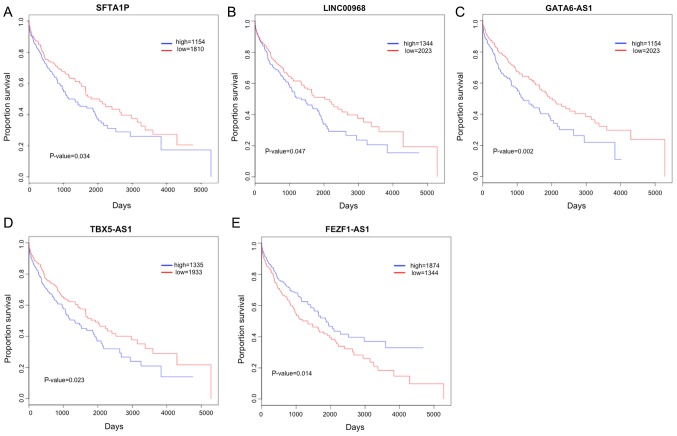
According to the median expression level of the identified DELs, samples were divided into high and low expression groups. KM survival analysis for the two groups indicated that the expression of 22 DELs (including SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 and FEZF1-AS1) correlated with the prognosis of patients with LUSC (Table I). In addition, the expression of SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 and FEZF1-AS1 were also identified in LUSC tissues and normal tissues using RT-qPCR. As illustrated in Fig. 3, the expression levels of SFTA1P, LINC00968, GATA6-AS1 and TBX5-AS1 were significantly increased in LUSC tissues, and FEZF1-AS1 expression was significantly decreased in LUSC tissues compared with that in normal tissues (P<0.01). The KM survival curves for SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 and FEZF1-AS1 are displayed in Fig. 4. High expressions of SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 predicted poor prognosis of LUSC, while low expression of FEZF1-AS1 predicted a poor prognosis of LUSC.
Table I.
Differentially expressed long non-coding RNAs correlating with the prognosis of lung squamous cell carcinoma.
| Gene | P-value |
|---|---|
| SFTA1P | 0.034 |
| LINC00968 | 0.047 |
| AC0796304 | 0.024 |
| TBX5-AS1 | 0.023 |
| RP11-434D91 | 0.036 |
| AC1337851 | 0.047 |
| GATA6-AS1 | 0.002 |
| RP11-613D138 | 0.013 |
| RP11-385J12 | 0.032 |
| CTD-2626G112 | 0.013 |
| RP11-174G61 | 0.037 |
| RP5-823G155 | 0.037 |
| RP11-108M123 | 0.011 |
| LINC00519 | 0.017 |
| CTB-113P194 | 0.024 |
| RP11-573D152 | 0.045 |
| CDIPT-AS1 | 0.016 |
| RP11-356K231 | 0.041 |
| FEZF1-AS1 | 0.014 |
| RP11-255A1121 | 0.015 |
| RP11-357H1417 | 0.017 |
| RP11-48O204 | 0.014 |
Figure 3.
Relative expression levels of long non-coding RNAs in tumor and adjacent, normal tissues. (A) SFTA1P, (B) LINC00968, (C) GATA6-AS1, (D) TBX5-AS1 and (E) FEZF1-AS1 in 37 paired LUSC tumor tissues and adjacent normal tissues were identified using reverse transcription-quantitative polymerase chain reaction. **P<0.01, ***P<0.001. SFTA1P, surfactant associated 1, pseudogene; LINC00968, long intergenic non-protein coding RNA 968; GATA6-AS1, GATA6 antisense RNA 1; TBX5-AS1, TBX5 antisense RNA 1; and FEZF1-AS1, FEZF1 antisense RNA 1.
Figure 4.
Kaplan-Meier survival curves for long non-coding RNA associations with patient survival. (A) SFTA1P, (B) LINC00968, (C) GATA6-AS1, (D), TBX5-AS1 and (E) FEZF1-AS1. Blue and red lines indicate high and low expression groups, respectively. SFTA1P, surfactant associated 1, pseudogene; LINC00968, long intergenic non-protein coding RNA 968; GATA6-AS1, GATA6 antisense RNA 1; TBX5-AS1, TBX5 antisense RNA 1; and FEZF1-AS1, FEZF1 antisense RNA 1.
Co-expression network and enrichment analysis
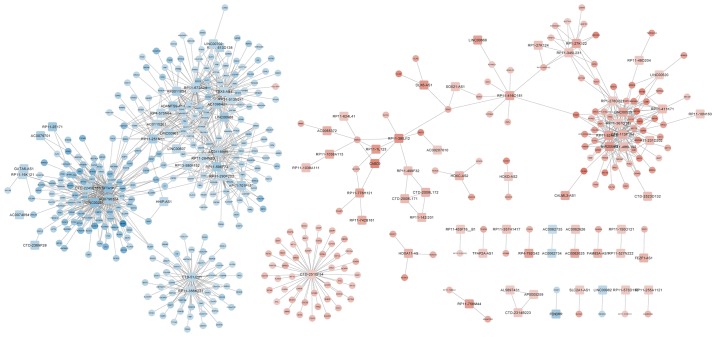
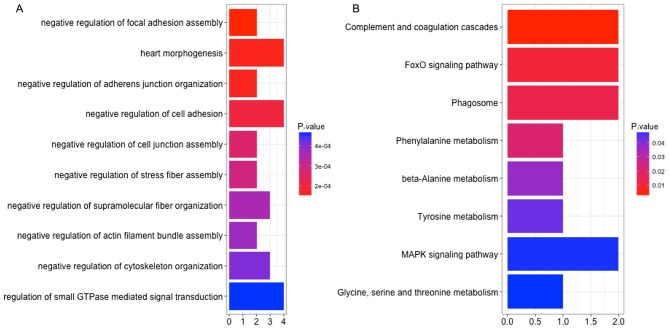
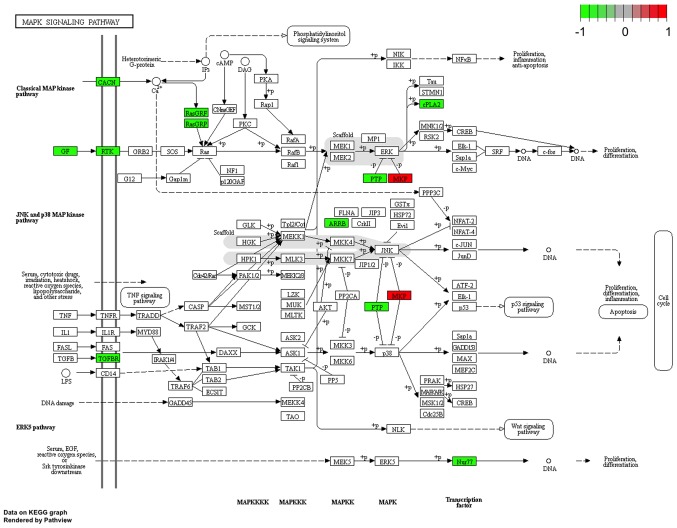
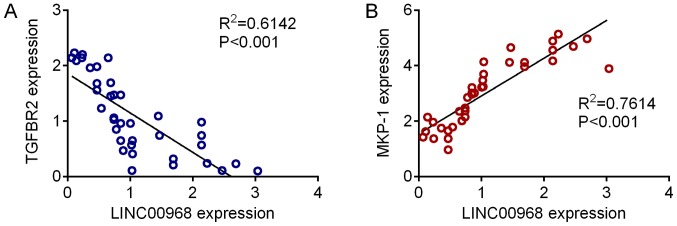
The construction of a DEL-DEG co-expression network revealed 1,096 co-expressed pairs, including 93 DELs (Fig. 5). Notably, lncRNA-SFTA1P was co-expressed with the majority of the other genes identified. A total of 8 prognosis-associated DELs (including LINC00968, SFTA1P and TBX5-AS1) were enriched. The GO terms and KEGG pathways outlining the enrichment of LINC00968 are presented in Fig. 6, and details of the MAPK signaling pathway are highlighted in Fig. 7. The expression levels of TGFBR2 and MKP-1, which are involved in MAPK signaling in 37 LUSC tumor samples, were identified using RT-qPCR. Fig. 8 demonstrates that the expression of TGFBR2 negatively correlated with that of LINC00968 (P<0.001), whilst the expression of MKP-1 positively correlated with LINC0096 expression (P<0.001).
Figure 5.
Co-expression network of genes implicated in lung squamous cell carcinoma. Red and blue represent high and low gene expression, respectively. Squares indicate long non-coding RNA genes, and circles represent coding genes or other genes.
Figure 6.
(A) Functional terms and (B) pathway enrichment of long non-coding RNA long intergenic non-protein coding RNA 968. Horizontal and vertical coordinates represent the count of implicated genes and term/pathway names, respectively. Red and blue indicate low and high P-values, respectively.
Figure 7.
The mitogen-activated protein kinase signaling pathway in lung squamous cell carcinoma. Red and green represent upregulated and downregulated genes, respectively.
Figure 8.
Correlation between LINC00968 and TGFBR2/MKP-1 expression in LUSC. (A) Expression of TGFBR2 in LUSC tissues was identified by RT-qPCR analysis, and revealed a negative correlation with LINC00968 expression. (B) Expression of MPK-1 in LUSC tissues revealed a positive correlation with LINC00968 expression. LINC00968, long intergenic non-protein coding RNA 968; TGFBR2, TGF-β receptor type-2; MPK-1, mitogen-activated protein kinase 1; LUSC, lung squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
In the present study, a total of 1,305 DEGs between LUSC samples and control samples were selected (including 153 DELs, 1,109 DECs and 43 DEOs). Survival analysis revealed that 22 DELs (including SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 and FEZF1-AS1) were prognosis-associated lncRNAs for LUSC. Additionally, a DEL-DEG co-expression network involving 93 DELs was constructed. Enrichment analysis for the DECs co-expressed with the DELs suggested that only 8 prognosis-associated DELs (including LINC00968, SFTA1P and TBX5-AS1) were enriched.
SFTA1P and cancer susceptibility candidate 2 mediate the functions of tumor suppressor genes and oncogenes during the development of LUSC, which may be used in diagnosis, prognostic prediction and targeted therapy (24). Increased expression of SFTA1P results in apoptosis and promotes cisplatin sensitivity in LUSC cells, indicating that SFTA1P may be considered a promising diagnostic marker, and an indicator for cisplatin sensitivity in patients with LUSC (25). Downregulation of GATA6-AS1 results in adverse outcomes for patients with gastric cancer (26), and also serves a tumor suppressing role in CLL (27). GATA6 has a regulatory effect in alveolar epithelial cells (28,29), and during lung development, GATA6-AS1 expression has been associated with that of GATA6 (30). These reports suggest that SFTA1P and GATA6-AS1 may be implicated in the prognosis of LUSC.
TBX5 expression is significantly associated with the disease stage, lymph node status and histopathological type of NSCLC, and serves a tumor-suppressing role in the disease (31,32). FEZF1-AS1 overexpression is associated with disease stage, differentiation degree and lymph node metastasis of NSCLC, and thus FEZF1-AS1 may promote tumor formation and be considered as a therapeutic target for the disease (33). Furthermore, FEZF1-AS1 expression levels were significantly upregulated in lung adenocarcinoma (LAD) samples, which resulted in unfavorable prognosis. This suggests that FEZF1-AS1 dysregulation promotes the development and progression of LAD, and may therefore be used in LAD therapy (34). The overexpression of FEZF1-AS1 is apparent in LAD tissues and cell lines, which may affect the cell cycle and proliferation of LAD cells by inhibiting the expression of p57 (35). Therefore, TBX5-AS1 and FEZF1-AS1 may also be associated with the clinical outcome of patients with LUSC.
LINC00968 inhibition suppresses the growth, invasion and migration of NSCLC cells, and is therefore an notable molecular marker for the diagnosis, prognosis and treatment success of patients with NSCLC (36). By activating the p38 MAPK and c-Jun N-terminal kinase signaling pathways, and suppressing the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling pathway, treatment with platycodin-D results in autophagy in A549 and NCI-H460 cells, and serves as an alternative compound for the treatment of NSCLC (37). Through the extracellular signal-regulated protein kinase/MAPK signaling pathway, claudin-7 suppresses the migration and invasion of growth factor-stimulated lung cancer cells (38). Pathway enrichment analysis revealed that LINC00968 was enriched for the MAPK pathway, thus, it was speculated that LINC00968 may affect the survival of patients with LUSC by regulating MAPK signaling.
In conclusion, 1,305 DEGs were identified from LUSC samples relative to corresponding control samples. The lncRNAs-SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1 and FEZF1-AS1 may be associated with the prognosis of patients with LUSC. However, the effects of these key lncRNAs on patient survival rate requires further experimental verification.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant. no. 81460010), the Youth Science Foundation from the Science and Technology Department of Jiangxi Province (grant. no. 20171BAB215002 and 20171BAB215040), the Medical Scientific Research Foundation of Guangdong Province (grant. no. A2017155), the China Postdoctoral Science Foundation funded project (grant. no. 2017M622107) and the Postdoctoral Science Foundation of Jiangxi Province (grant. no. 2016KY51).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YX, ZL and XZ performed the literature search and data extraction, and analyzed the data. AX analyzed the data. SX and JL performed the literature search and data extraction. WZ and XZ conceived and designed the study, and modified the manuscript.
Ethics approval and consent to participate
The present study conformed to the guidelines of the 1975 Declaration of Helsinki, and was approved by the Institutional Ethical Review Committee of The Second Affiliated Hospital of Nanchang University. All patients enrolled in the study gave written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cataldo JK, Brodsky JL. Lung cancer stigma, anxiety, depression and symptom severity. Oncology. 2013;85:33–40. doi: 10.1159/000350834. [DOI] [PubMed] [Google Scholar]
- 3.McGuire S. World cancer report 2014. geneva, Switzerland: World health organization, international agency for research on cancer, WHO press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: Recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: Molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18:2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genets. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu FQ, Hu R, Wang MS. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2014;7:8881–8886. [PMC free article] [PubMed] [Google Scholar]
- 10.Zequn N, Xuemei Z, Wei L, Zongjuan M, Yujie Z, Yanli H, Yuping Z, Xia M, Wei W, Wenjing D, et al. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget. 2016;7:18219–18228. doi: 10.18632/oncotarget.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao QS, Li L, Zhang L, Meng XW, Li LL, Ge XF, Li ZP. Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20:3031–3034. [PubMed] [Google Scholar]
- 12.Chen J, Hu L, Wang J, Zhang F, Chen J, Xu G, Wang Y, Pan Q. Low expression LncRNA TUBA4B is a poor predictor of prognosis and regulates cell proliferation in non-small cell lung cancer. Pathol Oncol Res. 2017;23:265–270. doi: 10.1007/s12253-016-0089-y. [DOI] [PubMed] [Google Scholar]
- 13.Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37:4127–4134. doi: 10.1007/s13277-015-4261-x. [DOI] [PubMed] [Google Scholar]
- 14.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt LH, Görlich D, Spieker T, Rohde C, Schuler M, Mohr M, Humberg J, Sauer T, Thoenissen NH, Huge A, et al. Prognostic Impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9:1294–1304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 16.Servant N, Roméjon J, Gestraud P, La Rosa P, Lucotte G, Lair S, Bernard V, Zeitouni B, Coffin F, Jules-Clément G, et al. Bioinformatics for precision medicine in oncology: Principles and application to the SHIVA clinical trial. Front Genet. 2014;5:152. doi: 10.3389/fgene.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Springer; New York: 2005. Limma: Linear models for microarray data. [DOI] [Google Scholar]
- 20.Miettinen OS. Survival analysis: Up from Kaplan-Meier-Greenwood. Eur J Epidemiol. 2008;23:585–592. doi: 10.1007/s10654-008-9278-7. [DOI] [PubMed] [Google Scholar]
- 21.Cogill SB, Wang L. Co-expression network analysis of human lncRNAs and cancer genes. Cancer Inform. 2014;13(Suppl 5):S49–S59. doi: 10.4137/CIN.S14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 23.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang GQ, Ke ZP, Hu HB, Gu B. Co-expression network analysis of long noncoding RNAs (IncRNAs) and cancer genes revealsSFTA1P and CASC2abnormalities in lung squamous cell carcinoma. Cancer Biol Ther. 2017;18:115–122. doi: 10.1080/15384047.2017.1281494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Yin JY, He FZ, Huang MS, Zhu T, Gao YF, Chen YX, Zhou DB, Chen X, Sun LQ, et al. Long noncoding RNA SFTA1P promoted apoptosis and increased cisplatin chemosensitivity via regulating the hnRNP-U-GADD45A axis in lung squamous cell carcinoma. Oncotarget. 2017;8:97476–97489. doi: 10.18632/oncotarget.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ, et al. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. doi: 10.1038/emm.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang LQ, Wong KY, Li ZH, Chim CS. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget. 2016;7:82400–82410. doi: 10.18632/oncotarget.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou B, Francis TA, Yang H, Tseng W, Zhong Q, Frenkel B, Morrisey EE, Ann DK, Minoo P, Crandall ED, Borok Z. GATA-6 mediates transcriptional activation of aquaporin-5 through interactions with Sp1. Am J Physiol Cell Physiol. 2008;295:C1141–C1150. doi: 10.1152/ajpcell.00120.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, Yang J, DeMayo FJ, Whitsett JA, Parmacek MS, Morrisey EE. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma R, Yang Y, Tu Q, Hu K. Overexpression of T-box transcription factor 5 (TBX5) inhibits proliferation and invasion in non-small cell lung carcinoma cells. Oncol Res. 2017:1495–1504. doi: 10.3727/096504017X14883287513729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 33.Ghassemi S, Vejdovszky K, Sahin E, Ratzinger L, Schelch K, Mohr T, Peter-Vörösmarty B, Brankovic J, Lackner A, Leopoldi A, et al. FGF5 is expressed in melanoma and enhances malignancy in vitro and in vivo. Oncotarget. 2017;8:87750–87762. doi: 10.18632/oncotarget.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Zhao P, Han Y, Lu S. LincRNA FEZF1-AS1 is associated with prognosis in lung adenocarcinoma and promotes cell proliferation, migration and invasion. Onco Res. 2018;27:39–45. doi: 10.3727/096504018X15199482824130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S, Chen S, Ma Y, Yang B, Liu Y. LincRNA FEZF1-AS1 contributes to the proliferation of LAD cells by silencing p57 expression. Oncotarget. 2017;8:103004–103013. doi: 10.18632/oncotarget.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Zhou J, Xu YJ, Hu HB. Long noncoding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J Cell Physiol. 2018;233:3397–3406. doi: 10.1002/jcp.26186. [DOI] [PubMed] [Google Scholar]
- 37.Zhao R, Chen M, Jiang Z, Zhao F, Xi B, Zhang X, Fu H, Zhou K. Platycodin-D induced autophagy in non-small cell lung cancer cells via PI3K/Akt/mTOR and MAPK signaling pathways. J Cancer. 2015;6:623–631. doi: 10.7150/jca.11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317:1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.