Abstract
The poor therapeutic effect of the current treatments for malignant glioma may be attributed to glioma stem cells (GSCs), which have been demonstrated to divide symmetrically. All-trans retinoic acid (ATRA)-induced differentiation is considered to target GSCs and has been reported to have the capability of eradicating cancer stem cells in specific malignancies. The aim of the present study was to investigate the effects of ATRA on the division mode of GSCs isolated from the U87MG glioblastoma cell line of unknown origin. The expressions of the GSC markers CD133 and nestin were detected using immunocytochemistry to identify GSCs. In addition, the differentiation potency of these GSCs was observed by detecting the expression of glial fibrillary acidic protein, β-tubulin III and galactosylceramidase using immunofluorescent staining. The Numb protein distribution was analyzed in two daughter cells following a GSC division. The results of the present study demonstrated that Numb protein is symmetrically segregated into two daughter cells during GSC division. Furthermore, the present study demonstrated that treatment with ATRA increased the asymmetric cell division of GSCs. In conclusion, these results suggest a therapeutic effect from ATRA-induced asymmetric division of GSCs from the U87MG cell line.
Keywords: all-trans retinoic acid, asymmetric division, glioma stem cells, cell line U87
Introduction
Malignant gliomas, which are classified by the World Health Organization as grade III–IV tumors, are extremely lethal primary brain tumors with an incidence of 3–5/100,000 each year between 2000 and 2004 (1). At present, the maximal therapy for malignant gliomas comprises surgical resection followed by radiotherapy and concomitant adjuvant temozolomide (1). However, patients with malignant glioma exhibit a dismal prognosis (2), with a 5-year survival rate of <3% (3). This poor prognosis is due to therapeutic resistance and tumor recurrence following the combined treatment approach (4). Previously, numerous studies have identified that the resistance of malignant gliomas to current therapy may reside in a relatively small population of glioma stem cells (GSCs), which are hypothesized to substantially influence tumor initiation and serve a critical role in tumor propagation (5–9). The majority of GSCs have been revealed to divide through symmetric cell division and not through asymmetric cell division (10). Recent reports have determined that the frequency of asymmetric division of cancer stem cells is negatively correlated with their proliferative capacity (11,12). A previous study in mouse models have further demonstrated that decreased asymmetry in normal stem cells is associated with neoplastic transformation, while therapies that increase asymmetric division result in decreased numbers of resistant GSCs (13).
All-trans retinoic acid (ATRA)-based differentiation treatment has been observed to target the GSC population and induce therapy-sensitizing and antitumor effects (14,15). Although it has been demonstrated that ATRA-treatment alone is able to induce antitumor effects (5), to the best of our knowledge, it remains unknown whether and how the GSC division mode is affected by treatment with ATRA. Therefore, the present study used a single-cell-based method to observe the differentiation-inducing effects of ATRA on the cell division features of GSCs from the U87MG cell line.
Materials and methods
Culture of U87MG cells
The U87MG cell line (HTB-14; glioblastoma of unknown origin) was purchased from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen: Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2. The identity of the U87MG cell line was confirmed through comparison with the short tandem repeat profile (Amelogenin, CSF1PO, D13S317, D7S820, D5S818, D16S539, vWA, THO1 and TPOX). This U87MG cells were used in subsequent experiments up to the fifth passage.
Culture of U87MG GSCs
GSCs were isolated as described previously (16). U87MG cells were seeded onto plates at a density of 2×104 cells/well. The serum-free culture medium (termed tumor stem cell medium) consisted of DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.), B27 (1×; Gibco; Thermo Fisher Scientific, Inc.), recombinant human epidermal growth factor (20 ng/ml; Sigma-Aldrich, Merck, KGaA, Darmstadt, Germany) and basic fibroblast growth factor (20 ng/ml; Upstate Biotechnology, Inc., Lake Placid, NY, USA) without vincristine. Cultures were incubated in 5% CO2 at 37°C and half of the medium was replaced with DMEM every 2 days. When primary tumor spheres with diameters of 70–100 µm were visible under a light microscope, all of the culture medium was discarded and the wells were filled with 1 ml fresh serum-free tumor stem cell medium.
The primary tumor spheres were dissociated enzymatically using Accutase (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, into single cells and seeded into a 96-well plate (2–3×103 cells/well) with tumor stem cell medium (0.1 ml/well). The culture medium was changed every 3 days. The clonogenic efficacy was examined by calculating the percentage of the wells that contained spheres with >50 cells on day 14. Five fields of vision under a light microscope (×20 magnification) were randomly selected to count the number of spheres. Adherent non-sphere-forming cells and parental U87MG cells were used as controls for the capacity to form tumor cell clones. The morphology of the secondary spheres was examined microscopically. Five fields were randomly selected to examine the morphology of the secondary spheres under a light microscope (×100 magnification).
Immunostaining
GSCs were plated onto poly-L-lysine (Sigma-Aldrich; Merck, KGaA)-coated glass coverslips for 40 h at 37°C in DMEM containing 10% FBS. The cells were fixed with 4% paraformaldehyde (PFA) for 30 min at 4°C and then incubated with primary antibodies against nestin (mouse monoclonal IgG1; 1:200; cat. no. 60091; Stemcell Technologies, Inc., Vancouver, BC, Canada), β-tubulin III (mouse monoclonal IgG1; 1:1,000; cat. no. 60052; Stemcell Technologies, Inc.), glial fibrillary acidic protein (GFAP, rabbit polyclonal; 1:400; cat. no. Z0334; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA), galactosylceramidase (GALC, rabbit polyclonal; 1:20; cat. no. AB142; EMD Millipore, Billerica, MA, USA) or Numb (rabbit polyclonal; 1:100; cat. no. ab155415; Abcam, Cambridge, UK) overnight at 4°C. Appropriate secondary antibodies (Alexa Fluor® 594 donkey anti-mouse; 1:1,000; cat. no. A-21207; Alexa Fluor® 488 donkey anti-rabbit; 1:500; cat. no. A-21202; Alexa Fluor® 488 donkey anti-mouse; 1:400; cat. no. A-21206; and Alexa Fluor® 594 donkey anti-rabbit; 1:500; cat. no. A-21203; Molecular Probes; Thermo Fisher Scientific, Inc.) were then selected for section incubation at room temperature for 2 h. The cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich; Merck KGaA) for 5 min at room temperature for fluorescence microscopy (×200 magnification).
BrdU incorporation
Cells were cultured in DMEM/F12 medium containing 20 ng/ml EGF (Invitrogen; Thermo Fisher Scientific, Inc.). Once pairs were observed, 10 µg/ml bromodeoxyuridine (BrdU; Invitrogen; Thermo Fisher Scientific, Inc.) was added to the wells for 8–10 h. The washed cells were fixed in 4% PFA at 4°C for 30 min, stained for Numb (rabbit polyclonal; cat. no. ab155415; Abcam) overnight at 4°C, and then stained with anti-BrdU antibody (cat. no. ab6326; 1:250; Abcam). The cell nuclei were counterstained with DAPI for 5 min at room temperature for fluorescence microscopy (×200 magnification).
Flow cytometric analysis
For flow cytometric analysis, expanded GSCs were collected and evaluated on an FC500 flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). The GSCs were stained with PE-conjugated CD133 clone AC133 antibody (cat. no. 130-113-670; Miltenyi Biotec, GmbH, Bergisch Gladbach, Germany) at a dilution of 1:50 in PBS with 0.5% BSA containing 20% FcR Blocking Reagent (Miltenyi Biotec, GmbH) at 4°C for 10 min. The control was the non-stained cells in the same buffer. Data were analyzed using FlowJo software version 7.6.1 (Tree Star, Inc., Ashland, OR, USA).
Mitotic image analysis
The fluorescence immunostaining intensity was evaluated for each daughter cell to determine the degree of asymmetric distribution of detected molecules between the two daughter cells. The condensed DNA was revealed via staining with DAPI and the morphology of the phase-contrasted image was used to identify the dividing daughter cells. Using Image-Pro Plus v. 6.2 (Media Cybernetics, Inc., Rockville, MD, USA) with the polygon tool, the integrated fluorescent signals of each defined daughter cell in a dividing pair were determined for the stained markers and DNA.
Differentiation assay
U87MG GSC spheres and adherent non-sphere-forming cells were plated on a sterile 24-well glass slide coated with poly-L-lysine (Sigma-Aldrich, Merck KGaA) in DMEM/F12 neurosphere culture medium. These cells were fed with FBS-supplemented culture medium, and coverslips were stained for GFAP, GALC and β-tubulin III as described above on day 2. The nuclei were visualized using DAPI. Quantification of the positive cells stained with each antibody was evaluated as a percentage of the total number of cells counted under five random fluorescence microscope fields (×200 magnification).
ATRA-treatment
The cells in the culture medium were fed with 10% FBS (vehicle group) or FBS combined with ATRA (Sigma-Aldrich; dissolved in DMSO) at 10 nmol/l (ATRA group) (5).
Pair assay
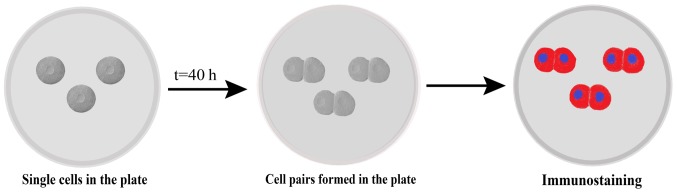
Cells were subjected to a pair assay as described previously (17). In brief, cells were plated at a density of 2,000–3,000 cells/well for 40 h and the cell pairs were observed using time-lapse microscopy, fixed with 4% PFA at 4°C for 30 min, and subjected to immunostaining with Numb primary and secondary antibodies as described above. Nuclei were stained with DAPI, and a total of 200 cell pairs were analyzed using a fluorescence microscope (×200 magnification; Fig. 1).
Figure 1.
Schematic representation of pair assay. Single cells plated under proliferative conditions enter mitosis and form cell pairs, which were fixed and immunostained. Cell pairs in which proteins partitioned to one daughter cell were considered asymmetrical.
Statistical analysis
Since paired daughter cells should have equal DAPI, the percentage deviation of the DAPI distribution was calculated in order to quantify the asymmetry, according to the protocol outlined in a previous study (12). For a minimum of 100 cells, the mean distribution of DAPI was calculated to be 6.848% and the standard deviation was 5.501. Then, the asymmetry cutoff was set to be a difference of >21% between paired daughter cells, based on the >99% confidence interval for DAPI. Based on the 21% cutoff value, each cell division was defined as either symmetrical or asymmetrical.
All experiments were repeated in triplicate, and the data are presented as the mean ± standard deviation. The data were processed using SPSS 22.0 statistical software (IBM Corp., Armonk, NY, USA). Unpaired Student's t-test was used to determine the significance between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Detection of tumor sphere forming cells with self-renewal capability
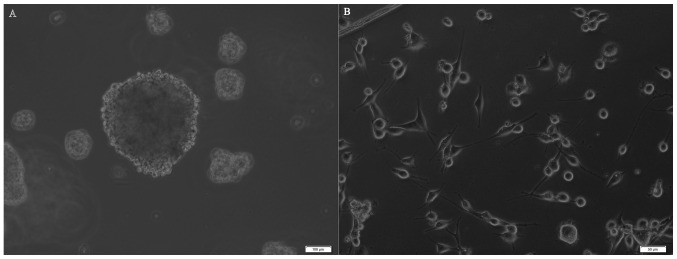
U87MG cells were seeded on plates at a density of 2×104/well with progressive increases in the serum-free neural stem cell culture medium. After 5–6 days, 30–50 small spheres per well (each containing 10–20 cells) were observed. At 2 weeks, these spheres had increased their diameters by 10 to 30-fold (Fig. 2A). Numerous adherent non-sphere-forming cells were observed in the bottoms of the wells (Fig. 2B), and remained viable during an 8-week period; however, they did not generate new spheres. Cells in tumor spheres could be passaged for many generations in serum-free neural stem cell culture medium.
Figure 2.
Morphology of tumor spheres. (A) Primary tumor spheres formed in the medium without vincristine. Scale bar, 100 µm. (B) Adherent non-sphere-forming cells. Scale bar, 50 µm.
Expression of neural stem cell markers
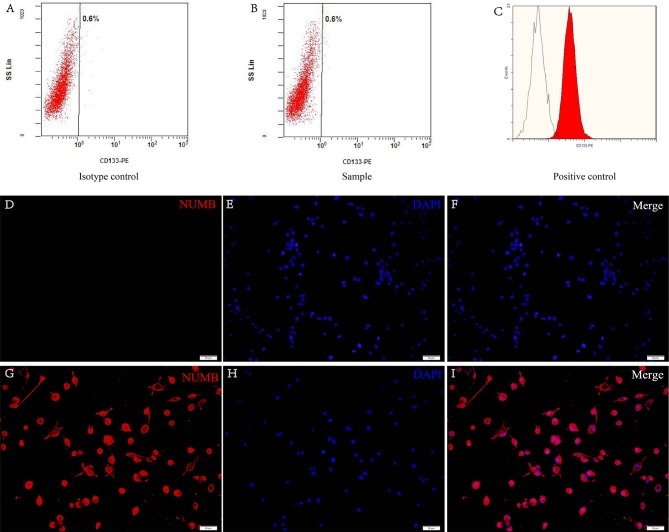
CD133 and nestin are cell surface markers for neural stem cells and are the widely accepted markers for brain GSCs (11). Flow cytometric analysis revealed that these cells had no presence of CD133 on the surface (Fig. 3A and B). The retinoblastoma cell line was used as a positive control (Fig. 3C). The data demonstrated that nestin was positive in the cells from the tumor secondary spheres (Fig. 3D-I).
Figure 3.
Expression of neural stem cell markers. Characterization of the flow-sorted CD133-negative populations. Flow cytometric analysis demonstrated the (A) isotype control and the (B) CD133-negative cells of the samples that were sorted by FACS. (C) The retinoblastoma cell line was used as a positive control. (D-I) The expression of nestin on neurospheres under fluorescence microscopy. (D-F) Negative expression of nestin in adherent cells. (G-H) Positive expression of nestin in glioma stem cells. Nestin (red) and DAPI (blue). Scale bars, 50 µm. FACS, fluorescence activated cell sorting.
Multipotency in the differentiation of secondary spheres
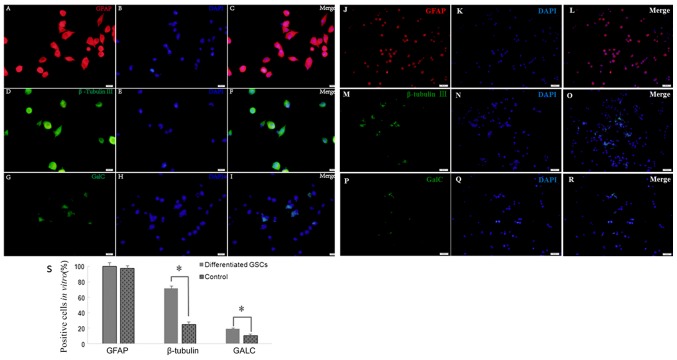
An important feature of GSCs are their differentiation capability. Following culture in medium with 10% FBS for 2 weeks, the cells differentiated from tumor secondary spheres were stained positive for GFAP (99.65±7.42 vs. 97.37±6.06%; P>0.05), β-tubulin III (71.25±3.45 vs. 25.33±3.18%; P<0.05) and GALC (19.23±1.41 vs. 10.87±2.01%; P<0.05) compared with the control cells (Fig. 4). Therefore, secondary spheres derived from the single parental cells of primary tumor spheres were able to differentiate into three neural cell lineages.
Figure 4.
Multipotency of neurosphere cell differentiation observed under a laser confocal scanning microscope. (A-I) Cells were stained for (A-C) GFAP (red), (D-F) β-tubulin III (green) and (G-I) GALC (green) after GSC differentiation. DAPI (blue) indicates nuclei. Scale bars, 20 µm. (J-R) GSCs were stained for (J-L) GFAP (red), (M-O) β-tubulin III (green) and (P-R) GALC as control. (S) Quantification of cells positively stained by each antibody. *P<0.05. Scale bars, 50 µm. GFAP, glial fibrillary acidic protein; GALC, galactosylceramidase.
Asymmetric distribution of Numb is associated with asymmetric GSC divisions
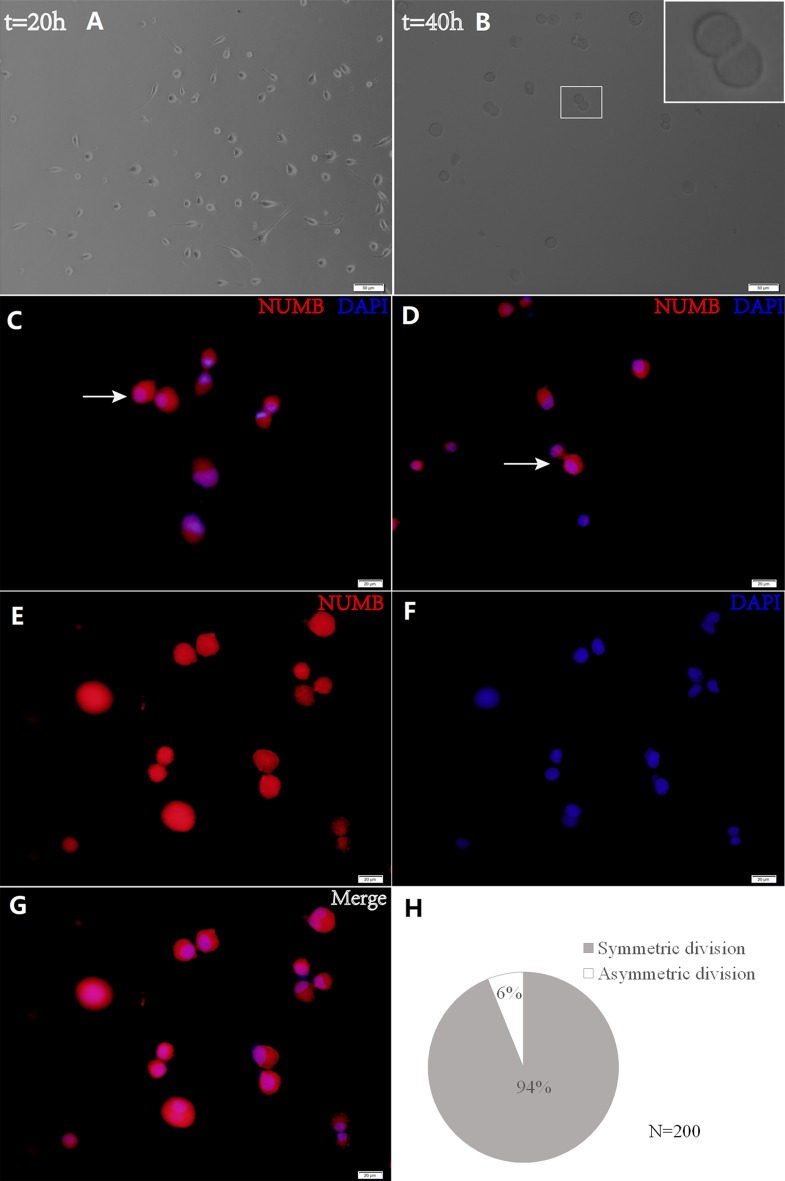
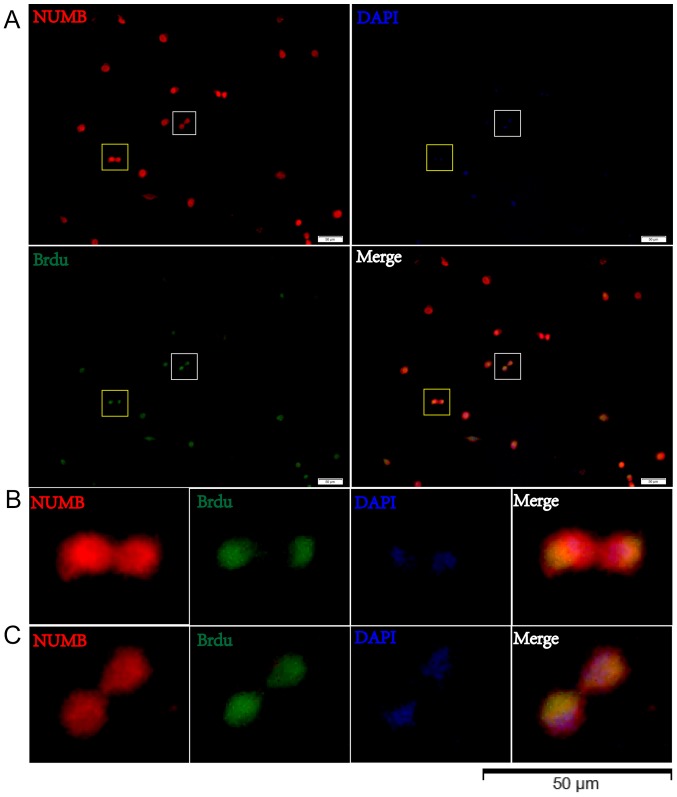
The present study utilized time-lapse lineage tracing to monitor GSC cell divisions at a single-cell resolution (Fig. 5). Cell divisions were recorded using time-lapse microscopy, and cells were subsequently fixed and stained for Numb. Following a 40-h culture, the present study identified the cells that had divided once and generated a cell pair (Fig. 5B). Through this analysis, the Numb distribution modes were detected in the daughter cells (Fig. 5C and D). Sister cells with symmetric Numb usually appeared similar. In order to quantify the asymmetry, asymmetrical division was defined as having occurred when the percent deviation of the stained marker between daughter cells was >21%. Using this criterion, it was discovered that the majority of pairs (93.91±1.59 vs. 6.09±1.57%) exhibited symmetry in the Numb distribution (Fig. 5H). These data demonstrate that GSCs are primarily maintained by symmetric, as opposed to asymmetric, cell divisions (Fig. 5E-G). Cells were then treated with BrdU for 8 h and the BrdU distribution was observed in relation to Numb expression by immunostaining in pair assays. The data revealed that BrdU incorporation in the two daughter cells was associated with Numb asymmetry (Fig. 6).
Figure 5.
Time-lapse lineage tracing detected cell division. Cell divisions were recorded using time-lapse video microscopy for (A) 20 h and (B) 40 h. Scale bars, 50 µm. The white box in (B) indicates a pair of daughter cells from a GSC, which is presented at a higher magnification (×800) in the inset. The representative cell pairs with Numb (C) symmetry and (D) asymmetry immunostained for Numb and DAPI (white arrow). (E-G) GSCs are primarily maintained by symmetric cell divisions. Scale bar, 20 µm. (H) Percentage of Numb distribution features (asymmetric or symmetric) observed from 200 pairs of cells when the GSCs had divided once. GSC, glioma stem cell.
Figure 6.
BrdU distribution in paired cells. (A) BrdU incorporation in two daughter cells is associated with Numb asymmetry. The white box in (A) indicates a pair of daughter cells from a GSC with asymmetric BrdU incorporation, which is presented at a higher magnification in (B) The yellow box in (A) indicates a pair of daughter cells from a GSC with symmetric BrdU incorporation, which is presented at a higher magnification in (C) GSC, glioma stem cell.
ATRA increased the asymmetric cell division of GSCs
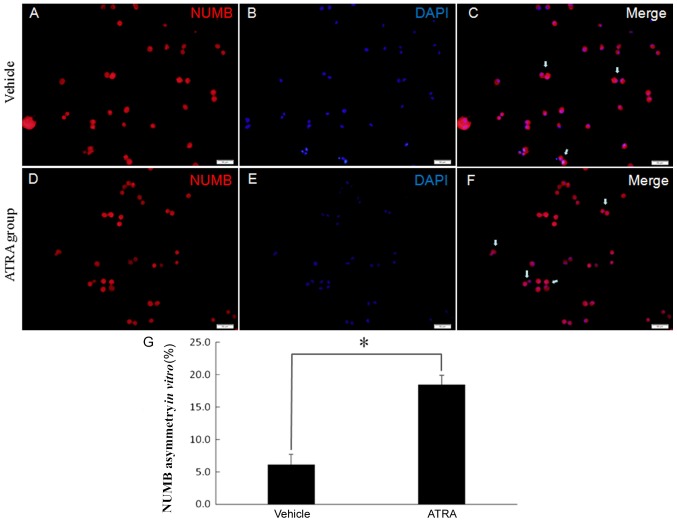
To investigate whether symmetric divisions are affected when ATRA is administered, the present study observed the division mode when ATRA was added to the culture. Neurospheres were grown in DMEM containing 10% FBS and 10 nmol/l ATRA to induce differentiation. Cell divisions were recorded for 40 h using time-lapse video microscopy. Pairs of daughter cells from single progenitor cells were identified, fixed and stained for Numb. There was a decrease in symmetrical stem cell expansion division incidence to 18.30±1.43% of the events in the ATRA-treated cells (Fig. 7). This suggests that the cancer stem cells were impaired in their ability to undergo a symmetric division.
Figure 7.
ATRA induces asymmetric cell divisions of GSCs. (A-F) Numb (red) distribution was used to represent GSC division mode in (A-C) vehicle group and (D-F) ATRA group. Cells were counterstained with DAPI (blue) to mark nuclei. White arrows indicate asymmetric pairs. Scale bars, 50 µm. (G) Quantification of the asymmetric cell pairs demonstrated that treatment with ATRA increased the fraction of cell pairs with an asymmetric Numb distribution. *P<0.05. GSC, glioma stem cell; ATRA, all-trans retinoic acid.
Discussion
The results of the present study demonstrated that the GSCs isolated from the U87MG cell line did not express CD133. Using single-cell-based analytical methods and quantitative immunofluorescence, the present study revealed that Numb was widely expressed in GSCs and symmetrically segregated into two daughter cells during the division of GSCs. The present study, to the best of our knowledge, is the first to demonstrate, that the Numb symmetrical distribution proportion was decreased following ATRA-based differentiation treatment.
GSCs have been identified in numerous primary culture GBM cells and malignant glioma cell lines (5,18–21). A previous study has confirmed that GSCs are the source of new tumor cells following the administration of temozolomide (22). The universal GSC marker remains a controversial topic due to high interpatient as well as intratumoral variability (8). Various surface markers, including CD133, nestin, Musashi-1, Nanog and platelet-derived growth factor receptor, have been reported to successfully identify GSC populations in vitro and in vivo (8,14). Additional stem cell marker detection would further support our conclusions. The present study analyzed CD133 expression using flow cytometry and identified that CD133 was negative in glioma cell spheres cells cultured from the U87MG cell line. This result differs from that of previous research, which reported that the majority of U87MG cells in the spheres were positive for CD133 (20). Further results using immunofluorescence revealed that the CD133-negative cell populations expressed nestin. In addition, the cell populations of the cultured tumor spheres were able to differentiate into cells positive for GFAP, β-tubulin III and GALC, which are representative markers of neuronal, astroglial and oligodendroglial cells (23). These results suggested successful induction of GSCs from the U87MG cell line. However, the lack of an exact evaluation of stemness/differentiation marker expression levels is a limitation of the present study. Although CD133 has been defined as a marker of glioma stem cells, an increasing amount of evidence has demonstrated that the use of CD133 as a unique glioma stem cell marker is insufficient to tag all GSCs. For example, fresh human glioma and gliomasphere cultures express CD133 at low and sometimes barely detectable levels (21). Secondly, CD133-positive and CD133-negative GSCs from cell lines and GBM tumors exhibited cancer stem cell properties (20,24). Thirdly, neither the expression of stemness genes nor the long-term self-renewal capacities of CD133-positive and CD133-negative cells were significantly different (25). Finally, CD133 negative cells were tumorigenic when implanted into rat brains (26). A previous study demonstrated that the levels of surface CD133 fluctuate during the cell cycle in GSCs (27), indicating that CD133 expression is likely a marker of certain stages of GSC division, rather than a constitutive marker of GSCs.
Lathia et al (10) examined a variety of molecules in GSCs and observed that only Numb and CD133 could be asymmetrically segregated. Since the results of the present study demonstrated that CD133 expression was negative in GSCs cultured from the U87MG glioblastoma of unknown origin cell line, the present study used Numb to analyze the GSC division mode. The data revealed that Numb protein was expressed in 99% of GSCs from the U87MG cell line. Using single-cell-based observations, the current study demonstrated that the Numb distribution was predominantly symmetric in the two daughter cells (94%) during GSC division. BrdU incorporation indicates the proliferative ability of cells that were actively replicating their DNA. The results of the present study demonstrated that the BrdU distribution in the two daughter cells was associated with Numb asymmetry. A limitation of the present study is that the exact level of BrdU in paired cells was not measured. In paraffin-embedded glioblastoma specimens, a previous study indicated that 85% of cells exhibited a symmetric pattern of Numb immunoreactivity (28). Numb is a so-called fate-determining molecule that promotes the differentiation of neural stem cells through antagonizing the notch and hedgehog signaling pathways (29,30). The function of Numb is critical for the occurrence of asymmetric cell division, and different expressions of Numb may indicate cell fate divergence (31). Previous studies have suggested that symmetric determinants exert pivotal functions in tumor initiation, as defects in either the function of fate determinants and regulators of asymmetric division, or the loss of asymmetric division may lead to tumor development (13,32). Although previous data demonstrated that the overexpression of Numb did not induce either differentiation of U87MG cells or alter their morphology, nor that the cell population doubling time was significantly affected (33), until now, numerous findings have demonstrated that Numb is associated with the GSC markers SRY (sex determining region Y)-box 2 and paired box protein Pax-6, as well as GSC survival, proliferation, aggressiveness and therapeutic resistance (12,28,34).
The present results demonstrated a decrease in symmetrical cell division incidence to 82% of the events in ATRA-treated GSCs from the U87MG cell line. ATRA is a metabolite of vitamin A that is able to induce complete remission in the majority of acute promyelocytic leukemia cases when administered in combination with light chemotherapy and/or arsenic trioxide (35). To date, the efficacy of ATRA to treat solid tumors remains poor (35). A previous study demonstrated that ATRA could reduce the proliferation and invasiveness of U87MG stem-like glioma cells in a dose-dependent manner and decrease tube formation and vascular endothelial growth factor secretion (36). When ATRA was administered in combination with mTOR and PI3K inhibition, there was a synergistic effect resulting in the minimum amount of cellular migration in GSCs (15). It has been revealed that ATRA-based differentiation targeted GSCs and induced GSCs to differentiate, which was mediated by the activation of ERK1/2 (37). The results of the present study demonstrated that differentiation treatment with ATRA induced asymmetric division in GSCs from the U87MG cell line. The underlying mechanism may be associated with the distribution of Numb (12,28,31). In a future study, we aim to investigate how ATRA affects the distributions of p53, E-cadherin and Notch in the paired GSCs in order to understand this underlying mechanism. Asymmetric division is a feature more closely associated with cancer stem cells from early-stage and well-differentiated tumors, while late stage tumors suppress asymmetric division and increase symmetric division (38). Therefore, the increase of asymmetric division following treatment with ATRA may be associated with the suppression of tumorigenesis in GSCs. However, from the present evidence it cannot be concluded that ATRA can be used as a glioblastoma treatment agent.
Although ATRA induces morphological differentiation of GSCs in vitro, one of the primary limitations in the present study is that it remains controversial as to whether the cell line U87MG is representative of highly malignant glioma. Therefore, further experiments on primary cell lines are required. Secondly, ATRA-metabolizing enzymes (39) and loss of retinoic acid receptors (40) may render cells irresponsive to ATRA effects. Additional studies are necessary in order to investigate how ATRA influences the GSC division mode in vivo. In conclusion, the results of the present study demonstrated that ATRA-induced asymmetric division of GSCs from the U87MG glioblastoma cell line of unknown origin.
Acknowledgements
Not applicable.
Funding
This study was supported by the Shaanxi Province Scientific and Technology Development Program (grant no. 2012SF2-03-05) and the Xi'an Jiaotong University Free Exploration Program (grant no. xjj2013059).
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
RW designed the study and performed the majority of experiments. CL performed the GSC cultures and immunostaining. RW was a major contributors to the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Second Hospital of Xi'an Jiaotong University (Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-κB in glioblastoma: A therapeutic approach. Sci Adv. 2016;2:e1501292. doi: 10.1126/sciadv.1501292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos B, Wan F, Farhadi M, Ernst A, Zeppernick F, Tagscherer KE, Ahmadi R, Lohr J, Dictus C, Gdynia G, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res. 2010;16:2715–2728. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundar SJ, Hsieh JK, Manjila S, Lathia JD, Sloan A. The role of cancer stem cells in glioblastoma. Neurosurg Focus. 2014;37:E6. doi: 10.3171/2014.9.FOCUS14494. [DOI] [PubMed] [Google Scholar]
- 8.Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119–1131. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olmez I, Shen W, McDonald H, Ozpolat B. Dedifferentiation of patient-derived glioblastoma multiforme cell lines results in a cancer stem cell-like state with mitogen-independent growth. J Cell Mol Med. 2015;19:1262–1272. doi: 10.1111/jcmm.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, Tsatsanis A Gallinger S, Dick JE. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 2012;21:777–792. doi: 10.1016/j.ccr.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. Nestin and CD133: Valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karsy M, Albert L, Tobias ME, Murali R, Jhanwar-Uniyal M. All-trans retinoic acid modulates cancer stem cells of glioblastoma multiforme in an MAPK-dependent manner. Anticancer Res. 2010;30:4915–4920. [PubMed] [Google Scholar]
- 16.Yu SC, Ping YF, Yi L, Zhou ZH, Chen JH, Yao XH, Gao L, Wang JM, Bian XW. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124–134. doi: 10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Goderie SK, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 18.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 19.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 20.Howard CM, Valluri J, Alberico A, Julien T, Mazagri R, Marsh R, Alastair H, Cortese A, Griswold M, Wang W, et al. Analysis of Chemopredictive assay for targeting cancer stem cells in glioblastoma patients. Transl Oncol. 2017;10:241–254. doi: 10.1016/j.tranon.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng X, Shen G, Yang X, Liu W. Most C6 cells are cancer stem cells: Evidence from clonal and population analyses. Cancer Res. 2007;67:3691–3697. doi: 10.1158/0008-5472.CAN-06-3912. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Richard JP, Wang SD, Rath P, Laterra J, Xia S. Regulation of glioblastoma multiforme stem-like cells by inhibitor of DNA binding proteins and oligodendroglial lineage-associated transcription factors. Cancer Sci. 2012;103:1028–1037. doi: 10.1111/j.1349-7006.2012.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 25.Clement V, Dutoit V, Marino D, Dietrich PY, Radovanovic I. Limits of CD133 as a marker of glioma self-renewing cells. Int J Cancer. 2009;125:244–248. doi: 10.1002/ijc.24352. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Sakariassen PØ, Tsinkalovsky O, Immervoll H, Bøe SO, Svendsen A, Prestegarden L, Røsland G, Thorsen F, Stuhr L, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 27.Barrantes-Freer A, Renovanz M, Eich M, Braukmann A, Sprang B, Spirin P, Pardo LA, Giese A, Kim EL. CD133 expression is not synonymous to immunoreactivity for AC133 and fluctuates throughout the cell cycle in glioma stem-like cells. PLoS One. 2015;10:e0130519. doi: 10.1371/journal.pone.0130519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Xing H, Kim TM, Jung Y, Huang W, Yang HW, Song S, Park PJ, Carroll RS, Johnson MD. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells. 2012;30:1313–1326. doi: 10.1002/stem.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Marcotullio L, Greco A, Mazzá D, Canettieri G, Pietrosanti L, Infante P, Coni S, Moretti M, De Smaele E, Ferretti E, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30:65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- 30.Beres BJ, George R, Lougher EJ, Barton M, Verrelli BC, McGlade CJ, Rawls JA, Wilson-Rawls J. Numb regulates Notch1, but not Notch3, during myogenesis. Mech Dev. 2011;128:247–257. doi: 10.1016/j.mod.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 32.Bajaj J, Zimdahl B, Reya T. Fearful symmetry: Subversion of asymmetric division in cancer development and progression. Cancer Res. 2015;75:792–797. doi: 10.1158/0008-5472.CAN-14-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Euskirchen P, Skaftnesmo KO, Huszthy PC, Brekkå N, Bjerkvig R, Jacobs AH, Miletic H. NUMB does not impair growth and differentiation status of experimental gliomas. Exp Cell Res. 2011;317:2864–2873. doi: 10.1016/j.yexcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Beier CP, Beier D. CD133 negative cancer stem cells in glioblastoma. Front Biosci (Elite Ed) 2011;3:701–710. doi: 10.2741/e280. [DOI] [PubMed] [Google Scholar]
- 35.Schenk T, Stengel S, Zelent A. Unlocking the potential of retinoic acid in anticancer therapy. Br J Cancer. 2014;111:2039–2045. doi: 10.1038/bjc.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling GQ, Liu YJ, Ke YQ, Chen L, Jiang XD, Jiang CL, Ye W. All-trans retinoic acid impairs the vasculogenic mimicry formation ability of U87 stem-like cells through promoting differentiation. Mol Med Rep. 2015;12:165–172. doi: 10.3892/mmr.2015.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman MD, Jeevan DS, Tobias M, Murali R, Jhanwar-Uniyal M. Targeting cancer stem cells in glioblastoma multiforme using mTOR inhibitors and the differentiating agent all-trans retinoic acid. Oncol Rep. 2013;30:1645–1650. doi: 10.3892/or.2013.2625. [DOI] [PubMed] [Google Scholar]
- 38.Bu P, Chen KY, Lipkin SM, Shen X. Asymmetric division: A marker for cancer stem cells in early stage tumors? Oncotarget. 2013;4:950–951. doi: 10.18632/oncotarget.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong JL, Taylor GA, Thomas HD, Boddy AV, Redfern CP, Veal GJ. Molecular targeting of retinoic acid metabolism in neuroblastoma: The role of the CYP26 inhibitor R116010 in vitro and in vivo. Br J Cancer. 2007;96:1675–1683. doi: 10.1038/sj.bjc.6603779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakravarti N, Lotan R, Diwan AH, Warneke CL, Johnson MM, Prieto VG. Decreased expression of retinoid receptors in melanoma: Entailment in tumorigenesis and prognosis. Clin Cancer Res. 2007;13:4817–4824. doi: 10.1158/1078-0432.CCR-06-3026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author upon reasonable request.