Abstract
Complete tumor removal during breast-conserving surgery remains challenging due to the lack of optimal intraoperative margin assessment techniques. Here, we use hyperspectral imaging for tumor detection in fresh breast tissue. We evaluated different wavelength ranges and two classification algorithms; a pixel-wise classification algorithm and a convolutional neural network that combines spectral and spatial information. The highest classification performance was obtained using the full wavelength range (450-1650 nm). Adding spatial information mainly improved the differentiation of tissue classes within the malignant and healthy classes. High sensitivity and specificity were accomplished, which offers potential for hyperspectral imaging as a margin assessment technique to improve surgical outcome.
1. Introduction
Breast-conserving surgery remains challenging due to the lack of clinically available intraoperative resection margin techniques. Therefore, in up to 37% of the women undergoing breast-conserving surgery, tumor is found in the resection margin of the resected specimen [1–5]. This is an indication of residual tumor, left behind in the patient, which increases the risks for developing a local recurrence and compromises long-term disease-specific survival [6]. Therefore, these patients often require additional treatment like a radiotherapy boost or a re-excision [7]. Currently, a pathologist, who evaluates the tissue under a microscope, assesses the resection margin a few days after surgery. As such, no direct feedback can be given to the surgeon during surgery.
To reduce the number of tumor-positive resection margins, multiple techniques for resection margin assessment during breast-conserving surgery have been proposed [8–13]. Margin assessment techniques that are currently available are frozen section analysis, imprint cytology, ultrasound, and specimen radiography [8–10,13]. However, none of them have made it to widespread successful clinical use. With frozen section analysis, tissue can be analyzed within 30 minutes with a sensitivity and specificity of 83% and 95% [8,10]. However, the main limitations of the technique are the need for a specialized pathologist, risk of false negatives, and the impracticability for analyzing the complete resection surface. With imprint cytology, an imprint is made of all six resection margins and analyzed by a specialist cytologist. With this technique, a diagnosis can be provided within 15 minutes. However, the sensitivity is limited to 72% due to errors in the interpretation of the data that are related to cytology expertise, the specimen surface irregularity, dryness and presence of atypical cells [8,10]. Ultrasound and specimen radiography are faster margin assessment techniques. However, both techniques are inferior to previously mentioned pathological techniques; ultrasound (sensitivity and specificity of 59% and 81%) has a limited role in detecting ductal carcinoma in situ (DCIS), a potential precursor of invasive carcinoma (IC). And specimen radiography (sensitivity and specificity of 53% and 84%) does not clearly improve the reoperation rate but is merely helpful in detecting microcalcifications [9,13,14].
A technique that offers great potential is hyperspectral (HS) imaging of diffusely reflected light. HS imaging is an optical imaging technique that can measure the entire resection margin within a limited amount of time, without tissue contact, and without the need for exogenous contrast agents [15]. HS imaging consists in capturing hundreds of images in narrow, contiguous and adjacent spectral bands over a wide spectral range. Thereby, a 3D hypercube is created that contains both spectral and spatial information of the imaged scene. HS imaging measures diffusely reflected light after it has undergone multiple scattering and absorption events within the tissue. Thereby, an ‘optical fingerprint’ of the tissue is obtained that reflects the composition and morphology of the tissue, which can be used for tissue analysis. Previous studies show the usefulness of HS imaging in noninvasive tissue analyses [16] and in the detection of cancer in ex vivo human tissue in head and neck [17,18], colon [19], skin [20], and breast tissue [21,22].
In a recent publication of our group, we showed promising results of classifying tumor in freshly excised breast tissue using an HS camera that operates in the near-infrared (NIR) wavelength range (∼900-1700 nm) [22]. The most challenging tissue types to differentiate were connective tissue and DCIS, which are in general the smallest tissue types: connective tissue often consists of small strands of collagen in and around a tumor or within adipose tissue, whereas DCIS is the precursor of IC and starts as a few premalignant cells in a glandular duct. This makes it difficult to measure a spot of only DCIS or only connective tissue. Especially with HS imaging, which measures tissue volumes in the range of square millimeters, the measured diffuse reflectance spectrum will represent a mixture of different tissue classes when the strands or DCIS pockets are smaller than the measured volume. HS data analysis can be performed using only the spectral information of the hypercube, i.e. considering each pixel as an individual measurement. However, HS data also contains valuable spatial and contextual information of the imaged scene. This spatial information can, when added to the classification methods, help to improve the detection of DCIS and connective tissue.
In this paper, spectral and spatial information is used to discriminate tumor from healthy tissue in broadband HS images obtained on fresh breast tissue slices. The novel contributions of this paper can be summarized as follows:
-
i
We perform measurements on fresh breast tissue slices, after gross-sectioning of the resection specimen. This allows us to create an extensive dataset with a high correlation with the gold standard, histopathologic assessment of the tissue under a microscope. With this dataset, algorithms can be developed that can directly be applied to the resection surface of unsliced resection specimen.
-
ii
We image all specimens with two HS cameras, covering the full wavelength range from 450-1650 nm. In previous HS research, this wavelength was limited to either the visual (VIS; 450-951 nm) or NIR wavelength (954-1650 nm) range. In Section 3.3, we demonstrate that this full spectral range (450-1650 nm) is required to obtain the highest discrimination between tumor and healthy tissue.
-
iii
We perform HS analysis with 1) a spectral algorithm that considers each pixel in the image as an individual measurement, and 2) a spectral-spatial algorithm that incorporates both the spectral and spatial information for classification. In Section 3.4, we demonstrate that adding spatial information to the classification algorithm improved the capability of HS imaging to differentiate different tissue classes within the malignant and healthy tissue classes.
-
iv
We made a clear distinction between 1) a dataset containing all pixels with a histopathologic label, and 2) a dataset containing only pixels of which we were certain that the histopathologic label represents the measured tissue type: the given histopathologic labels reflect just a single tissue class. However, at tissue transitions, the diffuse reflectance spectrum reflects a mixture of different tissue classes due to the diffuse nature of the reflected light. For these pixels, the histopathologic label cannot represent all measured tissue classes. Therefore, these pixels are excluded from the second dataset. This is further explained in Section 2.3.1 and demonstrated in Section 3.2.
-
v
The most important contribution of this paper is the introduction of a new approach, using HS imaging, for the discrimination of tumor and healthy tissue in ex vivo breast samples within 2 minutes. This approach offers great potential to be used during breast-conserving surgery for ex vivo resection margin assessment to detect tumor-positive resection margins.
The remainder of this paper is organized as follows. Section 2 describes the HS imaging setup, the data acquisition on fresh breast tissue slices, the correlation of HS measurements with histopathology, and the data preprocessing and analysis methods. The experimental results are presented in Section 3, followed by the discussion and conclusion in Section 4 and 5, respectively.
2. Materials and methods
2.1. Hyperspectral imaging setup
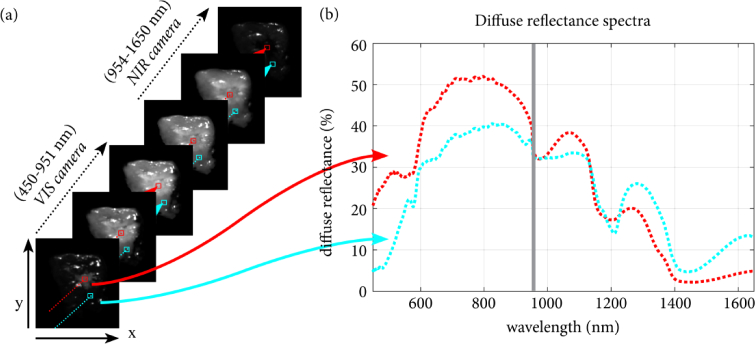
Hyperspectral images were measured with two push-broom HS imaging systems (Specim, Spectral Imaging Ltd., Finland) with a VIS camera (model: PFD-CL-65-V10E, lens: OLE 18.5 mm (Specim)) and a NIR camera (model: VLNIR CL-350-N17E, lens: OLES15 (Specim). These cameras operate respectively in the VIS (∼400-1000 nm, 384 wavelength bands, 3 nm increments) and NIR (∼900-1700 nm, 256 wavelength bands, 5 nm increments) wavelength range. Due to the low sensitivity of the sensors of the cameras at the edges of the spectral range, we only used wavelengths between 450 and 951 nm (318 wavelength bands) for the VIS camera. For the NIR camera, wavelengths between 954 and 1650 nm (210 wavelength bands) were used.
The tissue was placed under the camera on a translation stage, where it was illuminated by three halogen light sources (2900 K) under an angle of 45 degrees. By moving the translation stage, the tissue was imaged line-by-line so that the 3D hypercube was created. This hypercube contains both spectral and spatial information of the imaged scene, as shown in Fig. 1. Images acquired with the VIS camera (CMOS sensor with pixels) and the NIR camera (InGaAs sensor with pixels) had a spatial resolution of respectively 0.16 mm/pixel and 0.5 mm/pixel. The scanning speed for both cameras was adjusted to match the cameras spatial resolution of the imaged line. The imaging time required to acquire data of both the tissue sample (dimensions of the imaged scene: 12.5 cm × 18 cm) and two reference images (as described in Section 2.4) was 20 seconds for the NIR camera and 40 seconds for the VIS camera.
Fig. 1.
Hyperspectral data. a) The tissue was imaged with both the VIS and NIR camera. Thereby, two 3D hypercubes were created that contain both spectral and spatial information of the imaged scene. Therefore, each vector in the 3D HS image contains an entire spectrum over a broad wavelength range, as shown in (b). The red and cyan diffuse reflectance spectra shown in (b) correspond to the red and cyan selected pixels in (a). In this example, the VIS image was resized to match the resolution of the NIR image, as described in Section 2.4.
2.2. Data acquisition of breast tissue slices
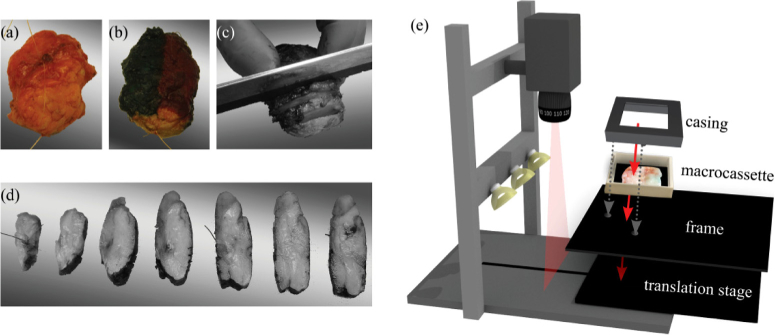
Measurements were performed on fresh ex vivo tissue from patients that had primary breast surgery at the Antoni van Leeuwenhoek hospital. This study was performed in compliance with the Declaration of Helsinki and approved by the Institutional Review Board of the Netherlands Cancer Institute/Antoni van Leeuwenhoek (Amsterdam, the Netherlands). According to Dutch law (WMO), no written informed consent from patients was required. After resection, the specimen was brought to the pathology department where it was inked and sliced according to standard protocol (Fig. 2(a-d)). In consultation with the pathologist, one tissue slice, that contained both healthy and tumor tissue, was placed in a macrocassette on top of black rubber and used for the optical measurements. The black rubber highly absorbs light from 400-1700 nm and prevents that any material underneath the tissue is measured. All tissue slices were at least 2 mm thick and imaged 4 times; 2 times with the VIS camera and 2 times with the NIR camera. For each camera, the tissue slice was rotated 180° by rotating the entire macrocassette, so that the tissue was illuminated from a different point of view. To ensure a reproducible location for each measurement, the macrocassette with the tissue was fixed with a casing on a frame with two holes that fitted the translational stage of both cameras (Fig. 2(e)). Both the casing and the frame were made of black polyoxymethylene that highly absorbs light from 400-1700 nm. After the optical measurements, which were performed within 10 minutes after collection of the tissue at the pathology department, the tissue was placed in formaldehyde and processed according to standard protocol.
Fig. 2.
Data acquisition of breast tissue slices. The breast specimen before (a) and after (b) inking with histopathologic ink. The specimen was sliced (c, d), and one slice was selected and placed in a macrocassette for optical measurements. The tissue slice was imaged with both HS imaging systems (e). To allow for a reproducible location for each measurement and an accurate registration between both cameras, the macrocassette with the tissue was fixed with a casing on a frame that fitted the translational stage of both systems.
2.3. Histopathologic annotation of breast tissue slices
A few days after surgery, the tissue slices were processed in hematoxylin and eosin (H&E) stained sections and analyzed by a pathologist. We used the histopathologic analysis of the tissue in these H&E stained sections as ground truth for the optical measurements. To correct for the tissue deformation that occurred during histopathologic processing of the tissue, the digitalized H&E sections were registered to the HS images using a white light image that was taken just before taking the HS image. With the annotations on the registered H&E sections, the whole HS image was annotated with four tissue classes, which were IC, DCIS, connective (including healthy glandular ducts), and adipose tissue. A detailed explanation of this annotation and registration process was described in our earlier publication [22].
2.3.1. Selection of two datasets: ALL and RIGHT dataset
However, even with a highly accurate registration of H&E sections to HS images, the tissue in the H&E annotations might not reflect the tissue that was optically measured. First, in the classified HS images, the size of each pixel was 0.5 mm × 0.5 mm. Due to this small size, the labeling of HS data was more sensitive to errors in the histopathologic registration. Second, the pathologist delineated tumor areas on the H&E sections. However, it might occur that within these delineations also healthy connective and adipose tissue was present. Third, the H&E sections represent a 2-dimensional section of the measured tissue whereas with HS imaging we measure a volume up to a few mm underneath the surface of the tissue. Fourth, the optical resolution of light is lower than the resolution of the HS images. Therefore, even though one pixel represents a tissue surface of 0.5 mm × 0.5 mm, the diffuse reflectance spectrum obtained with HS imaging originates from a much larger sampling volume. Therefore, the diffuse reflectance spectrum of a specific pixel might have been shaped by a different tissue type located at 1 mm distance.
As a result, the diffuse reflectance spectrum at one pixel might represent a mixture of different tissue types instead of the single tissue type provided by H&E annotations. Incorrect histopathologic annotations can decrease the classification performance of HS imaging. Therefore, we made a clear distinction between all pixels with a histopathologic label, the so-called ‘ALL’ (All histopathoLogy Labels) dataset, and pixels of which we are certain about the histopathologic label, the ‘RIGHT’ (RelIable Ground trutH annoTations) dataset.
Figure 3 shows the selection process of the RIGHT dataset. First, we manually selected adipose tissue (Fig. 3(d)) by thresholding all RGB channels in the H&E image (Fig. 3(a)): since the adipose content of the cells was washed away in the histopathologic processing, adipose cells were transparent on H&E sections and could be easily discriminated from the other tissue types. Second, we manually segmented the remaining tissue in the H&E image (Fig. 3(b)) to differentiate regions with high and low nuclei density by thresholding the red channel of the image: the H&E stain causes nuclei to stain dark blue, whereas amino acids and proteins turn red/pink. Since the nuclei density in connective tissue is low, this enabled us to select connective tissue (Fig. 3(e)). Third, we selected IC, DCIS and healthy glandular tissue using the annotations of the pathologist (Fig. 3(f)) and grouped the glandular tissue in the connective class. Finally, in the RIGHT dataset (Fig. e(i)), the edges of each tissue class area (at a distance of 1 mm from the edge) were removed because pixels close to these edges are likely to contain mixed spectra due to previously mentioned reasons.
Fig. 3.
Selection of regions that contain a single tissue class. From the original H&E image (a), first adipose tissue (d) was selected by thresholding all RGB channels so that the remaining tissue (b) contained the malignant tissue types, connective tissue, and healthy glandular ducts. By thresholding the red channel of the H&E image, a differentiation was made between tissue with a high and a low nuclei density. Since the nuclei density in connective tissue is low, this enabled us to select connective tissue (e). On the remaining tissue (c), we selected IC, DCIS and healthy glandular tissue using the annotations of the pathologist (f), and grouped the glandular tissue in the connective class. Finally, for each tissue class, the edges of a tissue class area (1 mm) were removed to remain only with the RIGHT dataset (i) from the ALL dataset (h). In addition, pixels that were contaminated with histopathology ink, as indicated with the arrows in (i) and the white light image (g), were removed from this RIGHT dataset.
2.4. Hyperspectral data preprocessing
All data analysis and tissue classification were performed using MATLAB 2018a (The Math Works Inc., Natick, Massachusetts, USA). Prior to classification, the HS data were preprocessed. First, raw HS data were converted from photon counts into normalized diffuse reflectance using:
| (1) |
where is the normalized diffuse reflectance (in percentage), the reference reflectance value of Spectralon (SRT-99-100, Lapsphere, Inc., Northern Sutton, New Hampshire, USA), x the location of the pixel in the imaged line and the wavelength band. and are the reference images acquired in addition to the tissue image (). The white reference image was acquired on Spectralon 99% and for the dark reference image, we closed the shutter of the camera. Prior to this normalization, we corrected for the slight non-linearity of the InGaAs sensor using a 3rd order polynomial [22].
Second, after segmenting the background from the tissue samples, pixels were excluded from analysis if they were over-illuminated or contaminated with pathology ink.
Third, HS images obtained with both cameras were registered using an affine registration algorithm based on the shape of the tissue in both images. To remain with the higher resolution of the VIS camera for the spectral-spatial algorithm, HS images obtained with the VIS camera were resized to match twice the resolution of the HS images obtained with the NIR camera. For the spectral algorithm, we further downsized the HS image obtained with the VIS camera to the resolution of the image obtained with the NIR camera.
Finally, the spectra obtained with each camera were pre-processed using standard normal variate (SNV) [23,24]. Thereby, spectral variability due to the oblique illumination and the nonflat surface of the tissue was eliminated by normalizing each individual spectrum to a mean of zero and a standard deviation of one.
2.5. Overview of classification algorithms
In this study, we first used a spectral classification algorithm to determine which wavelength range allows for the highest discrimination between tumor and healthy breast tissue with HS imaging. The wavelength ranges used were 1) visual (450-951 nm), 2) near-infrared (954-1650 nm) and 3) a combination of both (450-1650 nm). Second, we used the selected wavelength range as input for a classification algorithm that incorporates both spectral and spatial information. To allow for a good comparison between the different wavelength ranges and classification algorithms, the same training set (70% of the images) and test set (30% of the images) were used. Splitting of the whole dataset into a training and test set was performed randomly while keeping spectra from one patient together. To verify that the data partition was representative for the whole dataset, also a 7-fold cross-validation strategy was performed for the spectral classification algorithm and its results were compared to the results of the single data partition.
2.5.1. Spectral classification algorithm
First, a supervised classification model was developed that only incorporated the spectral information of the 3D hypercube, i.e. looks at each pixel individually without considering its surroundings. In the annotations of HS pixels with tissue types, it might occur that the diffuse reflectance spectrum in one pixel represents a mixture of different tissue types instead of the single tissue type provided by the H&E annotations. As the classification model will be affected by these incorrect histopathology annotations of the HS data, we only trained the model with the RIGHT pixels in the training set. As input for the spectral classification model, we used the wavelength range of the VIS camera, the NIR camera, and both cameras combined.
The SNV normalized RIGHT dataset was used to develop a supervised spectral classification model using Fisher's linear discriminant analysis (LDA). LDA finds a linear combination of wavelength bands that optimizes the separation between two classes. It maximizes a function that represents the difference between the means of two classes, normalized by a measure of the within-class variability [25]. Thereby, for each combination of two tissue classes (in total 6 combinations of two tissue types), LDA reduces the large number of wavelength bands to a single feature that most optimally separates the two tissue types. By setting a threshold between the two tissue classes for each feature, we obtained a multi-class classification algorithm. For each combination of tissue classes, the algorithm provides for each pixel the probability of the pixel belonging to a class. With pairwise coupling [26], those probabilities are combined so that finally, each pixel was labeled with one of the four tissue classes.
2.5.2. Spectral-spatial classification algorithm
Second, a deep neural network was used that incorporates both spectral and spatial information. Deep learning techniques usually require large training sets to achieve good performance. In addition, they require more pixels than only the RIGHT pixels to incorporate spatial information: in the RIGHT dataset, the contextual information of the complete imaged scene was altered. Therefore, at the edges of tissue areas in the RIGHT dataset (see Fig. 3(i)) some of the spatial information is missing. To increase the number of training samples and to keep the contextual information unaltered, all pixels with pathology labels (i.e. the ALL dataset) were used to train the algorithm. As input, we used one of the three wavelength ranges (VIS, NIR or a combination of both) that gave the highest performance for tissue discrimination using the spectral classification algorithm described in the previous subsection.
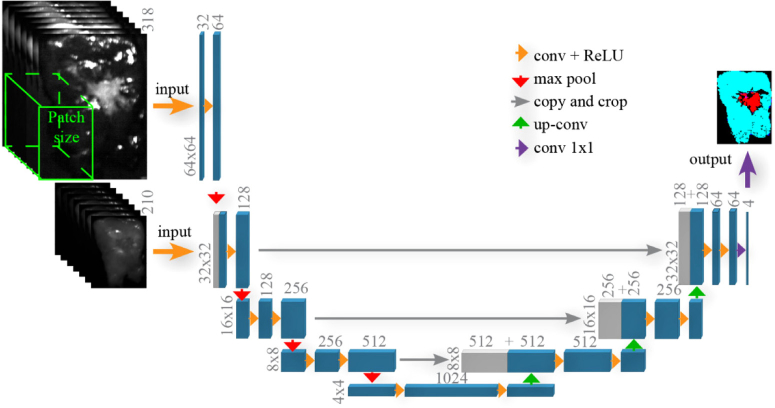
For the classification, we used a convolutional network architecture which was a modified version of U-Net [27]. U-Net allows for fast and precise segmentation of the HS images by using an architecture that consists of a contracting path to capture context and a symmetric expanding path that enables precise localization. Figure 4 shows a schematic overview of the multi-scale U-Net architecture. First, we start with the images obtained with the VIS camera that were twice the size of the images obtained with the NIR camera. We apply a multichannel feature web, which is a chain of 3D convolutional layers followed by a non-linear activation function (ReLU-rectified linear unit). Afterward, max pooling operators in regions were used, which reduced the size of the images and acted on each feature separately. In the second layer, the images obtained with the NIR camera are concatenated with the first layer output and used as inputs for this layer. After each max pooling operation, we increased the number of feature channels with a factor of two. To obtain segmentation results with a size similar to the HS NIR image, we used the expansion path. This path consisted of a sequence of up-convolutions and concatenation with high-resolution features. The up-convolution used a learned kernel to map each feature vector to the pixel output window, again followed by a non-linear activation function. The segmentation results were obtained by a 4-class softmax classifier at the end. A combination of cross entropy and Dice similarity coefficient was used to measure the loss value. The weights, , were included in the loss function to allow for reweighting of the strongly imbalanced classes:
| (2) |
where is the output of the softmax and denotes one-hot encoded ground truth, while runs over all samples and runs over the classes. controls the amount of the Dice term contribution in the loss function.
Fig. 4.
Architecture of U-Net for tissue segmentation.
Before feeding the images to the network, several pre-processing and data augmentation steps were applied. The first step consisted of cropping the borders of each image that did not contain tissue. Second, data augmentation steps included random scaling to of the initial size, horizontal /vertical flipping and random rotation between After applying these transformations, patches of pixels were extracted from the VIS camera. Next, from the NIR camera, corresponding patches of pixels were extracted. We used a Stochastic Gradient Descent optimization technique with a weight decay of 0.0001 and a momentum of 0.9. The learning rate was fixed to 0.001 for 100 epochs, then we decreased it to 0.0001 and trained the network for 200 more epochs. For the training and tuning model hyperparameters, the training set (described in Section 2.5) was randomly split into a training set (80% of the images) and a validation set (20% of the images) while keeping spectra from one patient together.
2.6. Performance metrics and statistical analysis
The classification performance of HS imaging on breast tissue was evaluated on the tissue slices that were used as test set. Clinically, differentiating tissue types within the healthy and tumor class is less relevant. Therefore, we evaluated the recall per tissue class, i.e. the percentage of pixels that were correctly classified as either tumor or healthy tissue. In addition, we calculated the Matthews Correlation Coefficient (MCC), true positive rate (sensitivity), true negative rate (specificity), positive predictive value (PPV) and negative predictive value (NPV). In these calculations, IC and DCIS pixels that were correctly classified as tumor were true positives (TP), and IC and DCIS pixels classified as healthy tissue were false negatives (FN). Connective and adipose pixels that were correctly classified as healthy tissue were true negatives (TN), whereas connective and adipose pixels classified as tumor were false positives (FP). MCC was used instead of accuracy since this performance metrics is able to handle the imbalance in measurements per tissue class in our dataset. MCC was calculated by [28,29]
| (3) |
and returns a value between and , which indicates no correlation and perfect correlation respectively. Therefore, a value of 0 indicates that the classification performance is no better than random prediction.
For the evaluation of a significant difference in the performance of the proposed tissue classification techniques, the paired nonparametric McNemar’s test was performed [30]. Thereby, the null hypothesis was tested that two algorithms disagree with the ground truth in the same way. P values < 0.05 are considered significant.
3. Experimental results
3.1. Data description
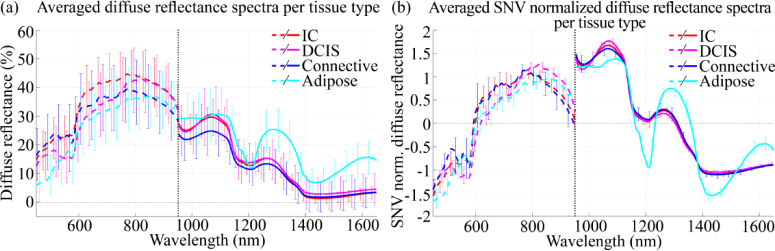
In total, 42 tissue slices from different patients were measured and divided into a training and test set. Table 1 shows the patient characteristics and the number of spectra per tissue class in each dataset. Figure 5(a) shows the average diffuse reflectance spectra for each tissue type. Two observations can be made: first, the standard deviations observed around the averaged diffuse reflectance spectra are large, and second, the spectra obtained with the VIS and NIR camera do not connect. Both observations are the result of the oblique illumination of the tissue in combination with the rough surface of the tissue, causing differences in illumination of the tissue and scatter nonspecific to the optical properties of the tissue measured. This is shown in Fig. 6(b); even when diffuse reflectance spectra are taken with the same camera, at the same location in the tissue slice, their intensity varied when the tissue was illuminated from a different point of view. This difference can be observed as a baseline shift of the diffuse reflectance spectrum, which caused the large standard deviations around the averaged spectra. Since both cameras have their own suspension system and light sources, the illumination angle of both cameras might differ slightly. Therefore, spectra obtained with both cameras did not connect. To correct for the differences in illumination and the nonspecific scatter, we used SNV normalization as a preprocessing step. As expected and shown in Fig. 5(b), this reduced the standard deviations around the averaged diffuse reflectance spectra. However, SNV was applied to the spectrum obtained with the VIS and NIR camera individually; therefore the SNV normalized spectra in Fig. 5(b) remained not connected.
Table 1. Data Description.
| Training set |
Test set |
||||
|---|---|---|---|---|---|
| ALL dataset | RIGHT dataset | ALL dataset | RIGHT dataset | ||
| Patient characteristics | Number | 29 (58 imagesa) | 13 (26 imagesa) | ||
| Age | 57 ± 11 | 58 ± 11 | |||
| Tissue class | IC | 13 (9,746) | 10 (2,854) | 11 (7,402) | 10 (3,200) |
| #patients (#spectra) | DCIS | 24 (11,694) | 5 (208) | 8 (1,483) | 3 (112) |
| Connective | 29 (69,612) | 10 (712) | 13 (15,731) | 5 (468) | |
| Adipose | 29 (133,809) | 29 (32,309) | 13 (59,720) | 13 (20,759) | |
| Total | 29 (224,861) | 29 (36,083) | 13 (84,336) | 13 (24,539) | |
Each tissue slice was imaged twice with each camera; the slice was rotated 180° so that it was illuminated from a different point of view. Both images from a tissue slice were assigned to the same set (training or test).
Fig. 5.
Averaged diffuse reflectance spectra for each tissue type in the test set before (a) and after (b) SNV normalization. The error bars indicate the standard deviation.
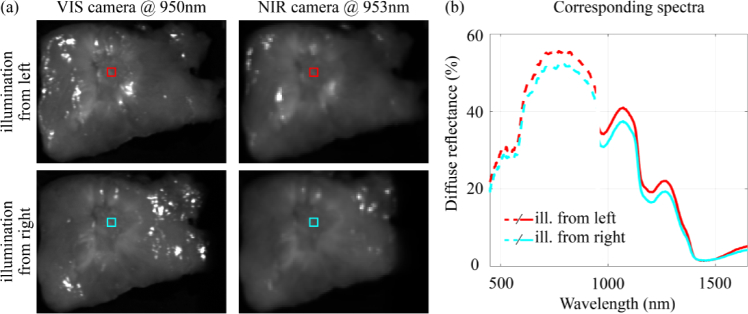
Fig. 6.
Intensity differences between spectra. (a) Hyperspectral images obtained with the VIS and NIR camera. In the top and bottom row, the tissue was illuminated from the left and the right, respectively. The colored squares in the HS images are located at the same position in the tissue and correspond to the diffuse reflectance spectra in (b). Spectra obtained with different cameras (VIS and NIR) did not connect due to differences in the measurement setup of both cameras in combination with the rough surface of the tissue slices. This might cause a spectral variability that can be observed as a baseline shift of the spectra. Even when diffuse reflectance spectra were taken with the same camera, at the same location in a tissue slice, their intensity varied when the tissue was illuminated from a different point of view.
Based on the SNV normalized spectra, shown in Fig. 5(b), adipose tissue differed the most from the other tissue classes in the NIR wavelength range, where the absorption of fat is most characteristic. For connective and the malignant classes, the spectral shape was more distinctive in the VIS wavelength range than in the NIR range.
3.2. ALL versus RIGHT dataset
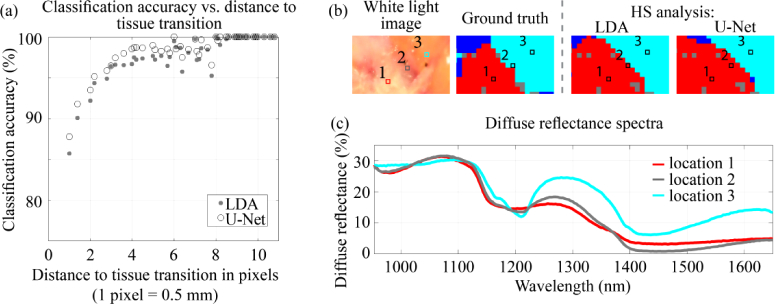
In this study, we made a clear distinction between all pixels with a histopathologic label, the ALL dataset, and pixels of which we are certain about the histopathologic label, the RIGHT dataset. In the ALL dataset, pixels that were classified incorrectly were mainly located at tissue transitions. This is shown in Fig. 7, which shows the classification accuracy of all pixels in the ALL dataset with respect to the distance to a tissue transition. At a tissue transition, the diffuse reflectance spectrum of a pixel will represent a mixture of optical properties of different tissue classes. Figure 7(b-c) show a representative example of a tissue transition and its corresponding spectrum; this spectrum (location 2) was neither equal to the spectrum taken in IC (location 1) or the spectrum taken in adipose tissue (location 3). Instead, it resembled a mixture of these two tissue types. Since we cannot be certain that the histopathologic label of pixels at a tissue transition represents the tissue measured optically, these pixels were excluded from the RIGHT dataset.
Fig. 7.
(a) The classification accuracy of all pixels in the ALL dataset with respect to the distance to a tissue transition. Both the VIS and NIR wavelength range were used as input for classification. (b) shows an example of a tissue slice with an IC-adipose tissue transition. The circles in the images (b) are taken in the middle of IC (location 1), in the middle of adipose tissue (location 3) and at the IC-adipose tissue transition (location 2). The three diffuse reflectance spectra (c) correspond to these circles. The colors in (b) represent IC (red), connective tissue (dark blue) and adipose tissue (cyan).
In this study, training of LDA and U-Net was performed on the RIGHT and ALL dataset, respectively (Table 2). Testing of the classification algorithms was performed on both datasets.
Table 2. Overview of Usage of RIGHT and ALL Dataset.
| Classification algorithm | Training set (70% of samples) | Test set (30% of samples) |
|---|---|---|
| LDA | RIGHT dataset | RIGHT dataset |
| RIGHT dataset | ALL dataset | |
| U-Net | ALL dataset | RIGHT dataset |
| ALL dataset | ALL dataset | |
3.3. Evaluation of optimal wavelength range
With the spectral classification algorithm, we evaluated which wavelength range allows for the highest discrimination between tumor and healthy tissue. As input, we used the RIGHT dataset, and the wavelength range of the VIS camera, the NIR camera, and both cameras combined. The results are shown in Table 3. The classification performance using only the VIS wavelength range was high with recalls above 91% for all tissue types. When using only the NIR wavelength range, these results were comparable for DCIS and adipose tissue but lower for IC and connective tissue. Especially for connective tissue, the NIR wavelength range performed worse with 21% of the pixels incorrectly classified as malignant tissue. By using both the VIS and NIR wavelength range, the highest recalls for both IC and DCIS were obtained. The recall for connective tissue was however 3 percentage point lower in comparison with the VIS range.
Table 3. Evaluation of Optimal Wavelength Range: Recall for each Tissue Type, Tumor and Healthy.
| VIS | NIR | VIS + NIR | |||
|---|---|---|---|---|---|
| #spectra | 450-950 nm | 953-1650 nm | 450-1650 nm | ||
| Tumor | 3,312 | 95.4% | 91.8% | 98.8% | |
| IC | 3,200 | 95.6% | 91.9% | 99.0% | |
| DCIS | 112 | 91.1% | 90.2% | 94.6% | |
| Healthy | 21,227 | 99.9% | 99.5% | 99.4% | |
| Connective | 468 | 97.0% | 79.1% | 94.0% | |
| Adipose | 20,759 | 100.0% | 99.9% | 99.5% | |
Recall = percentage of pixels that were correctly classified as either tumor or healthy tissue. Histopathologic assessment of the tissue was used as ground truth.
The results in Table 3 were obtained after a single data partition into a training set and a test set. To confirm that this data partition was representative for the whole dataset, also a 7-fold cross-validation strategy was performed. Using the 7-fold cross-validation strategy, the recall for tumor and healthy using the VIS + NIR wavelength range were respectively 98.7 ± 1.9 (mean ± standard deviation) and 99.3 ± 0.8 (mean ± standard deviation), and therefore comparable with the results shown in Table 3.
The recall for healthy tissue was comparable for all three wavelength ranges, whereas the recall for tumor varied and was the highest when both the VIS and NIR wavelength ranges were used. Based on these numbers, we considered the VIS + NIR range the wavelength range that gave the highest performance for tissue discrimination with LDA. Based on McNemar’s test, the performance of HS imaging using the VIS + NIR wavelength range was significantly different from both other wavelength ranges (VIS: p < 0.0001, odds ratio: 121.9; NIR: p < 0.0001, odds ratio: 238.6).
3.4. Spectral versus spectral-spatial classification results
3.4.1. Results on the RIGHT dataset
Table 4 and Table 5 show the performance metrics and the recall, respectively, of both the spectral classification algorithm, LDA, and the spectral-spatial classification algorithm, U-Net. On the RIGHT dataset, both the performance metrics (Table 4) and the recall for tumor and healthy tissue (Table 5) were high and similar using LDA and U-Net. The major differences between the classification algorithms were observed in recall per tissue type for DCIS and connective tissue: U-Net was capable of discriminating DCIS from healthy tissue with a recall of 100%, whereas LDA only achieved a recall of 94.6%. For connective tissue, the recall using LDA was higher (94%) than the recall using U-Net (85.4%).
Table 4. Performance Metrics averaged over Patients for the Discrimination of Tumor from Healthy Tissue (mean ± std.).
| Spectral: LDAa | Spectral-spatial: U-Neta | ||
|---|---|---|---|
| RIGHT dataset | MCC | 0.98 ± 0.03 | 0.98 ± 0.04 |
| Sensitivity | 0.98 ± 0.03 | 0.98 ± 0.04 | |
| Specificity | 0.99 ± 0.01 | 0.99 ± 0.01 | |
| PPV | 0.99 ± 0.01 | 0.99 ± 0.01 | |
| NPV | 0.98 ± 0.02 | 0.98 ± 0.04 | |
| ALL dataset | MCC | 0.70 ± 0.23 | 0.74 ± 0.25 |
| Sensitivity | 0.76 ± 0.24 | 0.80 ± 0.25 | |
| Specificity | 0.92 ± 0.15 | 0.93 ± 0.09 | |
| PPV | 0.90 ± 0.12 | 0.93 ± 0.09 | |
| NPV | 0.82 ± 0.11 | 0.88 ± 0.10 | |
MCC = Matthews Correlation Coefficient, PPV = Positive Predictive Value, NPV = Negative Predictive Value, P = Tumor, N = Healthy.
Bold values represent the highest performance metrics of LDA and U-Net.
LDA and U-Net were trained on respectively the RIGHT and the ALL dataset of the training set.
Table 5. Spectral versus Spectral-Spatial Classification Results: Recall for each Tissue Type, Tumor and Healthy.
| #spectra | Spectral: LDAa | Spectral-spatial: U-Neta | |||
|---|---|---|---|---|---|
| RIGHT dataset | Tumor | 3.312 | 98.8% | 98.1% | |
| IC | 3.200 | 99.0% | 98.1% | ||
| DCIS | 112 | 94.6% | 100.0% | ||
| Healthy | 21.227 | 99.9% | 99.6% | ||
| Connective | 468 | 94.0% | 85.4% | ||
| Adipose | 20.759 | 100.0% | 99.9% | ||
| ALL dataset | Tumor | 8.883 | 83.2% | 86.3% | |
| IC | 7.400 | 83.5% | 88.3% | ||
| DCIS | 1.483 | 81.8% | 76.3% | ||
| Healthy | 75.451 | 94.2% | 94.1% | ||
| Connective | 15.731 | 77.5% | 80.0% | ||
| Adipose | 59.720 | 98.6% | 97.8% | ||
Recall = percentage of pixels that were correctly classified as either tumor or healthy tissue. Histopathologic assessment of the tissue was used as ground truth.
LDA and U-Net were trained on respectively the RIGHT and the ALL dataset of the training set.
3.4.2. Results on the ALL dataset
Since U-Net was trained on all spectra with histopathologic labels, also the classification performance on the ALL dataset is shown. In general, the classification performance of both classification algorithms was lower on the ALL dataset than on the RIGHT dataset.
On the ALL dataset, there was a significant difference in performance between LDA and U-Net (p < 0.0001, odds ratio: 312.5). For the ALL dataset, all performance metrics were higher when they were classified with U-Net (Table 4). This includes the MCC, which is a measure that describes how often tumor is classified as tumor, and healthy tissue as healthy tissue. In addition, the recall of tumor was higher with U-Net (86.3%) than with LDA (83.2%), whereas the recall of healthy tissue was similar.
The recall in Table 5 represents the percentage of pixels that were correctly classified as either tumor or healthy tissue. Figure 8 shows the capability of both algorithms to discriminate different tissue classes within the malignant and healthy classes. As can be seen, U-Net was much better than LDA at differentiating DCIS from IC and connective tissue from adipose tissue. For connective tissue, for example, the percentage of connective pixels classified as healthy tissue was comparable (Table 5; LDA: 77.5%, U-Net: 80%). However, Fig. 8 shows that with LDA only 37% of the connective pixels were correctly classified as connective, in comparison with 56.5% using U-Net. The remaining pixels were classified as the other healthy tissue class, adipose tissue. This result is also illustrated in Fig. 9 by showing the classification result in three specimens. In the slice in the top row, both algorithms classified the DCIS pockets correctly as malignant. However, U-Net classified these pockets correctly as DCIS (Fig. 9(c)) whereas LDA classified them incorrectly as IC (Fig. 9(b)). Likewise, the smaller branches of connective tissue in the slice in the bottom two rows were detected with U-Net (Fig. 9(f, i)) but classified as adipose tissue with LDA (Fig. 9(e, h)) as indicated with the orange arrows.
Fig. 8.
For both classification algorithms, LDA and U-Net, the percentage of spectra in the ALL dataset classified as a tissue class within the malignant (IC + DCIS) and healthy (connective + adipose) tissue class. The entire bar corresponds to the percentage correctly classified as malignant or healthy tissue. The color in the bars corresponds to the tissue type as which the pixel was correctly classified with the classification algorithms: red = IC, magenta = DCIS, dark blue = connective, cyan = adipose.
Fig. 9.
The difference between pixel-based classification without (b, e, h) and with (c, f, i) adding contextual context in two tissue slices from the test set. a, d, g) shows the histopathology annotations. When adding contextual context, better differentiation between different tissue classes within the malignant (top row) and healthy classes (bottom two rows) can be made. The orange arrows point at smaller branches of connective tissue that were detected with U-Net (f) but classified as adipose tissue with LDA (e).
Therefore, adding spatial and contextual information to the classification algorithm improved the classification performance on the ALL dataset and improved the capability of differentiating different tissue classes within the malignant and healthy classes. On the RIGHT dataset, however, the classification performance was similar with and without adding textural structure to the classification algorithm.
4. Discussion
Obtaining tumor-negative resection margins after breast-conserving surgery remains challenging due to the lack of feedback during surgery. Diffuse reflectance HS imaging offers great potential to fill this gap as the entire resection margin can be imaged during surgery within a limited amount of time. This paper evaluates the classification performance of HS imaging for tumor detection in gross-sectioned fresh breast tissue slices. Specifically, we showed that the highest discrimination between tumor and healthy breast tissue was obtained when HS data over the full wavelength range of 450-1650 nm was used as input. In addition, the spatial information of the HS data contains valuable information that especially improves the classification performance on the ALL dataset and the capability of differentiating different tissue classes within the malignant and healthy tissue classes.
We showed that by using only the NIR wavelength range, high classification accuracies for IC, DCIS and adipose tissue could be obtained. However, the differentiation of connective tissue was more difficult as 21% of the spectra were incorrectly classified as malignant. These results are similar to our previously published work [22] and suggest that the amount of water, fat, and collagen, which are the main absorptions in the NIR wavelength range, are often similar in connective and malignant tissue. Previous research performing diffuse reflectance spectroscopy with fiber-optic probes shows that the chromophores in the NIR wavelength range allow for good discrimination between malignant and healthy tissue [31–34]. However, only a limited number of studies made a differentiation between connective and malignant tissue. In a study from Nachabé et al., connective tissue was reported to have a lower water content than IC as well as a lower collagen content than DCIS. Since the light absorption of both water and collagen is most characteristic in the NIR wavelength range, their study suggests that the NIR wavelength range is sufficient for discriminating connective from malignant tissue [34]. However, most studies show that chromophores in the VIS wavelength are required for the differentiation between connective and malignant tissue [31,35–37]. In our study, the classification performance indeed improved substantially to recalls above 94% for all four tissue types, including connective tissue, after adding HS data obtained in the VIS wavelength range to the HS data obtained in the NIR wavelength range.
To evaluate the performance of different wavelength ranges, we used the relatively simple classification algorithm LDA combined with pairwise coupling. This algorithm was trained using only pixels in the RIGHT dataset, i.e. pixels with a histopathologic label of which we were certain. U-Net, on the other hand, incorporates both spectral and spatial information in the classification. Therefore, all pixels in the HS image needed to be used as training data in order to keep the spatial and contextual information unaltered. As a result, also pixels with uncertain histopathologic labels were included in the training. Therefore, the classification performance on the RIGHT dataset could have been lower with U-Net than with LDA. However, the performance metrics were similar (Table 4) and only the recall for connective tissue was considerably lower using U-Net (Table 5). On the ALL dataset, we showed by using the McNemar’s test that the performances of LDA and U-Net were significantly different. However, this test does not determine whether one classification technique performs significantly better than the other. In Table 4, we did show that the performance metrics using U-Net was higher than when using LDA. In addition, the recall of tumor in Table 5 was higher with U-Net than with LDA. Based on these numbers, we considered U-Net to perform better on the ALL dataset than LDA. We would have expected that since, besides the fact that adding spatial information could improve the detection of smaller structures, also pixels were included in the training set that represented a mixture of different tissue types but were labeled with a single tissue class by histopathologic assessment. The latter increased the likelihood that pixels in the test set with a similar diffuse reflectance spectrum were classified with the same tissue class. In Fig. 9, we illustrated this by showing that smaller branches of connective tissue were detected with U-Net and missed with LDA.
The difference in performance metrics between the ALL and the RIGHT dataset was, as explained in Section 2.3.1, related to a difference in the data that we measured with the HS camera, and the information that histopathology is providing. Reducing the ALL dataset to the RIGHT dataset resulted in a large reduction of the number of spectra (remainder in RIGHT dataset per tissue class: IC 35%, DCIS 2.5%, connective tissue 1.4%, and adipose tissue 27%), especially for DCIS and connective tissue, which are smaller tissue classes in general. For the final purpose of this study, assessing resection margins of lumpectomy specimens, it is more likely that tumor-positive resection margins are caused by mixtures of malignant and healthy tissue instead of these RIGHT spectra. Nevertheless, the RIGHT dataset can be used to determine the maximum capability of HS imaging to differentiate malignant tissue types from healthy tissue types. On the RIGHT dataset, we report a sensitivity, specificity, PPV, and NPV all higher than 0.98 using either LDA or U-Net.
Thereby, the classification performance of HS imaging on the RIGHT dataset of the tissue slices was high in comparison with the performance reported for resection margin techniques that are currently available in the clinic (Table 6). For imprint cytology and frozen section analysis, a sensitivity of 72 and 83%, and a specificity of 97 and 95% was reported [10]. The classification performance of HS imaging on the ALL dataset was comparable to these numbers. However, imprint cytology and frozen section analysis add on average 10-30 minutes to operation times and are too time-consuming to analyze the whole resection surface. With HS imaging, the time required to image and analyze the entire resection surface can be much faster. In this study, each tissue slice was imaged in 1 minute (40 seconds with the VIS camera and 20 seconds with the NIR camera) and data analysis was performed within 45 seconds (30 seconds for preprocessing, and 2-3 seconds and 7-8 seconds for classifying the image using LDA and U-Net respectively). Therefore, with our approach, the surgeon would be able to get a diagnosis on the presence of tumor at a specific resection side within 2 minutes. This time is comparable to the other faster margin assessment techniques, ultrasound and specimen radiography [9,13]. However, the classification performance of those two techniques showed to be insufficient to clearly improve the reoperation rate [9,13,14].
Table 6. Comparison of Currently Available Margin Assessment Techniques and HS Imaging.
| Sensitivity | Specificity | Time required for diagnosis | ||
|---|---|---|---|---|
| Imprint Cytologya [10] | 72% | 97% | 27 min. | |
| Frozen Section Analysisa [10] | 83% | 95% | 13 min. | |
| Ultrasounda [9,12] | 59% | 81% | 3-6 min. | |
| Specimen Radiographya [9,14] | 53% | 84% | 1-2 min. | |
| Our approachb | RIGHT dataset | 98% | 99% | <2 min. |
| ALL dataset | 80% | 93% | ||
Classification performance reported in literature for resection margin assessment
Classification performance obtained on fresh tissue slices in this study
In this study, optimization of HS data acquisition and data analysis was out of the scope of this research: data were acquired with two separate imaging set-ups and data analysis was performed in MATLAB 2018a using an Intel Xeon CPU E3-1240 at 3.40 GHz. However, HS imaging for resection margin assessment does have the potential to be much faster by further development of the hardware by, for instance, combining both wavelength regions in a single camera, and optimization of the classification algorithm.
In the end, this study was performed to achieve our final goal: resection margin assessment with HS imaging on fresh lumpectomy specimen so that direct feedback can be given to the surgeon during surgery. However, in the current study, measurements are performed ex vivo on fresh tissue slices after inking and gross-sectioning of the resection specimen. This approach allowed us to obtain the highest possible correlation with histopathology since H&E sections are taken from the same surface as measured with the HS camera. However, our final goal with HS imaging is measuring lumpectomy specimens immediately after surgery, so that direct feedback on the resection margins can be given to the surgeon. Whether the developed classification algorithms in this study allows for the same high classification performance on the resection surface of lumpectomy specimens needs to be examined in a new clinical study. In such a study, fresh lumpectomy specimens can be imaged and analyzed immediately after surgery so that tumor-suspicious areas can be marked and retrieved on the specimen after histopathologic processing.
5. Conclusion
In summary, we have demonstrated that with HS imaging malignant tissue can be discriminated from healthy tissue with a sensitivity and a specificity of 0.98 and 0.99. Therefore, two HS cameras were required that together comprise a broad wavelength range from 450 to 1650 nm. Hyperspectral data analysis using this spectral range outperforms HS data analysis using wavelength ranges limited to either the VIS (450-951 nm) or NIR (954-1650 nm) alone. Adding spatial and contextual information to the classification algorithm especially improved the capability of HS imaging to differentiate different tissue classes within the malignant and healthy classes. As a result, smaller branches of connective tissue and DCIS, which were classified as adipose and IC respectively with the spectral algorithm, were detected with the spectral-spatial algorithm. With the algorithms developed in this study, it would be possible to, in the future, provide direct feedback on the resection margins to the surgeon during surgery. Therefore, the next step is to validate the technique as margin assessment technique during breast-conserving surgery.
Acknowledgment
The authors thank the NKI-AVL core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI-AVL biobank material, all surgeons and nurses from the Department of Surgery and all pathologist and pathologist assistants from the Department of Pathology for their assistance in collecting the specimens. The authors also thank K. Jóźwiak for help with statistical analyses. The Quadro P6000 GPU used for this research was donated by the NVIDIA Corporation.
Funding
KWF Kankerbestrijding10.13039/501100004622 (10747).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Alrahbi S., Chan P. M., Ho B. C., Seah M. D., Chen J. J., Tan E. Y., “Extent of margin involvement, lymphovascular invasion, and extensive intraductal component predict for residual disease after wide local excision for breast cancer,” Clin. Breast Cancer 15(3), 219–226 (2015). 10.1016/j.clbc.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 2.Merrill A. L., Coopey S. B., Tang R., McEvoy M. P., Specht M. C., Hughes K. S., Gadd M. A., Smith B. L., “Implications of new lumpectomy margin guidelines for breast-conserving surgery: Changes in reexcision rates and predicted rates of residual tumor,” Ann. Surg. Oncol. 23(3), 729–734 (2016). 10.1245/s10434-015-4916-2 [DOI] [PubMed] [Google Scholar]
- 3.Merrill A. L., Tang R., Plichta J. K., Rai U., Coopey S. B., McEvoy M. P., Hughes K. S., Specht M. C., Gadd M. A., Smith B. L., “Should new “no ink on tumor” lumpectomy margin guidelines be applied to ductal carcinoma in situ (DCIS)? A retrospective review using shaved cavity margins,” Ann. Surg. Oncol. 23(11), 3453–3458 (2016). 10.1245/s10434-016-5251-y [DOI] [PubMed] [Google Scholar]
- 4.Langhans L., Jensen M.-B., Talman M.-L. M., Vejborg I., Kroman N., Tvedskov T. F., “Reoperation rates in ductal carcinoma in situ vs invasive breast cancer after wire-guided breast-conserving surgery,” JAMA Surg. 152(4), 378–384 (2017). 10.1001/jamasurg.2016.4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart B. W., Wild C. P., World cancer report 2014, World Cancer Reports (IARC Press, 2014). [Google Scholar]
- 6.Vos E. L., Jager A., Verhoef C., Voogd A. C., Koppert L. B., “Overall survival in patients with a re-excision following breast conserving surgery compared to those without in a large population-based cohort,” Eur. J. Cancer 51(3), 282–291 (2015). 10.1016/j.ejca.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 7.Pleijhuis R. G., Graafland M., de Vries J., Bart J., de Jong J. S., van Dam G. M., “Obtaining adequate surgical margins in breast-conserving therapy for patients with early-stage breast cancer: Current modalities and future directions,” Ann. Surg. Oncol. 16(10), 2717–2730 (2009). 10.1245/s10434-009-0609-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating J. J., Fisher C., Batiste R., Singhal S., “Advances in intraoperative margin assessment for breast cancer,” Curr. Surg. Rep. 4(4), 15 (2016). 10.1007/s40137-016-0136-3 [DOI] [Google Scholar]
- 9.St John E. R., Al-Khudairi R., Ashrafian H., Athanasiou T., Takats Z., Hadjiminas D. J., Darzi A., Leff D. R., “Diagnostic accuracy of intraoperative techniques for margin assessment in breast cancer surgery: A meta-analysis,” Ann. Surg. 265(2), 300–310 (2017). 10.1097/SLA.0000000000001897 [DOI] [PubMed] [Google Scholar]
- 10.Esbona K., Li Z., Wilke L. G., “Intraoperative imprint cytology and frozen section pathology for margin assessment in breast conservation surgery: A systematic review,” Ann. Surg. Oncol. 19(10), 3236–3245 (2012). 10.1245/s10434-012-2492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Versteegden D., Keizer L., Schlooz-Vries M., Duijm L., Wauters C., Strobbe L., “Performance characteristics of specimen radiography for margin assessment for ductal carcinoma in situ: A systematic review,” Breast Cancer Res. Treat. 166(3), 669–679 (2017). 10.1007/s10549-017-4475-2 [DOI] [PubMed] [Google Scholar]
- 12.Maloney B. W., McClatchy D. M., Pogue B. W., Paulsen K. D., Wells W. A., Barth R. J., “Review of methods for intraoperative margin detection for breast conserving surgery,” J. Biomed. Opt. 23(10), 1 (2018). 10.1117/1.JBO.23.10.100901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler-Henderson K., Lee A. H., Price R. I., Waring K., “Intraoperative assessment of margins in breast conserving therapy: A systematic review,” The Breast 23(2), 112–119 (2014). 10.1016/j.breast.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 14.Kaufman C. S., Jacobson L., Bachman B. A., Kaufman L. B., Mahon C., Gambrell L.-J., Seymour R., Briscoe J., Aulisio K., Cunningham A., “Intraoperative digital specimen mammography: Rapid, accurate results expedite surgery,” Ann. Surg. Oncol. 14(4), 1478–1485 (2007). 10.1245/s10434-006-9126-5 [DOI] [PubMed] [Google Scholar]
- 15.Lu G., Fei B., “Medical hyperspectral imaging: A review,” J. Biomed. Opt. 19(1), 010901 (2014). 10.1117/1.JBO.19.1.010901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denstedt M., Bjorgan A., Milanič M., Randeberg L. L., “Wavelet based feature extraction and visualization in hyperspectral tissue characterization,” Biomed. Opt. Express 5(12), 4260–4280 (2014). 10.1364/BOE.5.004260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu G., Little J. V., Wang X., Griffith C. C., El-Deiry M., Chen A. Y., Fei B., “Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging,” Clin. Cancer Res. 23(18), 5426–5436 (2017). 10.1158/1078-0432.CCR-17-0906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fei B., Lu G., Wang X., Zhang H., Little J. V., Patel M. R., Griffith C. C., El-Diery M. W., Chen A. Y., “Label-free reflectance hyperspectral imaging for tumor margin assessment: A pilot study on surgical specimens of cancer patients,” J. Biomed. Opt. 22(8), 086009 (2017). 10.1117/1.JBO.22.8.086009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Z., Zhang A., Wang X., Sun Z., Wang M. D., Xie T., “In vivo use of hyperspectral imaging to develop a noncontact endoscopic diagnosis support system for malignant colorectal tumors,” J. Biomed. Opt. 21(1), 016001 (2016). 10.1117/1.JBO.21.1.016001 [DOI] [PubMed] [Google Scholar]
- 20.Neittaanmäki-Perttu N., Grönroos M., Tani T., Pölönen I., Ranki A., Saksela O., Snellman E., “Detecting field cancerization using a hyperspectral imaging system,” Lasers Surg. Med. 45(7), 410–417 (2013). 10.1002/lsm.22160 [DOI] [PubMed] [Google Scholar]
- 21.Pardo Franco A., Real Peña E., Krishnaswamy V., López Higuera J. M., Pogue B. W., Conde Portilla O. M., “Directional kernel density estimation for classification of breast tissue spectra,” IEEE Trans. Med. Imaging 36(1), 64–73 (2017). 10.1109/TMI.2016.2593948 [DOI] [PubMed] [Google Scholar]
- 22.Kho E., de Boer L. L., Van de Vijver K. K., van Duijnhoven F., Vrancken Peeters M.-J. T. F. D., Sterenborg H. J. C. M., Ruers T. J. M., “Hyperspectral imaging for resection margin assessment during surgery,” Clin. Cancer Res. 25(12), 3572–3580 (2019). 10.1158/1078-0432.CCR-18-2089 [DOI] [PubMed] [Google Scholar]
- 23.Barnes R., Dhanoa M. S., Lister S. J., “Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra,” Appl. Spectrosc. 43(5), 772–777 (1989). 10.1366/0003702894202201 [DOI] [Google Scholar]
- 24.Vidal M., Amigo J. M., “Pre-processing of hyperspectral images. Essential steps before image analysis,” Chemom. Intell. Lab. Syst. 117, 138–148 (2012). 10.1016/j.chemolab.2012.05.009 [DOI] [Google Scholar]
- 25.Fisher R. A., “The use of multiple measurements in taxonomic problems,” Ann. Eugen. 7(2), 179–188 (1936). 10.1111/j.1469-1809.1936.tb02137.x [DOI] [Google Scholar]
- 26.Wu T.-F., Lin C.-J., Weng R. C., “Probability estimates for multi-class classification by pairwise coupling,” J. Mach. Learn. Res. 5, 975–1005 (2004). [Google Scholar]
- 27.Ronneberger O., Fischer P., Brox T., “U-net: Convolutional networks for biomedical image segmentation,” in MICCAI, (Springer, 2015), 234–241. [Google Scholar]
- 28.Matthews B. W., “Comparison of the predicted and observed secondary structure of T4 phage lysozyme,” Biochim. Biophys. Acta, Protein Struct. 405(2), 442–451 (1975). 10.1016/0005-2795(75)90109-9 [DOI] [PubMed] [Google Scholar]
- 29.Boughorbel S., Jarray F., El-Anbari M., “Optimal classifier for imbalanced data using matthews correlation coefficient metric,” PLoS One 12(6), e0177678 (2017). 10.1371/journal.pone.0177678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietterich T. G., “Approximate statistical tests for comparing supervised classification learning algorithms,” Neural Comput. 10(7), 1895–1923 (1998). 10.1162/089976698300017197 [DOI] [PubMed] [Google Scholar]
- 31.Evers D., Nachabe R., Vranken Peeters M.-J., van der Hage J., Oldenburg H., Rutgers E., Lucassen G., Hendriks B. W., Wesseling J., Ruers T. M., “Diffuse reflectance spectroscopy: Towards clinical application in breast cancer,” Breast Cancer Res. Treat. 137(1), 155–165 (2013). 10.1007/s10549-012-2350-8 [DOI] [PubMed] [Google Scholar]
- 32.de Boer L. L., Molenkamp B., Bydlon T. M., Hendriks B. H. W., Wesseling J., Sterenborg H. J. C. M., Ruers T. J. M., “Fat/Water ratios measured with diffuse reflectance spectroscopy to detect breast tumor boundaries,” Breast Cancer Res. Treat. 152(3), 509–518 (2015). 10.1007/s10549-015-3487-z [DOI] [PubMed] [Google Scholar]
- 33.Taroni P., Paganoni A. M., Ieva F., Pifferi A., Quarto G., Abbate F., Cassano E., Cubeddu R., “Non-invasive optical estimate of tissue composition to differentiate malignant from benign breast lesions: A pilot study,” Sci. Rep. 7(1), 40683 (2017). 10.1038/srep40683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachabé R., Evers D. J., Hendriks B. H., Lucassen G. W., van der Voort M., Rutgers E. J., Peeters M.-J. V., Van der Hage J. A., Oldenburg H. S., Wesseling J., “Diagnosis of breast cancer using diffuse optical spectroscopy from 500 to 1600 nm: comparison of classification methods,” J. Biomed. Opt. 16(8), 087010 (2011). 10.1117/1.3611010 [DOI] [PubMed] [Google Scholar]
- 35.van Veen R. L., Amelink A., Menke-Pluymers M., van der Pol C., Sterenborg H. J., “Optical biopsy of breast tissue using differential path-length spectroscopy,” Phys. Med. Biol. 50(11), 2573–2581 (2005). 10.1088/0031-9155/50/11/009 [DOI] [PubMed] [Google Scholar]
- 36.Zhu C., Palmer G. M., Breslin T. M., Harter J., Ramanujam N., “Diagnosis of breast cancer using fluorescence and diffuse reflectance spectroscopy: A monte-carlo-model-based approach,” J. Biomed. Opt. 13(3), 034015 (2008). 10.1117/1.2931078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Boer L. L., Hendriks B. H. W., van Duijnhoven F., Vrancken Peeters-Baas M. J. T. F. D., Van de Vijver K. K., Loo C. E., Jóźwiak K., Sterenborg H. J. C. M., Ruers T. J. M., “Using DRS during breast conserving surgery: Identifying robust optical parameters and dealing with inter-patient variation,” Biomed. Opt. Express 7(12), 5188–5200 (2016). 10.1364/BOE.7.005188 [DOI] [PMC free article] [PubMed] [Google Scholar]