Abstract
The present study focused on exploring the inhibitory mechanism of microRNA (miR)-23a in endometrial cancer. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to investigate miR-23a expression in endometrial tissues and endometrial cancer cells. A colony formation assay using crystal violet staining was performed to compare cell proliferation, while wound-healing and Transwell assays were performed to compare cell migration and invasion. Subsequently, bioinformatics and a luciferase reporter gene assay were used to investigate the effect of miR-23a on sine oculis homeobox homolog 1 (SIX1) expression, and the biological function of SIX1 was analyzed. Additionally, a nude mouse tumorigenicity assay was performed to test the inhibitory effect of miR-23a and Taxol® therapy in endometrial cancer. Finally, immunohistochemistry and RT-qPCR were used to explore the association between miR-23a and SIX1 expression in endometrial cancer tissues. miR-23a was underexpressed in endometrial cancer tissues compared with in para-carcinoma tissues, and the overexpression of miR-23a inhibited proliferation and invasion of endometrial cancer cells. Furthermore, SIX1 was demonstrated to be a downstream target of miR-23a, and miR-23a reduced SIX1 expression. Additionally, SIX1 inversely promoted cell proliferation, migration and invasion. In addition, the effects of reduced cell proliferation and increased cell invasion following miR-23a overexpression could be reversed by adding SIX1 to in vitro culture. Furthermore, the inhibitory effect of miR-23a and Taxol therapy, which reduced SIX1 expression in endometrial cancer, was demonstrated in vivo. Finally, a negative association between miR-23a and SIX1 expression was demonstrated in endometrial cancer tissues. The results of the present study revealed that miR-23a may inhibit endometrial cancer development by targeting SIX1.
Keywords: microRNA-23a, endometrial cancer, sine oculis homeobox homolog 1
Introduction
Endometrial cancer has the highest mortality rate of malignant tumors in women in the USA, and the incidence rate is rising; 61,380 American women were diagnosed with endometrial cancer in 2017, and 10,920 were projected to succumb to the disease (1). Of all patients, ~80% are diagnosed at an early stage with an overall favorable prognosis; however, ~20% of patients will eventually succumb to the disease (1,2). Despite progress in the fields of integrated diagnosis and treatment, there are still some limitations, including disease biology, morbidity and mortality; in particular, the prognosis of endometrial carcinoma has not markedly improved (3–5). At present, the predominant treatment strategy for endometrial cancer is surgery combined with adjuvant chemotherapy. Taxol® is widely used as the most promising antitumor agent for women with endometrial cancer; however, some patients exhibit Taxol resistance, which results in cancer recurrence or metastasis (6).
MicroRNAs (miRNAs/miRs) are an abundant group of small endogenous non-coding RNA molecules (~22 nucleotides), which are single-stranded and bind to target mRNAs mainly at their 3′untranslated region (3′UTR) (7–9). Previous studies have revealed that miRNAs are involved in numerous types of cellular processes in various types of human cancer, including endometrial cancer (7–10). A study reported that the downregulation of miR-106b is associated with chemoresistance in endometrial cancer (11). Another study revealed that miR-218 is significantly downregulated in Taxol-resistant endometrial cancer cells compared with in non-drug-resistant cell lines, and that miR-218 may directly bind to the 3′UTR of the high mobility group box 1 (HMGB1) gene, which mediates autophagy and contributes to chemotherapy resistance in endometrial carcinoma in vitro (12). The results of this previous study revealed the effect of miR-218 on HMGB1-mediated cell autophagy during chemotherapy resistance in endometrial carcinoma cells (12). Additionally, it has been suggested that miR-194 could inhibit the epithelial-mesenchymal transition (EMT) of endometrial cancer cells by targeting oncogene BMI1 proto-oncogene polycomb ring finger, which regulates the expression levels of chemoresistance markers (SOX-2, Krüppel like factor 4 and mobility related protein-1) (13). Prior to the present study, our group used microarray analysis to identify that miR-23a expression was decreased and associated with chemoresistance in endometrial cancer (data not shown). These results were similar to the results of another study, which demonstrated that miR-23a expression is downregulated in endometrial cancer and miR-23a could directly downregulate human SMAD3 protein levels (14). Furthermore, it has been demonstrated that the overexpression of miR-23a could inhibit EMT by targeting SMAD3 in endometrial cancer (14). However, another study identified that overexpression of miR-23a could enhance the chemoresistance of colorectal cancer cells by targeting ATP binding cassette subfamily F member 1, and that miR-23a may promote cisplatin chemoresistance and protect against cisplatin-induced apoptosis through Twist in tongue squamous cell carcinoma cells (15). Thus far, there is only one study on the role of miR-23a in cancer (14), and the mechanism of miR-23a-inhibition in endometrial cancer remains unclear.
In the present study, the role of miR-23a in the development of endometrial cancer was examined. The results demonstrated that sine oculis homeobox homolog 1 (SIX1), a biomarker for carcinogenesis in human endometrial cancer, was a direct target of miR-23a. In the present study, miR-23a expression in endometrial cancer tissues was detected and the function of miR-23a was investigated in vitro, including a systematic analysis of miR-23a and its role in endometrial cancer development.
Materials and methods
Cell culture and reagents
Following approval by the Ethics Committee of The Secondary Hospital of Tianjin Medical University (Tianjin, China), 16 paired tissue sections were obtained from 16 female patients (age range, 35–50 years, mean age 38.7 years old) undergoing surgical resection at Tianjin Medical University General Hospital between July 2010 and May 2012. The patients or their families provided written informed consent for the use of these tissues. Human endometrial cancer cell lines, Ishikawa and HEC1B, were obtained from the American Type Culture Collection (Manassas, VA, USA). All cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), and 1% penicillin and 1% streptomycin at 37°C with 5% CO2.
RNA analysis
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from the cells, according to the manufacturer's protocol. A First Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China) was used to synthesize cDNA. The following temperature protocol was used: 30°C for 10 min, 42°C for 60 min and 95°C for 5 min. Mean values were used for calculations and β-actin was used as a loading control. The comparative detailed ΔCq method was utilized to analyze the results. SYBR Green PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to perform reverse transcription-quantitative (RT-q) PCR analysis (16): Pre-denature was performed at 94°C for 5 min; followed by 33 cycles of pre-denaturing at 94°C for 30 sec, 64°C for 30 sec, 72°C for 45 sec and 72°C for 10 min. The obtained PCR products were routinely subjected to agarose gel electrophoresis and scanned using an imaging system. The gray-scale ratio of target genes and internal parameters was used to represent the relative mRNA expression levels of each target gene. The levels of miR-23a was normalized to the U6 snRNA. Primer sequences used for RT-qPCR analysis were as follows: SIX1 forward, 5′AAGGAGAAGTCGAGGGGTGT-3′ and reverse 5′-TGCTTGTTGGAGGAGGAGTT-3′; miR-23a forward, 5′-CCTACTGTCGTCCCAAGACCT-3′ and reverse, 5′-GGGGCTCGTGCAGAAGAAT-3′; and β-actin forward, 5′-CTCCATCATGAAGTGTGACGTT-3′ and reverse, 5′-ATCTCCTTCTGCATCCTGTCAG-3′. The experiment was repeated three times, independently.
Reagents for the transient transfection assays
miR-23a mimics (5′-AUCACAUUGCCAGGGAUUUCC-3′), miR-23a mimic NC (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-23a antisense oligonucleotide (ASO; 5′-GUGGUAAUCCCUGGCAAUGUGAU-3′ and ASO-NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were obtained from Sangon Biotech Co., Ltd. (Shanghai, China) SIX1 small interfering (si)RNA and a pcDNA3.1 SIX1 overexpression plasmid were obtained from Guangzhou Ribobio Co., Ltd. (Guangzhou, China) (17,18). miR-23a mimics were transfected into Ishikawa cells, miR-23a ASO into HEC1B cells, and SIX1 siRNA and plasmid into Ishikawa and HEC1B cells. Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) was used for transfection, according to the manufacturer's protocol. The final concentration of ASO control (con) and miR-23a ASO used was 100 nM. SIX1 plasmids were obtained from OriGene Technologies, Inc. (Rockville, MD, USA).
The sequences were as follows: SIX1 siRNA forward 5′-GGAGCUCACAAGGCAAUAU-3′, and reverse 3′-CCUCGAGUGUUCCGUUAUA-5′; and SIX1 control siRNA forward 5′-GGAGUUCUCAAGGGAGUAU-3′, and reverse 3′-CCUCAAGAGUUCCCUCAUA-5′). Subsequent experimentation was performed at 48 h after transfection.
Colony formation assay
Cells (~400 cells/well) were seeded into 60-mm dishes and cultured for 10 days at 37°C with 5% CO2. Following fixation in methanol for 15 min at room temperature, the cells were stained with Giemsa for 10–30 min at room temperature. The number of colonies were counted under a light microscope. The experiment was performed in triplicate.
Western blotting
Whole cell extracts were prepared using cell lysis reagent (cat. no. C2978; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Total proteins were quantified using a bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.). A total of 50 µg protein was separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were blocked with 10% goat serum (dilution, 1:1,000; cat. no. ZLI-9022; OriGene Technologies, Inc.) at room temperature for 60 min and incubated with primary antibodies overnight at 4°C. The primary antibodies used included a rabbit polyclonal SIX1 antibody (1:500 dilution; cat. no. ab211359; Abcam, Cambridge, UK) and a mouse monoclonal β-actin antibody (1:2,500 dilution; cat. no. sc-8432; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). This was followed by incubation with a horseradish peroxidas-conjugated secondary antibody (cat. no. RI2341; polyclonal goat anti-rabbit/mouse; 1:5,000 dilution; Rockland Immunochemicals Inc., Limerick, PA, USA) at 37°C for 1 h. The visualization reagent used was Coomassie brilliant blue G-250 (cat. no. C8420; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The gray values were analyzed using Odyssey v3.0 software (Thermo Fisher Scientific, Inc.).
Scratch assay
A total of 1×105 cells/well were seeded in 6-well plates overnight. Subsequently, a sterile pipette tip was used to introduce a scratch in the middle of the well when the confluency had reached 100%. The migration of cells towards the center of the scratch was measured at the indicated time point (48 h). A light microscope was used to observe the cells and the migration was measured using a caliper by testing the scratch distance.
Transwell assay
Invasion assays were performed with an 8.0-µm pore inserts in a 24-well Transwell chamber plate (Costar; Corning Inc., Corning, NY, USA). For this assay, 2×105 cells were isolated and added to the upper chamber in serum-free RPMI-1640 medium, which was coated with Matrigel (BD Biosciences, San Jose, CA, USA). RPMI-1640 (500 µl) with 10% FBS was added to the lower chamber, followed by incubation for 24 h. Cells that had migrated to the bottom of the filter were stained fixed in 4% paraformaldehyde and stained with 0.1% crystal violet for 5 min at room temperature. The cells left on the lower side of each membrane were counted under a light microscope for five fields of view.
Bioinformatics analysis of miR-23a target genes
Putative miR-23a targets were predicted using several algorithms, including microRNA.org (http://www.microrna.org/microrna/getGeneForm.do), TargetScan (http://www.targetscan.org/) and miRanda (http://microrna.sanger.ac.uk/).
Dual-luciferase reporter assay
A Dual-Glo Luciferase Assay System (Promega Corporation, Madison, WI, USA) was used to detect luciferase activity, according to the manufacturer's protocol and as previously described (19). A total of 4×104 cells/well in 12-well plates were cultured without antibiotics for 12 h. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect cloned SIX1 wild-type 3′UTR and mutant 3′UTR target sequence. Renilla luciferase plasmids (pRL-SV40; Promega Corporation) were used as a control. After 48 h, a Dual Luciferase assay kit (Promega Corporation) was used to detect luciferase intensity. All data were normalized to Renilla luciferase expression. The wild-type miR-23a target site in SIX1 3′UTR was AAGUGUA of the SIX1 3′UTR region. The mutant miR-23a target site was AACUCUU. The two were designed and purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China).
Xenograft assays in vivo
The animal study protocols were approved by the Animal Experimentation Ethics Committee of The Secondary Hospital of Tianjin Medical University. All efforts were made to minimize suffering and relieve pain. The nude mice (n=20; age, 5–6 weeks; female; mean weight, 25 g) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice were housed in a pathogen-free animal facility at 25°C with a 12-h light/dark cycle, and randomly assigned to the control or experimental group (four mice per group) (20). HEC1B cells (8×106) were suspended in 0.1 ml DMEM, which was injected subcutaneously into the right flank of each mouse (20). The mice were divided into five groups and injected with control (transfected with vector), Taxol, miR-23a con, miR-23a or miR-23a+Taxol once every 3 or 4 days from day 14 (17). The final concentration of control, miR-23a con and miR-23a for each intradermal injection of HEC1B cells (0.01 mol) was 100 nM. The tumor volume was measured using Vernier calipers every 3 or 4 days. The maximum tumor size measured was 14 mm, and the maximum number of tumors observed in a single mouse was 20. The following formula was used for calculation of tumor volume: Tumor volume (mm3)=tumor length (mm) × tumor width (mm2/2). Only stable miR-23a mimics, not miR-23a ASO, and cell lines were used for the in vivo experiments.
Immunohistochemistry (IHC)
Following ethics committee approval, tumor samples were obtained from 16 patients (aged 31–49 years) undergoing surgical resection at Tianjin Medical University General Hospital between July 2010 and May 2012, and IHC diagnosis was performed in the hospital at the Department of Gynecology and Obstetrics. The specimens were fixed with 10% formaldehyde at room temperature for 24 h. Specimens were embedded in paraffin, sectioned (5-µm thick) and deparaffinized via the addition of alcohol at decreasing concentrations (100, 95, 85 and 75%) for 5 min per step. For antigen retrieval, a 96°C water-bath was used for antigen retrieval in 0.01 mol/l sodium citrate buffer (10 mM, pH 6.0) for 20 min. Subsequently, the sections were treated with 5 mM citrate buffer and 3% H2O2 for 15 min, 5% goat serum (cat. no. ZLI-9022; OriGene Technologies, Inc.) was used to block sections at 37°C for 30 min, which were then incubated with a SIX1 antibody (1:200 dilution; cat. no. ab211359; Abcam) for 12 h at 4°C. Subsequently, the sections were incubated at 37°C for 30 min with a secondary antibody (goat anti-rabbit immunoglobulin G; cat. no. ZB-2301; ZSGB-BIO; OriGene Technologies, Inc.). All sections were counterstained with hematoxylin at room temperature for 30 sec. The sections were observed under a light microscope (Olympus Corporation, Tokyo, Japan). The frequencies of positive cells were scored in degrees: 1 (low), <33; 2 (medium) 34–67%; and 3 (high) >67%. The cases were further classified into positive groups (2–5, low; >5, high) by the intensity and proportion of the cancer cells immunostained for SIX1 (21,22). PBS instead of the primary antibody was used for creating the negative control for IHC staining (data not shown).
Statistical analysis
All experiments were repeated at least three times, independently. Analyses were performed using SPSS v.22.0 software (IBM Corp, Armonk, NY, USA). A Student's t-test was used to compare between two groups. A two-way ANOVA followed by a post hoc Tukey's test was used to compare between three or more groups. All data are expressed as the means ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-23a expression is decreased in endometrial cancer tissues compared with in para-carcinoma tissues, and the overexpression of miR-23a may inhibit cell proliferation and invasion in Ishikawa and HEC1B cells
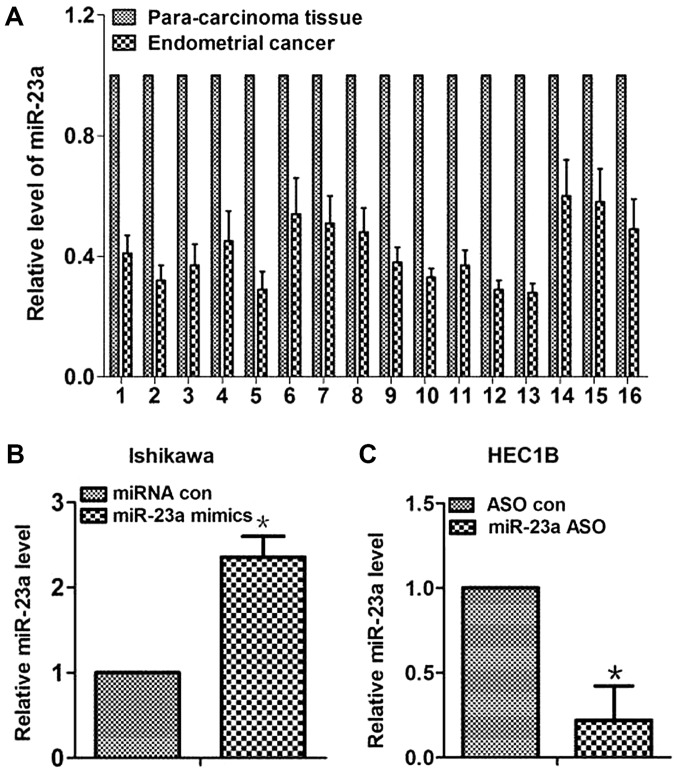
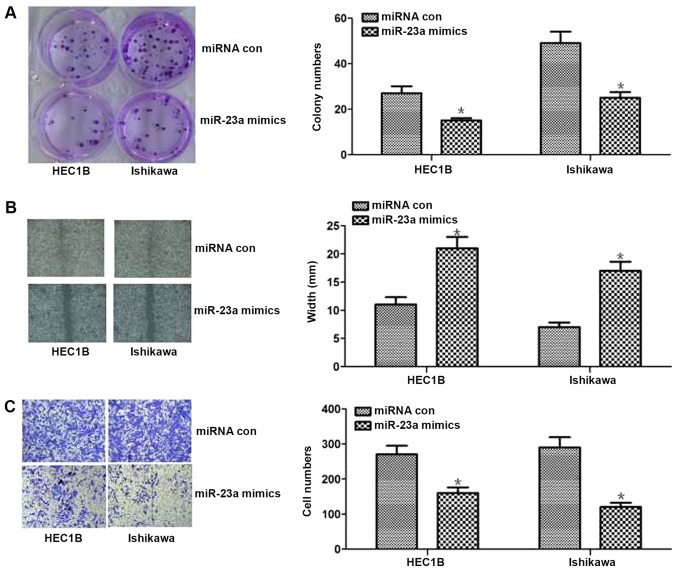
First, expression levels of miR-23a were measured in 16 pairs of human tissues. As shown in Fig. 1A, miR-23a expression levels were reduced by 42% on average in endometrial cancer tissues compared with in paired para-carcinoma tissues. To test the effect of miR-23a in endometrial cancer cells, miR-23a mimics were transfected into the Ishikawa and HEC1B cells to increase miR-23a expression (Fig. 1B and C). The results of cell function experiments revealed that increased miR-23a expression could inhibit proliferation and invasion in vitro (P<0.05; Fig. 2). The results of the colony formation assay revealed that proliferation was significantly decreased in miR-23a mimic-transfected cells compared with control cells (P<0.05; Fig. 2A). The results of the wound healing assay demonstrated significantly less wound healing in miR-23a mimic-transfected cells compared with the control cells (P<0.05; Fig. 2B). Additionally, the results of the Transwell assay revealed significantly lower numbers of invasive cells in the miR-23a mimic-transfected group compared with the control group (P<0.05; Fig. 2C). Overall, these results indicated that miR-23a expression was decreased in endometrial cancer tissues, and overexpression of miR-23 inhibited cell proliferation and invasion in endometrial cancer cells.
Figure 1.
Expression levels of miR-23a in 16 paired human endometrial cancer and para-carcinoma tissues. (A) miR-23a expression was reduced by 42% on average in endometrial cancer tissues compared with in paired para-carcinoma tissues. (B) miR-23a expression in the miR-23a mimics transfection and control groups for Ishikawa cells. (C) miR-23a expression in the miR-23a ASO transfection and control group in the HEC1B cells. *P<0.05. ASO, antisense oligonucleotide; con, control; miR-23a, microRNA-23a; miRNA, microRNA.
Figure 2.
Effect of miR-23a on the proliferation, migration and invasion of endometrial cancer cells in vitro. (A) Colony formation assay. Cells were cultured in 6-well plates and analyzed using a colony formation assay. After 10 days, cells were stained, images captured (left) and counted (right). The results of the colony formation assay revealed that proliferation was significantly decreased in miR-23a mimic-transfected cells compared with in the control cells in the two types of endometrial cancer cells. (B) Migration detected by wound healing assays. A significantly bigger wound width was observed in the treated group compared with in the control group in vitro. Magnification, ×200. (C) Invasion detected by Transwell assays. Significantly fewer invasive cells were observed in the treated groups compared with in the control groups in vitro. Magnification, ×200. *P<0.05 vs. con. con, control; miR-23a, microRNA-23a; miRNA, microRNA.
SIX1 is a direct target of miR-23a, and miR-23a downregulates SIX1 expression in endometrial cancer cells
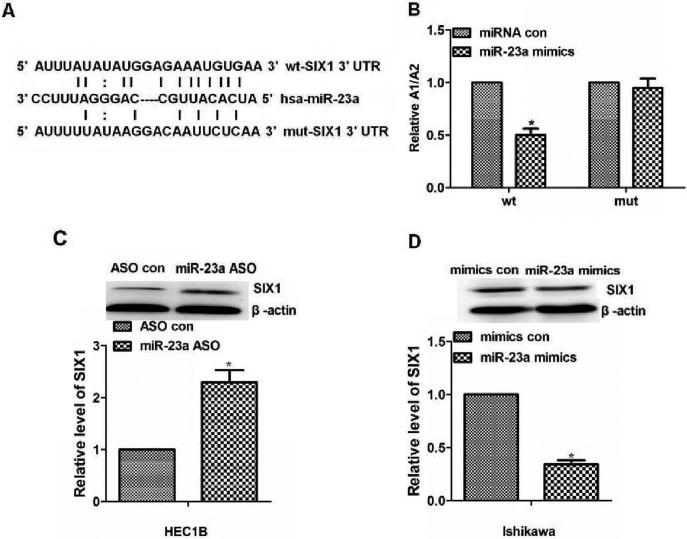
SIX1 was on the list of miR-23a targets suggested by microrna software. To test the possibility of a direct link between miR-23a and SIX1, the SIX1 3′UTR with either a wild-type or a mutant miR-23a target sequence downstream of the firefly luciferase gene was inserted (Fig. 3A). The pGL3-SIX1 3′UTR wild-type, pGL3-SIX1 3′UTR Mut and pGL3 constructs were individually transfected into HEC1B cells. The results indicated that miR-23a may directly bind to SIX1 3′UTR (P<0.05; Fig. 3B). In conclusion, the SIX1 gene may be a downstream post-transcriptional target of miR-23a.
Figure 3.
miR-23a downregulates SIX1 expression in vitro. (A) Bioinformatics results. The 3′UTR of SIX1 contains a potential miRNA-binding site for miR-23a. SIX1 UTR with either a wt or mut miR-23a target sequence downstream of the firefly luciferase gene was inserted into the pGL3-control vector to create the pGL3-SIX1 UTR wt or the pGL3-SIX1 UTR mut construct, respectively. (B) Luciferase reporter assay results. pGL3-SIX1 UTR WT, pGL3-SIX1 UTR Mut and pGL3 constructs were individually transfected into HEC1B cells. miR-23a significantly decreased the relative luciferase activity of the wild-type SIX1 3′UTR compared with the control, but miR-23a could not decrease the relative luciferase activity of the mutant SIX1 3′UTR. (C) SIX1 protein expression following different miR-23a treatments in endometrial cancer HEC1B cells, semi-quantified by western blotting. The SIX1 protein expression level was higher in the group treated with miR-23a ASO than that in the group treated with ASO con. (D) SIX1 protein expression following different miR-23a treatments in endometrial cancer Ishikawa cells, semi-quantified by western blotting. The SIX1 protein expression level was lower in the group treated with miR-23a mimics than that in the group treated with mimics con in the Ishikawa cells. *P<0.05, compared with wt miRNA con group in (B). 3′UTR, 3′untranslated region; ASO, antisense oligonucleotide; con, control; hsa, homo sapiens; miR-23a, microRNA-23a; miRNA, microRNA; mut, mutant; SIX1, sine oculis homeobox homolog 1; wt, wild-type.
To further investigate the mechanism of miR-23a in the chemoresistance of endometrial cancer via its ability to repress its downstream gene SIX1, SIX1 protein expression following different miR-23a treatment in two types of endometrial cancer cells was detected by western blotting. According to a previous study, SIX1 protein expression levels are high in Ishikawa cells and low in HEC1B cells (22). Consistent with the previous study, through the different miR-23a treatment in the two types of endometrial cancer cells, the results of the present study demonstrated that SIX1 protein expression was increased in HEC1B cells treated with miR-23a ASO compared with cells treated with ASO control (P<0.05; Fig. 3C). In addition, the present study revealed that the SIX1 protein expression level was lower in Ishikawa cells treated with miR-23a mimics than in cells treated with miRNA con (P<0.05; Fig. 3D). Overall, miR-23a may downregulate SIX1 expression in endometrial cancer cells.
SIX1 may inhibit cell proliferation, migration and invasion in endometrial cancer cells
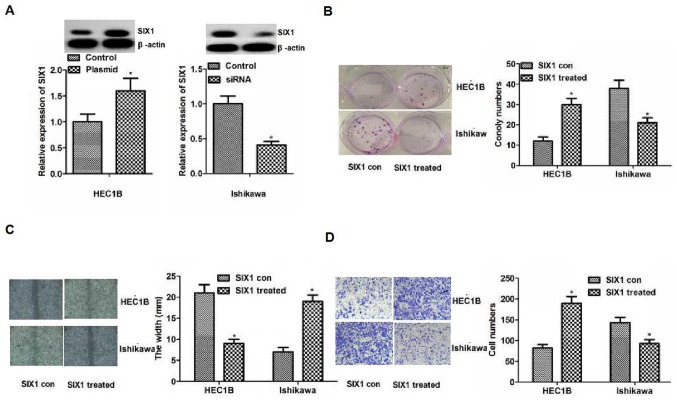
To investigate the influence of SIX1 on the biological behavior of endometrial cancer cells, SIX1 plasmid and siRNA were used to increase or decrease the expression levels of SIX1 in HEC1B or Ishikawa cells. Due to the differing levels of SIX1 expression in the two cell lines, the knockdown and overexpression experiments were performed in two different cell lines. As shown in Fig. 4A, the expression levels of SIX1 in HEC1B or Ishikawa cells were significantly increased or decreased, when treated with plasmid and siRNA, respectively (P<0.05). The results of the colony formation assay revealed that capacity for proliferation caused by the increased or reduced expression levels of SIX1 were significantly enhanced or weakened compared with the control in HEC1B or Ishikawa cells, respectively (P<0.05; Fig. 4B). The results of the wound healing assay indicated the significantly smaller or bigger width caused by the raised or reduced SIX1 than that in the control of HEC1B or Ishikawa cells, respectively (P<0.05; Fig. 4C). The results of the Transwell assay demonstrated significantly higher or lower numbers of invasive cells in association with increased or decreased expression levels of SIX1 compared with the control in HEC1B or Ishikawa cells (P<0.05; Fig. 4D). Overall, the results indicated that increased or decreased SIX1 expression promoted or inhibited cell proliferation, migration and invasion in vitro.
Figure 4.
Influence of SIX1 on the biological behavior of endometrial cancer cell lines. (A) SIX1 plasmid or siRNA were used to increase or reduce the protein expression levels of SIX1 in HEC1B or Ishikawa cells. Differences were detected using western blotting. Protein expression levels of SIX1 in HEC1B or Ishikawa cells were increased or reduced in the SIX1 plasmid or siRNA group compared with the control group, respectively. (B) Colony formation assay. The results of colony formation assay demonstrated that proliferation was significantly enhanced or weakened compared with the control in HEC1B or Ishikawa cells, respectively. Magnification, ×200. (C) Migration detected by wound healing assays. The results revealed significantly smaller or bigger width in the treated group compared with the control group in HEC1B or Ishikawa cells, respectively. (D) Invasion detected by Transwell assays. Significantly more or fewer invasive cells were observed in the treated group compared with in the control group in HEC1B or Ishikawa cells, respectively. Magnification, ×200. *P<0.05 vs. con. Con, control; siRNA, small interfering RNA; SIX1, sine oculis homeobox homolog 1; SIX1 treated, overexpression in the HEC1B cell line or knockdown in the Ishikawa cell line.
miR-23a inhibits the development of endometrial cancer by targeting SIX1
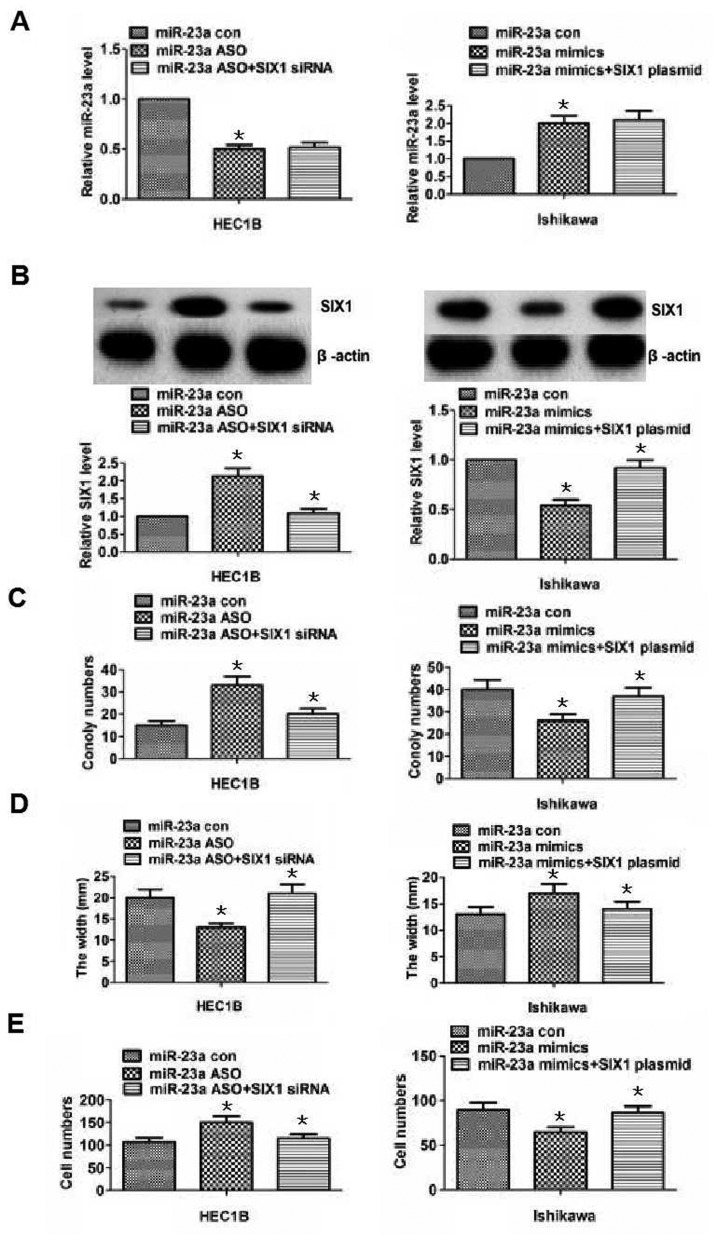
The aforementioned results revealed the possibility that miR-23a inhibited endometrial cancer cell proliferation, migration and invasion by downregulating SIX1. To further demonstrate this, cell proliferation, migration and invasion abilities were reversed by adding SIX1 siRNA or plasmid at the same time as performing knockdown or overexpression of miR-23a (Fig. 5). As shown in Fig. 5A, miR-23a expression was reduced in the miR-23a ASO group in HEC1B cells (P<0.05), while no significant difference in miR-23a expression was identified between the SIX1 siRNA + miR-23a ASO group and the miR-23a ASO group (P>0.05). Similarly, miR-23a expression was increased in Ishikawa cells treated with miR-23a mimics (P<0.05; Fig. 5A), while no significant difference was observed in the increase in miR-23a expression between the SIX1 plasmid + miR-23a mimics group and the miR-23a mimics group (P>0.05; Fig. 5A). The results demonstrated that the knockdown or overexpression of miR-23a was not altered by adding SIX1 siRNA or plasmid. Additionally, SIX1 expression was increased or decreased following the knockdown or overexpression of miR-23a in the miR-23a ASO or mimics group (P<0.05, respectively; Fig. 5B), and SIX1 expression levels were reduced or increased by adding SIX1 siRNA or plasmid in HEC1B or Ishikawa cells (P<0.05, respectively; Fig. 5B). The results demonstrated that miR-23a directly targeted SIX1 in endometrial cancer cells. Besides, the results of clone formation, wound healing and Transwell assays revealed that proliferation, migration and invasion abilities were strengthened or weakened, following the knockdown or overexpression of miR-23a in the miR-23a ASO or miR-23a mimics group in HEC1B or Ishikawa cells (P<0.05; Fig. 5C-E), but the altered proliferation, migration and invasion abilities following the knockdown or overexpression of miR-23a could be reversed by the downregulation or upregulation of SIX1 in HEC1B and Ishikawa cells (P<0.05; 5C-E). Overall, the aforementioned results indicated that miR-23a may promote the development of endometrial cancer by targeting SIX1.
Figure 5.
Alterations in proliferation, migration and invasion following miR-23a knockdown or overexpression are reversed by SIX1 downregulation or upregulation. (A) miR-23a expression was assessed using reverse transcription-quantitative polymerase chain reaction in HEC1B and Ishikawa cells for the differently treated groups. miR-23a expression was decreased or increased in the miR-23a ASO or mimic groups following the knockdown or the overexpression of miR-23a, compared with in the associated control groups. However, the decreased or increased miR-23a expression was not statistically different between the SIX1 siRNA + miR-23a ASO or SIX1 plasmid + miR-23a mimics groups compared with the miR-23a ASO or miR-23a mimics groups following the knockdown or overexpression of miR-23a in HEC1B or Ishikawa cells. (B) SIX1 protein expression was detected by western blotting in HEC1B or Ishikawa cells in the differently treated groups. SIX1 expression was increased or decreased in the miR-23a ASO or mimics following the knockdown or the overexpression of miR-23a compared with in the associated control groups. Additionally, SIX1 expression was significantly decreased or increased in the SIX1 siRNA + miR-23a ASO or SIX1 plasmid + miR-23a mimics groups compared with the miR-23a ASO or miR-23a mimics groups following the knockdown or overexpression of miR-23a in HEC1B or Ishikawa cells. (C) Results of the colony formation assays in HEC1B and Ishikawa cells. The number of colonies was increased or decreased in the miR-23a ASO or mimics group following the knockdown or the overexpression of miR-23a compared with the associated control group. Additionally, the number of colonies was significantly decreased or increased between the SIX1 siRNA + miR-23a ASO group or SIX1 plasmid + miR-23a mimics group compared with the miR-23a ASO group or miR-23a mimics group, following the knockdown or overexpression of miR-23a in HEC1B or Ishikawa cells, respectively. (D and E) Results of wound healing and Transwell assays for the different groups in HEC1B and Ishikawa cells. The results revealed that the migration and invasion abilities were altered in the miR-23a ASO and mimics groups following the knockdown or overexpression of miR-23a compared with in the associated control group. Additionally, migration and invasion were significantly altered in the SIX1 siRNA + miR-23a ASO group or SIX1 plasmid + miR-23a mimics group compared with in the miR-23a ASO group or miR-23a mimics group following the knockdown or overexpression of miR-23a in HEC1B and Ishikawa cells. *P<0.05, compared with the associated control group. ASO, antisense oligonucleotide; con, control; miR-23a, microRNA-23a; siRNA, small interfering RNA; SIX1, sine oculis homeobox homolog 1.
Inhibitory effect of miR-23a mimics and/or Taxol by reducing SIX1 expression in endometrial cancer in vivo
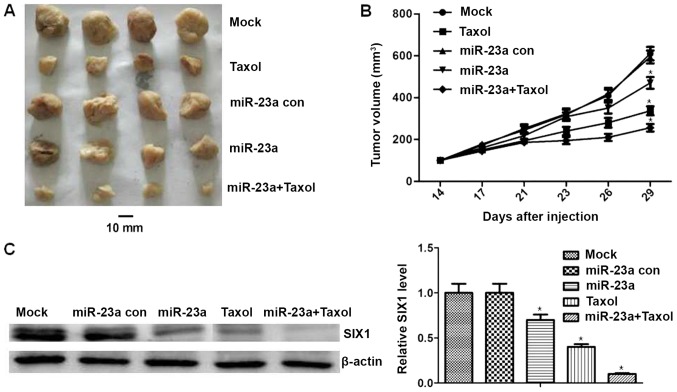
To verify the inhibitory effect of miR-23a mimics and/or Taxol and to determine whether miR-23a regulates SIX1 expression in vivo, Ishikawa cells were injected subcutaneously into the right flank of nude mice. The results revealed that the mean tumor volume in the Taxol group was significantly lower compared with in the control group (P<0.05; Fig. 6A and B). The mean tumor volume was significantly lower for the miR-23a mimics and miR-23a mimics + Taxol group compared with the mimics control group (P<0.05; Fig. 6A and B). Besides, the mean tumor volume was significantly lower in the miR-23a + Taxol group compared with in the Taxol group (P<0.05; Fig. 6A and B). In addition, SIX1 expression was assessed in mice tumors by western blotting. This revealed that the expression of SIX1 was decreased in the miR-23a mimics and/or Taxol groups (P<0.05; Fig. 6C). Additionally, compared with in the Taxol group, the expression levels of SIX1 in the miR-23a mimics + Taxol group were significantly decreased (P<0.05; Fig. 6C). There was no significant difference identified in the body weight of mice treated with miR-23a mimics and/or Taxol, none of the mice tested exhibited signs of other adverse effects, and no toxic effects were observed through blood counts or the observation of liver and renal function (data not shown). These results demonstrated the antitumor effects and safety of miR-23a mimics and/or Taxol. Overall, the aforementioned results indicated that miR-23a could inhibit chemoresistance of endometrial cancer in vivo by targeting SIX1.
Figure 6.
Inhibitory effect of miR-23a and Taxol by reducing SIX1 in vivo. (A and B) Mice tumor volumes in different groups. The average tumor volume in the miR-23a and miR-23a + Taxol groups was decreased significantly. Additionally, compared with the Taxol group, the average tumor volume in the miR-23a + Taxol group was decreased significantly. (C) Western blotting results indicating SIX1 expression in different groups. SIX1 expression was lower in the miR-23a and/or Taxol treated group than in the miR-23a con treated group. *P<0.05 vs. Mock. con, control; miR-23a, microRNA-23a; SIX1, sine oculis homeobox homolog 1.
Association between miR-23a and SIX1 expression in endometrial cancer tissues
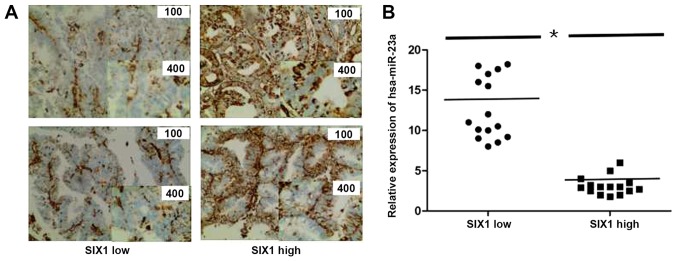
Finally, to explore the association between miR-23a and SIX1 in endometrial cancer tissues, their expression levels in 30 endometrial cancer cases were assessed. SIX1 expression was assessed by IHC, the 30 cases were divided into two groups: SIX1 low and high groups (n=15 for each group; Fig. 7A). Additionally, miR-23a expression was tested by RT-qPCR in the two groups, and a significant difference between the SIX1 low and high tissue samples was identified (P<0.05). Therefore, the results suggested a negative association between miR-23a and SIX1 expression in endometrial cancer tissues.
Figure 7.
Association between miR-23a and SIX1 expression in endometrial cancer tissues. (A) Immunohistochemistry results investigating SIX1 expression in 30 cases. Magnification, ×100 or ×400, as indicated. (B) Expression levels of miR-23a in endometrial cancer tissues according to RT-qPCR. A total of 30 endometrium samples (SIX1 low and high groups; n=15 for each group) were collected for RT-qPCR detection. The relative amount of miR-23a was normalized to the U6 small nuclear RNA. *P<0.05. miR-23a, microRNA-23a; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SIX1, sine oculis homeobox homolog 1.
Discussion
miRNAs serve different roles (promote or suppress cancer development) in cancer cellular processes, including in endometrial cancer (2,5–13). Previously, miR-23a has been linked to chemoresistance in cancer (10,12,15–17,19), and there have been three studies investigating the association between overexpression of miR-23a in cancer and chemoresistance (15,16,19). However, thus far, only one study investigated the association between miR-23a and endometrial cancer, and indicated that miR-23a is downregulated in cancer tissue and that miR-23a may inhibit EMT in endometrial endometrioid adenocarcinoma by targeting SMAD3 (14). In the present study, to investigate the function and mechanism of miR-23a, miR-23a was first demonstrated to be decreased in endometrial cancer tissues compared with in para-carcinoma tissues, and overexpression of miR-23a inhibited cell proliferation or invasion in Ishikawa cells and HEC1B cells. In addition, in the present study, overexpression of miR-23a inhibited proliferation, migration and invasion, which was different from the results reported in previous studies (15,16,19), and a possible explanation for this phenomenon was that this was due to the different types of cancer being investigated. Subsequently, it was identified that SIX1 was a target gene of miR-23a by different types of software, including TargetScan. Additionally, it was demonstrated that SIX1 was a functional target of miR-23a in endometrial cancer cells.
SIX1, a transcription factor, belongs to the SIX family of hemoproteins, and it has been reported to be less expressed in human normal tissue but expressed in mouse dental follicle, human periodontal ligament-derived, mouse skeletal muscle and cephalic neural crest cells; particularly, SIX1 overexpression has been reported to occur in several human types of cancer, including breast cancer, cervical cancer, ovarian cancer, oral squamous cell carcinoma, gastric cancer, hepatocellular carcinoma and pancreatic cancer, which leads to cancer cell proliferation, invasion and metastasis (21–38). Consistent with these results, the present study demonstrated that SIX1 promoted cell proliferation, migration and invasion in endometrial cancer. In addition, it has been reported that SIX1 upregulation induces cancer cell EMT (39–41). SIX1 upregulation leads to EMT via the activation of zinc-finger E-box binding homeobox1 (41). miR-23a could inhibit EMT in endometrial adenocarcinoma by targeting SMAD3 (14). This may provide clues for the association between miR-23a and SIX1. Additionally, SIX1 may mediate resistance to paclitaxel in breast cancer cells (27), which indicates that a similar mechanism may exist in endometrial cancer. Previous studies have identified an association between overexpression of miR-23a in cancer and chemoresistance (15,16,19). Particularly, a recent study demonstrated that SIX1 is overexpressed in endometrial carcinoma and promotes the malignant behavior of cancer cells via ERK and AKT signaling, the expression levels of SIX1 were high or low in Ishikawa or HEC1B cell lines (22), which is consistent with the results of the present study. Overall, the aforementioned studies and results revealed indirect evidence for the hypothesis that miR-23a may promote endometrial cancer development by targeting SIX1. In addition, the direct targeting association was further supported by the results of the luciferase reporter assay. Additionally, expression levels of miR-23a and SIX1 were assessed in 30 endometrial cancer cases, and this revealed a negative association between miR-23a and SIX1 expression in endometrial cancer tissues.
The present study demonstrated that miR-23a may be a suppressor gene in endometrial cancer cells, and alterations in the miR-23a-SIX1 interaction may be associated with the development of endometrial cancer. Furthermore, in contrast to the results of other studies, possibly due to different types of cancer and different perspectives, the results of the present study indicated that SIX1 was targeted by miR-23a, similar methods could be used to further explore the mechanism (24): For example, miR-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. The bioinformatics analysis, dual-luciferase reporter assay and the regulation results, with the same tendency of the associations between the upstream and downstream genes, indicated that SIX1 was a downstream target gene of miR-23a in vitro, and upregulation or downregulation of cell proliferation, migration and invasion, following the knockdown or overexpression of miR-23a, were reversed by adding SIX1 siRNA or plasmid in vitro.
As demonstrated in the present study, the inhibitory effect of miR-23a mimics, which reduces SIX1 expression in endometrial cancer, may be observed in vitro and in vivo. However, future studies should determine the aforementioned mechanism in various types of endometrial cancer cells in vitro and in vivo. Perhaps the association between genes known to be associated with endometrial cancer and the miR-23a-SIX1 interaction should be explored. A recent study indicated that it may be useful to investigate the association between miR-23a and SIX1 in the EMT and resistance mechanism (14). In conclusion, the results of the present study suggested that miR-23a may inhibit endometrial cancer cell development by targeting SIX1, and that miR-23a may serve as a therapeutic target for endometrial cancer.
Acknowledgements
Not applicable.
Funding
This work was supported by Tianjin Medical University Second Hospital Youth Fund (grant no. 2017ydey03).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HLL and JJS carried out the molecular biology experiments and drafted the manuscript. HM and SJL performed the animal experiments. NL, SJG and YS participated in the sequence alignment. HLL and JJS participated in the design of the study and performed the statistical analysis. YYX, ZYQ, YQW, FW, RMG and DL participated in the design and coordination of the study, and helped to draft the manuscript. FXX conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript
Ethics approval and consent to participate
All applicable international, national, and/or institutional guidelines for the care and use of human specimens and animals were followed. The animal study was carried out in accordance with the guidelines approved by the Animal Experimentation Ethics Committee of the Secondary Hospital of Tianjin Medical University. The protocol was approved by the committee, all surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering. The use of human samples was approved by the Ethics Committee of Secondary Hospital of Tianjin Medical University. The patients or their families provided written informed consent for the use of these tissues.
Patient consent for publication
The patients or parents provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Vogel RI, Pulver T, Heilmann W, Mooneyham A, Mullany S, Zhao X, Shahi M, Richter J, Klein M, Chen L, et al. USP14 is a predictor of recurrence in endometrial cancer and a molecular target for endometrial cancer treatment. Oncotarget. 2016;7:30962–30976. doi: 10.18632/oncotarget.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran AQ, Gehrig P. Recent advances in endometrial cancer. F1000Res. 2017;6:81. doi: 10.12688/f1000research.10020.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang S, Kang WD, Chung HH, Jeong DH, Seo SS, Lee JM, Lee JK, Kim JW, Kim SM, Park SY, Kim KT. Preoperative identification of a low-risk group for lymph node metastasis in endometrial cancer: A Korean gynecologic oncology group study. J Clin Oncol. 2012;30:1329–1334. doi: 10.1200/JCO.2011.38.2416. [DOI] [PubMed] [Google Scholar]
- 5.Wright JD, Burke WM, Wilde ET, Lewin SN, Charles AS, Kim JH, Goldman N, Neugut AI, Herzog TJ, Hershman DL. Comparative effectiveness of robotic versus laparoscopic hysterectomy for endometrial cancer. J Clin Oncol. 2012;30:783–791. doi: 10.1200/JCO.2011.36.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brasseur K, Gévry N, Asselin E. Chemoresistance and targeted therapies in ovarian and endometrial cancers. Oncotarget. 2017;8:4008–4042. doi: 10.18632/oncotarget.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer. 2016;15:48. doi: 10.1186/s12943-016-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Wang M, Wu J, Jie Z, Chang S, Shuang T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J Ovarian Res. 2015;8:23. doi: 10.1186/s13048-015-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Záveský L, Jandáková E, Turyna R, Langmeierová L, Weinberger V, Záveská Drábková L, Hůlková M, Hořínek A, Dušková D, Feyereisl J, et al. Evaluation of cell-free urine microRNAs expression for the use in diagnosis of ovarian and endometrial cancers. A pilot study. Pathol Oncol Res. 2015;21:1027–1035. doi: 10.1007/s12253-015-9914-y. [DOI] [PubMed] [Google Scholar]
- 12.Ran X, Yang J, Liu C, Zhou P, Xiao L, Zhang K. miR-218 inhibits HMGB1-mediated autophagy in endometrial carcinoma cells during chemotherapy. Int J Clin Exp Pathol. 2015;8:6617–6626. [PMC free article] [PubMed] [Google Scholar]
- 13.Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J, Sakuragi N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer. 2011;10:99. doi: 10.1186/1476-4598-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P, Wang C, Ma C, Wu Q, Zhang W, Lao G. MicroRNA-23a regulates epithelial-to-mesenchymal transition in endometrial endometrioid adenocarcinoma by targeting SMAD3. Cancer Cell Int. 2016;16:67. doi: 10.1186/s12935-016-0342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Li X, Liao D, Wang X, Wu Z, Nie J, Bai M, Fu X, Mei Q, Han W. Elevated microRNA-23a expression enhances the chemo-resistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr Protein Pept Sci. 2015;16:301–309. doi: 10.2174/138920371604150429153309. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Guan L, Hu X, Liu L, Xing Y, Zhou Z, Liang X, Yang Q, Jin S, Bao J, Gao H, et al. Bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci Rep. 2017;7:43716. doi: 10.1038/srep43716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Wang Y, Liang H, Fan Q, Zhu R, Cui J, Zhang W, Zen K, Zhang CY, Hou D, et al. miR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancer. Cell Death Dis. 2017;8:e3059. doi: 10.1038/cddis.2017.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang J, Yang F, Wang Y, Wang Y, Xue G, Mei Q, Wang F, Sun S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem. 2014;115:772–784. doi: 10.1002/jcb.24721. [DOI] [PubMed] [Google Scholar]
- 20.Sun KX, Jiao JW, Chen S, Liu BL, Zhao Y. MicroRNA-186 induces sensitivity of ovarian cancer cells to paclitaxel and cisplatin by targeting ABCB1. J Ovarian Res. 2015;8:80. doi: 10.1186/s13048-015-0207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z, Li G, Tang L, Li Y. SIX1 overexpression predicts poor prognosis and induces radioresistance through AKT signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:1071–1079. doi: 10.2147/OTT.S125330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin X, Li Y, Yang X. SIX1 is overexpressed in endometrial carcinoma and promotes the malignant behavior of cancer cells through ERK and AKT signaling. Oncol Lett. 2016;12:3435–3440. doi: 10.3892/ol.2016.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki T, Takahashi M, Yajima H, Mori Y, Kawakami K. Six1 is required for mouse dental follicle cell and human periodontal ligament-derived cell proliferation. Dev Growth Differ. 2016;58:530–545. doi: 10.1111/dgd.12291. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37:4105–4113. doi: 10.1007/s13277-015-4246-9. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Li L, Zhang XX, Wan DY, Xi BX, Hu Z, Ding WC, Zhu D, Wang XL, Wang W, et al. SIX1 promotes tumor lymphangiogenesis by coordinating TGFβ signals that increase expression of VEGF-C. Cancer Res. 2014;74:5597–5607. doi: 10.1158/0008-5472.CAN-13-3598. [DOI] [PubMed] [Google Scholar]
- 26.Feng GW, Dong LD, Shang WJ, Pang XL, Li JF, Liu L, Wang Y. HDAC5 promotes cell proliferation in human hepatocellular carcinoma by up-regulating Six1 expression. Eur Rev Med Pharmacol Sci. 2014;18:811–816. [PubMed] [Google Scholar]
- 27.Li Z, Tian T, Hu X, Zhang X, Nan F, Chang Y, Lv F, Zhang M. Six1 mediates resistance to paclitaxel in breast cancer cells. Biochem Biophys Res Commun. 2013;441:538–543. doi: 10.1016/j.bbrc.2013.10.131. [DOI] [PubMed] [Google Scholar]
- 28.Hetzler KL, Collins BC, Shanely RA, Sue H, Kostek MC. The homoeobox gene SIX1 alters myosin heavy chain isoform expression in mouse skeletal muscle. Acta Physiol (Oxf) 2014;210:415–428. doi: 10.1111/apha.12168. [DOI] [PubMed] [Google Scholar]
- 29.Garcez RC, Le Douarin NM, Creuzet SE. Combinatorial activity of Six1-2-4 genes in cephalic neural crest cells controls craniofacial and brain development. Cell Mol Life Sci. 2014;71:2149–2164. doi: 10.1007/s00018-013-1477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L, Li X, Li L, Ma W, Wu J, Zhang M. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato S, Ikeda K, Shioi G, Ochi H, Ogino H, Yajima H, Kawakami K. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344:158–171. doi: 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 32.Ng KT, Lee TK, Cheng Q, Wo JY, Sun CK, Guo DY, Lim ZX, Lo CM, Poon RT, Fan ST, Man K. Suppression of tumorigenesis and metastasis of hepatocellular carcinoma by shRNA interference targeting on homeoprotein Six1. Int J Cancer. 2010;127:859–872. doi: 10.1002/ijc.25105. [DOI] [PubMed] [Google Scholar]
- 33.Plant KE, Anderson E, Simecek N, Brown R, Forster S, Spinks J, Toms N, Gibson GG, Lyon J, Plant N. The neuroprotective action of the mood stabilizing drugs lithium chloride and sodium valproate is mediated through the up-regulation of the homeodomain protein Six1. Toxicol Appl Pharmacol. 2009;235:124–134. doi: 10.1016/j.taap.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66:1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 35.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Müller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci USA. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 37.Zheng XH, Liang PH, Guo JX, Zheng YR, Han J, Yu LL, Zhou YG, Li L. Expression and clinical implications of homeobox gene Six1 in cervical cancer cell lines and cervical epithelial tissues. Int J Gynecol Cancer. 2010;20:1587–1592. [PubMed] [Google Scholar]
- 38.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, Ford HL. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 39.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-b signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radisky DC. Defining a role for the homeoprotein Six1 in EMT and mammary tumorigenesis. J Clin Invest. 2009;119:2528–2531. doi: 10.1172/JCI40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono H, Imoto I, Kozaki K, Tsuda H, Matsui T, Kurasawa Y, Muramatsu T, Sugihara K, Inazawa J. SIX1 promotes epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation. Oncogene. 2012;31:4923–4934. doi: 10.1038/onc.2011.646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.