Abstract
Background
Female Aedes aegypti mosquitoes are vectors of arboviruses that cause diverse diseases of public health significance. Blood protein digestion by midgut proteases provides anautogenous mosquitoes with the nutrients essential for oocyte maturation and egg production. Midgut-specific miR-1174 affects the functions of the midgut through its target gene serine hydroxymethyltransferase (SHMT). However, less is known about SHMT-regulated processes in blood digestion by mosquitoes.
Methods
RNAi of SHMT was realized by injection of the double-stranded RNA at 16 h post-eclosion. The expression of SHMT at mRNA level and protein level was assayed by real-time PCR and Western blotting, respectively. Statistical analyses were performed with GraphPad7 using Student’s t-test.
Results
Here, we confirmed that digestion of blood was inhibited in SHMT RNAi-silenced female A. aegypti mosquitoes. Evidence is also presented that all SHMT-depleted female mosquitoes lost their flight ability and died within 48 h of a blood meal. Furthermore, most examined digestive enzymes responded differently in their transcriptional expression to RNAi depletion of SHMT, with some downregulated, some upregulated and some remaining stable. Phylogenetic analysis showed that transcriptional expression responses to SHMT silence were largely unrelated to the sequence similarity between these enzymes.
Conclusions
Overall, this research shows that SHMT was expressed at a low level in the midgut of Aedes aegypti mosquitoes, but blood-meal digestion was inhibited when SHMT was silenced. Transcriptional expressions of different digestive enzymes were affected in response to SHMT depletion, suggesting that SHMT is required for the blood-meal digestion in the midgut and targeting SHMT could provide an effective strategy for vector mosquito population control.
Keywords: Aedes aegypti, Serine hydroxymethyltransferase, Digestive enzyme, Midgut, Blood meal
Background
Female mosquitoes must take a blood meal from vertebrate hosts, including reptiles, birds and mammals, to obtain the proper nutrients for completion of the gonotrophic cycle [1, 2]. The blood-feeding behavior of the mosquito Aedes aegypti facilitates the transmission of the most prevalent arboviruses such as the yellow fever virus, dengue virus, chikungunya virus and Zika virus [3–9]. Blood-feeding in mosquitoes induces the production and secretion of many digestive enzymes in the midgut, causing decomposition of blood proteins into peptides and amino acids which serve as essential nutrition sources for vitellogenin biosynthesis and egg development [2, 10–12]. The major classes of digestive enzymes in blood-fed A. aegypti female mosquitoes are trypsins, chymotrypsins, aminopeptidases and carboxypeptidases [13, 14]. Over 300 serine protease-like genes were predicted in the A. aegypti genome, but only a few serine protease-like trypsins and chymotrypsins have been experimentally confirmed in the midgut [15, 16]. Chymotrypsin is induced by a blood meal in A. aegypti, and its protein levels and enzymatic activity remains high during protein digestion [17], but its role during blood-meal digestion remains undetermined. A chymotrypsin-like protease gene, JHA15, is transcriptionally activated by juvenile hormone (JH) in the newly emerged female adults but its silencing resulted in no clear phenotype in blood-meal digestion [18]. It is known that the digestion of blood in the midgut of A. aegypti mosquitoes can be divided into two phases, early phase one to three hours post-blood-meal (h PBM) and late phase 8–36 h PBM [19]. During the early phase of digestion, the Trypsin 3A1 Precursor AaET (AAEL007818) and female-specific chymotrypsin AaCHYMO (AAEL003060) accumulate [11, 17, 18], but Trypsin-like serine protease AaLT (AAEL013284) and Trypsin 5G1 Precursor Aa5G1 (AAEL013712) begin to be translated by 6–8 h PBM and reach maximal concentrations 24–36 h PBM [20, 21].
Previous work has shown that blood digestion is inhibited when serine hydroxymethyltransferase (SHMT) is depleted [22]. However, it remains undetermined whether and how SHMT regulates the digestive enzymes in the midgut of mosquitoes. As one evolutionarily conserved metabolic enzyme, SHMT catalyzes the reversible conversion of L-serine and tetrahydrofolate into glycine and 5,10-methylenetetrahydrofolate [23, 24]. The one-carbon units are raw materials for the synthesis of methylated DNA, and inhibition of SHMT activity blocks the biosynthesis of pyrimidine and purine, so SHMT is also an enzyme that links the metabolism of amino acids and nucleotides [25, 26]. SHMT serves as a scaffold protein essential for metabolic complex formation at sites of DNA replication initiation and thus may be even more important determinant of de novo thymidylate biosynthesis capacity than its catalytic activity [27]. In humans, SHMT exists in two isozymes in different cellular compartments, commonly termed cytosolic SHMT1 and mitochondrial SHMT2 [28, 29]. SHMT1 preferentially catalyzes serine synthesis, while SHMT2 participates in serine decomposition, thus forming a cyclic one-carbon unit flux between the cytosol and mitochondria [30, 31]. Although SHMT has been extensively studied in mammals and some other species, its role in insects is rarely mentioned. Here, our data suggest that SHMT controls the blood digestion via affecting transcriptional expression levels of diverse digestive enzymes in the midgut of mosquitoes.
Methods
Insect breeding and sample collection
The A. aegypti (NIH Rockefeller strain) colony was reared at 28 °C and 75% relative humidity under a 12 h light: 12 h dark photoperiod regime. Adult mosquitoes were provided a cotton pad soaked in water (0%) and a pad soaked in 10% sucrose solution before and after a blood meal. The adults were fed rat blood after 3 days of adult eclosion. The blood pellets were removed from the mosquitoes by pinching with forceps under a stereomicroscope before collecting samples. The whole-mount individuals of female A. aegypti were collected at 13 time points, namely 6, 12, 24, 36, 48 and 72 h post-eclosion (PE) and 1, 6, 12, 24, 36, 48 and 72 h PBM. Different tissues of adult female mosquitoes were collected before or after a blood meal, including the head (HD), midgut (MG), fat body (FB), ovary (OV) and leftover (LO). All samples were frozen with liquid nitrogen and were separately kept in 300 μl Trizol (for RNA extraction) and stored at – 80 °C.
Databases and phylogenetic analysis
All sequences of SHMT and digestive enzymes were downloaded from VectorBase at https://www.vectorbase.org/. Multi-sequence alignment and phylogenetic analysis were conducted in MEGA X with maximum likelihood method [32].
RNA extraction, cDNA synthesis and prokaryotic expression
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s manual. The first strand of cDNA was synthesized by PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan) using 1 μg of total RNA following the manufacturer’s manual. The protein coding region (CDS) of SHMT was amplified by PCR using PrimerSTAR Max DNA Polymerase (Takara Bio) with the following PCR program: 98 °C for 5 min; 30 cycles of 98 °C for 10 s, 60 °C for 20 s and 72 °C for 10 s; then 72 °C for 10 min. The primers were designed with Primer Premier 5 and synthesized at Sangon Biotech (Shanghai, China) (Additional file 1: Table S1). The PCR products were purified with an E.Z.N.A.Gel Extraction Kit (Omega Bio-tek, Norcross, GA, USA) following the manufacturer’s manual. After double digestion with restriction endonucleases SacI and NotI, the PCR products were cloned into a pET-28a His-tagged vector. The recombinant plasmids harboring the cloned SHMT CDS were then transformed into Escherichia coli BL21 (DE3) competent cells (Transgen Biotech, Beijing, China). The transformed recombinant strain E. coli BL21 (DE3) was inoculated into LB kanamycin-medium and grown for 2 h at 37 °C in a rotary shaker at 220× rpm until OD600 was 0.6. Then, prokaryotic expression was induced with 0.1 mmol/l of IPTG at 25 °C for 10 h.
Purification of recombinant protein and preparation of polyclonal antibody
The host E. coli BL21 (DE3) strains above were harvested by centrifugation at 4 °C and 12,000×g for 15 min, and were then suspended in binding buffer (20 mM Tris-HCl, 200 mM NaCl, pH 8.0). In order for the cells to rupture sufficiently, the suspended E. coli BL21(DE3) strains were repeatedly frozen and thawed in liquid nitrogen 3 times, and were then subjected to intermittent treatment with the ultrasonic cell-break method for 40 min. The recombinant protein SHMT-PA existed in the precipitation, while SHMT-PB was in the supernatant of ultrasound-treated bacterial solution. SHMT-PA precipitation and SHMP-PB were separately collected by centrifugation at 4 °C and 12,000×g, and SHMT-PA was then denatured in binding buffer (8 M Urea, 20 mM Tris-HCl, 200 mM NaCl, pH 8.0). Both the SHMT-PB supernatant and denatured SHMT-PA were separately filtered using a 0.45 μm filter membrane. The filtrate was slowly added to Ni ion affinity chromatography column (GE Healthcare, Stockholm, Sweden) with a flow rate of about 4 s/drop. The unbound proteins were rinsed with 40–50 ml of binding buffer (20 mM Tris-HCl, 200 mM NaCl, pH 8.0) with a flow rate of 3–4 s/drop. The His-tag recombinant proteins SHMT-PA and SHMT-PB were then eluted with 100 mM imidazole and 250 mM imidazole, respectively, followed by concentrating through centrifugation at 4 °C and 3,000×g in 10 kDa Millipore Amicon Ultra (Merck, Darmstadt, Germany). After purification by Ni-affinity chromatography, SHMT-PB was further purified by AKTA Purifier 100 (GE Healthcare), while SHMT-PA was refolded with gradient PBS buffer (6 M, 4 M, 2 M, 0 M Urea, pH 8.0) at 16 °C. The purified proteins were sent to Wuhan GeneCreate Biological Engineering Co., Ltd (Wuhan, China) to prepare the polyclonal antibodies.
Double-stranded RNA (dsRNA) synthesis and RNAi knockdown
After analyzing the functional domains of the SHMT with the online software SMART at http://smart.embl-heidelberg.de/, the primers used for SHMT RNAi were designed by Primer Premier 5 based on the sequence coding Pfam domain. The protein coding sequences of digestive enzymes were downloaded from VectorBase (https://www.vectorbase.org/organisms/aedes-aegypti) to design the primers for the synthesis of dsRNAs. All primers for RNAi are shown in Additional file 1: Table S1. The dsRNAs for SHMT and digestive enzyme genes were synthesized using MEGAscript RNAi Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s manual. To silence SHMT and digestive enzyme genes, mosquitoes at 16 h post-eclosion (PE) were injected with 500 ng of dsRNA of SHMT or digestive enzyme genes in 0.5 μl of nuclease-free water. To reveal the dose responses of mosquitoes, we injected different doses of dsSHMT, including 50, 100, 200, 400 and 800 ng. Transcript abundance was analyzed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), as described below. All dsRNA-injected mosquitoes were maintained on 10% (wt/vol) sucrose solution for 72 h and then a blood meal was given. The dsEGFP-injected mosquitoes served as a control. The whole body or tissues of mosquitoes were collected at expected time points for RNA extraction.
qRT-PCR analysis
The mRNA levels of SHMT and digestive enzyme genes were examined by means of qRT-PCR. All primers for qRT-PCR assay were designed with Primer Premier 5 and then synthesized at Sangon Biotech (Shanghai, China) (Additional file 1: Table S1). The cDNA was synthesized with 1μg of total RNA for each sample with Prime Script™ RT reagent kit with gDNA Eraser (Takara Bio) according to the manufacturer’s protocol, and was used as a template for qRT-PCR with SYBR® Premix Ex Taq™ II (Takara Bio). The qRT-PCR was performed in a 96-well plate on a real time PCR system, qTOWER2.0 (Analytik Jena, Jena, Germany). The ribosomal protein S7, which is stably expressed in mosquitoes before or after a blood meal [33], served as the baseline control. Unless otherwise stated, all qRT-PCR experiments in this study were performed in triplicate samples with at least three replicates. The relative mRNA expression levels were calculated by using the 2−ΔΔCT method [34].
Protein extraction and Western blotting analysis
Six female mosquitoes were collected as a whole for the total protein extraction of each sample. The total protein of each sample was prepared in NP-40 Lysis Buffer P0013 (Beyotime, Jiangsu, China). These six mosquitoes were fully ground in 100 μl of lysis buffer and were then incubated in an ice bath for 1 h. After centrifugation at 4 °C, 12,000×g for 15 min, the protein in the supernatant was then quantified with a BCA kit P0009 (Beyotime). Next, 50 ng of protein for each sample in 5× SDS loading buffer was denatured at 100 °C in a metal bath for 10 min, and was then loaded onto each gel well, followed by separation using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the proteins on the gel were transferred to the nitrocellulose/PVDF membrane (Roche, Basel, Switzerland) by a semi-dry membrane transfer instrument, Trans-Blot Turbo (Bio-Rad, Hercules, CA, USA). Membranes were blocked with 5% skimmed milk in 20 mM Tris-HCl containing 150 mM NaCl and 0.05% Tween 20 (TBST), then incubated with primary antibodies (1:10,000) against SHMT at room temperature for 2 h. After thoroughly washing with TBST (at least five times), horseradish peroxidase-conjugated secondary antibodies (Beyotime;1:20,000 dilution in TBST) was applied in incubation at room temperature for 1 h. After thoroughly washing with TBST (at least five times), the fluorescence signals were detected using an enhanced chemiluminescence detection kit (Thermo Fisher Scientific), and the photographs were obtained by a Chemiluminescence Imaging System (Clinx Science Instruments, Shanghai, China). The control Anti-TUBULIN (1:20,000) was purchased from Beyotime and the primary rabbit polyclonal anti-SHMT antibody was prepared by Wuhan Gene Create Biological Engineering Co. Ltd. (Wuhan, China).
Statistical analysis
Statistical analyses were performed using the GraphPad Prism v.7.0 (GraphPad Software, San Diego, CA, USA). The data are presented as means ± SEM of at least three independent samples. Statistical comparisons between groups were performed using one-way ANOVA followed by unpaired Student’s t-test. Differences of P < 0.05 were considered statistically significant, and differences of P < 0.01 were considered highly statistically significant.
Results
Sequence characteristics and expression patterns of A. aegypti serine hydroxymethyltransferase
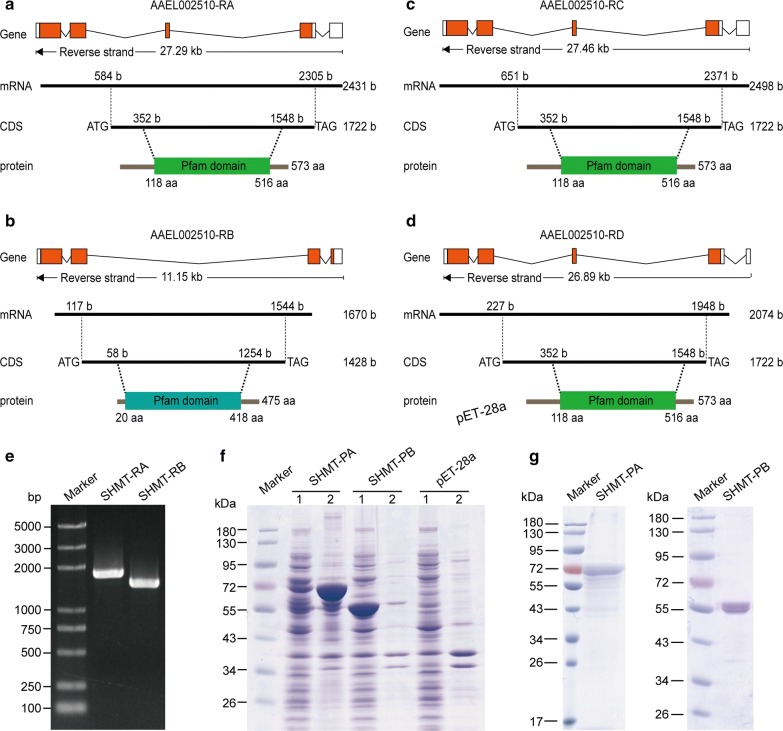
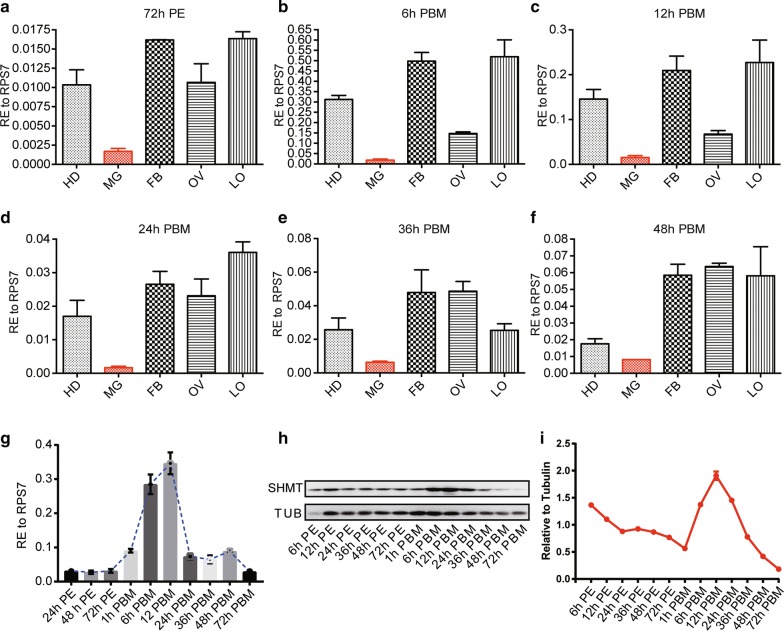
In the latest genome assembly for A. aegypti (AaegL5), SHMT is located on Chromosome 1:90, 394, 257-90, 421, 721, and has four alternative splicing forms (transcripts), namely AAEL002510-RA, AAEL002510-RB, AAEL002510-RC and AAEL002510-RD, but encodes two isoforms of protein with 573 aa and 475 aa (Fig. 1a–d). The short protein translated from AAEL002510-RB lacks 99 amino acids at the N-end and has seven amino acids different from the longer one encoded by the other three transcripts. The two kinds of protein-coding regions (CDS) were cloned into prokaryotic expression vector and expressed in bacteria (Fig. 1e–g), followed by purifying the proteins to prepare antibodies. We performed qRT-PCR to reveal the transcriptional expression patterns of SHMT in diverse tissues of female adults at different time points and found that it was highly expressed in the head, fat body, ovary and leftover tissues but expressed at very low levels in the midgut (Fig. 2a–f); the latter is consistent with our previous evidence [22]. Next, we confirmed that it was expressed at very low levels in the whole body of female mosquitoes before a blood meal (BBM) and quickly increased after a blood meal and even peaked at 12 h post-blood-meal (PBM), but swiftly decreased at 24 h PBM (Fig. 2g). We further examined its temporal expression at the protein level in the whole body (Fig. 2h). Quantified result of the Western blot showed that it was highly expressed at 6 h PE and decreased to basal levels at 1 h PBM, and then quickly increased to the peak level at 12 h PBM (Fig. 2i). Overall, SHMT was highly expressed at the early PE phase, then remained at low expression levels throughout the late PE phase but increased rapidly in both transcript and protein levels once the mosquitoes took a blood meal.
Fig. 1.
Gene structures, isoforms and prokaryotic expression patterns of A. aeSHMT. All sequences were downloaded from VectorBase at https://www.vectorbase.org/organisms/aedes-aegypti. a–d Schematic diagrams of isoforms A, B, C and D. The square box, black line and red box in the gene structure above represent the exon (mRNA), intron and protein coding region (CDS), respectively. These four SHMT isoforms correspond to four different lengths of mRNA and two different lengths of CDS or protein. The Pfam domain was generated by online software Smart at http://smart.embl-heidelberg.de/smart. e Cloning of the CDS for SHMT-RA and SHMT-RB. Two different lengths of CDS are shown in the 1% agarose gel as PCR products of SHMT-RA and SHMT-RB. f SDS-PAGE showing the SHMT proteins produced by prokaryotic expression. Lanes 1: detection of the supernatant; Lanes 2: detection of the precipitate. g SDS-PAGE showing the purified proteins used for the preparation of primary antibody. SHMT-PA was purified only through nickel affinity chromatography, while SHMT-PB was purified through nickel affinity chromatography and by AKTA Purifier 100 (GE Healthcare) (see “Methods”)
Fig. 2.
Expression profiles of SHMT mRNA and protein in A. aegypti. a–f mRNA levels of SHMT in diverse tissues at the following time points: a 72 h post-eclosion (PE); b 6 h post-blood-meal (PBM); c 12 h PBM; d 24 h PBM; e 36 h PBM; f 72 h PBM. g mRNA levels of SHMT in the whole body of adult females at diverse time points. h Western blotting analysis of temporal expression of SHMT in the whole body of adult females. i SHMT protein levels in h were quantified following normalization to TUBULIN. Nuclear RNA RPS7 was used as a loading control in all qRT-PCR assays (a–g). The qRT-PCR data are presented as mean ± SEM. TUBLIN was used as the loading control in western blot. Abbreviations: RE, relative expression; HD, head; MG, midgut; FB, fat body; OV, ovary; LO, leftover of the whole body
Knockdown of SHMT inhibits blood digestion and thus affects ovary development and oviposition
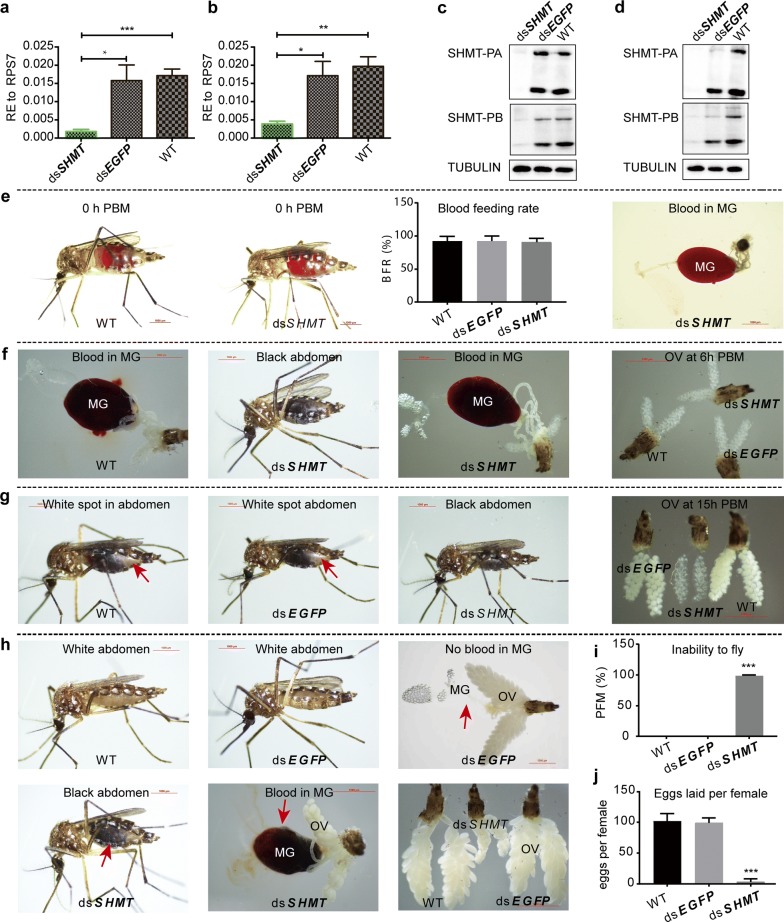
To reveal the function of SHMT, we injected 500 ng of dsSHMT in 0.5 μl of nuclease-free water into female adult mosquitoes at about 16 h PE. A blood meal was given at 72 h post-injection (h PIJ). The whole-mount individuals and tissues were collected before blood meal about 72 h PE (56 h PIJ) and 24 h PBM for assay by qRT-PCR and Western blot. The results showed that SHMT was remarkably decreased at both time points in the mRNA level (Fig. 3a, b) and protein level (Fig. 3c, d). Unlike miR-1174-depleted mosquitoes [22], SHMT-silenced mosquitoes displayed no abnormal phenotypes in blood-feeding, and the blood entered the midguts normally (Fig. 3e). At 6 h PBM, the blood in the midguts of SHMT-depleted mosquitoes normally turned dark brown as normal, and no obvious abnormal phenotypes could be observed in their ovaries (Fig. 3f). However, abnormal phenotypes were evident at about 15 h PBM (Fig. 3g). For example, the dorsal abdomens of the wild-type and dsEGFP controls showed a white spot (red arrow) due to the partial digestion of blood and normal growth of ovaries, whereas the abdomens of the SHMT-depleted mosquito were completely black, and the ovaries were about one half the size of the controls. At 48 h PBM, none of the wild-type (WT) or dsEGFP controls had blood left in the midguts (red arrow), their abdomens became white in appearance, and their ovaries were growing normally. By contrast, the SHMT-depleted mosquitoes still had a majority of undigested blood remaining, and the ovaries were not as developed nor were there as many as in the WT and the dsEGFP samples (Fig. 3h). More interestedly, all SHMT-depleted mosquitoes were not able to fly within 48 h PBM (Fig. 3i), and almost none of them laid eggs even 4 days after a blood meal (Fig. 3j). The knockdown efficiency of dsSHMT was confirmed by qRT-PCR (Fig. 3a, b), showing that 500 ng of dsSHMT could significantly silence this gene in the female mosquitoes. Furthermore, different dosages of dsSHMT (50, 100, 200, 400 and 800 ng) in 0.5 μl of nuclease-free water each were separately injected into female mosquitoes to compare the flightless time and oviposition (Additional file 2: Figure S1). When each mosquito was injected with 50 ng of dsSHMT, all of them could normally fly at 15 h PBM, 10% of them were not able to fly at 24 h PBM and nearly 40% of them were incapable of flying at 40 h PBM; when the injection dosage was increased to 800 ng for each female, about 10% of mosquitoes could not fly within 15 h PBM, and none of them could fly at 40 h PBM (Additional file 2: Figure S1a). The efficiency of different dosages of dsSHMT on oviposition was also obvious, i.e. the higher the dose of dsSHMT injected, the fewer eggs were laid (Additional file 2: Figure S1b).
Fig. 3.
Abnormal phenotypes caused by SHMT depletion. a qRT-PCR results showing RNAi of SHMT at 72 h PE. b qRT-PCR results showing RNAi of SHMT at 24 h PBM. c Western blotting analysis of SHMT RNAi at 72 h PE. d Western blotting analysis of SHMT RNAi at 24 h PBM. e RNAi of SHMT exerted no effect on blood-feeding. Blood meal was given at 3 d post-injection (PIJ) The wild-type, dsEGFP-injected and dsSHMT-injected mosquitoes normally sucked the blood which directly entered the midgut. f Phenotypes observed at 6 h PBM. The mosquitoes were dissected at 6 h PBM, showing the blood in the midguts of wild-type and dsSHMT samples as well as the ovaries of wild-type, dsEGFP and dsSHMT mosquitoes. g Phenotypes observed at 15 h PBM. A white spot appeared at the back of the abdomen for the wild-type and dsEGFP samples but the whole abdomen of dsSHMT was black. The red arrow shows the white spot in abdomens and growing ovaries. The ovaries are compared between wild-type, dsEGFP and dsSHMT mosquitoes at 15 h PBM, showing the developmentally delayed eggs in dsSHMT. h Phenotypes observed at 48 h PBM. The abdomens of wild-type and dsEGFP mosquitoes were all white. The wild-type and dsEGFP mosquitoes had no blood in their midguts and the ovaries developed normally. However, the abdomens of SHMT-depleted mosquitoes were still black in appearance and had undigested blood in the midgut. Ovaries of WT, dsEGFP and dsSHMT are compared at 48 h PBM, showing that the ovaries of dsSHMT were underdeveloped. i The proportion of flightless mosquitoes (PFM). j Number of eggs laid per female. The data are shown as the mean ± SEM. Abbreviations: dsSHMT, double-stranded RNA of SHMT; dsEGFP, double-stranded RNA of EGFP; PBM, post-blood-meal; WT, wild-type; MG, midgut; OV, ovary; BFR, blood-feeding rate; PFM, proportion of flightless mosquitoes. ***P < 0.001
Transcriptional expression of digestive enzymes in the wild-type A. aegypti mosquitoes
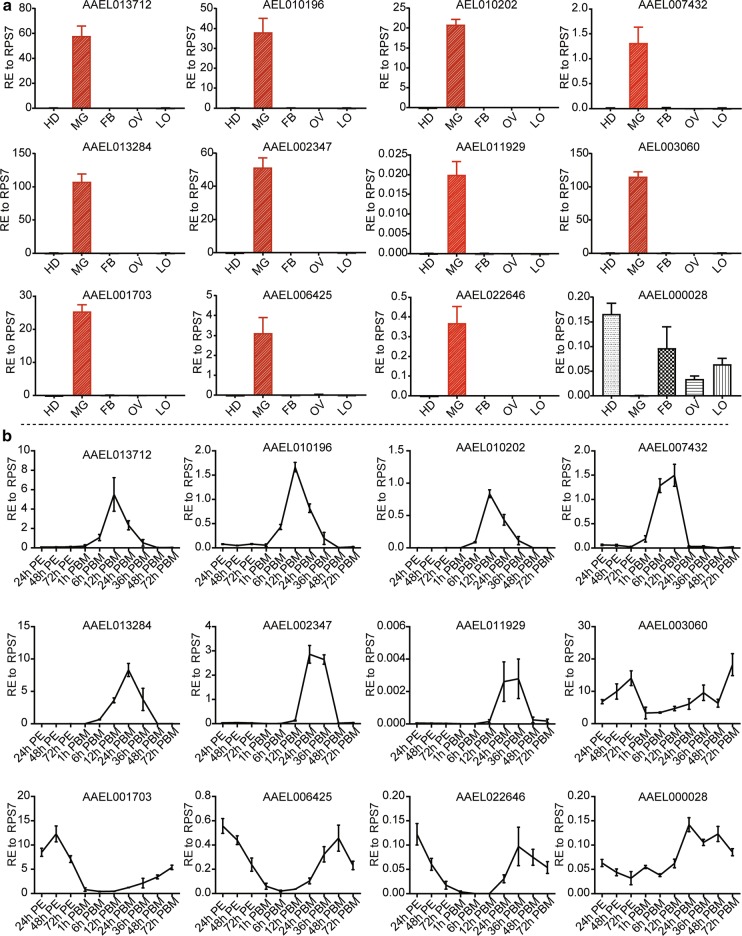
A large family of trypsin-like serine proteases is regarded as the main contributor to the digestion of the blood meal [35, 36]. So far, hundreds of serine protease-like genes have been predicted in the A. aegypti genome; however, only a few of them are known to be expressed in the midgut [15, 37–39]. Induced expression of AaET, AaSPVI, AaSPVII and AaLT proteins is not observed until 12 h PBM, with a peak level at 24 h PBM [39]. We speculate that inhibition of blood-meal digestion by SHMT RNAi was probably due to abnormal expression of digestive enzymes responding to SHMT depletion. To understand the potential effect of SHMT RNAi on these enzymes, it is necessary to determine the transcriptional expression patterns of digestive enzymes in wild-type mosquitoes. To this end, we examined the spatial expression of twelve digestive enzymes at 24 h PBM by qRT-PCR (see “Methods”), including three trypsins (AAEL013712, AAEL013284, AAEL006425), five chymotrypsins (AAEL002347, AAEL011929, AAEL003060, AAEL001703, AAEL022646) and four serine proteases (AAEL010196, AAEL010202, AAEL007432, AAEL000028) and found that the expression of eleven enzymes was limited to the midgut, while AAEL000028 was exclusively absent from the midgut (Fig. 4a). AAEL000028 is described as Serine Protease in NCBI (XM_001647815.3), and annotated as Clip-Domain Serine Protease family B in VectorBase. Next, we revealed their temporal expression patterns at a transcriptional level in adults from 24 h PE to 72 h PBM, and found roughly two modes of expression. The first group are those which were weakly expressed or not expressed before a blood meal, but were transcriptionally induced upon blood-feeding to peak levels at 12 or 24 h PBM and then quickly decreased to background levels, including one trypsin (AAEL013712, AAEL013284), three serine proteases (AAEL010196, AAEL010202, AAEL007432) and two chymotrypsins (AAEL002347, AAEL011929). The second group are those which were highly expressed before a blood meal, but dropped to baseline levels promptly after a blood meal and then dramatically returned to high levels at the late PBM phase, including one trypsin (AAEL006425), three chymotrypsins (AAEL003060, AAEL001703, AAEL022646), and one serine protease (AAEL000028) (Fig. 4b). In conclusion, transcriptional expression levels of digestive enzymes in the midguts of Aedes mosquitoes responded differently to the blood meal. Admittedly however, our data show mRNA expression profiles for 12 proteases without direct evidence to their involvement in blood-meal digestion. Nevertheless, protease genes for which mRNA expression decreased in response to the blood meal might also be involved in blood-meal digestion because mRNA levels could be high but disappear when translated to protein, especially after a blood meal.
Fig. 4.
Transcriptional expression profiles of digestive enzymes. Transcriptional expression profiles of 12 known digestive enzyme genes were examined by qRT-PCR (see “Methods”). a Spatial expression profiles of digestive enzyme genes at the transcriptional level at 24 h PBM. b Temporal expression profiles of digestive enzyme genes at the transcriptional level in adult mosquitoes. AAEL013712, AAEL013284, AAEL006425 are trypsins, AAEL002347, AAEL011929, AAEL003060, AAEL001703, AAEL022646 are chymotrypsins, AAEL010196, AAEL010202, AAEL007432, AAEL000028 are serine proteases. Data are presented as mean ± SEM. Abbreviations: HD, head; MG, midgut; FB, fat body; OV, ovary; LO, tissues left over from the whole body
Transcriptional expression of digestive enzymes responding to depletion of SHMT
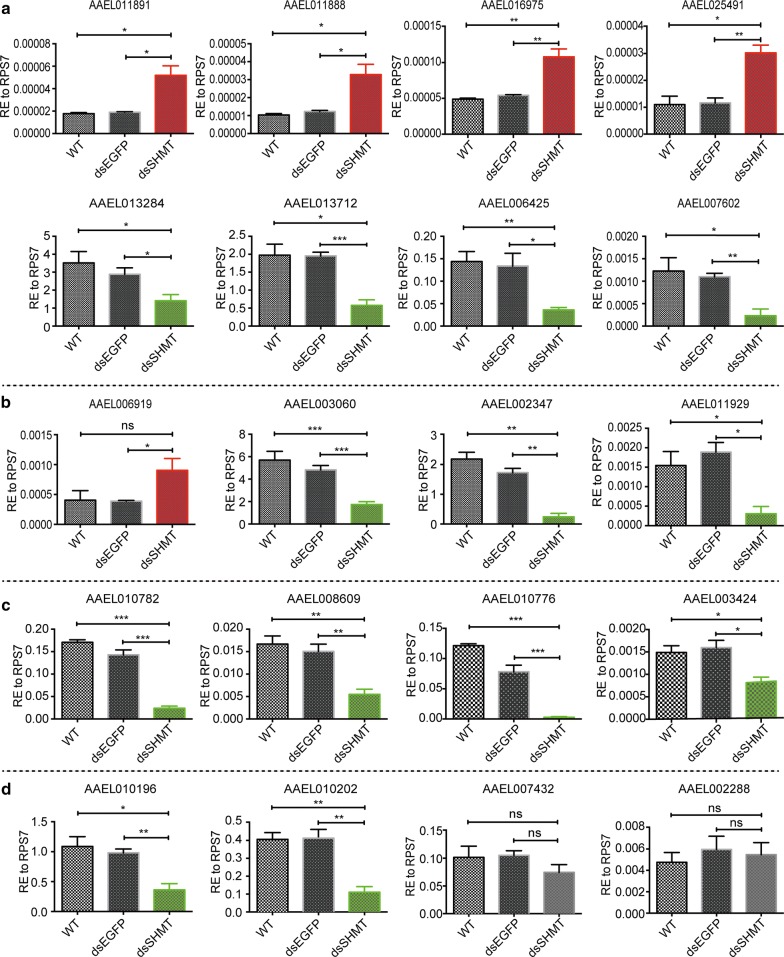
Next, we collected the SHMT-depleted whole-mount adult individuals at 24 h PBM and checked the transcriptional expressions of 74 digestive enzymes by qRT-PCR, including 26 trypsins, 26 chymotrypsins, 11 carboxypeptidases and 11 serine proteases (Additional file 1: Table S1 and Additional file 3: Table S2). The results showed that 6 trypsins were significantly increased (AAEL011891, AAEL011888, AAEL016975, AAEL025491, AAEL026329 and AAEL026347), 10 were significantly decreased (AAEL013284, AAEL013712, AAEL006425, AAEL007602, AAEL013703, AAEL007519, AAEL006903, AAEL004543, AAEL011553 and AAEL025114), and 10 remained unchanged (AAEL011889, AAEL006376, AAEL006403, AAEL004885, AAEL007818, AAEL009843, AAEL005611, AAEL024571, AAEL015638 and AAEL000203) (Fig. 5a and Additional file 2: Figure S2).
Fig. 5.
Transcriptional expression responses of digestive enzymes to SHMT RNAi. a Responses of trypsin to the depletion of SHMT by RNAi. b Responses of chymotrypsin to the depletion of SHMT by RNAi. c Responses of carboxyprotease to the depletion of SHMT by RNAi. d Responses of serine proteases to the depletion of SHMT by RNAi. The data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
Among these chymotrypsins, only AAEL006919 was upregulated, 8 were downregulated (AAEL003060, AAEL002347, AAEL011929, AAEL002360, AAEL002177, AAEL011920, AAEL009680 and AAEL024934) and 17 remained unchanged (AAEL011230, AAEL011916, AAEL015294, AAEL017475, AAEL001690, AAEL008784, AAEL006627, AAEL008782, AAEL001703, AAEL011917, AAEL024686, AAEL001690, AAEL004505, AAEL020852, AAEL006383, AAEL007938 and AAEL011922) (Fig. 5b and Additional file 2: Figure S3). Most of the examined carboxypeptidases have been reported to be involved in blood digestion [40]. When the SHMT gene was depleted, 6 carboxypeptidases were downregulated (AAEL010782, AAEL010776, AAEL003424, AAEL008609, AAEL001844 and AAEL001855), but 5 showed no significant response (AAEL001840, AAEL008600, AAEL001839, AAEL001863 and AAEL020960) (Fig. 5c and Additional file 2: Figure S4a). Serine proteases, which are characterized by their nucleophilic Ser residue at the active site [41], can be further divided into families and subfamilies, and function as the principle contributors in blood-meal digestion [15]. When SHMT was knocked down, the midgut serine proteases AAEL010196 (AaSPVI) and AAEL010202 (AaSPVII) were downregulated, while AAEL007432 (AaSPI), AAEL002288, AAEL000028, AAEL008767, AAEL012558 and AAEL013623 remained unchanged (Fig. 5d and Additional file 2: Figure S4b).
Overall, the digestive enzymes exhibited diverse responses to the depletion of SHMT at the transcriptional level. SHMT is known to be involved in the synthesis of dTMP [27, 42], a precursor of dTTP which is essential for nuclear and mitochondrial DNA replication [43]. So, SHMT depletion is probably affecting these processes, which in turn is affecting the expression of the proteases in the midgut in an indirect way. Along with regulation of midgut proteases at transcriptional and translational levels when SHMT is knocked down, given its known role in nuclear and mitochondrial DNA replication [27, 44–46], it is likely that DNA replication is also affected.
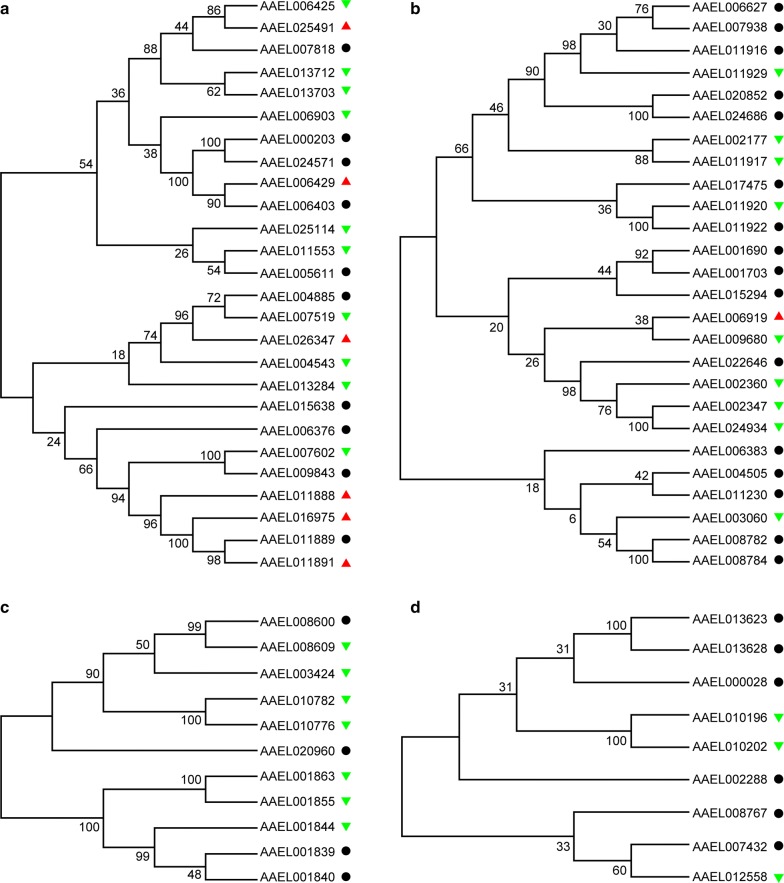
Phylogenetic analysis of A. aegypti digestive enzymes
To reveal whether the same expression responses of digestive enzymes are related to their sequence similarity, we downloaded the amino acid sequences of all examined digestive enzymes from VectorBase and performed phylogenetic analysis. For a better comparison, the expression changes are labeled on the phylogenetic tree, with the red triangle meaning upregulated expression, the green triangle representing downregulated expression and the black circle showing unchanged expression. Strikingly, the trypsins were obviously clustered into two large clades, and these three kinds of expression patterns could be observed in each clade (Fig. 6a). The closely clustered trypsins AAEL013712 (Trypsin 5G1 Precursor, Aa5G1) and AAEL013703 (Trypsin) with similar amino acid sequence (identity: 0.3941019) were both downregulated by SHMT RNAi. Meanwhile, mRNA expression levels remained unchanged for another sub-clustered AAEL000203 (trypsin-1) and AAEL024571 (trypsin-4), with similar amino acid sequence similarity (identity: 0.4790419) in the SHMT-depleted mosquitoes.
Fig. 6.
Maximum likelihood phylogenetic trees for digestive enzymes. a Trypsins. b Chymotrypsins. c Carboxyproteases. d Serine proteases. The red triangle indicates upregulation by SHMT RNAi, the green triangle indicates downregulation, and the black circle indicates unchanged expression
By contrast, some members which were clustered in the same sub-clade showed different or even opposite responses. For example, AAEL006429 (trypsin-7) was up-regulated while AAEL006403 (trypsin-7) remained unchanged; these two genes (identity: 0.2893401; Additional file 3: Table S2) are located on different chromosomes. Another pair of closely clustered trypsins AAEL006425 (trypsin-1) and AAEL025491 (trypsin-4-like) (identity: 0.4021448) showed opposite responses to SHMT depletion. All of these trypsins are encoded by different loci in the genome and actually have a low degree of homology with each other (Additional file 3: Table S2). Similar to trypsins, the chymotrypsins were organized into two large clades and the members within the same clade did not respond similarly to SHMT RNAi (Fig. 6b). A sub-clade member with low sequence similarity (identity: 0.2432432), AAEL006919 (chymotrypsin-1) was upregulated but AAEL009680 (chymotrypsin-2) was downregulated. Although assigned to a sub-clade with high sequence similarity (identity: 0.8248175), AAEL011920 was downregulated but AAEL011922 remained unchanged. However, most members within the same sub-clade with low or high sequence similarity showed exactly the same expression patterns, including AAEL006627 and AAEL007938 (identity: 0.7123746), AAEL020852 and AAEL024686 (identity: 0.9922780), AAEL002177 and AAEL011917 (identity: 0.3891892), AAEL001690 and AAEL001703 (identity: 0.2942708), AEL002347 and AAEL024934 (identity: 0.7894737), AAEL004505 and AAEL011230 (identity: 0.2708861), and AAEL008782 and AAEL008784 (identity: 0.4632768). Therefore, the expression of these chymotrypsins in response to SHMT depletion was not directly related to sequence similarity. The similar results could also be observed for carboxypeptidases (Fig. 6c) and serine proteases (Fig. 6d).
Discussion
Previous studies in other organisms have shown that SHMT mainly catalyzes the conversion of serine and tetrahydrofolate to glycine and 5,10-methyl tetrahydrofolate [23, 24]. Increased SHMT activity is associated with increased demand for DNA synthesis in rapidly proliferating cells [47]. In recent years, SHMT has been shown to be associated with various diseases and has emerged as an important biomarker and drug target [48, 49]. Our previous studies confirmed that SHMT is the target gene of mosquito-specific miR-1174. When SHMT is downregulated to a certain extent, the abnormal phenotype caused by the depletion of miR-1174 is partially rescued [22], but it remained undetermined whether and how SHMT regulates blood digestion, absorption and nutrient signaling in mosquitoes. In this study, we revealed that when the decrease of SHMT expression exceeded a certain limit, mosquitoes suffered from blockage of blood digestion, loss of flight and incapability of oviposition. Strikingly, these abnormal phenotypes began to appear in succession about 15 hours after a blood meal. Many factors could cause the failure of blood digestion; a likely possibility is that digestive enzymes are misregulated when SHMT is knocked down. At 48 h PBM, the majority of undigested blood remained in the midgut of SHMT-depleted mosquitoes, and the egg growth was severely inhibited when compared to that in the WT and the dsEGFP samples (Fig. 3h). It seems that blood-meal digestion occurred after dsSHMT treatment and enough nutrients could be provided for the formation of ovaries, whereas very few eggs were laid by SHMT-depleted mosquitoes (Fig. 3j). SHMT RNAi might affect the ovary development and oviposition via other processes like DNA replication in the ovary cells since SHMT is involved in the de novo synthesis of deoxythymidine monophosphate (dTMP) [23, 24], a precursor of deoxythymidine triphosphate (dTTP) [43]. In addition, SHMT knockdown recovered the miR-1174 depletion phenotype in mosquitoes [22], so SHMT might regulate the egg growth through miRNA regulatory pathway. However, additional evidence will be required for better explanation of the roles of SHMT in the ovary development and oviposition.
Previous studies showed that trypsin and chymotrypsin serine proteases produced by midgut epithelial cells might be the main contributors in the blood digestion of some hematophagous dipteran species [35, 36]. Furthermore, dsRNA-mediated knockdown of a single late phase serine protease, including AAEL013284 (AaLT), AAEL010196 (AaSPVI), and AAEL010202 (AaSPVII), significantly retards blood-meal digestion and decreases fecundity, while injection with a mix dsRNA of these protease genes does not show additive or synergistic effect on egg production [39]. All of these digestive enzymes were downregulated in SHMT-depleted mosquitoes (Additional file 3: Table S2), logically therefore, their absence might lead to impaired digestion and reproductive disorder. From the expression patterns observed, the digestive enzymes seemed to be negatively or positively regulated by SHMT at transcriptional level. We noted the fact that SHMT was expressed at low levels in the midgut but highly expressed in other tissues and organs. On the contrary, most digestive enzymes were exclusively upregulated in the midgut. Since the expression of SHMT and digestive enzymes is spatially segregated, how does SHMT regulate the digestive enzymes in midgut cells? SHMT is known to be involved in the formation of dTTP in DNA synthesis [27, 45, 46], then it seems more likely that transcription of digestive enzyme genes is being affected by SHMT depletion. However, further research is needed to better understand how SMHT RNAi is affecting these digestive enzymes.
The outcome of abnormal phenotype depended on the extent to which SHMT is downregulated, but in any case the abnormal phenotype did not appear until 15 hours after a blood meal. Previous studies demonstrated that some early digestive enzymes are quickly synthesized and secreted after a blood meal due to the stimulation of juvenile hormones; these early enzymes preliminarily digest blood and activate the expression of specific transcription factors, enabling some late digestive enzyme genes to be highly expressed 12 hours after a blood meal [50]. After female mosquitoes ingest blood from vertebrate hosts, exopeptidases and endopeptidases are required for digesting blood proteins in the midgut into amino acids, which female mosquitoes use to build yolk proteins. These proteases are not always present in the midgut, and their diverse expression patterns suggest that production of these enzymes is highly regulated in order to meet specific physiological demands at various stages [18]. It is known that the enzyme genes responsible for lipid metabolism in mosquitoes are downregulated at the late juvenile hormone-controlled PE phase and upregulated in the 20-hydroxyecdysone (20E)-mediated PBM phase [51]. SHMT might also negatively correlate with the pattern of JH titer while positively correlating with the 20E pulse, and accordingly, when SHMT is decreased, the hormone pathway will be blocked, thus resulting in abnormal expressions of various digestive enzymes. However, the evidence for violating these hypotheses remains to be revealed and the regulatory mechanism involved by SHMT will not be clearly established until then.
Conclusions
We examined the transcriptional expression profiles of SHMT before and after a blood meal and found that it was highly expressed in the head, fat body, ovary and leftover tissues, was expressed at very low levels in the midgut, and quickly increased in the whole body to the peak level at 12 h PBM. Western blotting analysis showed that the protein levels of SHMT also peaked at 12 h PBM in the whole body. Depletion of SHMT by RNAi caused severely abnormal phenotypes, including blockage of blood-meal digestion, loss of flight and incapability of oviposition. Strikingly, these abnormal phenotypes began to appear in succession about 15 h after a blood meal. Transcriptional expression of different diverse digestive enzymes was also affected in response to SHMT depletion, largely unrelated to the sequence similarity, indicating that SHMT might indirectly affect the transcriptional expression of digestive enzymes through its involvement in nuclear and mitochondrial DNA replication [27, 45, 46]. Therefore, we believe we have shown SHMT is required for blood-meal digestion in the midgut and targeting SHMT could provide an effective strategy for vector mosquito population control.
Supplementary information
Additional file 1: Table S1. All primers used in this study.
Additional file 2: Figure S1. Different doses of dsSHMT exerted different effect on the flight ability and oviposition of mosquitoes. Different doses of dsSHMT (50, 100, 200, 400 and 800 ng) dissolved in 0.5 μl of nuclease-free water were separately injected into mosquitoes at 16 h PE (see “Methods”). The phenotype of mosquitoes was examined at five time points post-blood-meal (PBM), namely 15, 17, 19, 24, 30 and 40 h PBM. a Effect on flight ability of mosquitoes. b Effect on the oviposition of mosquitoes. Figure S2. Transcriptional expression of trypsins responding to SHMT RNAi. The data are shown as the mean ± SEM. *P < 0.05; **P < 0.01; ns, not significant. Figure S3. Transcriptional expression of chymotrypsins responding to SHMT RNAi. All graphs have the same abscissa, i.e. three samples, WT, dsEGFP and dsSHMT. The data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ns, not significant. Figure S4. Transcriptional expression of carboxypeptidases and serine proteases responding to SHMT RNAi. a Transcriptional expression of carboxypeptidases. b Transcriptional expression of serine protease. The data are shown as the mean ± SEM. *P < 0.05.
Additional file 3: Table S2. Digestion enzymes of A. aegypti and their conservative analysis in different species of mosquitoes.
Acknowledgements
The authors would like to thank Professor Zhen Zou (Institute of Zoology, Chinese Academy of Sciences) and Professor Alexander S. Raikhel (Entomology Department, University of California, USA) for technical assistance and helpful discussions. We thank Professor Marian Goldsmith (University of Rhode Island Kingston, RI USA) for critical reading, kindly discussion and help with English.
Abbreviations
- SHMT
serine hydroxymethyltransferase
- PE
post-eclosion
- BBM
before a blood meal
- PBM
post-blood-meal
- PIJ
post-injection
- HD
head
- MG
midgut
- FB
fat body
- OV
ovary
- LO
leftover
- CDS
protein coding region
- WT
wild-type
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- 20E
20-hydroxyecdysone
- RE
relative expression
- dsSHMT
double-stranded RNA of SHMT
- dsEGFP
double-stranded RNA of EGFP
- BFR
blood-feeding rate
- PFM
proportion of flightless mosquitoes
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Authors’ contributions
XL and JY performed experiments, interpreted the data and contributed in writing. QP, XP and LX reared the mosquitoes and contributed in writing. SL conceived the research, designed and performed experiments and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31571334), State Key Programme of National Natural Science of China (31530071) and Chongqing Basic and Frontier Research Programme (cstc2014jcyjA00025).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
The experiments were carried out in accordance with the guidelines approved by the Southwest University Animal Care and Use Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuemei Li and Jinyu Yang contributed equally to this manuscript
Contributor Information
Xuemei Li, Email: lxm9812@163.com.
Jinyu Yang, Email: yang243917@163.com.
Qian Pu, Email: piaoqian1993@163.com.
Xinyue Peng, Email: xinyuepen@163.com.
Lili Xu, Email: xuli133324@163.com.
Shiping Liu, Email: lsp98668@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3714-2.
References
- 1.Briegel H. Physiological bases of mosquito ecology. J Vector Ecol. 2003;28:1–11. [PubMed] [Google Scholar]
- 2.Rascon AA, Jr, Gearin J, Isoe J, Miesfeld RL. In vitro activation and enzyme kinetic analysis of recombinant midgut serine proteases from the dengue vector mosquito Aedes aegypti. BMC Biochem. 2011;12:43. doi: 10.1186/1471-2091-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci AS, Morens DM. Zika virus in the Americas—yet another arbovirus threat. N Engl J Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 4.Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett AD, Higgs S. Yellow fever: a disease that has yet to be conquered. Annu Rev Entomol. 2007;52:209–229. doi: 10.1146/annurev.ento.52.110405.091454. [DOI] [PubMed] [Google Scholar]
- 6.Carrington LB, Tran BCN, Le NTH, Luong TTH, Nguyen TT, Nguyen PT, et al. Field- and clinically derived estimates of Wolbachia-mediated blocking of dengue virus transmission potential in Aedes aegypti mosquitoes. Proc Natl Acad Sci USA. 2018;115:361–366. doi: 10.1073/pnas.1715788115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mourya DT, Gokhale MD, Majumdar TD, Yadav PD, Kumar V, Mavale MS. Experimental Zika virus infection in Aedes aegypti: susceptibility, transmission and co-infection with dengue and chikungunya viruses. Indian J Med Res. 2018;147:88–96. doi: 10.4103/ijmr.IJMR_1142_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira-de-Lima VH, Lima-Camara TN. Natural vertical transmission of dengue virus in Aedes aegypti and Aedes albopictus: a systematic review. Parasites Vectors. 2018;11:77. doi: 10.1186/s13071-018-2643-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngoagouni C, Kamgang B, Kazanji M, Paupy C, Nakouné E. Potential of Aedes aegypti and Aedes albopictus populations in the Central African Republic to transmit enzootic chikungunya virus strains. Parasites Vectors. 2017;10:164. doi: 10.1186/s13071-017-2101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noriega FG, Edgar KA, Bechet R, Wells MA. Midgut exopeptidase activities in Aedes aegypti are induced by blood feeding. J Insect Physiol. 2002;48:205–212. doi: 10.1016/S0022-1910(01)00165-2. [DOI] [PubMed] [Google Scholar]
- 11.Noriega FG, Pennington JE, Barillas-Mury C, Wang XY, Wells MA. Aedes aegypti midgut early trypsin is post-transcriptionally regulated by blood feeding. Insect Mol Biol. 1996;5:25–29. doi: 10.1111/j.1365-2583.1996.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 12.Sanders HR, Evans AM, Ross LS, Gill SS. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem Mol Biol. 2003;33:1105–1122. doi: 10.1016/S0965-1748(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 13.Borovsky D. Biosynthesis and control of mosquito gut proteases. IUBMB Life. 2003;55:435–441. doi: 10.1080/15216540310001597721. [DOI] [PubMed] [Google Scholar]
- 14.Venancio TM, Cristofoletti PT, Ferreira C, Verjovski-Almeida S, Terra WR. The Aedes aegypti larval transcriptome: a comparative perspective with emphasis on trypsins and the domain structure of peritrophins. Insect Mol Biol. 2009;18:33–44. doi: 10.1111/j.1365-2583.2008.00845.x. [DOI] [PubMed] [Google Scholar]
- 15.Brackney DE, Isoe J, Black WC, IV, Zamora J, Foy BD, Miesfeld RL, Olson KE. Expression profiling and comparative analyses of seven midgut serine proteases from the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2010;56:736–744. doi: 10.1016/j.jinsphys.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saboia-Vahia L, Cuervo P, Borges-Veloso A, de Souza NP, Britto C, Dias-Lopes G, De Jesus JB. The midgut of Aedes albopictus females expresses active trypsin-like serine peptidases. Parasites Vectors. 2014;7:253. doi: 10.1186/1756-3305-7-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Q, Hall M, Noriega FG, Wells M. cDNA cloning and pattern of expression of an adult, female-specific chymotrypsin from Aedes aegypti midgut. Insect Biochem Mol Biol. 1997;27:283–289. doi: 10.1016/S0965-1748(97)00001-5. [DOI] [PubMed] [Google Scholar]
- 18.Bian G, Raikhel AS, Zhu J. Characterization of a juvenile hormone-regulated chymotrypsin-like serine protease gene in Aedes aegypti mosquito. Insect Biochem Mol Biol. 2008;38:190–200. doi: 10.1016/j.ibmb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felix CR, Betschart B, Billingsley PF, Freyvogel TA. Post-feeding induction of trypsin in the midgut of Aedes aegypti L. (Diptera, Culicidae) is separable into 2 cellular phases. Insect Biochem. 1991;21:197–203. doi: 10.1016/0020-1790(91)90050-O. [DOI] [Google Scholar]
- 20.Barillas-Mury C, Graf R, Hagedorn HH, Wells MA. cDNA and deduced amino acid sequence of a blood meal-induced trypsin from the mosquito Aedes aegypti. Insect Biochem. 1991;21:825–831. doi: 10.1016/0020-1790(91)90089-W. [DOI] [Google Scholar]
- 21.Kalhok SE, Tabak LM, Prosser DE, Brook W, Downe AE, White BN. Isolation, sequencing and characterization of two cDNA clones coding for trypsin-like enzymes from the midgut of Aedes aegypti. Insect Mol Biol. 1993;2:71–79. doi: 10.1111/j.1365-2583.1993.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci USA. 2014;111:14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szebenyi DM, Musayev FN, di Salvo ML, Safo MK, Schirch V. Serine hydroxymethyltransferase: role of glu75 and evidence that serine is cleaved by a retroaldol mechanism. Biochemistry. 2004;43:6865–6876. doi: 10.1021/bi049791y. [DOI] [PubMed] [Google Scholar]
- 24.Schirch V, Szebenyi DM. Serine hydroxymethyltransferase revisited. Curr Opin Chem Biol. 2005;9:482–487. doi: 10.1016/j.cbpa.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Winkler F, Kriebel M, Clever M, Groning S, Grosshans J. Essential function of the Serine Hydroxymethyl Transferase (SHMT) gene during rapid syncytial cell cycles in Drosophila. G3. 2017;7:2305–2314. doi: 10.1534/g3.117.043133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skibola CF, Smith MT, Hubbard A, Shane B, Roberts AC, Law GR, et al. Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood. 2002;99:3786–3791. doi: 10.1182/blood.V99.10.3786. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DD, Woeller CF, Chiang EP, Shane B, Stover PJ. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem. 2012;287:7051–7062. doi: 10.1074/jbc.M111.333120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tramonti A, Nardella C, di Salvo ML, Barile A, Cutruzzola F, Contestabile R. Human cytosolic and mitochondrial serine hydroxymethyltransferase isoforms in comparison: full kinetic characterization and substrate inhibition properties. Biochemistry. 2018;57:6984–6996. doi: 10.1021/acs.biochem.8b01074. [DOI] [PubMed] [Google Scholar]
- 29.Garrow TA, Brenner AA, Whitehead VM, Chen XN, Duncan RG, Korenberg JR, Shane B. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J Biol Chem. 1993;268:11910–11916. [PubMed] [Google Scholar]
- 30.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, et al. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Li M, Knyaz C, Tamura KMEGAX. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dana AN, Hong YS, Kern MK, Hillenmeyer ME, Harker BW, Lobo NF, et al. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genomics. 2005;6:5. doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 35.Wu DD, Wang GD, Irwin DM, Zhang YP. A profound role for the expansion of trypsin-like serine protease family in the evolution of hematophagy in mosquito. Mol Biol Evol. 2009;26:2333–2341. doi: 10.1093/molbev/msp139. [DOI] [PubMed] [Google Scholar]
- 36.Ramalho-Ortigao JM, Kamhawi S, Rowton ED, Ribeiro JM, Valenzuela JG. Cloning and characterization of trypsin- and chymotrypsin-like proteases from the midgut of the sand fly vector Phlebotomus papatasi. Insect Biochem Mol Biol. 2003;33:163–171. doi: 10.1016/S0965-1748(02)00187-X. [DOI] [PubMed] [Google Scholar]
- 37.Soares TS, Rodriguez Gonzalez BL, Torquato RJS, Lemos FJA, Costa-da-Silva AL, Capurro Guimaraes ML, Tanaka AS. Functional characterization of a serine protease inhibitor modulated in the infection of the Aedes aegypti with dengue virus. Biochimie. 2018;144:160–168. doi: 10.1016/j.biochi.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Torquato RJS, Lu S, Martins NH, Tanaka AS, Pereira PJB. High-resolution structure of a Kazal-type serine protease inhibitor from the dengue vector Aedes aegypti. Acta Crystallogr F Struct Biol Commun. 2017;73:469–475. doi: 10.1107/S2053230X17010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isoe J, Rascon AA, Jr, Kunz S, Miesfeld RL. Molecular genetic analysis of midgut serine proteases in Aedes aegypti mosquitoes. Insect Biochem Mol Biol. 2009;39:903–912. doi: 10.1016/j.ibmb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isoe J, Zamora J, Miesfeld RL. Molecular analysis of the Aedes aegypti carboxypeptidase gene family. Insect Biochem Mol Biol. 2009;39:68–73. doi: 10.1016/j.ibmb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blow DM. The tortuous story of Asp … His … Ser: structural analysis of alpha-chymotrypsin. Trends Biochem Sci. 1997;22:405–408. doi: 10.1016/S0968-0004(97)01115-8. [DOI] [PubMed] [Google Scholar]
- 42.Giardina G, Paone A, Tramonti A, Lucchi R, Marani M, Magnifico MC, et al. The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells. FEBS J. 2018;285:3238–3253. doi: 10.1111/febs.14610. [DOI] [PubMed] [Google Scholar]
- 43.Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci USA. 2011;108:15163–15168. doi: 10.1073/pnas.1103623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry C, Sastry R, Nasrallah IM, Stover PJ. Mimosine attenuates serine hydroxymethyltransferase transcription by chelating zinc. Implications for inhibition of DNA replication. J Biol Chem. 2005;280:396–400. doi: 10.1074/jbc.M410467200. [DOI] [PubMed] [Google Scholar]
- 45.Misselbeck K, Marchetti L, Field MS, Scotti M, Priami C, Stover PJ. A hybrid stochastic model of folate-mediated one-carbon metabolism: effect of the common C677T MTHFR variant on de novo thymidylate biosynthesis. Sci Rep. 2017;7:797. doi: 10.1038/s41598-017-00854-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Winkler F, Herde M, Witte CP, Grosshans J. A link between deoxyribonucleotide metabolites and embryonic cell-cycle control. Curr Biol. 2019;29(1187–1192):e3. doi: 10.1016/j.cub.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Renwick SB, Snell K, Baumann U. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998;6:1105–1116. doi: 10.1016/S0969-2126(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 48.Nonaka H, Nakanishi Y, Kuno S, Ota T, Mochidome K, Saito Y, et al. Design strategy for serine hydroxymethyltransferase probes based on retro-aldol-type reaction. Nat Commun. 2019;10:876. doi: 10.1038/s41467-019-08833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asano K, Suzuki T, Saito A, Wei FY, Ikeuchi Y, Numata T, et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018;46:1565–1583. doi: 10.1093/nar/gky068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barillas-Mury CV, Noriega FG, Wells MA. Early trypsin activity is part of the signal transduction system that activates transcription of the late trypsin gene in the midgut of the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1995;25:241–246. doi: 10.1016/0965-1748(94)00061-L. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Hou Y, Saha TT, Pei G, Raikhel AS, Zou Z. Hormone and receptor interplay in the regulation of mosquito lipid metabolism. Proc Natl Acad Sci USA. 2017;114:E2709–E2718. doi: 10.1073/pnas.1619326114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. All primers used in this study.
Additional file 2: Figure S1. Different doses of dsSHMT exerted different effect on the flight ability and oviposition of mosquitoes. Different doses of dsSHMT (50, 100, 200, 400 and 800 ng) dissolved in 0.5 μl of nuclease-free water were separately injected into mosquitoes at 16 h PE (see “Methods”). The phenotype of mosquitoes was examined at five time points post-blood-meal (PBM), namely 15, 17, 19, 24, 30 and 40 h PBM. a Effect on flight ability of mosquitoes. b Effect on the oviposition of mosquitoes. Figure S2. Transcriptional expression of trypsins responding to SHMT RNAi. The data are shown as the mean ± SEM. *P < 0.05; **P < 0.01; ns, not significant. Figure S3. Transcriptional expression of chymotrypsins responding to SHMT RNAi. All graphs have the same abscissa, i.e. three samples, WT, dsEGFP and dsSHMT. The data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ns, not significant. Figure S4. Transcriptional expression of carboxypeptidases and serine proteases responding to SHMT RNAi. a Transcriptional expression of carboxypeptidases. b Transcriptional expression of serine protease. The data are shown as the mean ± SEM. *P < 0.05.
Additional file 3: Table S2. Digestion enzymes of A. aegypti and their conservative analysis in different species of mosquitoes.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.