Abstract
Laboratory animal research was always crucial for scientific breakthroughs in the fields of medicine and biology. Animal trials offer insights into various disease mechanisms, genetics, drug therapy and the effect of different external factors onto living organisms. However, conducting animal trials is highly controversial. To ensure high ethical standards, a number of directives have been adopted in the European Union, which seek to replace, reduce and refine animal trials. Hence, severity assessment plays an important role in today’s laboratory animal research. Currently, severity of trials is assessed by highly rater dependent scoring systems. In this paper, we propose a method for unobtrusive, automated and contactless measurement of respiratory rate (RR) and heart rate (HR). We were able to extract RR and HR with an high agreement between our method and a contact-based reference method. The Root Mean Squared Error (RMSE) averaged 0.32 ± 0.11 breaths/min for RR and 1.28 ± 0.62 beats/min for HR in rats, respectively. In mice, the RMSE averaged 1.42 ± 0.97 breaths/min for RR and 1.36 ± 0.87 beats/min, respectively. In the future, these parameters can be used for new, objective scoring systems, which are not susceptible to inter-rater variability
1. Introduction
Most advances in modern medicine were made possible through animal trials [1,2]. With growing public consciousness, animal welfare became a more and more important factor in laboratory animal science [3,4]. In 1959 Russel and Burch promoted the so called 3R-Principles [5], in which the three ”R” stand for: replacement, reduction and refinement. They strive to replace animal trials as far as possible, reduce the quantity of animals used in scientific research and refine the trials, seeking for the most efficient and least severe procedures [6,7]. Since then, a number of regulations have been adopted to promote animal welfare, culminating in the EU Directive 2010/63/EU.
Today’s guidelines set a great value on severity assessments. Although it is an instrumental part of current animal experiments, there is still a lack of an objective tool, capable of accurately expressing the degree of severity [8,9]. Nowadays, severity assessment is performed, based on manual scoring systems. Most of the scores rely on unspecific parameters, like posture, fur condition [10] and grimace [11], which share a high inter-rater variability. While promising, rater independent severity measures, such as body weight and fecal corticosterone metabolites [12] are currently evaluated for severity assessment, they still require frequent handling of the animals for their collection. Thus, a truly continuous, unobtrusive and rater independent severity assessment requires other parameters.
Vital signs, like respiratory rate (RR) and heart rate (HR) are good candidates to monitor an animal’s state [13]. However, the current monitoring procedures are most often invasive [14] and require sedation. Such interventions cause distress to the animal and hence increase the experiments severity [15].
To obtain continuous vital signals in chronic experiments, telemetric sensors can be used. However, their surgical implantation is stressful [13,15], their size and volume also cause considerable discomfort and restrict the animal’s freedom of movement [16]. In addition, those sensors have a short lifespan, as the battery is implanted into the animal [17].
Ironically, by using the current vital signs monitoring techniques for severity assessment, animal welfare can be decreased, since they induce additional stress or pain [15]. Furthermore, experiment outcomes can be altered through additional manipulations [17].
In order to avoid this, animal friendly monitoring should be completely unobtrusive [13,14,18]. A few possible solutions have been proposed by using infrared thermography, [13,19], visual imaging [20] or capacitive sensing [14].
This paper presents a robust approach for monitoring breath- and heart rates, using price efficient visual cameras. Contactless vitals assessment using cameras isn’t a new idea. In fact, many approaches have been published for contactless vitals assessment in humans [21,22]. However the transferability of these approaches to animal test subjects is limited, as they require visible skin in order to work. Our algorithm, on the other hand, also works with furry animals because it is purely motion-based. Also, the pulse signal may be disturbed by a much larger respiratory signal, which is the case in small animals.
2. Materials and methods
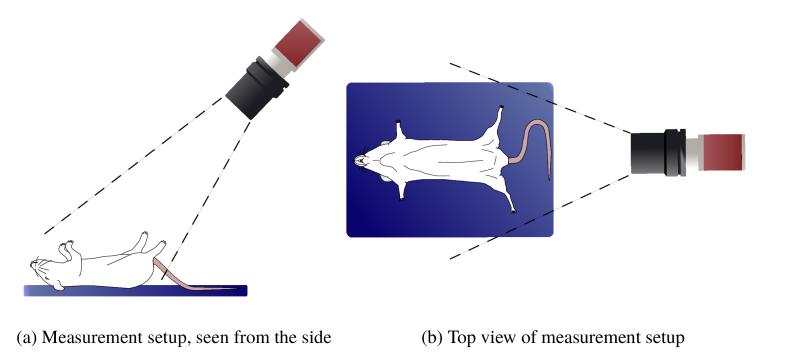
Color video of anesthetized mice and rats was recorded in cooperation with three different laboratories. A schematic overview of the recording setup is shown in Fig. 1 . The videos were imported into MATLAB (The MathWorks, Inc., Natick, Massachusetts, USA), which was used for the complete data analysis.
Fig. 1.
Graphical representation of the measurement setup
2.1. Animal detection
Inspired by Kumar et al. [23], regions on the animal, which are ideal for vitals extraction were automatically detected by performing a temporal pulsatility analysis on the video data. To reduce the computation time, the video frame size was reduced by calculating the mean values for patches of 5x5 pixels and converting the images to grayscale. For every resulting pixel the temporal power spectral density was computed, using a fast Fourier transform (FFT). The maximum spectral component in the frequency band of interest and the corresponding frequency were identified for every pixel:
| (1) |
The frequency band (where is the minimum and the maximum band frequency) was chosen, so that it contains all physiological breath rates of the recorded species (mice: [60, 230] breaths /min, rats: [60, 170] breaths /min. The noise floor was then computed by:
| (2) |
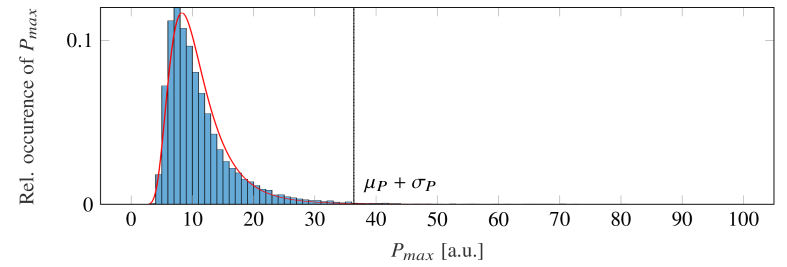
where and are the mean value and standard deviation of an inverse gamma distribution, fitted to the set , which includes all (see Fig. 2 ).
Fig. 2.
Probability density function (PDF) for the set of maximum spectral power of each pixel (blue bar graph). An inverse gamma distribution (red line graph), fitted to the set, is used to compute its mean value and standard deviation . The sum of both moments represents the spectral noise floor (dashed line).
Via a random sample consensus (RanSaC) approach [24], the frequency , which represents the most was computed. The pulsatility was then computed as the signal to noise ratio (SNR) of every pixel at :
| (3) |
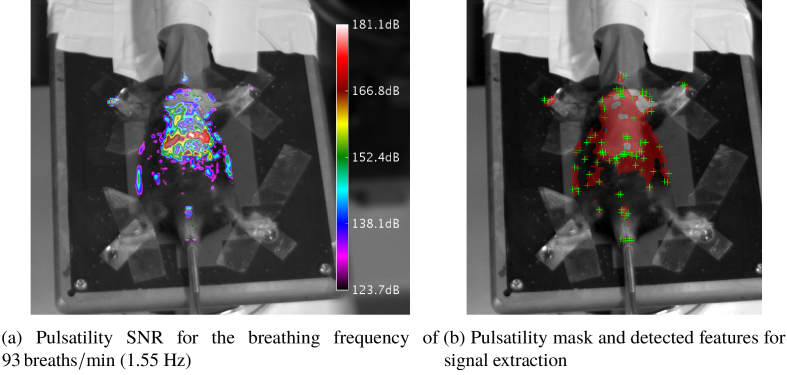
The parameter represents the half lobe width for the chosen window type and length. contains an image, which holds the SNR values of every pixel at the determined frequency (Fig. 3(a) ). Regions of high SNR were morphologically closed to obtain a mask for the animal (Fig. 3(b)).
Fig. 3.
Results of the pulsatility analysis overlayed onto grayscale image of animal for better visual reference (M3)
2.2. Signal extraction
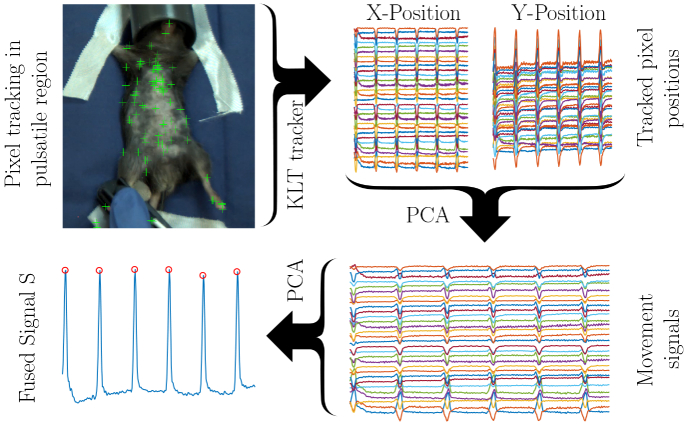
Within the mask, all minimum eigenvalue feature points were found, using the algorithm of Shi and Tomasi [25]. The fifty strongest feature points were tracked over the whole video, using the Kanade-Lucas-Tomasi (KLT) algorithm [26] to extract spatially redundant signals. The positions of the individually tracked points represent the raw signals.
2.3. Signal fusion
Firstly, all extracted signals were band-pass-filtered between 0.3 Hz and 15 Hz using a finite impulse response filter to remove baseline wander and high frequent noise. We chose 15 Hz as the upper border, because it is well above the maximum physiological heart rates of mice and rats (800 beats/min and 700 beats/min, respectively). The result is a set of redundant signals without baseline wander or high frequent noise. Different from the original, motion-based remote photoplethysmography (rPPG) algorithm by Balakrishnan et al. [27], we performed a principle component analysis (PCA) to extract the raw motion signal given its x- and y-components. A second PCA was performed to merge all positional signals into the signal . A schematic overview of the signal fusion algorithm can be found in Fig. 4 .
Fig. 4.
Schematic overview of the signal fusion algorithm (Visualization 1 (6.8MB, mp4) )
2.4. Respiratory rate extraction
features distinctive peaks in the time domain, which correspond to the respiratory activity of the animal. We used MATLABS’s findpeaks function to extract the locations of the breath-related spikes and computed the raw, non-equally sampled instantaneous RR signal:
| (4) |
Artifacts were removed by two threshold filters, which only allow RRs in the range between and and limit their rates of change to :
| (5) |
The resulting signal was interpolated onto an continuous grid with a sampling rate of 1 Hz and filtered with a 5 s median filter to yield the final RR.
2.5. Heart rate extraction
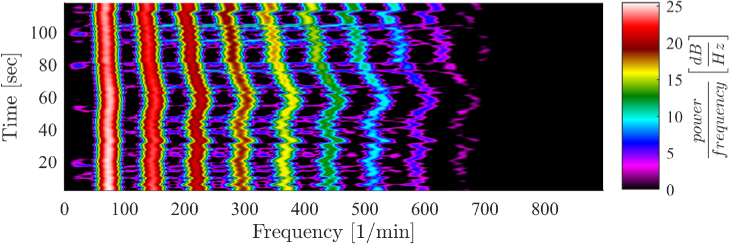
The impulse-like and relatively low frequent RR signal masks all higher frequency components, as can be seen in Fig. 5 . The spectrogram shows an exponentially decaying pattern of distinctive harmonics, which correspond to a RR of 60 breaths /min. These harmonics mask the high frequent heart rate signal that is about two orders of magnitude smaller than the RR signal.
Fig. 5.
Signal spectrum contaminated by harmonics of respiratory frequency
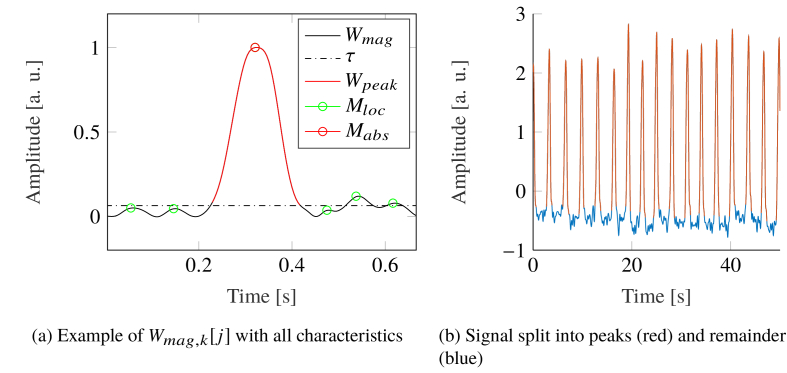
To reduce the influence of the RR, all breath related spikes were removed in the time domain. For this purpose, the continuous signal was split into multiple signals, only containing one breath period each. All these signals were de-trended and rectified independently, as given by:
| (6) |
Afterwards, the signals were normalized onto :
| (7) |
All local maxima – excluding the global maximum – were found. Their median value was taken as threshold. The signal around the global maximum, which was greater than this threshold was cut out (see Fig. 6(a) ). The gap was filled by using a shape-preserving piece-wise cubic spline algorithm (see Fig. 6(b)).
Fig. 6.
Spike removal from
The spike-free signal was used to compute the heart rate via a short-time FFT with length , after applying a hamming window and padding the signal with zeros. The window was shifted along the signal in steps of 1s (see Fig. 7 ). Using an adaptive bandpass filter, the HR was extracted from the signal spectrum within the frequency range of .
Fig. 7.
After removing the breath related spikes, the signal spectrogram unveils the HR
2.6. Experimental protocol
The presented study was conducted as part of the German Research Association (DFG) research unit project 2591 entitled “Severity Assessment in Animal Based Research”. The goal of this study was the development of algorithms for contactless vitals monitoring, which work in different lab-settings. All animals were anesthetized for scheduled interventions as part of different ongoing trials.
Six male WISTAR rats (Janvier S.A.S., Saint – Berthevin Cedex, France) weighing between 250-300 g were supplied by the Institute for Laboratory Animal Science and Experimental Surgery of University Hospital of RWTH Aachen University (R1-R6). Anesthesia was established with 5 vol% Isoflurane and 5 L min−1 of oxygen. Afterwards, it was maintained by reducing the anesthetic concentration to 2 vol% and the oxygen flow to 2 L min−1. The abdominal skin was shaved and a SHAM laparotomy was performed on all animals, during which 2 min of video were recorded. A lead II Electrocardiogram (ECG) was collected with needle electrodes by using an AD Instruments PowerLab data acquisition device. Reference heart- and breath rate were computed from the raw ECG signal. The study was approved by the governmental institution “Landesamt für Natur, Umwelt und Verbraucherschutz NRW” (Germany; 84-02.04.2017.A304).
Six male C57BL/6 N Crl mice aged 8 weeks, which previously underwent left coronary artery ligation to induce a myocardial infarction [28] were supplied from the Institute of Molecular and Translational Therapeutic Strategies of Hannover Medical School (M1-M6). The animals were anesthetized with 3 vol% Isoflurane at 0.8 L min−1 oxygen flow. Anesthesia was maintained by reducing the anesthetic agent to 0.8-2 vol%. The chest and abdominal areas were shaved to perform echocardiography on all animals. During six cardiography recordings, 2 min of video was captured, each. The reference ECG was acquired using the Vevo 2100 Imaging System (VisualSonics, Toronto, ON, Canada), which was also used for the echocardiographies. Both, RR and HR were recorded by capturing the screen of the ultrasound scanner and were later manually digitized. Approval for this study was granted by “Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit” (Germany; 33.12-42502-04-15/1978).
Five additional male mice (two healthy C57BL/6 mice and three colitis mice C57BL/6.129P2-Il10tm1Cgn) aged 14 weeks were provided from the Institute for Laboratory Animal Science of Hannover Medical School (M7-M11). The animals were anesthetized with 5 vol% Isoflurane at 6 L min−1 oxygen flow in an induction chamber. Afterwards, anesthesia was maintained with 1.5 - 2.5 vol% Isoflurane and an oxygen flow of 1 L min−1. The chest and abdominal areas were shaved, as required by the original protocol. During anesthesia, 10 min of video were recorded. For the HR reference a KentScientific PhysioSuite system, including a pulse oximetry unit was used. The study was again approved by “Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit” (Germany; 33.19-42502-04-15/1905)
All trials are summarized in Table 1 and were performed according to the EU directive 2010/63/EU and the German animal welfare law.
Table 1. Summarized data set description.
| Animal | Species | Strain | Intervention | Supplier |
|---|---|---|---|---|
| R1 - R6 | Rat | WISTAR | SHAM laparotomy | Institute for Laboratory Animal Science and Experimental Surgery, University Hospital RWTH Aachen |
| M1 - M6 | Mouse | C57BL/6 N Cr | Echo-cardiography | Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School |
| M7 - M8 | Mouse | C57BL/6 | Colitis monitoring | Institute for Laboratory Animal Science, Hannover Medical School |
| M9 - M11 | Mouse | C57BL/6 .129P2-Il10tm1Cgn | Colitis monitoring | Institute for Laboratory Animal Science, Hannover Medical School |
For the video recordings, an Mako G-223 B (Allied Vision GmbH, Stadtroda, Germany) camera with Bayer color filter and 12 bit pixel resolution, together with a KOWA LM12HC lens (Kowa Optimed GmbH, Düsseldorf, Germany) were, mounted on a tripod. The camera was set to a frame rate of 60 frames/sec and 640x1368 pixel resolution. The exposure time was set according to the ambient light conditions. No additional lighting was used and the frames were stored in an uncompressed, binary format. The camera was positioned according to Fig. 1. All raw video was converted to raw RGB footage using the debayering algorithm provided by the Allied Vision Vimba SDK.
The mice provided by the Institute for Laboratory Animal Science were used to develop the presented algorithms. The remaining mice and rats were used to evaluate the algorithms by comparing it with the recorded references through a statistical analysis.
3. Results
3.1. Animal detection
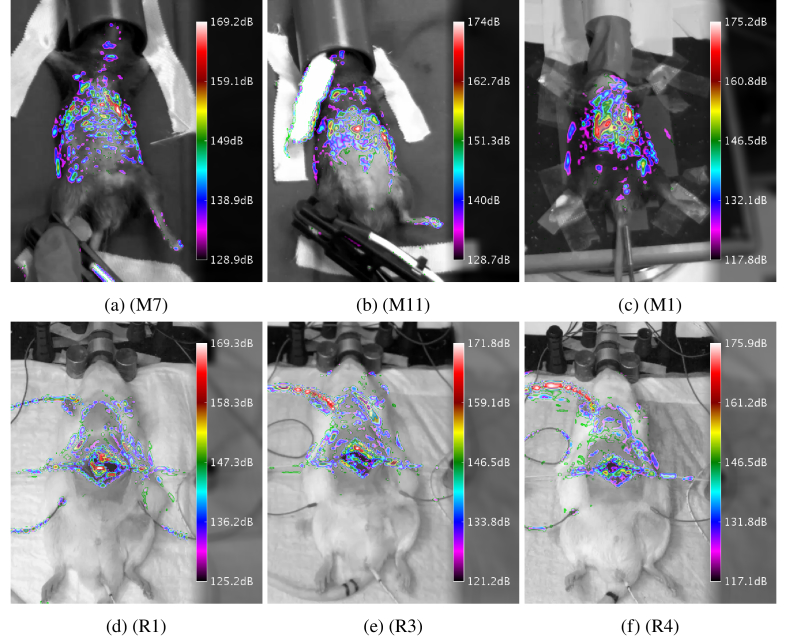
The animal detection was verified by comparing the computed pulsatility masks with a frame of the underlying video. Some representative results are shown in Fig. 8 . The masks have clear maxima in the chest- and abdominal regions, which corresponds well to the respiratory activity. Figures 8(e) and 8(f) show a maximum pulsatility located on a cable, which was attached to the animal, but not on the surface on the animal itself.
Fig. 8.
Results of the animal detection, using the pulsatility analysis for selected animals. Animals in top row are mice. The bottom row contains rats.
3.2. Respiratory rate extraction
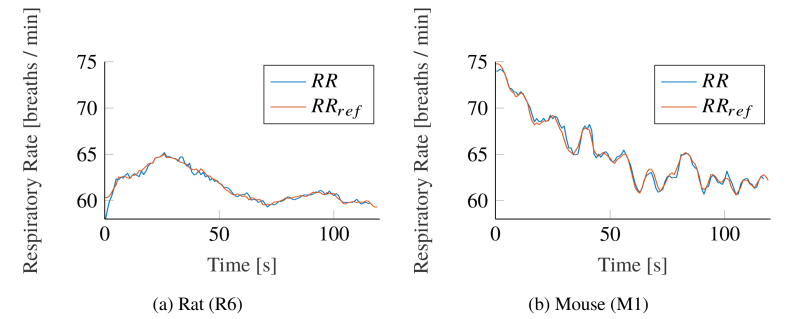
Figure 9 shows both, the result of our proposed algorithm and the reference RR for exemplary animals qualitatively. The accordance between both signals is good in rats and mice. While the mice showed larger variations in their RR (see Fig. 9(b)), the anesthetized rats breathed more constantly (see Fig. 9(a)).
Fig. 9.
Exemplary Respiratory Rate (RR) tracking comparison between the presented algorithm and the reference signal
Table 2 shows the quantitative performance of the presented algorithm for RR extraction in rats. The mean root mean squared error (RMSE) of breaths /min, minimal relative errors %) and high correlations with a mean of show a good accordance between the extracted signals and ECG reference.
Table 2. Results for respiratory rate estimation in videos from rats.
| Rodent ID | Mean RR (breaths/min) |
RMSE | (%) | (%) | Correlation | |
|---|---|---|---|---|---|---|
| IMG | REF | (breaths/min) | ||||
| R1 | 51.16 | 51.15 | 0.30 | 0.00 | 0.01 | 0.98 |
| R2 | 42.13 | 42.11 | 0.20 | 0.00 | 0.01 | 0.96 |
| R3 | 53.84 | 53.61 | 0.29 | 0.00 | 0.01 | 0.97 |
| R4 | 43.59 | 43.55 | 0.24 | 0.00 | 0.01 | 0.96 |
| R5 | 49.45 | 49.37 | 0.42 | 0.01 | 0.01 | 0.97 |
| R6 | 61.55 | 61.54 | 0.50 | 0.01 | 0.01 | 0.96 |
| Mean ± SD | 50.29±7.10 | 50.22±7.09 | 0.32±0.11 | 0.01±0.00 | 0.01±0.00 | 0.97±0.01 |
IMG: Image-derived, REF: Reference Measurement, : mean relative error, : 90th percentile of the relative errors
The results for mice are shown in Table 3. As represented, the mean RMSE averaged breaths /min and the mean relative error was %, which significantly differed from the observations in rats. The correlation of was comperable with the results for rats.
Table 3. Results for respiratory rate estimation in videos from mice.
| Rodent ID | Mean RR (breaths/min) |
RMSE | (%) | (%) | Correlation | |
|---|---|---|---|---|---|---|
| IMG | REF | (breaths/min) | ||||
| M1 | 112.6 | 112.9 | 1.68 | 1.25 | 2.42 | 0.86 |
| M2 | 65.00 | 65.07 | 0.85 | 0.99 | 2.08 | 0.97 |
| M3 | 92.04 | 92.39 | 1.52 | 1.26 | 2.59 | 0.88 |
| M4 | 161.2 | 160.2 | 0.40 | 0.64 | 2.12 | 0.96 |
| M5 | 79.66 | 79.70 | 0.94 | 0.92 | 1.90 | 0.97 |
| M6 | 73.44 | 74,84 | 3.15 | 2.59 | 5.84 | 0.87 |
| Mean±SD | 97.34±35.43 | 97.53±34.87 | 1.42±0.97 | 1.27±0.69 | 2.82±1.50 | 0.92±0.05 |
IMG: Image-derived, REF: Reference Measurement, : mean relative error, : 90th percentile of the relative errors
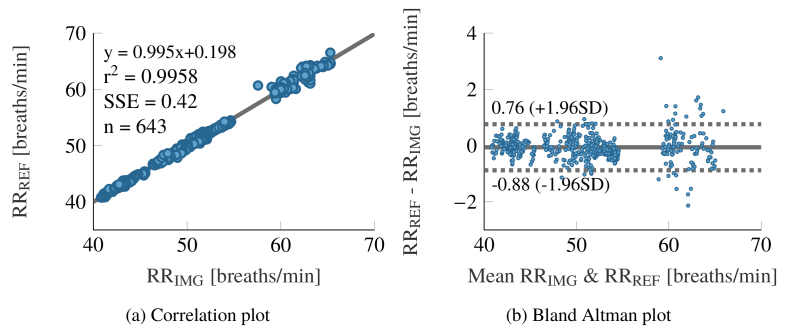
Figure 10 shows the statistical analysis for the RR signals extracted from all rats. The correlation plot 10a shows a good accordance between both signal sources. The sum squared error (SSE) of 0.42 breaths /min is sufficiently small. No bias can be observed in the Bland Altman plot (Fig. 10(b)), but the random errors appear to grow with frequency.
Fig. 10.
Statistical analysis between extracted and reference respiration rate. The plots comprise the data of all rats.
3.3. Heart rate extraction
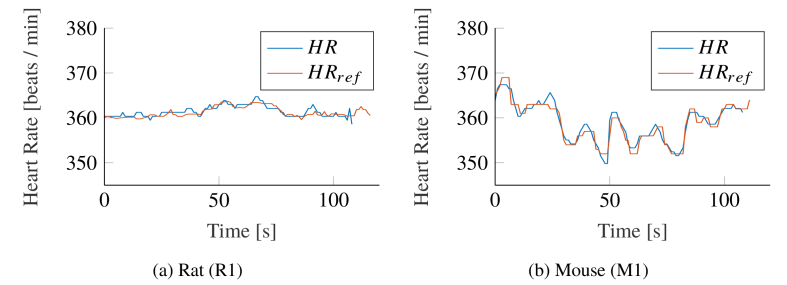
In Fig. 11 , we compare our algorithm with the reference HR for exemplary animals of both species qualitatively. Both signals show a good accordance, but while the HR in rats remained mostly constant during our measurements (see Fig. 11(a)), mice showed larger variations (Fig. 11(b)).
Fig. 11.
Exemplary Heart Rate (HR) tracking comparison between the presented algorithm and the reference signal
The quantitative performance of the HR extraction in rats is shown in Table 4. A mean RMSE of beats /min and mean relative errors of % are good. The mean correlation of 0.86 with a high standard deviation of 0.12 implies minor performance of the HR extraction algorithm in some cases. In fact, rat video R6 mainly accounts for the high deviation on its own, as its correlation coefficient of 0.64 is significantly lower than the rest.
Table 4. Results for heart rate estimation in videos from rats.
| Rodent ID | Mean HR (breaths/min) |
RMSE | (%) | (%) | Correlation | |
|---|---|---|---|---|---|---|
| IMG | REF | (breaths/min) | ||||
| R1 | 361.42 | 361.23 | 0.73 | 0.16 | 0.36 | 0.83 |
| R2 | 319,80 | 319,37 | 0.61 | 0.16 | 0.33 | 0.98 |
| R3 | 324.43 | 323.46 | 1.45 | 0.36 | 0.79 | 0.90 |
| R4 | 390.42 | 389.37 | 2.10 | 0.41 | 0.93 | 0.88 |
| R5 | 418.58 | 416.84 | 1.86 | 0.41 | 0.65 | 0.94 |
| R6 | 426.38 | 426.49 | 0.95 | 0.19 | 0.38 | 0.64 |
| Mean±SD | 373.51±45.94 | 372.79±45.87 | 1.28±0.62 | 0.28±0.13 | 0.57±0.25 | 0.86±0.12 |
IMG: Image-derived, REF: Reference Measurement, : mean relative error, : 90th percentile of the relative errors
For mice, the mean correlation is , which is better compared to the rats (see Table 5). The mean RMSE of beats /min and mean relative error of % is comparable to the results in rats.
Table 5. Results for heart rate estimation in videos from mice.
| Rodent ID | Mean HR (breaths/min) |
RMSE | (%) | (%) | Correlation | |
|---|---|---|---|---|---|---|
| IMG | REF | (breaths/min) | ||||
| M1 | 358.89 | 358.74 | 1.26 | 0.27 | 0.59 | 0.96 |
| M2 | 467.38 | 467.48 | 0.77 | 0.13 | 0.28 | 0.99 |
| M3 | 370.13 | 370.08 | 1.78 | 0.40 | 0.78 | 0.93 |
| M4 | 363.46 | 363.34 | 1.64 | 0.34 | 0.79 | 0.95 |
| M5 | 391.19 | 381.20 | 0.10 | 0.00 | 0.31 | 0.97 |
| M6 | 377.51 | 377.04 | 2.63 | 0.47 | 1.02 | 0.76 |
| Mean±SD | 386.43±40.53 | 386.31±40.62 | 1.36±0.87 | 0.27±0.18 | 0.63±0.29 | 0.93±0.09 |
IMG: Image-derived, REF: Reference Measurement, : mean relative error, : 90th percentile of the relative errors
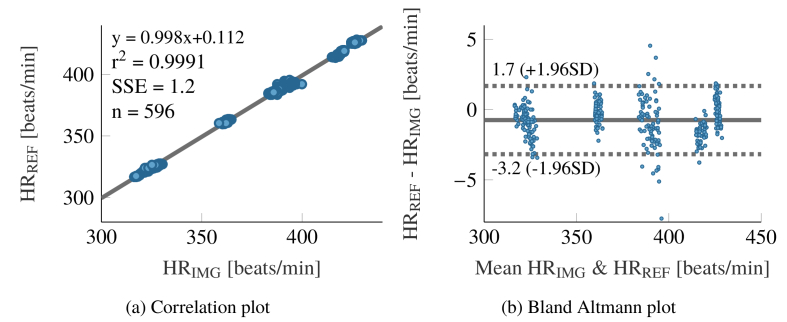
Figure 12 shows the statistical analysis of the HR computation for all recorded rats. The correlation plot 12a shows a good accordance between the computed- and reference HR. The value of 0.9991 is very good. A bias of −0.74 beats /min can be observed for the overall data set through the Bland Altman plot (Fig. 12(b)). The subsets corresponding to the different animals can clearly be seen, as their HR were almost constant over the recordings. All show different biases.
Fig. 12.
Statistical analysis between extracted and reference heart rate. The plots comprise the data of all rats.
4. Discussion
The presented research work aims to evaluate the possibility of automatically monitoring rodents during general anesthesia contactlessly and unobtrusively. Assessing important vital parameters such as RR and HR is important for laboratory animal science, since it delivers valuable information about the animal undergoing anesthesia. In most lab settings, only one researcher is responsible for both, the intervention, requiring anesthesia and the maintenance of the anesthesia itself. Providing an automated system, which is able to monitor the animals state and to warn in case of significant deviations from the norm, can improve the outcome of interventions, reduce the number of complications and thus reduce the number of animals needed for the research question.
Generally our results were promising. We proved the feasibility of an automated and contact-less approach for vitals monitoring in rodents. The evaluation of the presented algorithm revealed a few points, which should be discussed in the following:
-
•
In some videos, external hardware, such as cables were detected as regions of the animal’s body.
-
•
The errors in the RR detection of mice were significantly bigger than those for rats.
-
•
The computed HR didn’t correlate well with its reference in rat R6.
Cables, attached to the animals body can act like levers, which amplify the vital signals under certain conditions. In these situations the signal on the cables are larger than on the body itself. Thus, the developed algorithm treats the cable as part of the animal and extracts the vitals signal mainly from this region. This fact changes the form of the extracted waveforms, but not the retrieved HRs and RRs, so it is of no relevance for this work.
The significantly higher errors in the statistics for the extracted RR in mice M1-M6 can be explained with an unreliable reference signal. Since it was designed for the use on humans, the Vevo 2100 system had trouble in determining the correct RR of mice. This was notable through many physiologically not explainable signal jumps. The system extracts the breath rate from a lowpass filtered version of the ECG signal. Since the cutoff frequency of this filter is ideal for human- but to low for rodent RRs, we think that in fact our proposed method performed more reliable than the reference signal.
While the HR of the recorded mice fluctuated noticeable over the recording duration, the HR of most of the rats remained very stable and showed little to non fluctuation during the measurements. This can be verified by the distinct value clusters in Fig. 12(b) and explains bad correlations, as correlation is a standardized measure for joint variance. For signals with little fluctuations, the joint variance is also small. Because the HR of rat R6 was particular constant, the correlation value delivered no viable information about the algorithm performance in this case.
In summary, we can conclude that all presented results prove the performance of the described algorithm. Automated, contactless and unobtrusive monitoring of rodents during anesthesia is possible and viable, not only under certain conditions, but in most lab settings.
There exist a few devices, which are also able to assess rodent vitals during anesthesia. In fact, we used multiple systems to compare them against our method. The shortcoming of all those solutions is that they aren’t contact-free, which means additional hardware restraining the researcher even more in his already little space to work. Moreover, the contact-based methods did not perform better for RR monitoring than the presented approach. Therefore we think a contactless modality is greatly beneficial.
Zhao et. al [20] already proposed another possibility to extract HR and RR from animals in 2013. Using the videos color information in conjunction with a delay vector independent component analysis (ICA) signal separation approach [29], they were able to extract both, RR and HR rate from mice, fish and pigs.
The difference to our proposed approach is that they used the color signal, obtained from a region of interest around the tail to extract HR and a region of interest around the chest for the RR extraction in mice. This is not a feasible solution for our case, since we aim to use the developed algorithm not only in sedated, but also in alert rodents. In this case, the tail is hard to track or completely hidden beneath the cage litter, which makes HR extractions impossible.
Our method, on the other hand, extracts the vitals directly from the animals trunk. Here, the delay vector ICA approach would not be able to separate the raw signal into a RR-, a HR- and a noise related component. Our algorithm on the other hand is specifically designed for this case and handles it well. Thanks to this approach, it does not matter whether the animals have fur or are partially hidden beneath the cage litter. Furthermore, the tracking of a rodents trunk is easily possible [13].
5. Conclusion and outlook
This joint research work presents the possibility of camera-based vital signs monitoring in laboratory animal science. Our results can potentially be used to monitor and improve the well-being of animals, used for experimentation. The automated assessment of vitals, using unobtrusive, cost-effective hardware is not only an important step towards an objective severity assessment, which can contribute significantly to the refinement of animal trials. It can also be used to monitor animals during anesthesia and potentially prevent over dosage, which can lead to hypoxemia and death.
To reach this goal, the presented algorithm needs to be made real-time capable. An implementation in more efficient programming language, such as C++ and the use of available graphics acceleration for redundant tasks, like resampling and FFT computation should allow the algorithm to run in real-time on standard hardware.
Another important goal is to test and adjust the presented algorithm to work with alert, moving animals. Continuous, real-time vitals monitoring in the home cage is an important milestone to improve housing conditions and animal welfare, not only for laboratory animals, but for livestock, as well.
Ethics
The animal protocols used in this work were evaluated and approved by the governmental institutions:
-
1.
“Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit” (Germany; approval IDs: 33.12-42502-04-15/1978 for M1-M6 and 33.19-42502-04-15/1905 for M7-M11)
-
2.
“Landesamt für Natur, Umwelt und Verbraucherschutz NRW” (Germany; approval ID: 84-02.04.2017.A304 for R1-R6)
The trials were performed according to the EU directive 2010/63/EU and the German animal welfare law.
All videos were taken during ongoing projects. No animal was specifically handled or anesthetized for the purpose of video recording. The videos were taken during scheduled interventions.
Acknowledgments
The authors wish to acknowledge Anna Kümmecke for her help in conducting the studies and Christina Holländer for assisting in the evaluation process.
Acronyms
- DFG
German Research Association
- ECG
Electrocardiogram
- FFT
fast Fourier transform
- HR
heart rate
- ICA
independent component analysis
- KLT
Kanade-Lucas-Tomasi
- PCA
principle component analysis
- PPG
photoplethysmography
- RanSaC
random sample consensus
- RMSE
root mean squared error
- rPPG
remote photoplethysmography
- RR
respiratory rate
- SNR
signal to noise ratio
- SSE
sum squared error
Funding
Deutsche Forschungsgemeinschaft (DFG)10.13039/501100001659 (321137804, BL953/10-1, CZ215/3-1, TH903/22-1, TO 542/5-1).
Disclosures
JK and CBP: Docs in Clouds GmbH, camera-based vitals monitoring, Vaalser Str. 460 52074 Aachen, Germany (E).
MC: Docs in Clouds GmbH, camera-based vitals monitoring, Vaalser Str. 460 52074 Aachen, Germany (I,E).
TT: Cardior Pharmaceuticals GmbH, Feodor-Lynen-Str.15 30625 Hannover, Germany (I,E).
References
- 1.Fernandes M. R., Pedroso A. R., “Animal experimentation: A look into ethics, welfare and alternative methods,” Rev. Assoc. Med. Bras. 63(11), 923–928 (2017). 10.1590/1806-9282.63.11.923 [DOI] [PubMed] [Google Scholar]
- 2.Festing S., Wilkinson R., “The ethics of animal research. Talking Point on the use of animals in scientific research,” EMBO Rep. 8(6), 526–530 (2007). 10.1038/sj.embor.7400993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehinde E. O., “They See a Rat, We Seek a Cure for Diseases: The Current Status of Animal Experimentation in Medical Practice,” Med. Princ. Pract. 22(s1), 52–61 (2013). 10.1159/000355504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy N., “The Use of Animal as Models: Ethical Considerations,” Int. J. Stroke 7(5), 440–442 (2012). 10.1111/j.1747-4949.2012.00772.x [DOI] [PubMed] [Google Scholar]
- 5.Russel W., Burch R., The principle of humane experimental technique (Methuen, London, 1959). [Google Scholar]
- 6.Roelfsema P., Treue S., “Basic Neuroscience Research with Nonhuman Primates: A Small but Indispensable Component of Biomedical Research,” Neuron 82(6), 1200–1204 (2014). 10.1016/j.neuron.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 7.Hajar R., “Animal testing and medicine,” Hear. Views 12(1), 42 (2011). 10.4103/1995-705X.81548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleich A., Tolba R. H., “How can we assess their suffering? german research consortium aims at defining a severity assessment framework for laboratory animals,” Lab. Anim. 51(6), 667 (2017). 10.1177/0023677217733010 [DOI] [PubMed] [Google Scholar]
- 9.Häger C., Keubler L. M., Talbot S. R., Biernot S., Weegh N., Buchheister S., Buettner M., Glage S., Bleich A., “Running in the wheel: Defining individual severity levels in mice,” PLoS Biol. 16(10), e2006159 (2018). 10.1371/journal.pbio.2006159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunamaker E. A., Artwohl J. E., Anderson R. J., Fortman J. D., “Endpoint refinement for total body irradiation of C57bl/6 mice,” Comp. Med. 63, 22–28 (2013). [PMC free article] [PubMed] [Google Scholar]
- 11.Langford D. J., Bailey A. L., Chanda M. L., Clarke S. E., Drummond T. E., Echols S., Glick S., Ingrao J., Klassen-Ross T., LaCroix-Fralish M. L., Matsumiya L., Sorge R. E., Sotocinal S. G., Tabaka J. M., Wong D., M. van den Maagdenberg A. M. J., Ferrari M. D., Craig K. D., Mogil J. S., “Coding of facial expressions of pain in the laboratory mouse,” Nat. Methods 7(6), 447–449 (2010). 10.1038/nmeth.1455 [DOI] [PubMed] [Google Scholar]
- 12.Cinque C., Zinni M., Zuena A. R., Giuli C., Alema S. G., Catalani A., Casolini P., Cozzolino R., “Faecal corticosterone metabolite assessment in socially housed male and female Wistar rats,” Endocr. Connect. 7(2), 250–257 (2018). 10.1530/EC-17-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira C., Kunczik J., Zieglowski L., Tolba R., Abdelrahman A., Zechner D., Vollmar B., Janssen H., Thum T., Czaplik M., “Remote Welfare Monitoring of Rodents Using Thermal Imaging,” Sensors 18(11), 3653 (2018). 10.3390/s18113653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Sánchez C., Fraile J.-C., Pérez-Turiel J., Damm E., Schneider J., Zimmermann H., Schmitt D., Ihmig F., “Capacitive Sensing for Non-Invasive Breathing and Heart Monitoring in Non-Restrained, Non-Sedated Laboratory Mice,” Sensors 16(7), 1052 (2016). 10.3390/s16071052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesarovic N., Jirkof P., Rettich A., Arras M., “Implantation of Radiotelemetry Transmitters Yielding Data on ECG, Heart Rate, Core Body Temperature and Activity in Free-moving Laboratory Mice,” J. Visualized Exp. 57, e3260 (2011). 10.3791/3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helwig B. G., Ward J. A., Blaha M. D., Leon L. R., “Effect of intraperitoneal radiotelemetry instrumentation on voluntary wheel running and surgical recovery in mice,” J. Am. Assoc. for Lab. Animal Sci. JAALAS 51, 600–608 (2012). [PMC free article] [PubMed] [Google Scholar]
- 17.Niemeyer J. E., “Telemetry for small animal physiology,” Lab Anim 45(7), 255–257 (2016). 10.1038/laban.1048 [DOI] [PubMed] [Google Scholar]
- 18.Mutlu K., Rabell J. E., Martin del Olmo P., Haesler S., “IR thermography-based monitoring of respiration phase without image segmentation,” J. Neurosci. Methods 301, 1–8 (2018). 10.1016/j.jneumeth.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 19.Vainer B. G., “A Novel High-Resolution Method for the Respiration Rate and Breathing Waveforms Remote Monitoring,” Ann. Biomed. Eng. 46(7), 960–971 (2018). 10.1007/s10439-018-2018-6 [DOI] [PubMed] [Google Scholar]
- 20.Zhao F., Li M., Qian Y., Tsien J. Z., “Remote measurements of heart and respiration rates for telemedicine,” PLoS One 8(10), e71384 (2013). 10.1371/journal.pone.0071384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDuff D. J., Estepp J. R., Piasecki A. M., Blackford E. B., A survey of remote optical photoplethysmographic imaging methods, in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), (IEEE, Milan, 2015), pp. 6398–6404. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y., Thakor N., “Photoplethysmography Revisited: From Contact to Noncontact, From Point to Imaging,” IEEE Trans. Biomed. Eng. 63(3), 463–477 (2016). 10.1109/TBME.2015.2476337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar M., Veeraraghavan A., Sabharwal A., “DistancePPG: Robust non-contact vital signs monitoring using a camera,” Biomed. Opt. Express 6(5), 1565 (2015). 10.1364/BOE.6.001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischler M. A., Bolles R. C., “Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography,” Commun. ACM 24(6), 381–395 (1981). 10.1145/358669.358692 [DOI] [Google Scholar]
- 25.Shi J., Tomasi C., “Good features to track” (1994), pp. 593–600.
- 26.Tomasi C., Kanade T., “Detection and tracking of point features,” Tech. rep., International Journal of Computer Vision (1991).
- 27.Balakrishnan G., Durand F., Guttag J., “Detecting pulse from head motions in video,”, in 2013 IEEE Conference on Computer Vision and Pattern Recognition, (IEEE, 2013). [Google Scholar]
- 28.Frantz S., Hu K., Bayer B., Gerondakis S., Strotmann J., Adamek A., Ertl G., Bauersachs J., “Absence of NF-κb subunit p50 improves heart failure after myocardial infarction,” FASEB J. 20(11), 1918–1920 (2006). 10.1096/fj.05-5133fje [DOI] [PubMed] [Google Scholar]
- 29.McSharry P. E., Clifford G. D., “A comparison of nonlinear noise reduction and independent component analysis using a realistic dynamical model of the electrocardiogram,” in Fluctuations and Noise in Biological, Biophysical, and Biomedical Systems II, Abbott D., Bezrukov S. M., Der A., Sanchez A., eds. (SPIE, 2004). [Google Scholar]