Abstract
Objective:
This study tests associations of DNA methylation-based (DNAm) measures of epigenetic age acceleration (EAA) with cross-sectional and longitudinal depressive symptoms in an urban sample of middle-aged adults.
Methods:
White and African–American adult participants in the Healthy Aging in Neighborhoods of Diversity across the Life Span study for whom DNA samples were analyzed (baseline age: 30–65 years) we included. We estimated three DNAm based EAA measures: (1) universal epigenetic age acceleration (AgeAccel); (2) intrinsic epigenetic age acceleration (IEAA); and (3) extrinsic epigenetic age acceleration (EEAA). Depressive symptoms were assessed using the 20-item Center for Epidemiological Studies-Depression scale total and sub-domain scores at baseline (2004–2009) and follow-up visits (2009–2013). Linear mixed-effects regression models were conducted, adjusting potentially confounding covariates, selection bias and multiple testing (N = 329 participants, ~ 52% men, k = 1.9 observations/participant, mean follow-up time ~ 4.7 years).
Results:
None of the epigenetic age acceleration measures were associated with total depressive symptom scores at baseline or over time. IEAA – a measure of cellular epigenetic age acceleration irrespective of white blood cell composition – was cross-sectionally associated with decrement in “positive affect” in the total population (γ011 ± SE = −0.090 ± 0.030, P = 0.003, Cohen’s D: −0.16) and among Whites (γ011 ± SE = −0.135 ± 0.048, P = 0.005, Cohen’s D: −0.23), after correction for multiple testing. Baseline “positive affect” was similarly associated with AgeAccel.
Limitations:
Limitations included small sample size, weak-moderate effects and measurement error.
Conclusions:
IEAA and AgeAccel, two measures of EAA using Horvath algorithm, were linked to a reduced “positive affect”, overall and among Whites. Future studies are needed to replicate our findings and test bidirectional relationships.
Keywords: Depressive symptoms, Epigenetic Age acceleration, Health disparities, Adults
1. Introduction
The global burden of major depressive disorder (MDD) is currently estimated at 350 million people (Smith, 2014). This chronic condition is ranked second worldwide in years lost due to disability (Smith, 2014; Uchida et al., 2018). Despite its public health importance, researchers have yet to uncover the causes of MDD and its associated elevation in depressive symptoms. With a heritability not exceeding 37% (Uchida et al., 2018), MDD may indeed be a product of gene and environment interactions, with stressful life events as a key environmental factor based on previous epidemiological evidence (Uchida et al., 2018). Changes in neuronal plasticity triggers adaptation to chronic stress and other environmental modifications (West and Greenberg, 2011). In fact, neuronal synaptic structure is constantly modified in response to the need for neuronal plasticity (Uchida et al., 2018). The former is dependent on de novo gene expression, which is regulated through various epigenetic mechanisms, including DNA methylation (DNAm), covalent histone modifications and non-coding RNAs (Uchida et al., 2018). Those epigenetic mechanisms have the unique characteristics of altering gene expression through chromatin structural changes without modifying DNA sequence per se (Nestler, 2014).
DNAm has been associated with psychopathology, including post-traumatic stress (Conrad et al., 2018; Mehta et al., 2017; Parade et al., 2017) and major depressive disorder (Bustamante et al., 2018; Han et al., 2018; Li et al., 2018; Saavedra et al., 2016), as well as cognitive aging (Chouliaras et al., 2018; Levine et al., 2015; Marioni et al., 2018, 2015b; McCartney et al., 2018; Starnawska et al., 2017). With the help of the Horvath and Hannum “epigenetic clocks” well-established epi-genetic age algorithms, DNAm can be utilized to estimate biological aging at the cellular level (Wolf et al., 2019). Despite differences in those algorithms and loci, both approaches produce clocks that are strongly associated with chronological age (Wolf et al., 2019). Generally speaking, an epigenetic age acceleration, or a faster “epigenetic clock” has been linked to age-related health decline, including a higher mortality risk (Chen et al., 2016; Marioni et al., 2015a; Perna et al., 2016) and faster rates of cognitive decline (Chouliaras et al., 2018; Levine et al., 2015; Marioni et al., 2018, 2015b; McCartney et al., 2018; Starnawska et al., 2017). However, only a few epidemiological studies have directly linked epigenetic clocks or DNAm in general to MDD (Bustamante et al., 2018; Han et al., 2018; Li et al., 2018; Saavedra et al., 2016) and only one has indirectly examined its association with elevated depressive symptoms, by testing pathways between socioeconomic disadvantage and epigenetic cellular aging (Austin et al., 2018). In fact, according to the Research Domain Criteria (RDoC) approach, “which encourages studies to focus on the neurobiological mechanisms and core aspects of behavior rather than to rely on traditional diagnostic categories” (such as MDD), examining epigenetic aging in relation to domains of depressive symptoms is of great importance (Katahira and Yamashita, 2017). Moreover, previous studies have reported higher rates of epigenetic aging among men compared to women and that DNAm levels also differ by race/ethnicity in several tissues including blood, saliva and brain (Horvath et al., 2016). Moreover, differences in depressive symptoms by sex and race have also been detected (Beydoun et al., 2016). Thus, it is important to uncover the relationship between epigenetic age acceleration and depressive symptoms while stratifying by sex and race.
In the present study, we test relationships of 3 DNAm-based “epigenetic clocks” with cross-sectional and longitudinal elevation in depressive symptoms in a socio-economically diverse sample of White and African–American middle-aged adults. We hypothesize that a baseline epigenetic age acceleration predicts higher baseline depressive symptoms or faster increase in those symptoms over time. Finally, we also test whether those key relationships of interest differ across those two socio-demographic factors.
2. Methods
2.1. Study design
HANDLS was initiated in 2004 as a prospective cohort study focused on disparities pertaining to cardiovascular disease and cognitive aging. Using an area probability sampling strategy, an ethnically and socioeconomically diverse sample of urban adults was recruited in HANDLS. Middle-aged African American and White adults (baseline age: 30–64 years) residing in urban areas were sampled with widely ranging household incomes (above and below poverty). Thirteen Baltimore city neighborhoods were selected to define primary sampling units (Evans et al., 2010). The current study analyzed data from visit 1 (2004–2009) in addition to the initial follow-up examination (visit 2: 2009–2013), with follow-up time between waves ranging between 1 year and ~ 8 years, mean ± SD of 4.64 ± 0.93 years. HANDLS collected data using several cognitive tests at the two waves of data; a sub-sample of visit 1 included DNAm data from which three epigenetic clocks reflecting accelerated aging were estimated. Written informed consent was obtained from all study participants who were provided with a booklet and a video explaining key study procedures. The study protocol was approved by the National Institute on Environmental Health Sciences Institutional Review Board of the National Institutes of Health.
2.2. Participants
The HANDLS consisted of N1=3720 participants (30–65 years, AA and Whites, Phase I, visit 1). During Phase II of visit 1 (Medical Research Vehicle (MRV) baseline visit), in-depth examinations were performed including a fasting blood draw, a physical examination, a DEXA scan, an EKG, a 24-h dietary recall and an assessment of depressive symptoms severity. A second 24-h dietary recall telephone interview was completed for most participants with one 24-h recall, 3–10 days following the MRV visit. The average of those two dietary recalls was computed to evaluate dietary intakes. Subsequently, epi-genetic analyses were performed using frozen peripheral blood mono-nuclear cells (PBMC) on a sub-sample of Whites and AA participants. The participant flowchart is detailed in Figure S1. In this study, we included participants who had complete “epigenetic clock” data (visit 1: N 2b=470) who additionally had data on depressive symptoms scores at either visit (visit 1: N3=465). The final analytic sample (N4=329) excluded participants with missing data on several covariates, including dietary, self-reported chronic conditions, use of non-steroidal anti-inflammatory drugs (NSAIDs), measured body mass index (BMI) among others. Using a probit model with a binary outcome (1=selected, 0=unselected) and with predictors being the key socio-demographic variables, it was determined that the selected group differed from the remaining HANDLS participants by being older, less likely to be male and less likely to be African-American or to fall in the above poverty income category. Adjustment for sample selectivity was done using a 2-stage Heckman selection model, as described later.
2.3. Depressive symptoms
At each visit, depressive symptoms were measured using the original version of the 20-item Center of Epidemiological Studies-Depression (CES-D), a self-reported symptom rating scale assessing affective and depressed mood (Radloff, 1977) with suitable psychometric properties in various studies of older adults (Beekman et al., 1997). A total CES-D (CES-Dtotal) score ≥16 reflects elevated depressive symptoms (EDS) (Beydoun et al., 2016). CES-Dtotal consists of meaningful domains that exhibit invariant factor structure between the National Health and Nutrition Examination Survey I and pilot HANDLS data (Nguyen et al., 2004). Our hypotheses were tested using the total score and domain-specific CES-D scores: (1) Somatic complaints (e.g., poor sleep, poor appetite); (2) Depressive affect (e.g., feeling sad); (3) Positive affect (e.g., having positive thoughts) and (4) Interpersonal problems (e.g., having trouble in social settings) (Nguyen et al., 2004). The raw sub-scores were used by summing up the scores on symptoms that were shown to fall under each domain. Details regarding which items (scored between 0 and 3) are used to construct each domain are previously described (Nguyen et al., 2004).
2.4. DNA methylation and epigenetic clocks
A random sample of 508 participants was identified to examine DNA methylation (DNAm), based on a factorial design defined across sex, race and poverty status and available DNA samples. Further, 250 ng of DNA was extracted from blood and treated with sodium bisulfite Zymo EZ-96 DNA Methylation kit as suggested in manufacturer’s protocol (Zymo Research, Orange, CA, USA). The Zymo DNA methylation kit allows DNA bisulfite conversion directly from blood without the prerequisite for DNA purification. It completes both DNA denaturation and bisulfite conversion processes in a single step. Genome-wide DNAm was measured utilizing the Illumina Infinium MethylationEPIC BeadChip (Illumina Inc., San Diego, CA, USA). Of initial 508 participants, a total of 487 had DNAm measures, and quality control was carried out on 12 technical replicates and performed at sample and probe levels. Furthermore, 17 samples were excluded because they were outliers, had poor quality methylation values (i.e., a mean detection p value ≥ 0.01) or an evidence of sex mismatch between self-report and methylation prediction. In terms of probe, we excluded those of low quality (mean detection p value ≥ 0.01), with overlapping single nucleotide polymorphisms (minor allele frequency cut-off = 0.05), cross-hybridizing probes, and probes mapping to sex chromosomes. To identify an optimal method for DNAm data normalization, we compared performance levels of different commonly utilized data normalization and pre-processing algorithms in terms of their reduction in technical variations, by using DNAm measured in technical replicates. Selected algorithms were the following: Illumina Genome Studio, normal-exponential out-of-band (NOOB) (Triche et al., 2013), stratified quantile normalization (quantile) (Touleimat and Tost, 2012), and subset-quantile within array normalization (SWAN) (Maksimovic et al., 2012). Because it yielded the lowest probe variance and highest correlation between technical replicates, NOOB method was chosen for DNAm data normalization and background correction in this study. Using DNAm data, proportions of multiple white blood cell types (granulocytes, natural killer cells, monocytes, B cells, CD8+ naïve T cells, CD4+ T cells, exhausted CD8+ T cells (CD8+CD28–CD45RA–), plasmablasts, and the number (count) of naïve CD8+ T cells (CD8+CD45RA+CCR7+)) were estimated (Houseman et al., 2012).
2.5. DNA methylation age (DNAm age) prediction and epigenetic age acceleration (EAA) measures
DNAm age was calculated using the Horvath (Horvath, 2013) and Hannum (Hannum et al., 2013) methods, both of which rely on methylation beta values of 353 and 71 CpG sites, respectively, while applying the epigenetic clock algorithm. We selected participants with variable genetic ancestries. Algorithms were trained and validated while using DNA derived from different tissues that include blood DNA. The DNAm age and epigenetic age acceleration estimation process is available from Horvath’s laboratory (https://dnamage.genetics.ucla.edu/home). In brief, the Horvath method predicts age while being agnostic to tissue type or DNA cell source. In contrast, Hannum method was developed based on blood DNAm. Universal epigenetic age acceleration (AgeAccel or “Epigenetic clock1”) are the residuals obtained from regressing DNAm age-predicted by the Horvath algorithm on chronological age, with positive residual value suggesting faster aging and negative value reflecting a slower aging. Moreover, two additional epigenetic age acceleration (EAA) measures were used, reflecting intrinsic and extrinsic epigenetic age acceleration – IEAA (“Epigenetic clock 2) and EEAA (“Epigenetic clock 3”), respectively. Believed to be a measure of cellular epigenetic age acceleration irrespective of white blood cell composition, IEAA is the residual from regressing DNAm age (predicted by the Horvath algorithm) on chronological age and white blood cell proportions (naive CD8+ T cells, exhausted CD8+ T cells, plasmablasts, CD4+ T cells, natural killer cells, monocytes, and granulocytes). On the other hand, using the Hannum algorithm, EEAA based on the DNAm age and is believed to be a measure of epigenetic age acceleration combined with changes in white blood cell proportions, and may indicate immune system cell aging (immunosenescence) (Chen et al., 2016).
2.6. Covariates
2.6.1. Sociodemographic, lifestyle, and health-related potential confounders
All regression models were adjusted for sociodemographic factors, age, sex, race (White vs. African American), educational attainment categories (0 ≤ High School (HS); 1=HS and 2 ≥ HS) and poverty status (below vs. above 125% the federal poverty line). Poverty status was categorized as such by using the US Census Bureau poverty thresholds for 2004 (Bureau, 2004) relying on income, and total family size including children under age 18 years. Furthermore, all analyses were adjusted for measured body mass index (kg/m2), current drug use (“opiates, marijuana or cocaine”=1 vs. not=0) and current smoking status (0: “never or former smoker” vs. 1 “current smoker”) without evaluating exposure-covariate associations. These models were further adjusted for visit 1 self-reported history of type 2 diabetes, hypertension, dyslipidemia, cardiovascular disease (stroke, congestive heart failure, non-fatal myocardial infarction or atrial fibrillation), auto-immune disease (multiple sclerosis, systemic lupus, gout, rheumatoid arthritis, psoriasis, thyroid disorder and Crohn’s disease) and use of NSAIDs (prescription and over-the-counter) over the past two weeks, as was done previously (Bettcher et al., 2012; Gimeno et al., 2009).
2.6.2. Dietary potential confounders
For all exposures, dietary covariates were considered as potential confounders if they were linked to depression based on previous studies; these included vitamins B-6, folate and B-12, total carotenoids (α-carotene, β-carotene, β-cryptoxanthin, lutein+zeaxanthin, lycopene), vitamin C and α-tocopherol (all divided by total energy intake and expressed per 1000 kcal) and ratio of n-3 PUFA:n-6 PUFA, as was done in previous studies (Beydoun et al., 2015a). To emulate multivariable nutrient density model, energy intake was entered as a covariate (Willet, 1998). The Healthy Eating Index (HEI-2010) total score, A measure of overall dietary quality, (http://appliedresearch.cancer.gov/tools/hei/tools.html and http://handls.nih.gov/06Coll-dataDoc.htm) was also considered. Noteworthy is the inclusion of alcohol intake in component 12 of HEI-2010, a dietary factor known to influence DNA methylation and epigenetic aging (Rosen et al., 2018).
2.7. Statistical analysis
Stata 15.0 (StataCorp, College Station, TX) was used to conduct all analyses (STATA, 2017). First, baseline characteristics, including covariates and exposures, were compared by sex, race and EDS status (based on mean score across waves), using t-tests and ANOVA for continuous variables and χ2 tests for categorical variables. Second, several mixed-effects regression models on continuous CES-D total or on domain-specific score(s) were conducted to test associations with 3 “epigenetic clock” measures, while controlling for potential confounders. Sex- and race-specific associations were examined by adding interaction terms to multivariable mixed-effects regressions and stratifying by sex and race, separately. The methodology used is outlined in Supplemental Method 1 (Blackwell et al., 2006).
Non-random selection of participants from the initial HANDLS sample (n = 3720) may cause bias due to systematic differences in baseline characteristics including age, sex, race and socio-economic status between final analytic excluded samples. A 2-stage Heckman selection process accounted for this potential bias in our final regression models. At a first stage, a probit model with binary outcome variable coded as selected=1 vs. unselected=0 was constructed from which an inverse mills ratio (derived from the predicted probability of being selected, conditional on the covariates baseline age, sex, race, poverty status and education) was estimated. At a second stage, this inverse mills ratio was entered into each mixed-effects regression model as a covariate, as previously done (Beydoun et al., 2013). An inverse mills ratio was computed for the sample with “epigenetic clock” measures.
A type I error of 0.05 was used, with 0.05 < p-values < 0.10 judged as borderline significant for main effects and 2-way interaction terms (Selvin, 2004) before family-wise Bonferroni correction for multiple testing (Hochberg and Tamhane, 1987), assuming each of total CES-D and sub-domains of CES-D are distinctive outcomes, while the 3 exposures that are conceptually related. This approach was adopted in several previous studies (Beydoun et al., 2015a, 2015b). Accounting for 3 exposures, type I error was reduced to 0.05/3 = 0.0165 for main effects and for interaction terms for the mixed-effects regression models. 3-way interaction terms were deemed statistically significant at an α-error level of 0.05. Raw p-values were presented and annotated for significance upon correction for multiple testing. Those significant findings were illustrated using predictive margins from mixed-effects regression models. Moreover, Cohen’s D was estimated by transforming the related outcome (e.g., CES-D total score or domains of CES-D) and the key exposures (e.g., Epigenetic clocks 1–3) into standardized z-scores. Effect sizes were then obtained and determined to be weak if below 0.20 and medium/moderate if between 0.2 and 0.8 and strong if above 0.80. Finally, a sensitivity analysis was conducted for models that passed correction for multiple testing for at least parameter in the full model, whereby a series of reduced models were carried out and compared to the full model. Specifically, a crude model with only the inverse mills ratio (Model 0), followed by a model adding age, sex and race (Model 1), a third model adding all socio-demographic and socioeconomic factors (Model 2). We then ran models that adjusted for lifestyle factors (i.e., smoking, drug use, dietary factors). In addition to the socio-demographic and socio-economic factors in Model 1 (Model3) and a final model included health-related factors (BMI, co-morbid conditions, NSAIDs) to Model 1 (Model 4).
3. Results
Based on descriptive findings outlined in Table 1, EEAA (“epigenetic clock 3”) was higher among men compared to women (+1.35 vs. −1.15, P = 0.0002) and higher among Whites compared to African-Americans (+2.26 vs. −1.85, P < 0.001), reflecting faster age acceleration that includes immunosenescence. On the other hand, women had higher CES-D scores based on mean scores across waves (16.9 vs.14.3, P = 0.020). Other notable differences include lower educational attainment among African–Americans, a lower proportion above poverty or employed among depressed individuals. Moreover, depressed individuals were likely to report hypertension and autoimmune conditions. The latter was also more frequently reported among women compared to men. While energy intake was higher on average among men, adjusting for it, micronutrient intakes differed by sex (total carotenoids, vitamin C, vitamin E, n3 PUFA: n6 PUFA), race (vitamin C) and depression status (HEI-2010).
Table 1.
Characteristics of HANDLS study participants by sex, race and EDS status [based on CES-D score (mean across waves)]a.
| By sex | Pb | Race | Pb | EDS status | Pb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Men | Women | Men vs. | Whites | African-Americans | Whites vs. | EDS− | EDS+ | EDS− vs. | |
| women | African-Americans | EDS+ | |||||||
| % or Mean ± SEM | |||||||||
| (n = 171) | (n = 158) | (n = 160) | (n = 169) | (n = 179) | (n = 150) | ||||
| Depressive Symptoms | |||||||||
| CES-D, Mean ± SEM | 14.3 ± 0.73 | GO | 0.020 | 16.2 ± 0.8 | 14.9 ± 0.8 | 0.23 | 7.82 ± 0.31 | 24.7 ± 0.62 | <0.001 |
| Epigenetic clock | (n = 171) | (n = 158) | (n = 160) | (n = 169) | (n = 179) | (n = 150) | |||
| Epigenetic clock 1: AgeAccel | + 0.45 ± 0.36 | −0.36 ± 0.37 | 0.12 | −0.13 ± 0.35 | +0.24 ± 0.37 | 0.47 | + 0.12 ± 0.35 | −0.00 ± 0.38 | 0.81 |
| Epigenetic clock 2: IEAA | +0.14 ± 0.34 | −0.09 ± 0.36 | 0.65 | −0.16 ± 0.35 | 0.21 ± 0.36 | 0.47 | −0.00 ± 0.34 | + 0.07 ± 0.37 | 0.88 |
| Epigenetic clock 3: EEAA | + 1.35 ± 0.47 | −1.15 ± 0.46 | 0.0002 | 2.26 ± 0.38 | −1.85 ± 0.51 | <0.001 | + 0.27 ± 0.46 | −0.00 ± 0.50 | 0.68 |
| Sociodemographic characteristics | (n = 171) | (n = 158) | (n = 169) | (n = 160) | 0.84 | (n = 179) | (n = 150) | 0.83 | |
| Age (y), Mean ± SEM | 48.9 ± 0.7 | 48.9 ± 0.7 | 0.99 | 49.0 ± 0.7 | 48.8 ± 0.7 | 48.8 ± 0.7 | 49.1 ± 0.7 | ||
| Sex, % men | 51.3 | 52.7 | 0.79 | 34.8 | 26.9 | 0.12 | |||
| African-American, % | 52.1 | 50.6 | 0.80 | 53.6 | 48.7 | 0.37 | |||
| Education, % | 0.031 | 0.14 | |||||||
| <HS | 9.9 | 7.6 | 0.37 | 5.3 | 12.5 | 8.4 | 9.3 | ||
| HS | 63.7 | 59.4 | 61.0 | 62.5 | 57.4 | 66.7 | |||
| >HS | 26.3 | 32.9 | 33.7 | 25.0 | 34.1 | 24.0 | |||
| PIR≥125%, % | 48.0 | 50.0 | 0.71 | 49.4 | 48.5 | 0.88 | 58.1 | 38.0 | <0.001 |
| Employed, % | 0.31 | <0.001 | <0.001 | ||||||
| Yes | 49.1 | 44.3 | 48.3 | 47.3 | 55.9 | 36.0 | |||
| Missing | 14.0 | 20.3 | 25.6 | 8.9 | 17.3 | 16.7 | |||
| Lifestyle and health-related factors | |||||||||
| Current smoking status, % | 0.16 | 0.29 | |||||||
| Currently smoking | 50.9 | 43.0 | 45.0 | 49.1 | 43.6 | 51.3 | 0.083 | ||
| Missing | 0.6 | 2.5 | 0.6 | 2.4 | 0.6 | 2.7 | |||
| Current use of illicit drugs, % | <0.001 | 0.23 | |||||||
| Used any type | 61.4 | 37.3 | 45.0 | 54.4 | 48.6 | 51.3 | 0.65 | ||
| Missing | 1.2 | 1.3 | 1.3 | 1.2 | 1.7 | 0.7 | |||
| (n = 171) | (n = 158) | (n = 169) | (n = 160) | (n = 179) | (n = 150) | ||||
| Body mass index, kg/m2; Mean ± SEM | 28.7 ± 0.50 | 31.1 ± 0.65 | 0.0048 | 29.9 ± 0.6 | 29.8 ± 0.6 | 0.89 | 29.3 ± 0.5 | 30.5 ± 0.6 | 0.13 |
| Co-morbid conditions and NSAIDs | (n = 171) | (n = 158) | (n = 169) | (n = 160) | (n = 179) | (n = 150) | |||
| Diabetes, % | 12.3 | 12.0 | 0.94 | 14.3 | 10.1 | 0.23 | 8.9 | 16.0 | 0.051 |
| Hypertension,% | 35.0 | 37.3 | 0.67 | 33.1 | 39.1 | 0.26 | 29.6 | 44.0 | 0.007 |
| Dyslipidemia,% | 20.5 | 23.4 | 0.52 | 25.0 | 18.9 | 0.18 | 19.0 | 25.3 | 0.17 |
| Cardiovascular diseased, % | 12.9 | 12.7 | 0.96 | 10.0 | 15.4 | 0.14 | 11.2 | 14.7 | 0.34 |
| Inflammatory conditionse, % | 8.2 | 18.4 | 0.006 | 11.3 | 14.8 | 0.34 | 8.4 | 18.7 | 0.006 |
| NSAIDSf, % | 24.0 | 15.8 | 0.065 | 23.8 | 16.6 | 0.10 | 19.0 | 21.3 | 0.60 |
| Dietary factors, daily intakes | (n = 171) | (n = 158) | (n = 160) | (n = 169) | (n = 179) | (n = 150) | |||
| Energy, kcal | 2402 ± 83 | 1706 ± 54 | <0.001 | 2034 ± 65 | 2099 ± 85 | 0.55 | 2142 ± 74 | 1980 ± 80 | 0.13 |
| Total carotenoids, mg/1000 kcal | 2998 ± 293 | 4006 ± 381 | 0.035 | 3145 ± 269 | 3801 ± 390 | 0.17 | 3557 ± 315 | 3393 ± 368 | 0.74 |
| Vitamin A, RE/1000 kcal | 309 ± 51 | 355 ± 71 | 0.59 | 286 ± 17 | 375 ± 82 | 0.30 | 319 ± 48 | 347 ± 75 | 0.16 |
| Vitamin C, mg/1000 kcal | 30.1 ± 2.9 | 40.4 ± 4.2 | 0.044 | 28.9 ± 2.4 | 40.9 ± 4.3 | 0.018 | 38.3 ± 2.9 | 31.2 ± 4.4 | 0.16 |
| Vitamin E, mg/1000 kcal | 2.8 ± 0.1 | 3.4 ± 0.2 | 0.007 | 3.00 ± 0.13 | 3.19 ± 0.20 | 0.44 | 3.31 ± 0.19 | 2.86 ± 0.12 | 0.06 |
| Vitamin B-6, mg/1000 kcal | 0.91 ± 0.04 | 0.89 ± 0.05 | 0.75 | 0.89 ± 0.04 | 0.91 ± 0.04 | 0.81 | 0.92 ± 0.04 | 0.87 ± 0.04 | 0.44 |
| Vitamin B-12, μg/1000 kcal | 3.27 ± 0.54 | 3.28 ± 0.71 | 0.99 | 2.70 ± 0.15 | 3.82 ± 0.85 | 0.20 | 3.00 ± 0.51 | 3.60 ± 0.76 | 0.50 |
| Folate, μg/1000 kcal | 175.9 ± 7.7 | 200.3 ± 10.2 | 0.055 | 197.4 ± 9.2 | 178.4 ± 8.8 | 0.14 | 189.3 ± 8.4 | 185.7 ± 9.7 | 0.77 |
| n3 PUFA:n6 PUFA ratioc | 0.108 ± 0.002 | 0.127 ± 0.009 | 0.033 | 0.121 ± 0.007 | 0.114 ± 0.005 | 0.44 | 0.113 ± 0.006 | 0.121 ± 0.006 | 0.37 |
| Healthy Eating Index-2010 | 39.7 ± 0.82 | 41.8 ± 1.00 | 0.094 | 40.1 ± 1.00 | 41.2 ± 0.84 | 0.39 | 42.6 ± 0.9 | 38.4 ± 0.8 | 0.0012 |
Abbreviations: AA=arachidonic acid; ALA=α-linolenic acid; CES-D=Center for Epidemiologic Studies-Depression scale; DHA=docosahexaenoic acid; DPA=docosapentaenoic acid; EDS=elevated depressive symptoms; EPA=eicosapentaenoic acid; HANDLS=Healthy Aging in Neighborhoods of Diversity Across the Lifespan; HDL-C]High-Density Lipoprotein-Cholesterol; HS=High School; LA=linoleic acid; n3=omega-3; n6=omega-6; PIR=Poverty Income Ratio; PUFA=polyunsaturated fatty acids; SEM=standard error of the mean; TC=total cholesterol.
Values are percent or Mean ± SEM or % ± SE.
P-value was based on independent samples t-test when row variable is continuous and χ2 test when row variable is categorical.
n3 PUFA included DHA+EPA+n3DPA +ALA. n6 PUFA included AA+LA.
Cardiovascular disease include self-reported stroke, congestive heart failure, non-fatal myocardial infarction or atrial fibrillation.
Inflammatory conditions include multiple sclerosis, systemic lupus, gout, rheumatoid arthritis, psoriasis, thyroid disorder and Crohn’s disease.
Non-steroidal anti-inflammatory drugs (NSAIDS) include over the counter and prescription drugs in that category.
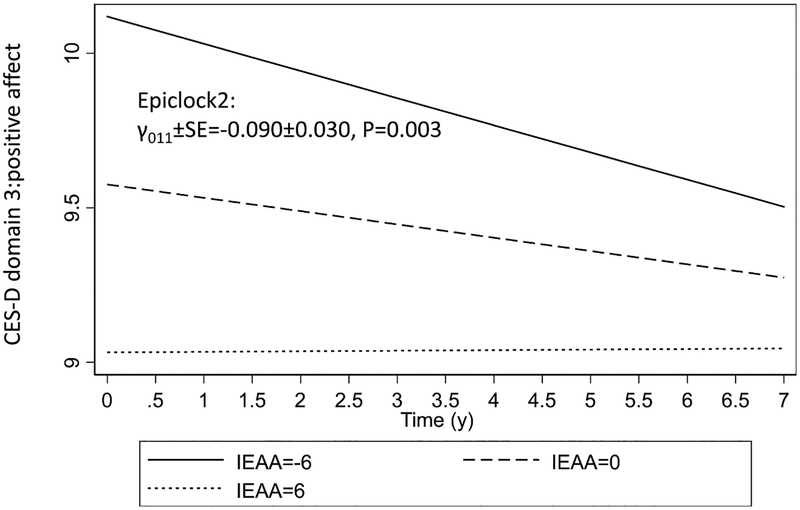
Table 2 displays findings from the linear mixed-effects regression models for depressive symptoms as predicted by the three epigenetic clock exposures, adjusting for key confounders both at the levels of the intercept and the slope. After adjustment for multiple testing, none of the epigenetic clock of accelerated aging were associated with baseline or rate of change in the total CES-D score. However, “epigenetic clock2” or IEAA which is measured using the Horvath algorithm while adjusting for while blood cell count, was inversely associated with baseline CES-D domain 3, which reflects “positive affect” (higher score → lower depressive symptoms), both in the total population (γ011 ± SE=−0.090 ± 0.030, P = 0.003; Cohen’s D: −0.16) and among Whites (γ011 ± SE=−0.135 ± 0.048, P = 0.005, Cohen’s D: −0.23). This association in the total population is illustrated in Fig. 1 showing no divergence in the trajectories but rather a significant difference in baseline positive affect at increasing levels of epigenetic clock 2. Moreover, baseline “epigenetic clock 1” (Horvath algorithm, AgeAccel) had a similar inverse relationship with the positive affect domain of the CES-D at baseline, both in the total population (γ011 ± SE=−0.071 ± 0.030, P = 0.016; Cohen’s D: −0.13) and among Whites (γ011 ± SE=−0.012 ± 0.047, P = 0.011; Cohen’s D:−0.21). Other associations deemed non-significant after correction for multiple testing showed some inconsistencies across sex and race, and between cross-sectional and longitudinal effects. Thus, even though associations were generally weak, they were stronger among Whites compared to the overall population. In fact, in the sensitivity analysis, the crude model as well as models 1–3 (adding socio-demographic factors, lifestyle and health-related factors) retained statistical significance to a greater extent among Whites as opposed to the total population. For instance, IEAA (“epigenetic clock 2”) was associated with lower positive affect among Whites in all models, particularly those adjusting for all socio-demographic and socio-economic factors in addition to health-related and/or dietary factors (data not shown).
Table 2.
Analysis of baseline epigenetic clock measures and CES-D scores (total population, sex- and race-stratified), mixed-effects linear regression analysis, HANDLS study, 2004–2013.
| Total population | Men | Women | Whites | African-Americans | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| γ ± SEE | p-value | γ ± SEE | p-value | γ ± SEE | p-value | γ ± SEE | p-value | γ ± SEE | p-value | ||||
| CES-D total score | N = 329 | N’ = 626 | N = 171 | N’ = 321 | N = 158 | N’ = 305 | N = 160 | N’ = 302 | N = 169 | N’ = 324 | |||
| Model 1: Epigenetic clock 1 | |||||||||||||
| Epiclock1 (γ011 for π0i) | +0.125 ± 0.131 | 0.34 | +0.117 ± 0.159 | 0.46 | +0.05 ± 0.20 | 0.81 | +0.28 ± 0.20 | 0.15 | −0.063 ± 0.173 | 0.72 | |||
| Epiclock1 × Time (γ111 for π1i) | +0.017 ± 0.028 | 0.54 | +0.026 ± 0.036 | 0.46 | 0.04 ± 0.04 | 0.36 | +0.035 ± 0.040 | 0.39 | −0.013 ± 0.035 | 0.71 | |||
| Model 2: Epigenetic clock 2 | |||||||||||||
| Epiclock2 (γ011 for π0i) | +0.166 ± 0.134 | 0.22 | +0.185 ± 0.166 | 0.26 | +0.051 ± 0.204 | 0.80 | +0.310 ± 0.200 | 0.12 | −0.028 ± 0.180 | 0.87 | |||
| Epiclock2 × Time (γ111 for π1i) | +0.021 ± 0.028 | 0.45 | +0.023 ± 0.037 | 0.54 | +0.048 ± 0.040 | 0.24 | +0.027 ± 0.041 | 0.51 | +0.002 ± 0.036 | 0.97 | |||
| Model 3: Epigenetic clock 3 | |||||||||||||
| Epiclock3 (γ011 for π0i) | −0.054 ± 0.108 | 0.61 | +0.016 ± 0.132 | 0.91 | −0.179 ± 0.168 | 0.29 | +0.025 ± 0.19 | 0.90 | −0.082 ± 0.127 | 0.52 | |||
| Epiclock3 × Time (γ111 for π1i) | +0.011 ± 0.023 | 0.62 | +0.034 ± 0.028 | 0.23 | −0.002 ± 0.035 | 0.96 | +0.051 ± 0.040 | 0.20 | −0.008 ± 0.026 | 0.76 | |||
| CES-D domain 1: somatic complaints | |||||||||||||
| Model 1: Epigenetic clock 1 | |||||||||||||
| Epiclock1 (γ011 for π0i) | −0.007 ± 0.051 | 0.89 | −0.011 ± 0.060 | 0.86 | −0.023 ± 0.077 | 0.77 | +0.060 ± 0.075 | 0.42 | −0.078 ± 0.069 | 0.26 | |||
| Epiclock1 × Time (γ111 for π1i) | +0.008 ± 0.011 | 0.51 | +0.024 ± 0.015 | 0.10 | +0.003 ± 0.016 | 0.88 | +0.006 ± 0.017 | 0.74 | +0.010 ± 0.015 | 0.53 | |||
| Model 2: Epigenetic clock 2 | |||||||||||||
| Epiclock2 (γ011 for π0i) | +0.001 ± 0.051 | 0.99 | +0.006 ± 0.062 | 0.92 | −0.030 ± 0.078 | 0.70 | +0.073 ± 0.077 | 0.34 | −0.084 ± 0.072 | 0.24 | |||
| Epiclock2 × Time (γ111 for π1i) | +0.011 ± 0.011 | 0.36 | +0.026 ± 0.152 | 0.089 | +0.007 ± 0.017 | 0.67 | +0.009 ± 0.017 | 0.62 | +0.015 ± 0.016 | 0.33 | |||
| Model 3: Epigenetic clock 3 | |||||||||||||
| Epiclock3 (γ011 for π0i) | −0.013 ± 0.042 | 0.76 | +0.037 ± 0.051 | 0.46 | −0.078 ± 0.064 | 0.22 | −0.001 ± 0.073 | 0.99 | −0.009 ± 0.051 | 0.87 | |||
| Epiclock3 × Time (γ111 for π1i) | +0.002 ± 0.010 | 0.84 | +0.005 ± 0.012 | 0.65 | −0.000 ± 0.014 | 0.98 | +0.005 ± 0.017 | 0.78 | +0.001 ± 0.012 | 0.96 | |||
| CES-D domain 2: depressed affect | |||||||||||||
| Model 1: Epigenetic clock 1 | |||||||||||||
| Epiclock1 (γ011 for π0i) | +0.038 ± 0.058 | 0.51 | +0.066 ± 0.071 | 0.35 | −0.037 ± 0.921 | 0.69 | +0.071 ± 0.083 | 0.39 | −0.004 ± 0.079 | 0.96 | |||
| Epiclock1 × Time (γ111 for π1i) | +0.012 ± 0.013 | 0.36 | +0.010 ± 0.016 | 0.53 | +0.031 ± 0.191 | 0.10 | +0.019 ± 0.017 | 0.27 | −0.003 ± 0.018 | 0.86 | |||
| Model 2: Epigenetic clock 2 | |||||||||||||
| Epiclock2 (γ011 for π0i) | +0.054 ± 0.059 | 0.36 | +0.094 ± 0.073 | 0.20 | −0.033 ± 0.092 | 0.72 | +0.079 ± 0.084 | 0.35 | +0.019 ± 0.082 | 0.82 | |||
| Epiclock2 × Time (γ111 for π1i) | +0.013 ± 0.013 | 0.29 | +0.009 ± 0.017 | 0.59 | +0.037 ± 0.019 | 0.058 | +0.015 ± 0.018 | 0.38 | +0.003 ± 0.018 | 0.89 | |||
| Model 3: Epigenetic clock 3 | |||||||||||||
| Epiclock3 (γ011 for π0i) | −0.008 ± 0.048 | 0.87 | +0.026 ± 0.06 | 0.66 | −0.066 ± 0.077 | 0.39 | +0.017 ± 0.081 | 0.84 | −0.020 ± 0.058 | 0.72 | |||
| Epiclock3 × Time (γ111 for π1i) | +0.002 ± 0.011 | 0.89 | +0.009 ± 0.013 | 0.49 | +0.001 ± 0.017 | 0.94 | +0.024 ± 0.017 | 0.16 | −0.008 ± 0.013 | 0.54 | |||
| CES-D domain 3: positive affect | |||||||||||||
| Model 1: Epigenetic clock 1 | |||||||||||||
| Epiclock1 (γ011 for π0i) | −0.071 ± 0.030 | 0.016d | −0.077 ± 0.039 | 0.051 | −0.046 ± 0.043 | 0.29 | −0.012 ± 0.047 | 0.011d | −0.028 ± 0.033 | 0.40 | |||
| Epiclock1 × Time (γ111 for π1i) | +0.006 ± 0.007 | 0.38 | +0.017 ± 0.009 | 0.047 | −0.006 ± 0.010 | 0.59 | −0.001 ± 0.011 | 0.95 | +0.018 ± 0.008 | 0.031 | |||
| Model 2: Epigenetic clock 2 | |||||||||||||
| Epiclock2 (γ011 for π0i) | −0.090 ± 0.030 | 0.003d | −0.096 ± 0.019 | 0.019 | −0.067 ± 0.044 | 0.13 | −0.135 ± 0.048 | 0.005d | −0.047 ± 0.035 | 0.18 | |||
| Epiclock2 × Time (γ111 for π1i) | +0.007 ± 0.007 | 0.29 | +0.021 ± 0.009 | 0.023 | −0.006 ± 0.010 | 0.59 | +0.005 ± 0.011 | 0.68 | +0.016 ± 0.008 | 0.062 | |||
| Model 3: Epigenetic clock 3 | |||||||||||||
| Epiclock3 (γ011 for π0i) | +0.047 ± 0.024 | 0.054 | +0.040 ± 0.033 | 0.22 | +0.071 ± 0.036 | 0.049 | +0.032 ± 0.046 | 0.48 | +0.043 ± 0.025 | 0.081 | |||
| Epiclock3 × Time (γ111 for π1i) | −0.007 ± 0.006 | 0.25 | −0.010 ± 0.007 | 0.12 | −0.002 ± 0.009 | 0.81 | −0.014 ± 0.011 | 0.17 | +0.000 ± 0.006 | 0.99 | |||
| CES-D domain 4: Interpersonal problems | |||||||||||||
| Model 1: Epigenetic clock 1 | |||||||||||||
| Epiclock1 (γ011 for π0i) | +0.017 ± 0.015 | 0.28 | +0.001 ± 0.021 | 0.98 | +0.029 ± 0.022 | 0.19 | +0.029 ± 0.022 | 0.19 | −0.016 ± 0.022 | 0.46 | |||
| Epiclock1 × Time (γ111 for π1i) | +0.005 ± 0.004 | 0.20 | +0.006 ± 0.005 | 0.31 | +0.004 ± 0.005 | 0.44 | +0.008 ± 0.006 | 0.18 | +0.000 ± 0.0050 | 0.93 | |||
| Model 2: Epigenetic clock 2 | |||||||||||||
| Epiclock2 (γ011 for π0i) | +0.014 ± 0.016 | 0.36 | +0.007 ± 0.022 | 0.74 | +0.088 ± 0.064 | 0.17 | +0.027 ± 0.022 | 0.22 | −0.018 ± 0.023 | 0.44 | |||
| Epiclock2 × Time (γ111 for π1i) | +0.005 ± 0.004 | 0.16 | +0.005 ± 0.006 | 0.39 | 0.017 ± 0.022 | 0.44 | +0.005 ± 0.006 | 0.36 | +0.003 ± 0.005 | 0.58 | |||
| Model 3: Epigenetic clock 3 | |||||||||||||
| Epiclock3 (γ011 for π0i) | +0.011 ± 0.013 | 0.41 | −0.003 ± 0.018 | 0.87 | +0.031 ± 0.018 | 0.094 | +0.031 ± 0.021 | 0.14 | −0.012 ± 0.016 | 0.44 | |||
| Epiclock3 × Time (γ111 for π1i) | +0.001 ± 0.003 | 0.68 | +0.007 ± 0.004 | 0.13 | −0.004 ± 0.004 | 0.26 | +0.011 ± 0.006c | 0.062 | −0.001 ± 0.004 | 0.72 | |||
Abbreviations: CES-D=Center for Epidemiologic Studies-Depression scale; Epiclock=Epigenetic clock (See methods for definition of each); HANDLS=Healthy Aging in Neighborhoods of Diversity Across the Lifespan; HS=High School; n3=omega-3; n6=omega-6; PUFA=polyunsaturated fatty acids; SEE=standard error of the estimate.
Models were further adjusted for other covariates (main effects and interaction with time). Time at baseline visit was set to zero. Covariates considered as potential confounders included: baseline age was centered at 50y, sex, race, PIR, education, employment status, total energy intake at 2000 kcal/d, total carotenoid intake at 3 mg/1000 kcal/d, vitamin C intake at 30 mg/1000 kcal/d, vitamin A intake at 300 RE/1000 kcal/d, vitamin E at 3 mg/1000 kcal/d, vitamin B-6 at 0.8 mg/1000, vitamin B-12 at 3 μg/1000 kcal/d, folate at 170 μg/1000 kcal/d, n-3 PUFA:n-6 PUFA at 0.11, Healthy Eating Index-2010 was centered at 42, body mass index at 30, co-morbid conditions (diabetes, hypertension, dyslipidemia, CVD, inflammatory conditions) and use of NSAIDs. All these covariates were entered in models with each epigenetic clock exposures. P-value presented a raw values prior to correction for multiple testing.
N=number of participants in the analysis; N’=total number of visits included in the analysis. Findings that were significant at a type I error of 0.05 are bolded, while those that are marginally significant (P<0.10) are bolded and italicized.
In a separate model with interaction of epigenetic clock exposures by (sex/race) by TIME, including all other terms in the current model, p<0.05 for null hypothesis that this interaction term is=0.
P < 0.0165 for exposure main (exposure) or interaction term effects (exposure×TIME).
Fig. 1.
Predictive margins for positive affect by Time, across levels of epigenetic clock 2, total population: Mixed-effects linear regression models.
4. Discussion
This study comprehensively tested the relationship between DNAm epigenetic age acceleration and depressive symptoms in a prospective bi-racial cohort of urban adults. Our findings indicated that in the total population and among Whites, there was a cross-sectional relationship between two measures of epigenetic age acceleration utilizing the Horvath algorithm and the domain of positive affect, indicating that accelerated aging may influence this specific domain of depressive symptoms in an adverse manner. No longitudinal associations were detected in our present analyses, indicating that this relationship was for the most part a contemporaneous one, whereby epigenetic age acceleration can trigger depressive symptoms or vice versa. Nevertheless, reverse causality whereby CES-D total and domain-specific scores can alter the trajectory of any of the three DNAm epigenetic clock measure cannot be ruled out.
Previously, methods such as candidate gene approaches and methylome-wide association studies (MWAS) were used to study MDD-associated and stress-induced alterations in DNA methylation (Pishva et al., 2017). Herein, we tested the associations of three DNAm measures of EAA in a socio-economically diverse sample of White and African–American middle-aged adults which may provide a clue for MDD biomarker identification. Previous reports have shown the epi-genetic aging in individuals with Werner’s syndrome (Maierhofer et al., 2017), HIV infection (Chen et al., 2019), Post-traumatic Stress Disorder (Verhoeven et al., 2018), cognitive impairment (White et al., 2017) and frailty (Breitling et al., 2016).
Only a few studies have previously examined the relationship between epigenetic aging and MDD. One key study detected no significant age or Post-Mortem Interval differences between MDD cases and controls, though this difference was found between suicide cases and controls (Bustamante et al., 2018). In this study, they have used the publicly available dataset which is a cross-sectional study containing the DNAm patterns associated with glial and neuronal cell types in 58 post-mortem brain prefrontal cortex tissue samples collected from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders and the University of Maryland, Baltimore (Bustamante et al., 2018). Among the 58 (30 females, 28 males) tissue samples they have selected for their study, 29 were with MDD with an age group around 32.5 ± 15.9 years. They showed that 4 probes for Interleukin 1 Receptor Accessory Protein-Like 1 (IL1RAPL1) i.e., cg06927864, cg18230558, cg20350671, and cg26791231 has higher methylation in MDD cases compared to the controls. Limitations included the use of postmortem brain tissue and small sample size with a resulting reduced statistical power to detect meaningful differences between MDD cases and controls. In our present study, we overcome these pitfalls by selecting a larger sample size (N = 329) and by implementing stringent statistical procedures.
In contrast to Bustamante et al. study, Han et al. reported significantly higher epigenetic aging in patients with MDD compared to controls (Han et al., 2018). The study participants selected by Han et al. group were from the Netherlands Study of Depression and Anxiety (NESDA), which is an ongoing longitudinal multicenter cohort study designed to investigate the long-term course and consequences of depressive and anxiety related disorders (Han et al., 2018). Among the cohort samples of 1130 participants, they selected the samples with no lifetime psychiatric disorders and low depressive symptoms with a score <14 as controls (N = 319) and samples with a score ≥14 as MDD (N = 811) based on the Inventory of Depressive Symptomology with a follow up of 4 years (Han et al., 2018). The mean age of their selected sample controls was 41.6 years and MDD samples was 41.5 years (Han et al., 2018). Their results suggested that higher epigenetic aging in MDD may be driven largely by severity of illness (Han et al., 2018). They did not identify any additional relationships between higher epigenetic aging and cumulative clinical characteristics (Han et al., 2018). Our findings of a cross-sectional association between two epi-genetic clocks and lower positive affect was most robust among Whites. Despite that neither one of those two epigenetic clocks differed by race, we found that being White was associated with a reduction in positive affect by 0.28 SD compared to AAs, even after adjusting for age, sex, and poverty status (P = 0.007). Thus, White urban adults may be more affected by epigenetic age acceleration due to their reduced level of positive affect at baseline.
While our understanding of the pathophysiology of depression has been dominated by theories such as the monoamine hypothesis for decades, it is not without some significant limitations. In addition, hypothalamic–pituitary–adrenal (HPA) axis dysfunctions (Anacker et al., 2011), inflammation and neuroimmune processes (Miller) have also been linked to the pathophysiology of numerous mood disorders, including depression (Prins et al.). Inflammatory connection to depressive symptoms has been explained using nitrooxidative (NOS) mechanisms in one study by Luca et al. (insert citation). NOS stress in brain aging could be a result of: (a) oxidative DNA damage, primarily affecting mitochondrial DNA (mtDNA); (b) oxidation of polyunsaturated fatty acids leading to increased production of reactive oxygen species (ROS) and; (c) activation of microglia; also a source of free radicals-prolonged activation of which leads to oxidative damage and neuronal cell death. In short, increased systemic inflammation and impaired antioxidant defense mechanisms expose brain cells to increased oxidative stress, resulting in chronic physiological alterations underlying aging and depression (Luca et al., 2013). Recent studies have shifted the direction towards epigenetic mechanisms, particularly histone modification and DNAm, affecting depression in human subjects or depression-like symptoms in animal models (Massart et al.). A recent study showed that age-associated epigenetic upregulation of the FK506 binding protein 5 (FKBP5) may increase the risk for PTSD and MDD in mouse models (Sabbagh et al., 2014). They showed that the progressive FKBP5 demethylation occurs with age in wild-type mice thereby explaining the mechanism by which FKBP5 levels alter throughout the life. Their findings explicitly suggested that aging acts as an important epigenetic entity interacting at the early stage life events thereby making a person vulnerable to depression and other disorders (Sabbagh et al., 2014). Our study indicates that epigenetic aging using DNAm biomarkers is specifically linked to one aspect of depressive symptoms, namely positive affect, and was not associated with other domains of the CES-D. This reinforces the need for the RDoC approach as recommended by the National Institute on Mental Health, to examine biological markers in relation to continuous symptoms or groups of symptoms (e.g., domains) as opposed to classifying people based on diagnostic criteria that often produces highly heterogeneous cases of a mental condition (Katahira and Yamashita, 2017).
Social and environmental cues earlier in life moderate epigenetic programming and result in subsequent adaptive responses to changing landscapes. Any insult to the estimated trajectory will presumably result in progressive maladaptation and an increased risk of developing numerous diseases. Since DNA methylation is susceptible to environmental changes (Swanson et al.), it is not unusual to observe early environmental manipulation in mood-related disorders, as demonstrated by Meaney and Szyf in post-natal maternal interactions (Szyf). This is further supported by Weaver and colleagues, who showed that maternal behavior alters DNA methylation and chromatin structure in rats, suggesting long-term and reversible effects of maternal care in the offspring (Weaver et al.). Maternal depression in women with high burdens of depressive symptoms before pregnancy and antenatally were significantly associated with child’s lower epigenetic gestational age at birth, where lower epigenetic age was an indicator of higher mental adversities later in life (Suarez et al.). DNA methylation has been studied extensively in relation to the embryonic brain. In mammals, DNA methylation occurs predominantly at CpG islands and involves DNA methyltransferases (DNMTs) to carry out desired modifications (Babenko et al.). Loss of DNMT1 action in humans, for example, through specific mutations cause neurodegeneration in the form of hereditary sensory neuropathy with dementia and hearing loss (Babenko et al.).
Our study has several strengths. First, we used a longitudinal design to ascertain temporality of those relationships and stratifying by sociodemographics relevant to epigenetic age acceleration. In addition to using a well-validated scale of depressive symptomology, sub-domains were also investigated in order to separate somatic complaints from other domains such as depressed affect, positive affect and interpersonal problems. Those sub-domains had factorial invariance in national data (Nguyen et al., 2004). Our analyses used multivariable regression models such as mixed-effects linear regression that adjusted for sample selectivity and allowed us to use a more complete set of data while assuming missingness at random. Finally, we used a standard and readily available blood-based DNAm markers of epigenetic aging which can be replicated in future studies.
Nevertheless, some study limitations should be noted. First, although our models were adjusted for a wealth of potentially confounding covariates, causality cannot be inferred given the observational nature of the study and the possible role played by residual confounding. Notably, an adequate measure for anti-depressant use was not available at the time of this analysis, nor was an accurate measure of MDD history at visit 1. In fact, MDD history was not made consistently available in our study sample which used a proxy for elevated depressive symptoms (CES-D score >16) previously shown to be associated with MDD (Wada et al., 2007). Second, outcome measures were only repeated up to twice over an average follow-up of 5 years, our overall sample was of moderate size and while stratification by race was warranted, pooled analysis may introduce a bias in terms of population structure. This allows room for improvement in larger studies with 3 or more timepoints that could be carried out in the near future which would mirror true change in depressive symptomology as opposed to random fluctuation and would allow more adequate stratum-specific sample sizes that would detect smaller effects. Third, selective non-participation could bias the main associations of interest. However, this bias was minimized by using a 2-stage Heckman selection process that was applied to the multiple linear regression models. Fourth, exposure measurement can affect our conclusion given the multiplicity of potential techniques that can be used to assess DNAm, the wide range of possible tissues that can be targeted such as blood and brain tissue, and the difficult task to define a “normal” epigenetic profile (Mill and Petronis, 2007). In fact, level of blood DNA methylation may not necessarily reflect its level in the central nervous system, the target tissue of interest. Fifth, relationships between epigenetic age acceleration and depressive symptoms can be bi-directional. Given the current lack of follow-up data on epigenetic age acceleration, this hypothesis can be tested in a comparable future study. Sixth, our findings with positive affect may be due to chance and the standardized association implies a weak to moderate effect detected only among Whites. Finally, while the CES-D reliably measures depressive symptoms and acts as an important screening tool, it faces important limitations as a diagnostic test for major depressive disorder (Carleton et al., 2013).
In our study, EAA and AgeAccel, two measures of epigenetic age acceleration relying on the Horvath algorithm, were linked to a reduced level of “positive affect” in the complete sample and among Whites. Further longitudinal studies are needed to replicate our findings, while uncovering potential bi-directional relationships and future mechanistic studies are required to determine the specific pathways behind this association.
Supplementary Material
Acknowledgment
This work was fully supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging, NIA/NIH/IRP.
Sources of funding
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations:
- AA
African Americans
- AgeAccel
age acceleration
- BMI
body mass index
- CES-Dtotal
Center for Epidemiologic Studies Depression total score
- CESD
Center for Epidemiologic Studies Depression
- CHD
coronary heart disease
- DNAm
DNA methylation
- DNMTs
DNA methyltransferases
- EDS
elevated depressive symptoms
- EAA
epigenetic age acceleration
- EEAA
extrinsic epigenetic age acceleration
- FKBP5
FK506 binding protein 5
- (HANDLS) study
Healthy Aging in Neighborhoods of Diversity across the Life Span
- (HEI-2010) total score
Healthy Eating Index
- HS
High School
- HIV
Human Immunodeficiency Virus
- IL1RAPL1
Interleukin 1 Receptor Accessory Protein-Like 1
- IEAA
intrinsic epigenetic age acceleration
- MDD
major depressive disorder
- NESDA
Netherlands Study of Depression and Anxiety
- NSAIDs
non-steroidal anti-inflammatory drugs
- n-3 PUFA
Omega-3 polyunsaturated fatty acids
- n-6 PUFA
Omega-6 polyunsaturated fatty acids
- PBMC
Peripheral blood mononuclear cells
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The views expressed in this article are those of the author(s) and do not reflect the official policy of the Department of the Army/Navy/Air Force, Department of Defense, or the U.S. Government.
Conflict of interest
All authors declare no conflict of interest.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jad.2019.06.032.
References
- Anacker C, Zunszain PA, Carvalho LA, Pariante CM, 2011. The glucocorticoid receptor: pivot of depression and of antidepressant treatment? Psychoneuroendocrinology 36 (3), 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MK, Chen E, Ross KM, McEwen LM, Maclsaac JL, Kobor MS, Miller GE, 2018. Early-life socioeconomic disadvantage, not current, predicts accelerated epigenetic aging of monocytes. Psychoneuroendocrinology 97, 131–134. [DOI] [PubMed] [Google Scholar]
- Babenko O, Kovalchuk I, Metz GA, Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health [DOI] [PubMed]
- Beekman AT, Deeg DJ, Van Limbeek J, Braam AW, De Vries MZ, Van Tilburg W, 1997. Criterion validity of the Center for Epidemiologic Studies Depression Scale (CES-D): results from a community-based sample of older subjects in the Netherlands. Psychol. Med 27, 231–235. [DOI] [PubMed] [Google Scholar]
- Bettcher BM, Wilheim R, Rigby T, Green R, Miller JW, Racine CA, Yaffe K, Miller BL, Kramer JH, 2012. C-reactive protein is related to memory and medial temporal brain volume in older adults. Brain Behav. Immun 26, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beydoun MA, Beydoun HA, Dore GA, Fanelli-Kuczmarski MT, Evans MK, Zonderman AB, 2015a. Total serum cholesterol, atherogenic indices and their longitudinal association with depressive symptoms among US adults. Transl. Psychiatry 5, e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beydoun MA, Beydoun HA, Kitner-Triolo MH, Kaufman JS, Evans MK,Zonderman AB, 2013. Thyroid hormones are associated with cognitive function: moderation by sex, race, and depressive symptoms. J. Clin. Endocrinol. Metab 98, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beydoun MA, Fanelli-Kuczmarski MT, Shaked D, Dore GA, Beydoun HA, Rostant OS, Evans MK, Zonderman AB, 2016. Alternative pathway analyses indicate bidirectional relations between depressive symptoms, diet quality, and central adiposity in a sample of urban US adults. J. Nutr 146, 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beydoun MA, Fanelli Kuczmarski MT, Beydoun HA, Rostant OS, Evans MK, Zonderman AB, 2015b. Associations of the ratios of n-3 to n-6 dietary fatty acids with longitudinal changes in depressive symptoms among US women. Am. J. Epidemiol 181, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E, de Leon CF, Miller GE, 2006. Applying mixed regression models to the analysis of repeated-measures data in psychosomatic medicine. Psychosom. Med 68, 870–878. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Saum KU, Perna L, Schottker B, Holleczek B, Brenner H, 2016. Frailty is associated with the epigenetic clock but not with telomere length in a german cohort. Clin. Epigenetics 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau UC, 2004. US Census Bureau, social, economic, and housing statistics division[Accessed January, 2016] Poverty Thresholds, 2004. 2014.
- Bustamante AC, Armstrong DL, Uddin M, 2018. Epigenetic profiles associated with major depression in the human brain. Psychiatry Res 260, 439–442. [DOI] [PubMed] [Google Scholar]
- Carleton RN, Thibodeau MA, Teale MJ, Welch PG, Abrams MP, Robinson T, Asmundson GJ, 2013. The center for epidemiologic studies depression scale: a review with a theoretical and empirical examination of item content and factor structure. PLOS One 8, e58067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S, 2016. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 8, 1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang S, Pan X, Hu X, Zhang YH, Yuan F, Huang T, Cai YD, 2019. HIV infection alters the human epigenetic landscape. Gene Ther 26 (1–2), 29–39. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Pishva E, Haapakoski R, Zsoldos E, Mahmood A, Filippini N,Burrage J, Mill J, Kivimaki M, Lunnon K, Ebmeier KP, 2018. Peripheral DNA methylation, cognitive decline and brain aging: pilot findings from the Whitehall II imaging study. Epigenomics 10, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D, Wilker S, Schneider A, Karabatsiakis A, Pfeiffer A, Kolassa S, Freytag V, Vukojevic V, Vogler C, Milnik A, Papassotiropoulos A, de Quervain JFD, Elbert T, Kolassa IT, 2018. Integrated genetic, epigenetic, and gene set enrichment analyses identify NOTCH as a potential mediator for PTSD risk after trauma: results from two independent African cohorts. Psychophysiology e13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MK, Lepkowski JM, Powe NR, LaVeist T, Kuczmarski MF, Zonderman AB, 2010. Healthy aging in neighborhoods of diversity across the life span (HANDLS): overcoming barriers to implementing a longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethn. Dis 20, 267–275. [PMC free article] [PubMed] [Google Scholar]
- Gimeno D, Kivimaki M, Brunner EJ, Elovainio M, De Vogli R, Steptoe A, Kumari M, Lowe GD, Rumley A, Marmot MG, Ferrie JE, 2009. Associations of C-reactive protein and interleukin-6 with cognitive symptoms of depression: 12-year follow-up of the Whitehall II study. Psychol. Med 39, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, Zhao M, Kumar G, Xie LY, Jansen R, Milaneschi Y, Dean B, Aberg KA, van den Oord E, Penninx B, 2018. Epigenetic aging in major depressive disorder. Am. J. Psychiatry 175, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K, 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Tamhane AC, 1987. Multiple comparison procedures Wiley, New York. [Google Scholar]
- Horvath S, 2013. DNA methylation age of human tissues and cell types. Genome Biol 14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, Jamieson BD, Sun D, Li S, Chen W, Quintana-Murci L, Fagny M, Kobor MS, Tsao PS, Reiner AP, Edlefsen KL, Absher D, Assimes TL, 2016. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 17, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira K, Yamashita Y, 2017. A theoretical framework for evaluating psychiatric research strategies. Comput. Psychiatry 1, 184–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S, 2015. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and alzheimer’s disease related cognitive functioning. Aging (Albany NY) 7, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He Y, Ma X, Chen X, 2018. Epigenetic age analysis of brain in major depressive disorder. Psychiatry Res 269, 621–624. [DOI] [PubMed] [Google Scholar]
- Luca M, Luca A, Calandra C, 2013. Accelerated aging in major depression: the role of nitro-oxidative stress. Oxid. Med. Cell. Longev 2013, 230797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maierhofer A, Flunkert J, Oshima J, Martin GM, Haaf T, Horvath S, 2017. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY) 9, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic J, Gordon L, Oshlack A, 2012. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 beadchips. Genome Biol 13, R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, McRae AF, Bressler J, Colicino E, Hannon E, Li S, Prada D, Smith JA, Trevisi L, Tsai PC, Vojinovic D, Simino J, Levy D, Liu C, Mendelson M, Satizabal CL, Yang Q, Jhun MA, Kardia SLR, Zhao W, Bandinelli S, Ferrucci L, Hernandez DG, Singleton AB, Harris SE, Starr JM, Kiel DP, McLean RR, Just AC, Schwartz J, Spiro A 3rd, Vokonas P, Amin N, Ikram MA, Uitterlinden AG, van Meurs JBJ, Spector TD, Steves C, Baccarelli AA, Bell JT, van Duijn CM, Fornage M, Hsu YH, Mill J, Mosley TH, Seshadri S, Deary IJ, 2018. Meta-analysis of epigenome-wide association studies of cognitive abilities. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ, 2015a. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 16, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, Corley J, Taylor A, Murphy L, Starr JM, Horvath S, Visscher PM, Wray NR, Deary IJ, 2015b. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth cohort 1936. Int. J. Epidemiol 44, 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Mongeau R Lanfumey L, Beyond the monoaminergic hypothesis: neuroplasticity and epigenetic changes in a transgenic mouse model of depression [DOI] [PMC free article] [PubMed]
- McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A, Murray AD, Whalley HC, Porteous DJ, McIntosh AM, Evans KL, Deary IJ, Marioni RE, 2018. Investigating the relationship between DNA methylation age acceleration and risk factors for alzheimer’s disease. Alzheimers Dement. (Amst) 10, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Bruenig D, Carrillo-Roa T, Lawford B, Harvey W, Morris CP, Smith AK, Binder EB, Young RM, Voisey J, 2017. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta Psychiatr. Scand 136, 493–505. [DOI] [PubMed] [Google Scholar]
- Mill J, Petronis A, 2007. Molecular studies of major depressive disorder: the epigenetic perspective. Mol. Psychiatry 12, 799–814. [DOI] [PubMed] [Google Scholar]
- Miller AH, Depression and immunity: a role for T cells? [DOI] [PMC free article] [PubMed]
- Nestler EJ, 2014. Epigenetic mechanisms of depression. JAMA Psychiatry 71, 454–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Kitner-Triolo M, Evans MK, Zonderman AB, 2004. Factorial invariance of the CES-D in low socioeconomic status African Americans compared with a nationally representative sample. Psychiatry Res 126, 177–187. [DOI] [PubMed] [Google Scholar]
- Parade SH, Novick AM, Parent J, Seifer R, Klaver SJ, Marsit CJ, Gobin AP, Yang BZ, Tyrka AR, 2017. Stress exposure and psychopathology alter methylation of the serotonin receptor 2A (HTR2A) gene in preschoolers. Dev. Psychopathol 29, 1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H, 2016. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a german case cohort. Clin. Epigenetics 8, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishva E, Rutten BPF, van den Hove D, 2017. DNA methylation in major depressive disorder. Adv. Exp. Med. Biol 978, 185–196. [DOI] [PubMed] [Google Scholar]
- Prins J, Olivier B Korte SM, Triple reuptake inhibitors for treating subtypes of major depressive disorder: the monoamine hypothesis revisited [DOI] [PubMed]
- Radloff L, 1977. The CES-D scale: a self-report depression scale for research in the general population. Appl. Psychol. Meas 1 (3), 385–401. [Google Scholar]
- Rosen AD, Robertson KD, Hlady RA, Muench C, Lee J, Philibert R, Horvath S, Kaminsky ZA, Lohoff FW, 2018. DNA methylation age is accelerated in alcohol dependence. Transl. Psychiatry 8, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra K, Molina-Marquez AM, Saavedra N, Zambrano T, Salazar LA, 2016. Epigenetic modifications of major depressive disorder. Int. J. Mol. Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh JJ, O’Leary JC 3rd, Blair LJ, Klengel T, Nordhues BA, Fontaine SN, Binder EB, Dickey CA, 2014. Age-associated epigenetic upregulation of the FKBP5 gene selectively impairs stress resiliency. PLoS ONE 9, e107241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin S, 2004. Statistical Analysis of Epidemiologic Data, 3rd ed Oxford UniversityPress. [Google Scholar]
- Smith K, 2014. Mental health: a world of depression. Nature 515, 181. [DOI] [PubMed] [Google Scholar]
- Starnawska A, Tan Q, Lenart A, McGue M, Mors O, Borglum AD, Christensen K, Nyegaard M, Christiansen L, 2017. Blood DNA methylation age is not associated with cognitive functioning in middle-aged monozygotic twins. Neurobiol. Aging 50, 60–63. [DOI] [PubMed] [Google Scholar]
- STATA, 2017. Statistics/Data Analysis: Release 15.0 Stata Corporation, Texas. [Google Scholar]
- Suarez A, Lahti J, Czamara D, Lahti-Pulkkinen M, Knight AK, Girchenko P,Hamalainen E, Kajantie E, Lipsanen J, Laivuori H, Villa PM, Reynolds RM, Smith AK, Binder EB, Raikkonen K, The epigenetic clock at birth: associations with maternal antenatal depression and child psychiatric problems [DOI] [PMC free article] [PubMed]
- Swanson JM, Entringer S, Buss C, Wadhwa PD, Developmental origins of health and disease: environmental exposures [DOI] [PMC free article] [PubMed]
- Szyf M, The early life environment and the epigenome [DOI] [PubMed]
- Touleimat N, Tost J, 2012. Complete pipeline for Infinium((R)) human methylation 450 K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 4, 325–341. [DOI] [PubMed] [Google Scholar]
- Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD, 2013. Low-level processing of illumina infinium DNA methylation beadarrays. Nucleic Acids. Res 41, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Yamagata H, Seki T, Watanabe Y, 2018. Epigenetic mechanisms of major depression: targeting neuronal plasticity. Psychiatry Clin. Neurosci 72, 212–227. [DOI] [PubMed] [Google Scholar]
- Verhoeven JE, Yang R, Wolkowitz OM, Bersani FS, Lindqvist D, Mellon SH, Yehuda R, Flory JD, Lin J, Abu-Amara D, Makotkine I, Marmar C, Jett M, Hammamieh R, 2018. Epigenetic age in male combat-exposed war Veterans: associations with posttraumatic stress disorder status. Mol. Neuropsychiatry 4, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Tanaka K, Theriault G, Satoh T, Mimura M, Miyaoka H, Aizawa Y, 2007. Validity of the center for epidemiologic studies depression scale as a screening instrument of major depressive disorder among japanese workers. Am. J. Ind. Med 50, 8–12. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ, Epigenetic programming by maternal behavior [DOI] [PubMed]
- West AE, Greenberg ME, 2011. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold. Spring Harb. Perspect. Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CC, Yang HS, Yu L, Chibnik LB, Dawe RJ, Yang J, Klein HU, Felsky D, Ramos-Miguel A, Arfanakis K, Honer WG, Sperling RA, Schneider JA, Bennett DA, De Jager PL, 2017. Identification of genes associated with dissociation of cognitive performance and neuropathological burden: multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med 14, e1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet WC, 1998. Nutritional Epidemiology, 2nd ed Oxford University Press, New York. [Google Scholar]
- Wolf EJ, Logue MW, Morrison FG, Wilcox ES, Stone A, Schichman SA, McGlinchey RE, Milberg WP, Miller MW, 2019. Posttraumatic psychopathology and the pace of the epigenetic clock: a longitudinal investigation. Psychol. Med 49(5), 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.