Abstract
The use of mRNAs as biomarkers serves to diagnose, treat, as well as aid the prognosis of cancer. The present study involved an analysis of mRNAs in the cell cycle at the G2 and G3 tumor grades for the prognosis of ovarian serous cystadenocarcinoma (OSC) using 364 clinical samples (G2:G3=42:322). Statistics aided the identification of NPFFR2, XPNPEP2 and CELA3B; the 3-mRNA model that allows for classification of patients into high- and low-risk groups using a median value of 0.9580745. The rates of survival varied (P=0.00149) and the independent detection of stratification of the risk of this disease was validated with success using the 3-mRNA signature, which was demonstrated to be more successful than the weight model. This approach was revealed to provide the prognosis of grade G2 and G3 in patients with OSC compared with factors used traditionally. Compared with traditional factors, this 3-mRNA model was demonstrated to be the only and independent prognostic factor for patients with G2 and G3 stage OSC. A literature survey was also performed in the present study in order to assess the role of the 3 genes and indirectly prove their effectiveness. The establishment of this new genetic model will enhance prospective prognosis and treatment for patients with OSC.
Keywords: ovarian serous cystadenocarcinoma, mRNA, prognosis, overall survival, The Cancer Genome Atlas
Introduction
Ovarian cancer adversely affects female health worldwide and is one of the major causes of mortality of women with severe gynecological issues (1). Ovarian cancer primarily presents as tumors of the epithelial cells that grow from the surface epithelial cells of the ovary, and are histologically classified into four main subtypes: Mucinous, endometrioid, serous and clear cells. The most common histologic subtype in epithelial ovarian cancer is the OSC, representing 75–80% of all cases worldwide (2,3). OSC is a common female genital cancer. These types of cancer are either asymptomatic or have similar symptoms to other benign gynecological diseases, until the tumor has metastasized on the surface of the peritoneum and then it will be diagnosed. Thus, the majority of patients are diagnosed when the disease has reached an advanced stage. Furthermore, due to the current absence of effective treatment options, patients with the disease experience extremely poor overall survival rate (OS), and only 45% 5-year relative survival rate of all stages (4). Therefore, identification and validation of prognostic biomarkers to predict OSC outcomes are of high clinical value.
There is an urgent need to identify new, highly sensitive and specific biomarkers for improved diagnosis and targeted therapies (5). The biomarkers bestow early detection as well as predict a poor prognosis (6,7). A number of studies have used certain genes as cancer prognostic biomarkers with significant success in patients with ovarian cancer. For instance, Liu et al (8) reported overexpression of TRIM44 in ovarian cancer, and revealed a close association with lower rates of overall- and disease-free survival. TRIM44 can be used as an independent marker to predict poor prognosis in ovarian cancer. Lee et al (9) discovered that another protein, CENPK, was overexpressed in ovarian cancer cells, and proved its direct association with a poor prognosis in patients with ovarian cancer. Furthermore, incorporating CENPK with the tumor markers CA125 or HE4 can increase the sensitivity of CA125 or HE4 for predicting ECO outcomes (9). In addition, ASAP1, MAGE-A9 and keratin 17 have been linked to poor prognosis and, hence, their utility as a prognostic indicator in human ovarian cancer (10–12). Thus, while the biomarkers are of great clinical value for predicting outcomes for patients with ovarian cancer, there is very limited prognostic value to a single candidate biomarker. This could be attributed to inconsistent sample collection, detection methods and small sample sizes (11).
Despite an increasing number of studies focusing on malignant tumors, the exact underlying molecular mechanisms remain unclear. For the majority of tumors, the treatment effect and prognosis are not ideal. In clinical practice, the histological grade provides an important prognosis for tumors, which aids in assessing the tumor behavior (13) and is most commonly used for the prognosis of hepatocellular carcinoma, breast cancer and mucinous appendiceal adenocarcinoma (13–15). The aggressive potential of the tumor is defined by histological grades as: G1, well-differentiated and the least aggressive (slow growing); G2, moderately differentiated; G3, highly proliferative and most aggressive but poorly differentiated; G4, undifferentiated (16). A statistically significant difference was observed by Overman et al (17) between the apparent diffusion coefficient and pure diffusion coefficient among the different histological grades. They also found that in the mucinous appendiceal adenocarcinoma with peritoneal metastasis, the G3 group values were lower than those of the G2 group (17). While studying hepatocellular carcinoma (HCC), Granata et al (18) demonstrated that within the HCC groups, the perfusion fraction values of G1, G2 and G3 histological grades were significantly different (18). Similarly, Grotz et al (16) demonstrated that in mucinous appendiceal adenocarcinoma (MAA), the G2 clinical behavior is distinctly different from that in G1 and G3, with remarkably different cancer-specific survival in stage IV G2 and G3 of MAA. Although OSC is an extremely common form of ovarian cancer, no article has reported the utility of multiple biomarkers in determining the associated histological grade to the best of our knowledge. With this aim, the current study investigated the utility of multiple biomarkers to assess the OS rate of OSC and determined the prognostic biomarkers for assessing poor prognosis and disease progression.
Materials and methods
Dataset for patients with OSC
Patient clinical and cognate data for pre-processed transcript mRNA (as of September 2018) were sourced from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The information includes intact data on mRNA expression and clinical characteristics (including age, ethnicity, sex, tumor type and histological grade, and status and time of survival).
Screening of differentially expressed mRNAs between G2 and G3 of OSC
Data from a total of 364 patients with OSC were downloaded from TCGA, as per screening criteria, transcript data and basic clinical data (Table I), including 42 in G2 and 322 in G3. The differentially expressed mRNAs in G2 and G3 grades were distinguished using edgeR version 3.22.5 (bioinf.wehi.edu.au/edgeR) in R (19), and the threshold values were |log 2-fold change |≥2 and false discovery rate <0.05. The edgeR package automatically deletes outliers when analyzing differences, thus reducing the impact of sample differences and making the analytical data more reliable.
Table I.
Summary of clinical characteristics of the patients with ovarian serous cystadenocarcinoma included in the present study.
| Patients (n=364) | ||
|---|---|---|
| Characteristic | n | % |
| Age, years | ||
| 30–59 | 191 | 52.47 |
| 60–87 | 173 | 47.53 |
| Sex | ||
| Female | 364 | 100 |
| Ethnicity | ||
| Caucasian | 316 | 86.81 |
| Black or African American | 23 | 6.32 |
| Asian | 11 | 3.02 |
| Unknown | 14 | 3.85 |
| Tumor grade | ||
| G2 | 42 | 11.54 |
| G3 | 322 | 88.46 |
| Patient status | ||
| Alive | 166 | 45.60 |
| Succumbed | 198 | 54.40 |
Analysis of survival and the prognostic model based on mRNA status
The association between patient OS rate and differentially expressed genes was assessed using the univariate Cox package for univariate Cox proportional hazard regression analysis, and those with P<0.01 were used as candidate variables. A multi-gene prediction model was then established simultaneously, and appropriate prognostic information for validation comparisons was downloaded from Kaplan-Meier plotter (kmplot.com/analysis/). Based on the mean of expression, the cases were divided into a high and low expression groups. Furthermore, the multivariate Cox regression model was performed to assess the prognostic value of the differentially expressed mRNAs (DEMs). Based on the mean expression levels of the gene, the cases were classified into high and low expression groups. A score for prognosis risk was established to predict OS based on a linear combination of the expression level multiplied by the multivariate Cox regression model (β) derived regression coefficient, applying the formula: Risk score=exp DEM1 × β DEM1 + exp DEM2 × β DEM2 + … exp DEMn × β DEMn).
Risk classification and receiver operating characteristic (ROC) curve
The risk scores of 364 patients were determined as per the multi-gene model, and then grouped into high- and low-risk groups based on the median value. The survival status and risk scores of high- and low-risk patients were compared using the Survival_Graph and Point_Graph ggplot version 2.3.0.0 (ggplot2.tidyverse.org). The areas under the curve (AUC) of the predictive model were evaluated computationally to estimate the sensitivity and specificity of mRNA prognosis, and plotted using the Survival_ROC package version 1.0.3 (cran.r-project.org/web/packages/survivalROC/index.html).
Prediction of independent survival time based on the 3-mRNA signature prognosis from other clinical variables
Multivariate Cox analyses were performed to determine the independence of the 3-mRNA signature from other clinical factors (age, race and grade) of patients with OSC, using OS as the dependent variable. Furthermore, stratification analysis was carried out on clinical features mentioned in Cox regression analysis, to assess if there is any prognostic value of the 3-mRNA signature within the same clinical factor.
Statistical analysis
The data were derived from TCGA, processed by edgeR package to obtain differentially expressed genes, and the integration of differential genes with clinical data was performed in the command prompt of Windows 10 (Microsoft Corporation). The multivariate Cox package was used to establish a multi-gene survival model and generate the model's survival curve and the ROC package was utilized to predict the survival rate of patients with G2 and G3 grade OSC. Following the construction of the 3-gene model, the 3-gene data (selected from the differentially expressed genes) were entered into GraphPad Prism software (version 5.0; GraphPad Software, Inc.) to determine whether the three mRNAs were prognostically significant for the G2 and G3 grades of OSC and a Mann-Whitney U test was used to assess the data. The univariate Cox package Survival version 2.43–3 (github.com/therneau/survival) was used to obtain the survival curve of each gene. The heat map was generated using the pheatmap package version 1.0.10 (cran.r-project.org/web/packages/pheatmap/), and the Survival_Graph package was used to draw the survival scatter plot and the Point_Graph package to plot the survival score curve. The relevant gene survival curves were downloaded from the KM plotter website. The editing and splicing of images in the text was performed using Photoshop CS6 (Adobe Systems Europe, Ltd.), and the data processing and data packet applications were based on R. Language. P<0.05 was considered to indicate a statistically significant result.
Results
OSC in G2 and G3 exhibit differentially expressed mRNAs
As per the selection criteria, a total of 144 differentially expressed genes (104 upregulated and 40 downregulated) were analyzed in order to identify the OSC G2 and G3 grades.
Association between the 3-mRNA signature with comprehensive survival of patients with OSC
To shortlist prognosis-associated mRNA from differential genes, the association between the expression of each differentially expressed gene and overall patient survival was analyzed via univariate Cox regression analysis, and 3 mRNAs were identified (P<0.01; Table II). Then, through multivariate Cox regression analysis, these 3 mRNAs (Table II) were used as a predictive model (P<0.015), which is linearly associated with the level of corresponding mRNA expression. For the gene predictive model in the multivariate Cox regression analysis, the risk score was calculated as 0.0817 × NPFFR2 + 0.0976 × XPNPEP2-0.1304 × CELA3B. The risk scores were positively associated with NPFFR2 and XPNPEP2, indicating that enhanced expression of these two mRNAs in patients with OSC have shorter OS. In addition, CELA3B was inversely associated with risk scores, suggesting that it may be a potential protective gene for patients with OSC, and its high expression indicates longer OS.
Table II.
3-mRNA risk score signature.
| Gene symbol | Coefficient | Univariate P-value | Multivariate P-value |
|---|---|---|---|
| NPFFR2 | 0.0817 | 0.009221 | 0.011 |
| XPNPEP2 | 0.0976 | 0.007264 | 0.010 |
| CELA3B | −0.1304 | 0.009284 | 0.013 |
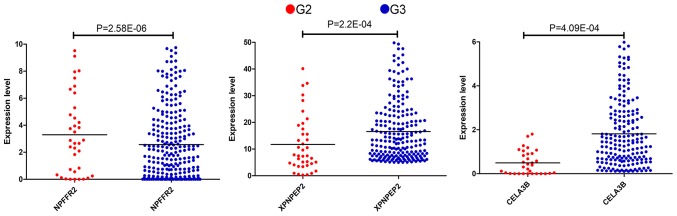
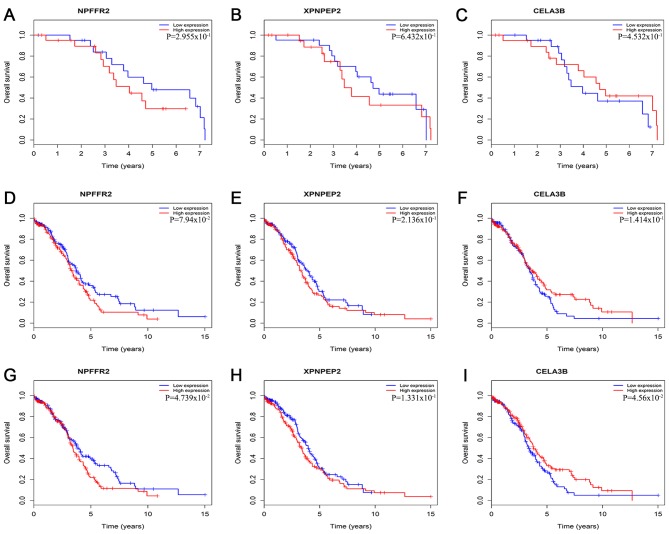
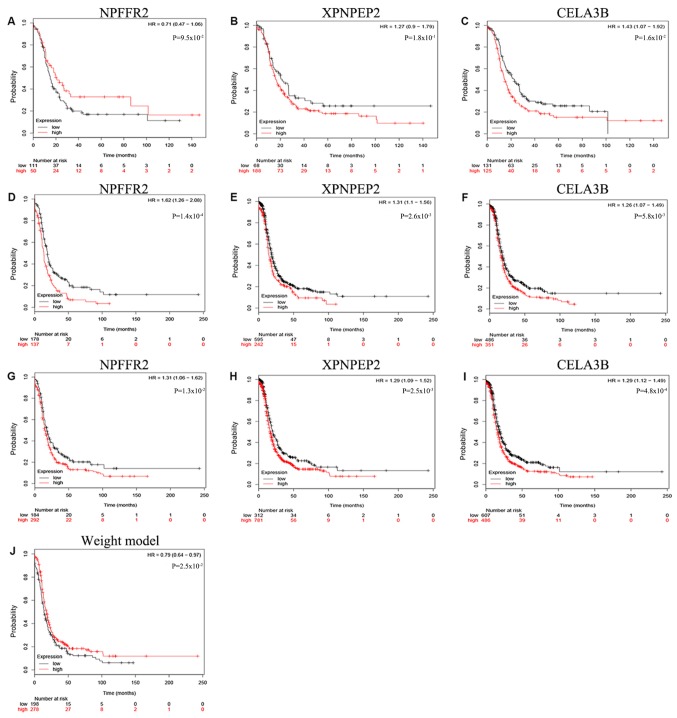
Thus, according to the Cox regression analysis, the three mRNAs were prognostically significant for the G2 and G3 grades of OSC (Fig. 1). The enhanced expression of NPFFR2 in G2 phase and a reduced expression of XPNPEP2 and CELA3B in the sample were statistically significant (P<0.001). NPFFR2 and CELA3B determined to be independent prognostic factors, whereas XPNPEP2 was not. Furthermore, the Kaplan-Meier curves demonstrate statistically significant differences in OS (P<0.05) of NPFFR2 and CELA3B (Fig. 2). High expression of NPFFR2 (Fig. 2A, D and G), and XPNPEP2 (Fig. 2B, E and H) was associated with a lower overall survival rate, and similarly, lower CELA3B expression was associated with a lower overall survival rate (Fig. 2F and I), in agreement with the results of univariate analysis.
Figure 1.
Comparison of expression levels of the three genes in G2 group with those in G3 group.
Figure 2.
Kaplan-Meier survival curves presenting the association between overall survival and the three genes. Based on the mean gene expression level, the cases were classified into high and low expression group. (A) NPFFR2 in G2. (B) XPNPEP2 in G2. (C) CELA3B in G2. (D) NPFFR2 in G3. (E) XPNPEP2 in G3. (F) CELA3B in G3. (G) NPFFR2 in G2 and G3. (H) XPNPEP2 in G2 and G3. (I) CELA3B in G2 and G3.
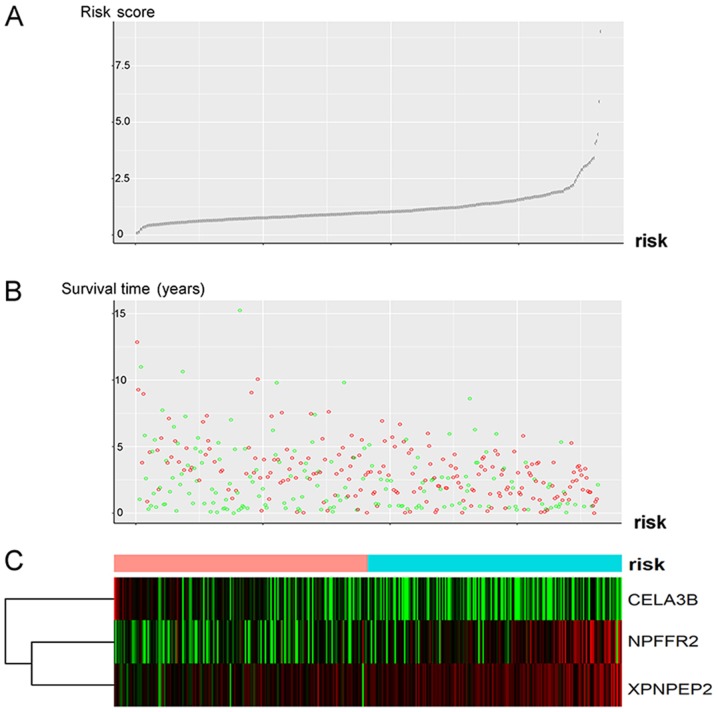
The risk scores of these patients with OSC could be divided into low- and high-risk groups according to the median value of the risk score which was 0.9580745 (Fig. 3C). This group visualizes the survival score and survival status of patients with OSC (Fig. 3A and B). Patients with higher risk scores were observed to have higher NPFFR2 and XPNPEP2 and lower CELA3B expressions. The majority of the risk scores were <2.5, and their survival time was <7.5 months (Fig. 3A and B); however, as the risk score increased, the survival time and the survival rate decreased (Fig. 3A and B).
Figure 3.
Analysis of the risk score of the differentially expressed 3-mRNA signature of G2 and G3 grade of OSC. (A) Curve representing the risk score of the 3-gene signature. (B) Risk score distribution representing status and survival time of the patients. Green circles represent patients who were alive, and red circles represent patients that succumbed. (C) Heatmap of the three genes from all patients. Color transition from green to red indicates increasing expression from low to high, respectively. The color bar indicates the risk score curve individual inflection point, for classifying the patients with OSC into the low-risk (light red), or high-risk group (blue). (The abscissa of Part A and Part B is the case sequence of the color bars according to Part C). OSC, ovarian serous cystadenocarcinoma.
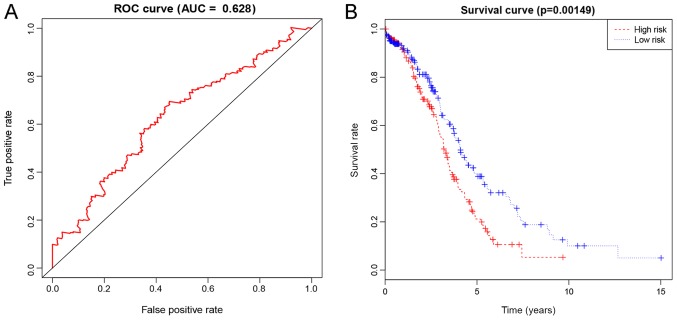
Based on the 5-year ROC AUC of 0.628 (Fig. 4A), the risk score was used to predict the survival rate of G2 and G3 grade patients with OSC. Furthermore, the Kaplan-Meier curve of the 3-mRNA signature further validated that the high-risk group had notably shorter survival times compared with the low-risk group (P=0.00149, Fig. 4B).
Figure 4.
Representation of the differentially expressed 3-mRNA signature of G2 and G3 grade of OSC in terms of prognostic performance. (A) The risk-score prognostic performance presented as the time-dependent ROC curve for the prediction of 5-year survival. (B) The risk score for the overall survival in terms of the Kaplan-Meier survival analysis. OSC, ovarian serous cystadenocarcinoma; ROC, receiver operating characteristic.
The Kaplan-Meier curve for NPFFR2 indicated that there was no significant difference when the OS rate of the patients with G2 grade were compared with others (P>0.05; Fig. 5). In addition, the Kaplan-Meier curve of the 3-mRNA signature further validated that patients in the high-risk group had significantly shorter survival times compared with the low-risk group (P=0.025; Fig. 5J). The role of NPFFR2 appears to change from being a protective factor in G2 to a risk factor in G3, making it a risk factor overall (Fig. 5A, D and G); although the other two biomarkers, XPNPEP2 and CELA3B, were risk factors in both G2 and G3 (Fig. 5B, E and H, and C, F and I), thus confirming the 3-mRNA model.
Figure 5.
Association between the three genes and overall survival as demonstrated via Kaplan-Meier plotter curves. Based on the mean of expression, the cases were divided into a high and low expression groups. (A) NPFFR2 in G2. (B) XPNPEP2 in G2. (C) CELA3B in G2. (D) NPFFR2 in G3. (E) XPNPEP2 in G3. (F) CELA3B in G3. (G) NPFFR2 in G2 and G3. (H) XPNPEP2 in G2 and G3. (I) CELA3B in G2 and G3. (J) the 3-mRNA signature in G2 and G3 (gene weight model).
3-mRNA signature is prognostically independent of other clinical factors
The 3-mRNA risk scoring model exhibited predictive power independent of other clinical factors [hazard ratio (HR), 1.442; 95% confidence interval [CI] 1.081–1.924; P=0.013; Table III]. Age (P=0.119) and histological grade (P=0.128) were not considered significant factors. It was observed that in terms of age, ethnicity, histological grade and risk score HR, values for age, histological grade and risk scores corresponded with greater susceptibility to cancer (HR can help determine if the factor is protective or detrimental; Table III). Among different races, Caucasian individuals may have been less susceptible, as the P-value was close to 0.05, and the HR value is 0.669. Therefore, race may be a protective factor. The histological grades and the corresponding age were considered risk factors (HR>1), which authenticates this scoring model. Although, whether ethnicity serves as a protective factor requires further investigation. P<0.05 for each of the 3 mRNAs in the signature, suggested that this model was highly specific. Thus, according to these results, the 3-mRNA signature used in the present study may serve as a predictor of other clinical factors.
Table III.
Patient overall survival in terms of multivariate Cox regression analysis.
| Variables | HR | 95% CI | P-value |
|---|---|---|---|
| Age, years (≤60 vs. >60) | 1.251 | 0.944–1.658 | 0.119 |
| Ethnicity (Caucasian vs. others) | 0.669 | 0.436–1.026 | 0.066 |
| Grade (G2 vs. G3) | 1.386 | 0.911–2.108 | 0.128 |
| RS (Low vs. high) | 1.442 | 1.081–1.924 | 0.013 |
HR, hazard ratio; CI, confidence interval; RS, 3-mRNA risk score.
Discussion
Nearly 90% of all ovarian cancers present as OSC, making it the most common type of epithelial ovarian cancer (20). OSC results in greater mortality rates than any other type of cancer of the female reproductive system (21). According to the Global Cancer Statistics, in 2016 there were ~230,000 cases of female ovarian cancer diagnosed, and 150,000 women succumbed to the disease (21). While studying ovarian cancer, focus should be paid to the differences in gene expression between normal and tumor tissues (22), characterizing differences between histological subtypes (23,24) and marking differences between tumors with invasive and low malignant potential (25,26). The present study focuses on the moderate differentiation of G2 tumors while transitioning from G2 to G3, which are the most invasive and poorly differentiated tumor grades. To achieve good curative effect, the most important prerequisite is accurate tumor staging in the patients with ovarian cancer. A number of studies have used gene expression for the prognosis of OSC, which exhibit high sensitivity and specificity, and may be clinically significant (27,28). The G2 and G3 grades are crucial indicators for prognosis in OSC as the metastatic rate of ovarian cancer is closely associated with histological grade. A cancer tissue with a lower degree of differentiation is more likely to metastasize, leading to worse prognosis. Hence, the differentiation between G2 and G3 is helpful in evaluating patient prognosis. Owing to similarly hypothesized prognoses, the majority of studies combine G2 and G3, whereas others do not support this amalgamation, suggesting that the differentiated G2 have distinct clinical behavior and outcome from that of G1 and G3. The data from the present study demonstrate differential clinical manifestations of G2 and G3, with different prognoses and different treatment options, including the scope of surgery and the course of chemotherapy.
In the present study, three different genes (NPFFR2, XPNPEP2 and CELA3B) were obtained that distinguished G2 and G3 grades of patients with OSC, and established a 3-mRNA signature. The OS rate of patients with G2 and G3 grade OSC was predicted based on the high-differential low-risk group. To the best of our knowledge, this is the first genetic prediction model based on the histological grading of patients with OSC. The differentially expressed mRNAs were obtained from 364 patients with OSC whose clinical information was available on TCGA (42 cases in G2 stage and 322 cases in G3 stage). These differentially expressed mRNAs were assessed through single factor and stepwise multivariate Cox analyses and a linear prediction model was built. It was observed that the high- and low-risk patient groups were significantly different, in addition to high sensitivity and specificity as determined through the ROC curve (AUC, 0.628; Fig. 4A). The multivariate Cox regression analysis indicated that the 3-mRNA signature can aid in prognosis, independent of the traditional clinical factors such as age, ethnicity and histological grade. Furthermore, the KM plotter confirmed that OS based on NPFFR2 and CELA3B were significant in all patients (G2+G3) and that the 3-mRNA signature was therefore greatly different from the gene weight model. The expression of NPFFR2 is inversely proportional to cancer malignancy. A similar pattern is observed in the pathological grading and staging of head and neck squamous cell carcinoma; the degree of malignancy was directly proportional to that of NPFFR2 methylation (29). NPFFR2 is also associated with the activity of hypothalamic-pituitary-adrenal axis (30). In terms of stress-associated hormones and released neurotransmitters, quality has an adverse effect on stress-induced tumor progression and cancer treatment (31), which indirectly indicate that the progression of cancer (increased histological grade, etc.) is indirectly affected by NPFFR2. As a proline hydrolase, XPNPEP2, also known as aminopeptidase P or APP2, hydrolyzes numerous biologically active peptides, such as XPNPEP2, which inactivates bradykinin (32). XPNPEP2 serves a vital role in the ovarian breakdown and follicular dysplasia induced by hexavalent chromium in rat germ cells (33). XPNPEP2 overexpression is also observed in cervical cancer tissues, and is associated with pathological staging, lymph node metastasis and poor OS (34), which coincides with the results from the present study findings in the XPNPEP2 study in OSC, in renal clear cell carcinoma (CCRCC) (35) and in advanced gastric cancer (36).
Human CELA3B is a product of gene duplication. In CELA3B, the 241st residue is polymorphic (p.A241 G), and the differences may lead to the risk of chronic pancreatitis (37) and capsular fibrosis islets (38). While capsular fibrosis also conforms to histological changes in OSC, in type 2 diabetes, CELA3B has been revealed to be associated with microvascular ischemia (39), which may explain the cause of OSC rupture. Therefore, NPFFR2, XPNPEP2 and CELA3B can be associated with the occurrence of OSC and the associated prognosis.
There were also some limitations to the present study. The number of patients in G2 and G3 were not equal. The pathological grades of OSC are G1, G2, G3 and G4. However, limited data were available on TCGA for the G1 and G4 stages of OSC (<5 cases each) and no data for adjacent tissues. If the sample size is too small, errors occur in the statistical analyses, and thus should not be included within the study. Updates to the OSC data on TCGA will be assessed in future studies once they become available. The clinical data of OSC are incomplete, and the Tumor-Node-Metastasis (TNM) stages were unknown. While TNM is the standardized staging method for malignant tumor progression (4), it also plays an important role in guiding the prognosis and treatment of tumors. In the multi-factor analysis, if the influence of TNM staging on OS is also assessed then, through database verification, testing the feasibility of the gene model can be improved. Another high-risk factor for ovarian cancer is age, and it should be assessed in greater detail; the small amount of age groups in the present study may be one of the most important shortcomings of the present study. Due to the small number of age groups, the results may have demonstrated larger errors, therefore age should be divided into additional groups for more accurate comparisons. Furthermore, there are large inconsistencies between the case survival and the database survival analyses. More than 3,000 patients with ovarian cancer are usually included in the KM plotter database, but the number of cases in the present study, obtained from TCGA, was much lower, causing inconsistencies in the results. The fact that some of the data in Figs. 2 and 5 do not match may have been due these restrictions. While the model is feasible according to the moderate area of the AUC curve, it is not highly sensitive and specific, and so may only be suitable for the initial screening of disease. Whilst the sensitivity was high compared to other factors (age, histological grade and ethnicity), when the data was compared independently (amongst itself), the absolute sensitivity was not high. To the best of our knowledge, the genetic model involving the three genes (NPFFR2, XPNPEP2 and CELA3B) signature has not been studied in other tumors, making it difficult to prove that they have roles in OSC. Thus, a combination of clinical experience and other auxiliary examinations is required in order to make a correct diagnosis.
The present study established a genetic model using three genes, NPFFR2, XPNPEP2 and CELA3B, for identifying G2 and G3 grades of OSC through statistical analysis. The model is different from traditional prognosis and clinical identification methods. The role of these three genes was further analyzed and compared against the published literature. This newly established genetic model has the potential for prognosis of patients with OSC.
Acknowledgements
The authors would like to acknowledge Professor Hong Wang (Hong Wang, Pediatric Surgery II Ward, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi) for her support.
Funding
The present study was supported by the Scientific Research Project of Guangxi Provincial Health and Family Planning Commission (grant no. Z20180900).
Availability of data and materials
The datasets generated and analyzed during the current study are available in the TCGA repository, https://portal.gdc.cancer.gov/.
Authors' contributions
JZ designed the study. CS and YL revised the experimental design. JZ and YY acquired, analyzed and interpreted the data. CW validated the experimental data. CS and YL verified the results of the experiment. JZ, YY and CW wrote the manuscript and revised it for important intellectual content.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escalona RM, Chan E, Kannourakis G, Findlay JK, Ahmed N. The many facets of metzincins and their endogenous inhibitors: Perspectives on ovarian cancer progression. Int J Mol Sci. 2018;19(pii):E450. doi: 10.3390/ijms19020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang S. Gastric metastasis of ovarian serous cystadenocarcinoma. Int Med Case Rep J. 2018;11:201–204. doi: 10.2147/IMCRJ.S171985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cress RD, Chen YS, Morris CR, Petersen M, Leiserowitz GS. Characteristics of long-term survivors of epithelial ovarian cancer. Obstet Gynecol. 2015;126:491–497. doi: 10.1097/AOG.0000000000000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Qiu J, Ye C, Yang D, Gao L, Su Y, Tang X, Xu N, Zhang D, Xiong L, et al. ROR1 expression correlated with poor clinical outcome in human ovarian cancer. Sci Rep. 2014;4:5811. doi: 10.1038/srep05811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal SK, Figlin RA, Reckamp K. Targeted therapies for non-small cell lung cancer: An evolving landscape. Mol Cancer Ther. 2010;9:1931–1944. doi: 10.1158/1535-7163.MCT-10-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly K, Huang C. Biological agents in non-small cell lung cancer: A review of recent advances and clinical results with a focus on epidermal growth factor receptor and vascular endothelial growth factor. J Thorac Oncol. 2008;3:664–673. doi: 10.1097/JTO.0b013e3181758141. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Yin H, Ji H, Zhu J, Ma R. Overexpression of TRIM44 is an independent marker for predicting poor prognosis in epithelial ovarian cancer. Exp Ther Med. 2018;16:3034–3040. doi: 10.3892/etm.2018.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YC, Huang CC, Lin DY, Chang WC, Lee KH. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ. 2015;3:e1386. doi: 10.7717/peerj.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Wang C, Zhang Y, Jia L, Huang J. Overexpression of MAGE-A9 is predictive of poor prognosis in epithelial ovarian cancer. Sci Rep. 2015;5:12104. doi: 10.1038/srep12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou T, Yang C, Tong C, Zhang H, Xiao J, Li J. Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int J Clin Exp Pathol. 2013;7:280–287. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YF, Lang HY, Yuan J, Wang J, Wang R, Zhang XH, Zhang J, Zhao T, Li YR, Liu JY, et al. Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol. 2013;34:1685–1689. doi: 10.1007/s13277-013-0703-5. [DOI] [PubMed] [Google Scholar]
- 13.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010;12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoog P, Ohlsson M, Fernö M, Rydén L, Borrebaeck CAK, Wingren C. Tumor tissue protein signatures reflect histological grade of breast cancer. PLoS One. 2017;12:e0179775. doi: 10.1371/journal.pone.0179775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu SC, Liu YH, Wei Y, Li LL, Dou SW, Sun TY, Shi DP. Intravoxel incoherent motion diffusion-weighted magnetic resonance imaging for predicting histological grade of hepatocellular carcinoma: Comparison with conventional diffusion-weighted imaging. World J Gastroenterol. 2018;24:929–940. doi: 10.3748/wjg.v24.i8.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grotz TE, Royal RE, Mansfield PF, Overman MJ, Mann GN, Robinson KA, Beaty KA, Rafeeq S, Matamoros A, Taggart MW, Fournier KF. Stratification of outcomes for mucinous appendiceal adenocarcinoma with peritoneal metastasis by histological grade. World J Gastrointest Oncol. 2017;9:354–362. doi: 10.4251/wjgo.v9.i9.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overman MJ, Fournier K, Hu CY, Eng C, Taggart M, Royal R, Mansfield P, Chang GJ. Improving the AJCC/TNM staging for adenocarcinomas of the appendix: The prognostic impact of histological grade. Ann Surg. 2013;257:1072–1078. doi: 10.1097/SLA.0b013e318269d680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granata V, Fusco R, Catalano O, Guarino B, Granata F, Tatangelo F, Avallone A, Piccirillo M, Palaia R, Izzo F, Petrillo A. Intravoxel incoherent motion (IVIM) in diffusion-weighted imaging (DWI) for Hepatocellular carcinoma: Correlation with histologic grade. Oncotarget. 2016;7:79357–79364. doi: 10.18632/oncotarget.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team 2012. R, corp-author. R Foundation for Statistical Computing; Vienna, Austria: A language and environment for statistical computing. ISBN 3-900051-07-0. [Google Scholar]
- 20.Cancer Genome Atlas Research Network, corp-author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 22.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 24.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, et al. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.e03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 26.Gilks CB, Vanderhyden BC, Zhu S, van de Rijn M, Longacre TA. Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol Oncol. 2005;96:684–694. doi: 10.1016/j.ygyno.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Xu M, Gao H, Guo JC, Guo YL, Zou M, Wu XF. Two protein-coding genes act as a novel clinical signature to predict prognosis in patients with ovarian serous cystadenocarcinoma. Oncol Lett. 2018;15:3669–3675. doi: 10.3892/ol.2018.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu LW, Zhang Q, Guo W, Qian K, Wang Q. A Five-gene expression signature predicts clinical outcome of ovarian serous cystadenocarcinoma. Biomed Res Int. 2016;2016:6945304. doi: 10.1155/2016/6945304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misawa K, Imai A, Mochizuki D, Misawa Y, Endo S, Hosokawa S, Ishikawa R, Mima M, Shinmura K, Kanazawa T, Mineta H. Genes encoding neuropeptide receptors are epigenetic markers in patients with head and neck cancer: A site-specific analysis. Oncotarget. 2017;8:76318–76328. doi: 10.18632/oncotarget.19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YT, Yu YL, Hong WC, Yeh TS, Chen TC, Chen JC. NPFFR2 activates the HPA axis and induces anxiogenic effects in rodents. Int J Mol Sci. 2017;18(pii):E1810. doi: 10.3390/ijms18081810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin KJ, Lee YJ, Yang YR, Park S, Suh PG, Follo MY, Cocco L, Ryu SH. Molecular mechanisms underlying psychological stress and cancer. Curr Pharm Des. 2016;22:2389–2402. doi: 10.2174/1381612822666160226144025. [DOI] [PubMed] [Google Scholar]
- 32.Hui Y, Wu Y, Tormey CA. The development of a novel molecular assay examining the role of aminopeptidase P polymorphisms in acute hypotensive transfusion reactions. Arch Pathol Lab Med. 2013;137:96–99. doi: 10.5858/arpa.2011-0466-OA. [DOI] [PubMed] [Google Scholar]
- 33.Banu SK, Stanley JA, Sivakumar KK, Arosh JA, Barhoumi R, Burghardt RC. Identifying a novel role for X-prolyl aminopeptidase (Xpnpep) 2 in CrVI-induced adverse effects on germ cell nest breakdown and follicle development in rats. Biol Reprod. 2015;92:67. doi: 10.1095/biolreprod.114.125708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng T, Wei R, Jiang G, Zhou Y, Lv M, Dai Y, Yuan Y, Luo D, Ma D, Li F, Xi L. XPNPEP2 is overexpressed in cervical cancer and promotes cervical cancer metastasis. Tumour Biol. 2017;39:1010428317717122. doi: 10.1177/1010428317717122. [DOI] [PubMed] [Google Scholar]
- 35.Drendel V, Heckelmann B, Chen CY, Weisser J, Espadas G, Schell C, Sabido E, Werner M, Jilg CA, Schilling O. Proteome profiling of clear cell renal cell carcinoma in von Hippel-Lindau patients highlights upregulation of Xaa-Pro aminopeptidase-1, an anti-proliferative and anti-migratory exoprotease. Oncotarget. 2017;8:100066–100078. doi: 10.18632/oncotarget.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeung HC, Rha SY, Kim HK, Lim HY, Kim S, Kim SY, Gong SJ, Park CH, Ahn JB, Noh SH, Chung HC. Multi-institutional phase II study of S-1 monotherapy in advanced gastric cancer with pharmacokinetic and pharmacogenomic evaluations. Oncologist. 2007;12:543–554. doi: 10.1634/theoncologist.12-5-543. [DOI] [PubMed] [Google Scholar]
- 37.Párniczky A, Hegyi E, Tóth AZ, Szücs Á, Szentesi A, Vincze Á, Izbéki F, Németh BC, Hegyi P, Sahin-Tóth M. Genetic analysis of human chymotrypsin-like Elastases 3A and 3B (CELA3A and CELA3B) to assess the role of complex formation between proelastases and procarboxypeptidases in chronic pancreatitis. Int J Mol Sci. 2016;17:2148. doi: 10.3390/ijms17122148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X, Yi Y, Xie W, Liang B, Winter MC, He N, Liu X, Luo M, Yang Y, Ode KL, et al. CFTR influences beta cell function and insulin secretion through non-cell autonomous exocrine-derived factors. Endocrinology. 2017;158:3325–3338. doi: 10.1210/en.2017-00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han D, Moon S, Kim H, Choi SE, Lee SJ, Park KS, Jun H, Kang Y, Kim Y. Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. J Proteome Res. 2011;10:564–577. doi: 10.1021/pr100759a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the TCGA repository, https://portal.gdc.cancer.gov/.